Background: Hox hydrogenase (HoxEFUYH) is present in some bacteria and cyanobacteria.
Results: In Synechocystis, HoxYH and HoxEFU form subcomplexes that can be disrupted in Hox subunit mutants.
Conclusion: Hox assembly is modular and most reliant on the presence of HoxF and -U, consistent with effects on Hox protein abundance and activity.
Significance: Subcomplexes of Hox hydrogenase may represent steps in complex assembly and maturation.
Keywords: Bacterial Metabolism, Cyanobacteria, Enzyme Inactivation, Enzyme Processing, Enzyme Purification, Hydrogenase, Hox Hydrogenase, Hydrogenase Assembly, Hydrogenase Function, Hydrogenase Maturation
Abstract
Hydrogenases are metalloenzymes that catalyze 2H+ + 2e− ↔ H2. A multisubunit, bidirectional [NiFe]-hydrogenase has been identified and characterized in a number of bacteria, including cyanobacteria, where it is hypothesized to function as an electron valve, balancing reductant in the cell. In cyanobacteria, this Hox hydrogenase consists of five proteins in two functional moieties: a hydrogenase moiety (HoxYH) with homology to heterodimeric [NiFe]-hydrogenases and a diaphorase moiety (HoxEFU) with homology to NuoEFG of respiratory Complex I, linking NAD(P)H ↔ NAD(P)+ as a source/sink for electrons. Here, we present an extensive study of Hox hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803. We identify the presence of HoxEFUYH, HoxFUYH, HoxEFU, HoxFU, and HoxYH subcomplexes as well as association of the immature, unprocessed large subunit (HoxH) with other Hox subunits and unidentified factors, providing a basis for understanding Hox maturation and assembly. The analysis of mutants containing individual and combined hox gene deletions in a common parental strain reveals apparent alterations in subunit abundance and highlights an essential role for HoxF and HoxU in complex/subcomplex association. In addition, analysis of individual and combined hox mutant phenotypes in a single strain background provides a clear view of the function of each subunit in hydrogenase activity and presents evidence that its physiological function is more complicated than previously reported, with no outward defects apparent in growth or photosynthesis under various growth conditions.
Introduction
Hydrogenase enzymes are a unique and diverse family of metalloenzymes widely distributed throughout Archaea, Prokaryotes, and some unicellular Eukaryotes that catalyze the reduction/oxidation of H+/H2. These enzymes are classified by their metal-containing active sites and include [Fe]-hydrogenases, [FeFe]-hydrogenases, and [NiFe]-hydrogenases (1). The [NiFe]-hydrogenases are minimally heterodimeric, consisting of a large catalytic subunit and a small subunit containing at least one [FeS] cluster that functions in electron transfer to and from the large subunit (1). The maturation of [NiFe]-hydrogenases involves at least six maturation proteins (HypABCDEF) essential for the assembly of the [NiFe] active site. Some [NiFe]-hydrogenases additionally require a specialized protease that cleaves the large subunit's C terminus, a step required for hydrogenase function (2). Research in Escherichia coli suggests that the hydrogenase small subunit only associates with the large subunit after it is fully processed (3).
Among the diverse hydrogenases, the bidirectional [NiFe]-hydrogenase (Hox) in cyanobacteria is of great interest for basic biological study as well as for the development of solar hydrogen production technologies (4–6). NAD(P)-linked Hox hydrogenases have been identified and characterized in cyanobacteria (4, 5), the Gram-positive bacterium Rhodococcus opacus (7, 8), the Gram-negative bacterium Ralstonia eutropha (6), and the purple sulfur photosynthetic bacteria Thiocapsa roseopersicina and Allochromatium vinosum (9–12). These Hox hydrogenases are multimeric with at least four related subunits expressed from a single operon. HoxH and HoxY form the hydrogenase moiety and are homologous to the large and small subunits of prototypical heterodimeric [NiFe]-hydrogenases, respectively (1). The [FeS] cluster-containing subunits HoxF, HoxU, and, in some organisms, HoxE form the diaphorase moiety (13, 14) that catalyzes the oxidation/reduction of NAD(P)H/NAD(P)+ (via FMN and NAD binding sites in HoxF) coupled to the hydrogenase moiety (HoxYH) (10, 12–15). The R. eutropha Hox hydrogenase does not contain HoxE and instead harbors an unrelated fifth subunit, HoxI, which functions in linkage to NADPH (16).
Purification of the Hox hydrogenase has been performed in some of the organisms it has been characterized in with varied results. In R. eutropha, the purified Hox complex is HoxFUYHI2, although HoxI (not present in other characterized Hox hydrogenases) dissociates easily upon more stringent purification conditions (which is why the complex was initially characterized as HoxFUYH) (16). Other purifications in R. eutropha utilizing gene knockouts and ectopic expression demonstrate that HoxYH and HoxFU subcomplexes can be isolated, although these subcomplexes were reportedly unstable (13). In A. vinosum (12) and the cyanobacterium Gleocapsa alpicola CALU 743 (17), attempted purifications of native complex resulted in isolation of only HoxYH, whereas in T. roseopersicina (18) and Synechocystis (19, 20), intact HoxEFUYH complexes were purified. Purification of the Synechocystis Hox hydrogenase by Schmitz et al. (19) resulted in a dimer of a 1:1:1:1:1 HoxE/F/U/Y/H complex via a series of column chromatography steps, whereas Germer et al. (20) purified a complex with a 0.2:2:2:1:1 HoxE/F/U/Y/H ratio using a StrepII-tagged HoxF, raising questions about Hox complex assembly and association in vivo.
The physiological role of Hox hydrogenase varies, depending on the organism in which it is present, but it generally functions in anoxic/micro-oxic conditions due to a reversible inactivation by O2 (1). In T. roseopersicina, Hox hydrogenase catalyzes H2 production under dark, fermentative conditions and in light when thiosulfate is present and alternatively functions in H2 uptake in light when O2 is absent (11). The Hox hydrogenase of R. eutropha (soluble hydrogenase) functions in H2 oxidation linked to the regeneration of NADH in support of carbon fixation (6). In cyanobacteria, Hox hydrogenase is expressed under both anaerobic and aerobic conditions (4, 21) but is only active under dark, fermentative conditions and in the transition from dark to light prior to inhibition by O2 generated during photosynthesis (22–24). Respiration and nitrate assimilation mutants exhibit increased H2 photoevolution rates under low O2 conditions, whereas defects in photosynthetic reactions/ratios have been reported in Hox hydrogenase mutants (15, 22–26). Therefore, it is hypothesized that the Hox hydrogenase functions as an electron valve for cells by H2 production/oxidation in response to changes in redox states (25, 26). HoxEFU are the only homologs for NuoEFG of respiratory complex I in cyanobacteria, but there is no evidence to date that these diaphorase subunits play any role in respiration (14, 27–30).
In this work, we generated a series of Hox hydrogenase mutants in a common parental strain, deleting individual hox genes or combinations of hox genes in the unicellular cyanobacterium Synechocystis sp. PCC 6803. This comprehensive collection of hox mutants enabled us to perform systematic studies of mutation effects on complex/subcomplex formation and composition, subunit abundance, and hydrogenase activity. In addition, we also provide data for growth, photosynthesis, and fermentation in a hox operon deletion mutant compared with wild type (WT), revealing that some previously reported hox mutant phenotypes may have been due to differences in strain backgrounds (31, 32).
EXPERIMENTAL PROCEDURES
Strain Background/Construction
All hox mutants were generated in a Synechocystis sp. PCC 6803 glucose-tolerant strain from Teruo Ogawa, who also provided the whole operon deletion, hox−::hygromycin resistance. The hoxH− deletion was constructed in a pSMART LC-Amp vector (Lucigen, Inc.) containing a 3.6-kb region spanning hoxY and hoxH cut with SspI (removing 816 bp of hoxH) to insert the kanamycin resistance cassette from pUC4K. All remaining hox deletion strains were created by two-step fusion PCR (33) for open reading frame (ORF) replacement, combining ∼600-bp 5′- and 3′-flanking regions for each hox gene ORF replaced with the following antibiotic resistance gene ORFs: hoxE−::aac3ia (gentamicin resistance), hoxF−::ermC (erythromycin resistance), hoxU−:: aph (kanamycin resistance), and hoxY−:: dfra17 (spectinomycin resistance). An slr0168 neutral site integration vector containing a plastocyanin (petE) promoter for ectopic gene expression (34) was altered by the addition of a 225-bp fragment from pETDuet containing the T7 terminator into the PpuMI restriction site. PCR primers to amplify HoxYH or HoxH from genomic DNA were constructed with sequence encoding His6/HRV3C protease site at the N terminus and 5′- and 3′-specific SapI sites for insertion behind the petE promoter following restriction digestion and ligation. The pPSBA2KS vector for the integration behind the psbAII promoter (35) was altered by removal of a SalI site via partial digest and blunting to allow for retention of the kanamycin resistance gene. PCR primers were designed for amplification and insertion of an N-terminal His6-tagged hoxE gene between NdeI and SalI sites. Transformations were conducted by incubating ∼1 μg of linear purified fusion PCR products or targeted integration vectors with 200 μl of cells (adjusted to OD730 = 2.5 from cell cultures at OD730 = 0.2–0.5) for 6 h, followed by the addition of 2 ml of BG11, 24-h outgrowth in culture tubes under standard growth conditions, and plating of 200 μl on BG11 plates with antibiotics as required for selection (200 μg/ml hygromycin or kanamycin, 100 μg/ml gentamicin, erythromycin, spectinomycin, or chloramphenicol).
Strain Growth
Cultures were inoculated into BG11 medium (ATCC medium 616) supplemented with 3 μm NiCl2, 20 mm TES (unbuffered), 100 mm NaHCO3, and antibiotics as required (one-half concentrations as used on plates listed above) at an initial OD730 = 0.05 and were grown by shaking in culture flasks with 5% CO2 under 50 microeinsteins2 (μE) m−2 s−1 continuous light from cool white fluorescent bulbs. Cultures were generally grown to logarithmic/linear growth phase (OD730 = 0.2–0.8) for analysis.
FPLC Analysis of Synechocystis sp. PCC 6803 WT Soluble Extract
Soluble extract was prepared from a WT 50-ml liquid culture at an OD730 > 1 (stationary phase). Cells were harvested, resuspended, and washed twice in ACA buffer (750 mm ϵ-amino caproic acid, 50 mm BisTris/HCl, pH 7.0, 0.5 mm EDTA). Approximately 200 μl of glass beads (150–212 μm; Sigma-Aldrich) were added to the cell suspension (∼500 μl), and cells were broken using a Disruptor Genie digital multiplace vortexer (Scientific Industries) at 4 °C for 5 min, setting 3,000, followed by centrifugation for 10 min at maximum speed in a microcentrifuge. Subsequently, 2 μl of DNase I (10,000 units/ml, Thermo Scientific) was added, and the sample was spun again in an ultracentrifuge at 100,000 × g for 30 min at 4 °C. For FPLC, the resulting supernatant was normalized to OD650 = 10 (typically, the OD650 of a 1:20 dilution was measured). To solubilize any remaining thylakoid membrane fragments, β-dodecyl-d-maltoside was added to a final concentration of 0.5% (w/v) from a 10% (w/v) stock solution. 100 μl of this soluble extract was loaded onto a Superdex 200 10/300 size exclusion column (GE Healthcare) connected to an Akta Purifier FPLC system (GE Healthcare). The sample was run at a flow rate of 0.5 ml min−1 with 50 mm Tris, pH 7.5, 100 mm NaCl, and 0.03% (w/v) β-dodecyl-d-maltoside as the buffer. 0.5-ml fractions were collected by a Frac-950 fraction collector (GE Healthcare). A high molecular weight marker mix (GE Healthcare) was run as a control for relative protein sizes in the fractions (supplemental Fig. 1).
Protein Preparations, Two-dimensional Blue Native/SDS-PAGE, Affinity Purification, and Western Blotting
Soluble extracts for two-dimensional blue native/SDS-PAGE (BN/SDS-PAGE) were prepared as described for FPLC analysis above for WT and hox mutant strains, and the extracts were normalized to an A650 = 0.6. β-Dodecyl-d-maltoside was added to a final concentration of 0.5% (w/v), and the same volume of Coomassie loading solution (750 mm ϵ-amino caproic acid, 5% (w/v) Coomassie-G) was added to the sample. Subsequently, 20 μl of the sample was loaded per lane and run on a 12% (w/v) polyacrylamide BN PAGE first dimension gel. The protein complexes in the first dimension gel strips were denatured for 1 h in solubilization buffer (66 mm Na2CO3, 2% (w/v) SDS, 2% (v/v) β-mercaptoethanol, and 4 m urea) prior to being layered on 17.5% (w/v) polyacrylamide, 6 m urea two-dimensional SDS-polyacrylamide gels (36). The resultant two-dimensional gels were either Coomassie-stained, silver-stained (37), or electroblotted onto nitrocellulose membranes.
For WT and hox mutant one-dimensional Western blots and affinity purifications, cells were resuspended in 20 mm phosphate buffer, pH 7.4, plus protease inhibitors (Fermentas) and broken by bead disruption as described above. Protein concentrations were determined by Bradford assay (Fermentas), and levels of each sample were adjusted to ∼100 μg/ml. For purification of His6-tagged proteins/complexes, Co2+ beads (Pierce) were added to cleared lysates and incubated for 1 h at 4 °C, and washes were performed using 5 times bead volume with 20 mm phosphate buffer, pH 7.4, with or without 0.5 m NaCl through reloadable columns (Pierce). Beads were resuspended in a 50% slurry in buffer. To rule out nonspecific binding of Hox proteins to beads, untagged WT samples were also subjected to the same regime, and a lack of Hox proteins in bead samples was verified by Western blot (data not shown). Samples for SDS-PAGE were boiled in 1× Laemmli sample buffer (Bio-Rad), and 10 μl of each was loaded onto precast TGX Stain-free Any kDa gels (Bio-Rad) and transferred onto a PVDF membrane (Bio-Rad). For Western blotting, the SnapID system (Millipore) was used according to the manufacturer's instructions. Membranes were blocked in 5% BSA, 1× PBST, and primary and secondary antibodies were diluted in 1% BSA, 1× PBST. Polyclonal rabbit primary antibodies developed for this study were generated using ∼20-amino acid C-terminal peptides (α-HoxE, -F, -U, -Y, and -H (C-terminal); α-HoxF and α-HoxY from these preparations are not shown but were used in quantitation of one-dimensional Western blots) and a HoxH internal peptide (amino acids 300–318) that allows recognition of both processed and unprocessed HoxH (YenZym, San Francisco, CA). α-HoxY and α-HoxF antibodies were also generated in rabbit using full-length E. coli expressed protein (SeqLab). α-Rps1 (Agrisera; not shown) and α-PsaD antibodies (38) were used as loading controls for normalization of band intensities for relative protein level quantitation. Secondary incubation was performed with Clean Blot HRP (Pierce), and Cell Biosciences ChemiWest or Pierce Dura Chemiluminescent reagents were used for signal development. Images and quantitation analysis were processed using a Cell Biosciences FluoroCam Q Gel imaging system.
Hydrogenase Assays
Cells grown to similar logarithmic growth densities (OD730 = 0.2–0.8) were concentrated to OD730 = 2.5, and 200 μl was added to 600 μl of assay mixture containing Triton X-100 (0.05%, w/v), phosphate buffer (50 mm, pH 7), and either methyl viologen (MV; 1 mm final) or NADH (1 mm final) in 2-ml HPLC vials sealed with anaerobic septa. The mixture was sparged with argon for 10 min, 100 μl of anaerobic sodium dithionite (10 mm final) was added, and the mixture was incubated with shaking for 30 min at 37 °C. The reaction was stopped by the addition of 100 μl of 20% (w/v) trichloroacetic acid, and 100 μl of the 1-ml headspace was analyzed using a gas chromatograph (Agilent 1100) equipped with a Molecular Sieve 5A column, a thermal conductivity detector, and argon as the carrier gas. To measure in vivo H2 production, cells were spun down and resuspended to OD730 = 2.5 in 2 ml of BG11 containing 5 mm glucose, placed in a 15-ml culture tube, sealed with anaerobic septum, sparged with argon for 30 min, and incubated at 30 °C in the dark for 24 h. A 100-μl sample from the 13-ml headspace was analyzed using a gas chromatograph as above.
Determination of Doubling Times
Log phase precultures (10 ml) of WT and hox− strains were used to inoculate 400-ml capacity Photobioreactor culture vessels (FMT 400, Photon System Instruments) without the addition of antibiotic to growth medium. Illumination parameters were programmed as listed in Table 1. Optical density was measured every 1–5 min at wavelength maximum 680 nm and recorded digitally, along with pH and O2 concentration (not shown). Culture vessels were maintained at 30 °C and continuously bubbled at a rate of ∼250 ml/min in air or argon supplemented with 2% CO2. Optical density values versus time were fitted to a single exponential function to determine doubling times.
TABLE 1.
Doubling times of WT and hox− cultures
| Growth condition | Doubling time |
|
|---|---|---|
| WT | hox− | |
| h | ||
| 24 h light, 65 μE m−2 s−1 | ||
| Aerobic: Bubbled with 2% CO2 in air | 19.5 ± 1.2 | 21.0 ± 1.2 |
| 3 min dark; 3 min light, 1000 μE m−2 s−1 | ||
| Aerobic: Bubbled with 2% CO2 in air | 8.9 ± 0.8 | 10.7 ± 1.3 |
| Microoxic: Bubbled with 2% CO2 in argon | 11.5 ± 1.3 | 11.7 ± 2.0 |
| 15 min dark; 3 min light, 1200 μE m−2 s−1 | ||
| Anaerobic: Bubbled with 2% CO2 in argon | 87 ± 8 | 79 ± 8 |
Fluorescence Measurements
Dark-adapted quantum efficiency (Fv/Fm) and effective quantum efficiency under illumination (ΔF/Fm′) of WT and hox− strain suspensions from log phase cultures were measured in multiwell plates with a closed FluorCam (FC 800-C/1010, Photon System Instruments) and used to calculate effective electron transport rates. ΔF/Fm′ was determined for cells under continuous illumination for at least 3 min at each light intensity with dark adaption between experiments. Cells were dark-adapted for at least 5 min prior to measurement and were measured in biological and technical triplicate (minimum number of samples per type = 9).
RESULTS
Composition of Hox Hydrogenase Protein Complexes
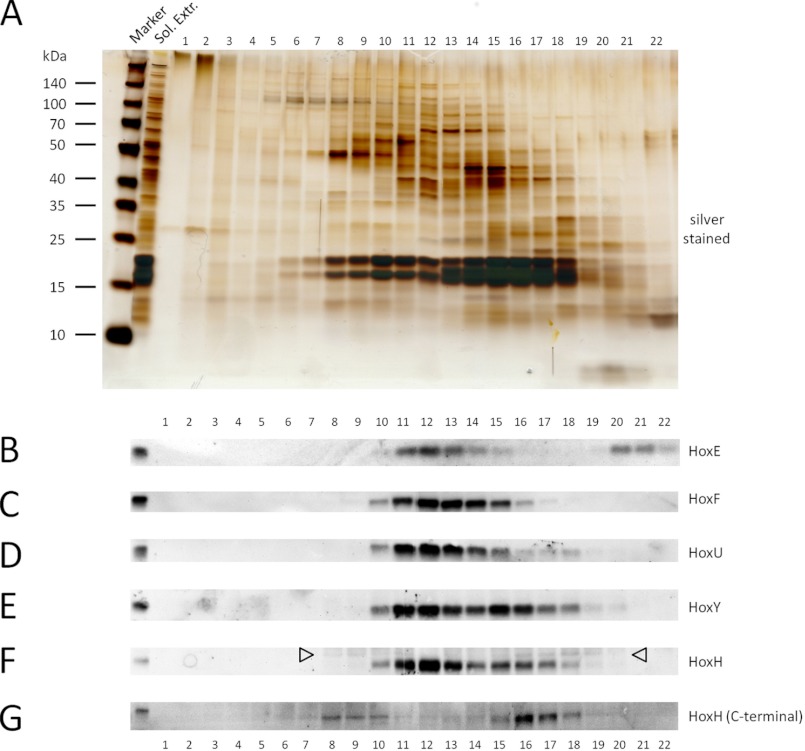
Reported subunit composition and stoichiometry of purified Hox hydrogenase varies in the literature (12, 16–20), raising questions about its in vivo structure. Therefore, we assessed fractionation of the Hox hydrogenase complex by performing size exclusion FPLC analysis of Synechocystis WT soluble extracts followed by Western blotting with Hox subunit-specific antibodies (Fig. 1). As expected, all five subunits, HoxE (18.8 kDa), HoxF (57.8 kDa), HoxU (26.2 kDa), HoxY (20.0 kDa), and HoxH (52.9 kDa), clearly co-fractionate (fractions 10–14). The band intensities of the diaphorase (HoxEFU) and hydrogenase (HoxYH) subunits fluctuate in a related manner, consistent with HoxYH and HoxEFU subcomplexes observed in purifications of other Hox hydrogenases (12, 16, 17). The presence of what is probably monomeric HoxE (fractions 20–22) suggests that HoxE may dissociate relatively easily from the complex. Interestingly, unprocessed, immature HoxH co-fractionates with other Hox proteins, including processed HoxH (fractions 10 and 16–18), implying association of unprocessed HoxH with other complex subunits prior to maturation and full complex assembly (Fig. 1, HoxH (open arrows) and HoxH (C-terminal) blots). In addition, unprocessed HoxH is clearly present in higher molecular weight complexes (fractions 8 and 9), revealing its association with other unknown proteins (possibly maturation factors) prior to its cleavage.
FIGURE 1.
FPLC and Western blot analysis of WT Synechocystis sp. PCC 6803 Hox hydrogenase. The crude soluble fraction of WT Synechocystis sp. PCC 6803 was separated by FPLC, and the resulting fractions were analyzed by Western blotting. A, silver-stained TGX Any kDa gradient SDS-polyacrylamide gel (Bio-Rad) of starting material (Sol. Extr.) and fractions 1–22 (for the chromatogram at A280 and A420 see supplemental Fig. 1). B–G, Western blotting of FPLC fractions with the indicated Hox subunit-specific antibodies. Arrowheads in F indicate the signal for unprocessed HoxH.
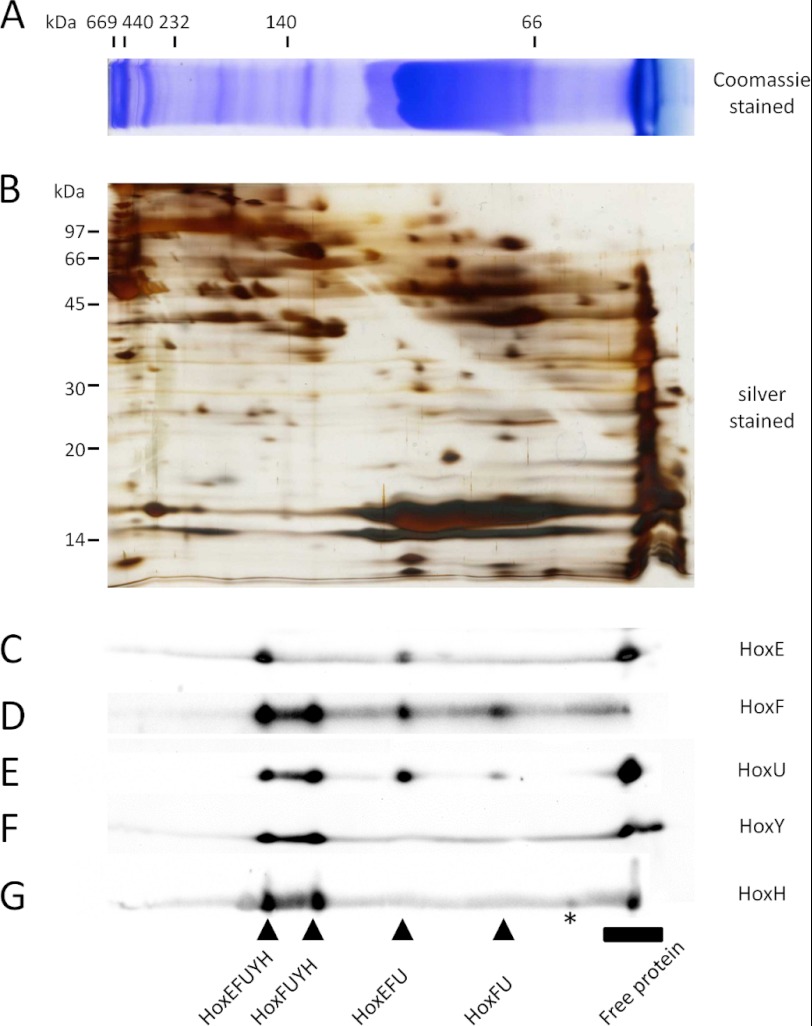
Results from size exclusion FPLC imply the presence of Hox complexes and possible subcomplexes, but co-fractionation does not provide enough evidence to confirm subunit association. To verify Hox protein complex/subcomplex associations in soluble extracts, we performed two-dimensional BN/SDS-PAGE followed by Western blotting (Fig. 2). Using this approach, we were able to identify a complex containing all five Hox subunits, again consistent with the composition of the purified pentameric complex reported in the literature (19, 20). The apparent molecular mass of this complex is ∼160 kDa, close to the calculated mass based on the predicted sizes of the individual subunits (175 kDa). We note that no dimer of this pentameric hydrogenase protein complex could be reliably detected in our analysis despite previous reports (19). However, we were able to detect the presence of HoxFUYH, HoxEFU, and HoxFU subcomplexes. We also failed to reliably detect a HoxYH subcomplex, yet there was a weak antibody signal for the HoxH subunit with a higher apparent molecular mass than that of the free HoxH protein alone (Fig. 2G, asterisk), suggesting the association of HoxH with another protein. Monomeric subunits were also detected in the low molecular mass region of the Western blots, with the HoxE, HoxH, and HoxU subunits all accumulating to significant levels when compared with levels of each associated with subcomplexes. We were unable to consistently detect unprocessed HoxH in any of these subcomplexes, suggesting that its possible association may only be transient. Nevertheless, these data reveal for the first time the presence of various subcomplexes of Synechocystis Hox hydrogenase in vivo.
FIGURE 2.
Two-dimensional BN/SDS-PAGE and Western blot analysis of WT Synechocystis sp. PCC 6803 Hox hydrogenase. A, the soluble fraction of WT Synechocystis sp. PCC 6803 was separated on a 12% (w/v) polyacrylamide BN-polyacrylamide gel followed by two-dimensional separation on a 17.5% (w/v) polyacrylamide SDS-polyacrylamide gel (B) and analysis by Western blotting with the indicated Hox subunit-specific antibodies (C–G). The first dimension BN-polyacrylamide gel was Coomassie-stained, whereas the second dimension SDS-polyacrylamide gel was silver-stained.
Subunit Abundance in Hox Hydrogenase Mutants
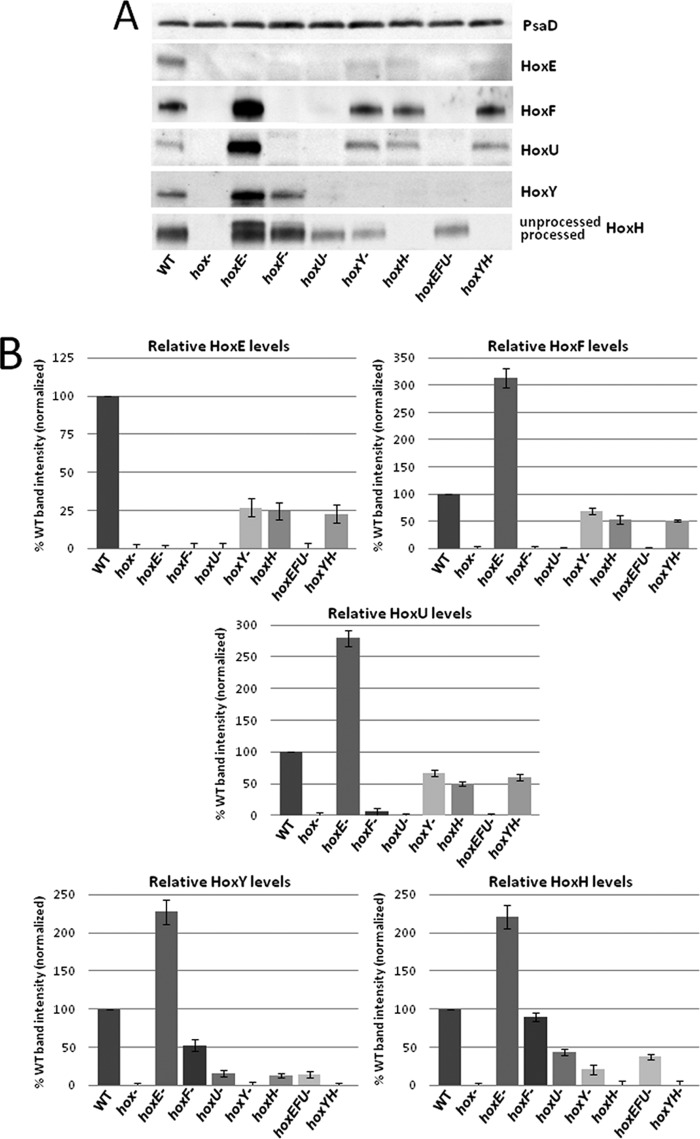
The detection of Hox subcomplexes in the WT strain by FPLC and two-dimensional BN/SDS-PAGE analyses prompted us to construct mutants of each hox gene alone and in combination in the same parental WT strain (supplemental Fig. 2) to further examine any relationship between complex/subcomplex associations and relative subunit abundance by Western blotting. All of the mutants except for hox− (whole operon deletion constructed by T. Ogawa) and hoxH− (see “Experimental Procedures” and supplemental Fig. 2) were created by ORF replacement with similar size antibiotic resistance genes to avoid changes in operon transcription from additional promoters in antibiotic resistance cassettes or large alterations in sequence size. As expected, no detectable Western signal for Hox subunits was apparent when the respective gene was disrupted, confirming full segregation of gene deletions (cyanobacteria carry multiple genome copies) (Fig. 3A). Interestingly, deletion of hoxE, the first gene in the operon, led to a 2–3-fold increase in the remaining Hox protein subunits (Fig. 3B). The increase in Hox protein levels in this mutant is probably linked to the first 190 bp of the hoxE ORF, whose absence results in increased hox transcript levels and possible enhancement in the rate of translation due to a decrease in minimum free energy in the hoxE− mutant mRNA (see “Discussion” and supplemental Fig. 3, A–D). The overexpression of HoxH in the hoxE− mutant leads to a discernible increase in levels of unprocessed HoxH compared with WT and other hox mutants (Fig. 3A and supplemental Fig. 4), consistent with increased unprocessed large subunit when the hydrogenase is overexpressed without additional maturation factors in previous studies (our unpublished results3 for R. eutropha soluble hydrogenase and Refs. 13 and 20). All other hox mutants exhibited decreased levels of remaining complex subunits (Fig. 3B). Levels of HoxH and HoxY are decreased to 20 and 10% of WT in the respective hoxY− and hoxH− mutants, whereas HoxE, HoxF, and HoxU levels are less than 5% of WT in hoxF− and hoxU− mutants, highlighting interdependencies for HoxYH and HoxEFU subcomplex protein abundance. In addition, HoxY and HoxH levels are decreased to 15–90% of WT in HoxEFU subcomplex mutants (most notably in hoxU− mutants), and HoxE, HoxF, and HoxU levels are decreased to 25–70% of WT in HoxYH subcomplex mutants, revealing interdependencies in subunit abundance across subcomplexes as well.
FIGURE 3.
One-dimensional SDS-PAGE and Western blot analysis of Synechocystis sp. PCC 6803 hox mutants. A, whole cell lysates of WT and individual/combined hox mutants were run on TGX Any kDa gradient SDS-polyacrylamide gels (Bio-Rad), transferred to PVDF, and immunoblotted with Hox subunit-specific antibodies and PsaD and/or Rps1 (not shown) as loading controls. B, relative levels of Hox subunits in each mutant, presented as a percentage of WT. Data represent quantitation of multiple Western blot analyses with error bars depicting variation between separate analyses.
Hox Subunit Association in Individual Subunit hox Mutants
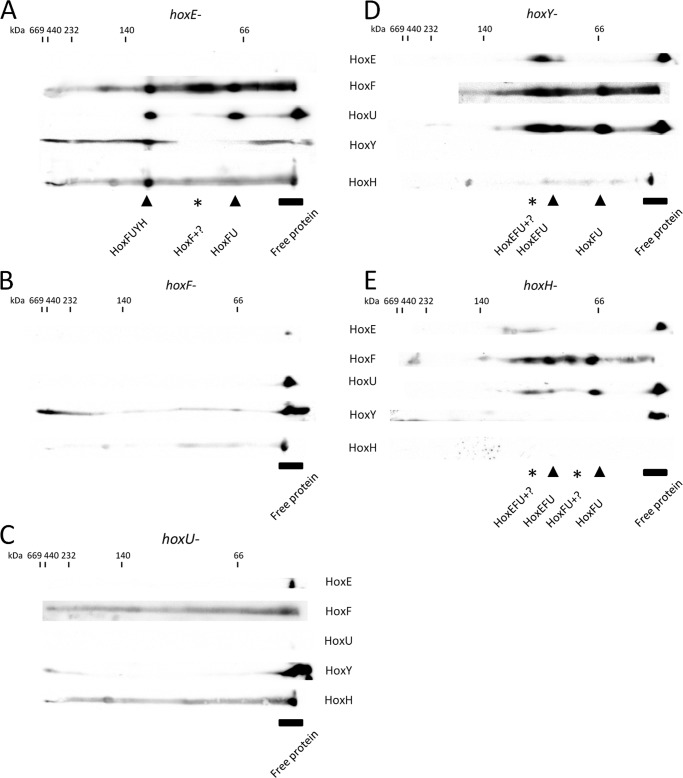
Although changes in transcription could be responsible for altered subunit abundance observed in hox mutants, the absence of complex subunits could also lead to instability of the remaining subunits because of an inability to form stable Hox complexes/subcomplexes. To assess the association of the remaining Hox subunits when one of the five subunits is absent, we again performed two-dimensional BN/SDS-PAGE and Western blotting of soluble extracts from single-subunit hox mutants (Fig. 4). No notable differences in total protein were observed between the WT and hox mutant strains by Coomassie-stained one-dimensional BN or silver-stained two-dimensional SDS-polyacrylamide gels (data not shown). In the hoxE− strain, HoxFUYH and HoxFU subcomplexes accumulated (Fig. 4A), consistent with the observation of those subcomplexes in WT (Fig. 2). Despite the increased levels of HoxY and HoxH in our hoxE− strain (Fig. 3A), we still were not able to observe the expected HoxYH subcomplex (Fig. 4A). Interestingly, none of the Hox subcomplexes are detected in hoxF− or hoxU− strains, with the remaining Hox proteins observed only as unassembled monomers, indicating that HoxF and HoxU are necessary for stable assembly/maintenance of the full Hox complex and Hox subcomplexes (Fig. 4, B and C). In contrast, the diaphorase subunits are still able to assemble into HoxEFU and HoxFU subcomplexes in hoxY− or hoxH− strains (Fig. 4, D and E), confirming that HoxY and HoxH are not required for the stable association of diaphorase subcomplexes.
FIGURE 4.
Two-dimensional-BN/SDS-PAGE and Western blot analysis of Synechocystis sp. PCC 6803 hox mutants. Shown are two-dimensional immunoblots (as described in the legend to Fig. 2) for hoxE− (A), hoxF− (B), hoxU− (C), hoxY− (D), and hoxH− (E) deletion mutant strains probed with the indicated Hox subunit-specific antibodies.
Pull-down Analysis of Hox Subunit Associations in Hox Hydrogenase Subcomplexes
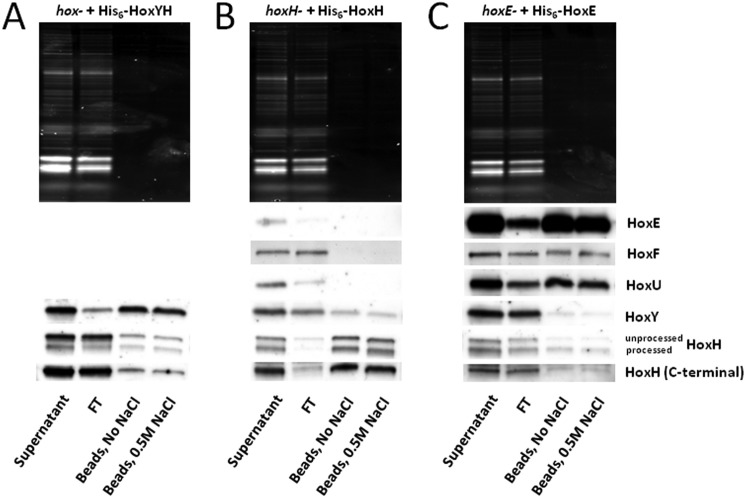
Additional mutant strains were constructed to ectopically express His6-tagged Hox subunits to further probe subcomplex associations: 1) HoxYH, tagged on HoxY (hox− + His6-HoxYH); 2) HoxEFUYH, tagged on HoxH (hoxH− + His6-HoxH); and 3) HoxEFUYH, tagged on HoxE (hoxE− + His6-HoxE). Overexpression of HoxH by ectopic expression (Fig. 5, A and B) or in the hoxE− strain background (Fig. 5C) results in clear unprocessed and processed HoxH signal (Fig. 5, A–C, upper and lower bands, respectively, in HoxH blots and in the HoxH (C-terminal) blot), increasing the likelihood that we would see unprocessed HoxH in bead fractions if it can indeed bind complex subunits, as suggested in our FPLC analysis (Fig. 1, F and G). After ensuring that the metal-containing active site of HoxH could not bind to Co2+ beads nonspecifically (data not shown; see “Experimental Procedures”), we performed pull-down assays of soluble lysates from each of these strains to verify subunit association on beads following washes with and without 0.5 m NaCl (Fig. 5). When His6-HoxY and HoxH are co-expressed, HoxH remains bound to HoxY under all conditions, as shown by Western blotting of the bead fraction (Fig. 5A), confirming their ability to form a tightly associated HoxYH subcomplex. When a His6-tagged HoxH is used to pull down the Hox complex, only HoxY and HoxH are detectably bound to beads, regardless of whether or not salt was included in the wash (Fig. 5B), validating the presence of a HoxYH subcomplex in soluble extracts. Alternatively, when a His6-tagged HoxE is used to pull down the Hox complex, the diaphorase subunits HoxF and HoxU are preferentially associated with the tagged HoxE, whereas only low levels of HoxY and HoxH are present in the bead fraction (Fig. 5C). Interestingly, we observed both processed and unprocessed HoxH associated with HoxY (Fig. 5A) as well as full complex (Fig. 5C). Overall, these data suggest that 1) HoxY and -H bind to each other more strongly than to the diaphorase components, 2) the diaphorase components bind to each other more strongly than to either HoxY or HoxH, and 3) unprocessed HoxH is able to bind HoxY and associate with the full complex.
FIGURE 5.
His6 pull-down of Synechocystis sp. PCC 6803 Hox subcomplexes. hox− mutant expressing His6-HoxYH (A), hoxH− mutant expressing His6-HoxH (B), and hoxE− mutant expressing His6-HoxE (C) were used for pull-down on Co2+ beads to determine subunit association. Supernatant, flow-through (FT) following bead incubation, pooled washes with/without 0.5 m NaCl, and boiled bead samples from each pull-down were run on TGX Any kDa gradient SDS-polyacrylamide gels (Bio-Rad), transferred to PVDF, and immunoblotted using Hox subunit-specific antibodies.
Physiological Function of Hox Hydrogenase
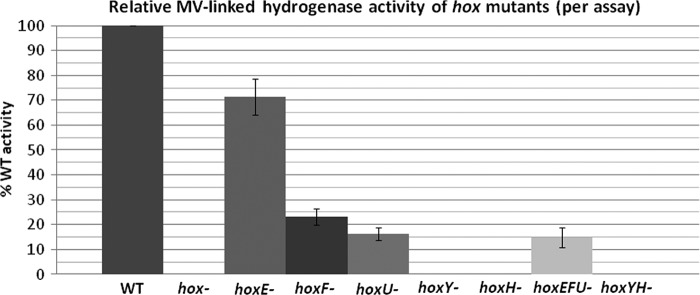
Various phenotypic and activity studies have been reported for hoxH− (25, 26, 39), hoxYH− (40), hoxE− (39), hoxU− (Anacystis nidulans) (14), and hoxEF− (15, 39) mutants as well as purified complex (19) in cyanobacteria. These studies are difficult to compare because of differences in experimental conditions and/or strain backgrounds. In this study, all of the individual and combined hox mutants we created are in the same species and strain background, providing a platform for detailed physiological comparison of hydrogenase activity and growth. Hydrogenase activities of the various hox mutants were similar to previously published data (14, 15, 25, 26, 39, 40) (Fig. 6 and supplemental Table 1), confirming that the full complex is necessary for in vivo activity and linkage to NAD(P)H and that HoxYH is the minimal hydrogenase with MV as the electron donor. The relative MV-linked activity in diaphorase subunit mutants is consistent with increased levels of unprocessed HoxH (hoxE−; Fig. 3 and supplemental Fig. 4) or decreased levels of HoxY and HoxH proteins and/or the lack of detectable HoxYH complex by two-dimensional BN/SDS-PAGE Western blot (hoxF− and hoxU−; Figs. 3 and 4).
FIGURE 6.
Relative hydrogenase activity of Synechocystis sp. PCC 6803 hox mutants. Whole cells of Synechocystis sp. PCC 6803 WT and hox− mutant strains were analyzed for hydrogenase activity with the exogenous electron donor MV. The graph shows MV-linked activities of hox mutants relative to WT with assay variation depicted by error bars. Rates (nmol of H2 ml−1 min−1) are presented in supplemental Table 1.
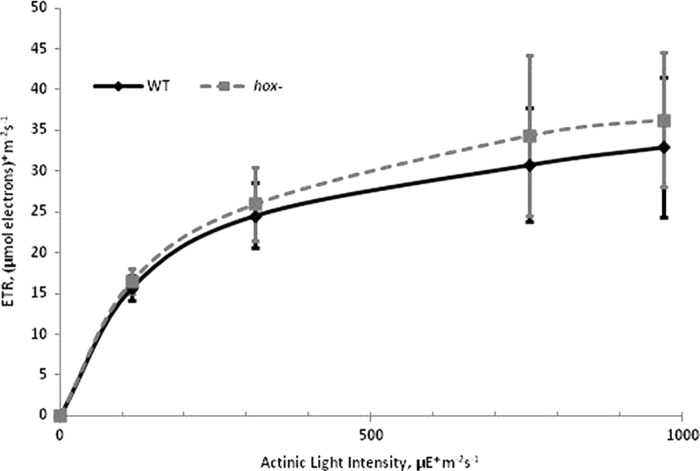
Previous research on a hoxH− mutant in Synechocystis suggested a slight growth defect when compared with WT cells (25), yet no growth difference was detected in a separate report of a hoxYH− mutant (40). This inconsistency prompted us to examine WT and hox− strains of Synechocystis under different illumination regimes in an attempt to compare growth under conditions of continuous or quickly cycled (on/off) illumination (Table 1). We hypothesized that quick transitions from darkness to bright illumination may require the Hox hydrogenase to function as an electron valve, a role suggested by previous studies (25, 26), to maintain normal growth rates (25). However, we did not observe a significant difference in doubling times for cultures under either continuous illumination or light/dark cycling conditions with varying durations of dark period (Table 1), consistent with results for similar continuous and light/dark conditions for the hoxYH− mutant (40). Likewise, we observed no difference in fluorescence-based electron transport rate (ETR) measurements under long term illumination (on a scale of several min) at light intensities between 100 and 1000 μE m−2 s−1 for WT versus hox− cultures (Fig. 7). We further quantified dark-adapted variable fluorescence (Fv/Fm) for cells grown under both lower light (65 μE m−2 s−1) and high light (700 μE m−2 s−1) and observed no significant difference between WT and hox− strains under either light regime (Table 2). This is contrary to the findings reported by Antal et al. (15), in which light sensitivity and a 20–30% lower Fv/Fm was measured in a hoxEF− mutant of Synechocystis sp. PCC 6803. In addition, the reported light sensitivity for this hoxEF− mutation was no longer apparent when transferred to our strain background (supplemental Fig. 5).
FIGURE 7.
Fluorescence-based ETR measurements for Synechocystis sp. PCC 6803 WT and hox− strains grown under 50 μE m−2 s−1 continuous illumination. Variable fluorescence parameters of dilute whole cell suspensions were collected after 3 min of actinic light adaptation at varying intensities. ETR = ΔF/Fm′ × (actinic light intensity) × 0.42. Error bars represent standard deviations of three biological replicates.
TABLE 2.
Dark-adapted quantum efficiency (Fv/Fm) of WT and hox− cultures grown under continuous illumination
| Illumination during growth | WT | hox− |
|---|---|---|
| μE m−2 s−1 | ||
| 65 | 0.49 ± 0.02 | 0.50 ± 0.02 |
| 700 | 0.54 ± 0.01 | 0.55 ± 0.01 |
DISCUSSION
Previous studies of the Hox hydrogenase are somewhat variable and fragmented, focused on only one or two subunit mutants (sometimes in different WT strain backgrounds), leaving open questions as to its composition, relative association, and physiological function(s) in Synechocystis. Herein, we shed light on these questions by the analysis of Hox subunit mutants, alone or in combination, generated in a common WT strain to ensure that phenotypes observed are solely due to the hox mutation and not differences in strain background. Using these strains, we provide comprehensive data on Hox subunit associations, enzyme activities in the absence of individual Hox subunits or subcomplexes, and effects of hox mutations on growth and photosynthesis, providing a sound platform to uncover the assembly and physiological role of Hox hydrogenase in future studies.
Hox Complex/Subcomplex Association in Synechocystis
In this work, we present multiple lines of evidence to support the presence of Hox subcomplexes in Synechocystis. Our pull-down experiment using a His6-tagged HoxE to pull down the full complex resulted in a better recovery of diaphorase proteins (HoxE, HoxF, and HoxU) than hydrogenase proteins (HoxY and HoxH) when comparing protein levels of each subunit on beads versus initial lysate (Fig. 5C). This result is consistent with the 2:1 diaphorase/hydrogenase ratio (with the exception of HoxE; discussed below) of Germer et al. (20) when HoxF was tagged for complex purification. Our FPLC and two-dimensional BN/SDS-PAGE analyses of WT soluble extracts (Figs. 1 and 2) uncovered the presence of HoxEFUYH, HoxFUYH, HoxEFU, and HoxFU complexes, but the HoxYH subcomplex was only detectable in the pull-down experiments (Fig. 5). Our inability to clearly see HoxYH subcomplex association by FPLC and two-dimensional BN methods (Figs. 3B and 4 (B and C)) can possibly be attributed to its instability in the absence of the diaphorase subunits. MV-linked hydrogenase activity measurements of purified His6-HoxYH samples decreased considerably during the course of purification (data not shown), consistent with the instability and loss of activity of purified HoxYH subcomplexes in R. eutropha (13).
Hox Complex Purification and Implications for Localization
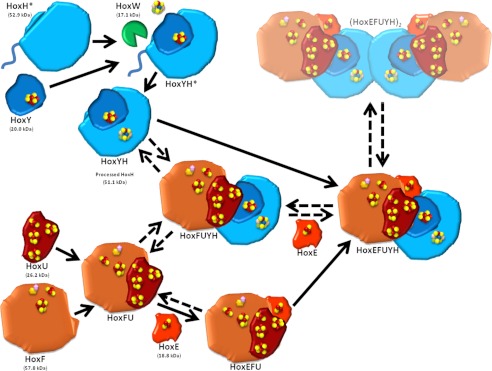
Data from FPLC and two-dimensional BN/SDS-PAGE analyses suggest that HoxE may dissociate relatively easily from Hox complexes and subcomplexes, as indicated by the large proportion of this protein present in monomeric form and the presence of HoxFUYH and HoxFU subcomplexes (Figs. 1 and 2). More HoxE protein is detected in lysed whole cell versus soluble protein preparations, and it is the only Hox protein detectable in pellet preparations (data not shown), consistent with the decreased ratio of HoxE (0.2) associated with the purified Hox hydrogenase complex reported by Germer et al. (20). Despite the clear localization of this complex in soluble fractions in R. eutropha (6), cyanobacterial Hox hydrogenase has been localized, at least in part, to the membrane (42–44), and others have suggested a role for HoxE in anchoring the complex to the membrane in Synechocystis (19). We postulate that this membrane association may be required for the association of the complex as a dimer of a pentamer reported by Schmitz et al. when purifications were made from an initial 30–50% ammonium sulfate precipitation of whole cell lysates (19). In contrast, Germer et al. (20) spun down lysates at high speed prior to a 20% ammonium sulfate addition, centrifugation, and passage of the supernatant through a Streptactin column, whereas we utilized ultracentrifugation of lysates, both of which would probably result in a loss of any complex associated with the membrane (monomer or dimer of Hox complex) and decreased abundance of HoxE. Collectively, these findings suggest that the various subforms of the Hox hydrogenase may become dissociated upon cell breakage and sample preparation or may exist transiently during complex formation, as depicted in our proposed model of assembly (Fig. 8).
FIGURE 8.
Model for Hox hydrogenase association in Synechocystis sp. PCC 6803. Unprocessed HoxH (HoxH*; containing an additional 16-amino acid C terminus) binds HoxY (containing a [4F-4S] cluster) and undergoes assembly and insertion of its [NiFe] active site and cleavage of its C terminus by the protease HoxW, resulting in a hydrogenase subcomplex (HoxYH). Alternatively, HoxF (containing NAD and FMN binding sites and [2Fe-2S] and [4Fe-4S] clusters) associates with HoxU (containing one [2Fe-2S] and three [4Fe-4S] clusters) and HoxE (containing a [2Fe-2S] cluster), forming the diaphorase subcomplex (HoxEFU). The presence and formation of HoxFU and HoxFUYH subcomplexes may represent steps in assembly, but may also formally be due to a loss of HoxE during preparation/extraction strategies. Purification by Schmitz et al. resulted in detection of a dimer of the HoxEFUYH pentamer ((HoxEFUYH)2), which is depicted here despite our inability to detect this dimer in our preparations. We hypothesize that this dimerization may be through HoxH, allowing association of unprocessed HoxH with the full complex.
Hox Complex Association and Implications for Maturation and Assembly
The maturation and assembly of Hox hydrogenase requires six Hyp proteins (HypABCDEF) for biosynthesis of the hydrogenase active site (2, 13, 45–48) and an endopeptidase (HoxW) that cleaves the C terminus of HoxH, a step essential for proper folding and final assembly of the active site (49). Unprocessed HoxH band intensities become more pronounced in our hoxE− mutant and its complement, a phenomenon that is apparently due to increased transcription and/or translation of the hox operon, resulting in increased levels of HoxH (Figs. 3 and 5C and supplemental Figs. 3 and 4; discussed below). Processing of these increased levels of HoxH in the hoxE− strain is probably limited by the availability of maturation factors, previously addressed by Germer et al. (20) by co-overexpression of the Synechocystis hox operon using the psbAII promoter and ectopic expression of additional maturation factors from Nostoc sp. 7120. We were able to detect the co-elution (Fig. 1) and association (Fig. 5) of unprocessed HoxH with other Hox proteins. We were unable to detect this association by two-dimensional BN/SDS-PAGE due to the inability to clearly separate processed and unprocessed HoxH signals and the presence of nonspecific background signals with the α-HoxH (C-terminal) antibody not apparent in one-dimensional analyses (data not shown). Nevertheless, we were able to detect unprocessed HoxH in the purified HoxYH subcomplex as well as in the full complex using pull-down assays (Fig. 5). The E. coli model for [NiFe]-hydrogenase assembly assumes the production of a holo-catalytic subunit prior to its association with the electron-relaying small subunit based on the lack of detection of unprocessed large subunit with the small subunit in purifications (2, 3). In contrast, our results suggest that HoxY can associate with an unprocessed HoxH prior to its maturation (Fig. 5A). The association of the unprocessed form of HoxH with the full complex is also clearly visible in pull-down experiments (Fig. 5C). We hypothesize that HoxY (and possibly other Hox subunits) may play a role in efficient maturation and processing of HoxH. Alternatively, it is also feasible that HoxH may dimerize, resulting in the association of unprocessed HoxH with full complex or HoxYH subcomplexes already containing processed HoxH (Fig. 8). The events leading to the assembly and maturation of the Hox hydrogenase are largely unknown, and the results here reveal a novel, yet to be identified mechanism of assembly that warrants further studies.
Hox Subunit Abundance and Complex/Subcomplex Assembly
Analysis of hox deletion strains provides insight into the relative abundance and association of the remaining Hox subunits when other subunits are absent. Various studies have demonstrated that the hox operon is transcribed as a single transcript (50–54). Our results confirm expression of all five proteins (Figs. 2 and 3) and suggest a relative balance between the expression of Hox subunits and levels of maturation factors because processing of HoxH and hydrogenase activity are affected negatively by Hox protein overexpression (hoxE− or ectopic expression of HoxH; Figs. 3, 5, and 6). Decreased Hox subunit stability is reported in the absence of other hydrogenase subunits in R. eutropha (13). In agreement, we observe a decrease in relative abundance of subunits of the hydrogenase (HoxYH) and diaphorase (HoxEFU) subcomplexes in Hox subunit deletion strains, with the exception of HoxE (not present in R. eutropha), whose absence actually results in an increase in other Hox protein levels (Fig. 3, A and B; discussed below). Most notably, the relative abundance of remaining Hox subunits (Fig. 3) and resultant hydrogenase activity with MV (Fig. 6) are greatly diminished in strains lacking HoxU. Moreover, despite higher relative protein abundance for remaining Hox subunits in hoxE− strains (Fig. 3), detectable subcomplexes are no longer observed in strains lacking either HoxF or HoxU (Fig. 4, B and C), suggesting that HoxF and HoxU are essential for complex and subcomplex assembly. These observations could explain why we do not observe detectable amounts of the HoxYH subcomplex in our two-dimensional BN/SDS-PAGE analyses (Figs. 1 and 4). Although the loss of HoxY or HoxH negatively affects the abundance of diaphorase subunits (Fig. 3), there remains a clear presence of the subforms HoxEFU/FU in hoxY− and hoxH− strains (Fig. 4, D and E), suggesting that the assembled diaphorase subcomplex may be relatively more stable than that of HoxYH and less dependent on the HoxYH subcomplex for assembly and stability.
Interestingly, although levels of HoxE are nearly undetectable in hoxF− and hoxU− mutant strains and are greatly reduced in strains devoid of HoxY and HoxH, a loss of HoxE does not result in any discernible instability of other complex components (Fig. 3) or a loss in the ability to form HoxFUYH and HoxFU subcomplexes (Fig. 4A). Instead, we observe at least a 2-fold increase in the levels of the remaining Hox proteins in the hoxE− strain when compared with levels present in WT (Fig. 3). Mutations of hoxE generated by Howitt and Vermaas (30) (supplemental Fig. 3B) and Aubert-Jousett et al. (39) did not exhibit increased Hox protein levels. Whereas our construct was made by ORF replacement of hoxE (522-bp) with a gentamicin resistance gene of nearly identical size (aac3ia; 534 bp), these previously published mutants were constructed using restriction digestion and insertion of a chloramphenicol resistance cassette, leaving 190 and 183 bp of the N-terminal nucleotide sequence unaffected, respectively (30, 39) (supplemental Fig. 3A). The increased Hox protein levels related to removal of this sequence in our hoxE− mutant could be a result of increased rates of transcription and/or translation of the remaining genes in the hox operon. Quantitative RT-PCR analysis of WT, hoxE−, and hoxEF− (mutation from Ref. 30 in our strain background) reveal 11- and 3-fold increases in relative 3′ hoxF and hoxH target sequences, respectively (supplemental Fig. 3C). There is precedence for higher levels of hoxE and hoxF transcript relative to that of downstream hoxU, hox Y, and hoxH transcripts (53, 55), which could possibly result from the presence of an antisense RNA just upstream of hoxU (56). Because full but not partial deletion of hoxE results in increased hox transcript levels, there may be some regulatory region in the first 190 nucleotides of hoxE that affects transcript stability and/or rate of transcription. In addition, this sequence could also affect rates of translation. Indeed, the online tool RNAfold (57) predicts a decrease in minimal free energy folding for the operon mRNA with aac3ia versus hoxE when sequences are input and analyzed (supplemental Fig. 3D).
Synechocystis Hox Hydrogenase Function
Previous activity measurements of purified Hox complexes and various hox mutants in Synechocystis and other cyanobacteria demonstrate activity linked to NAD(P)H, in vivo and in vitro, only with full complex, with the minimal functional hydrogenase, HoxYH, exhibiting activity only in the presence of reduced MV (14, 15, 39, 40). Relative hydrogenase activities of the hox mutants we constructed correlate well with calculated relative HoxH (processed) and HoxY protein present in each strain (Figs. 3 and 6 and supplemental Fig. 4). In addition, the substantially decreased MV-linked activity in mutants lacking HoxF and/or HoxU highlights their role in the association of the hydrogenase subcomplex as well as the full complex (Figs. 4 and 6).
In a study by Antal et al. (15), the hoxEF− mutant constructed by Howitt and Vermaas (30) exhibited both light sensitivity and photosynthetic defects when compared with their WT strain, which they interpreted as photosystem II (PSII) photoinhibition due to a lack of hydrogenase activity. We acquired the same hoxEF− strain and observed similar light sensitivity. However, when we generated the same hoxEF− mutation in our WT strain background, we no longer observed any light sensitivity when compared with the parental strain (supplemental Fig. 5), suggesting that the defects observed in the published study were not a result of the hoxEF− mutation. Appel et al. (25) also showed a slight growth defect and a difference in PSII/PSI fluorescence kinetics upon illumination under saturating red light in dark-adapted WT and hoxH− samples. In this study, we compared a hox− strain derived from the same WT for comparison and observed no defects in growth or light-adapted photosynthesis under many different conditions (Fig. 7 and Table 1), although we did observe a reproducible difference in PSII fluorescence kinetics under saturated red actinic light similar to that reported by Appel et al. (25) (data not shown). It is important to note that whereas growth and ETR measurements were taken from time scales on the order of minutes, the PSII fluorescence kinetic measurement happens on the order of seconds. Taken together, our data do not support the hypothesis that the Hox hydrogenase plays a role in photosynthesis or in protection against PSII photoinhibition over extended time scales (on the order of minutes to days). Nevertheless, we cannot exclude the possibility that Hox hydrogenase might play a role as an electron valve during photosynthesis under very transient (on the order of seconds) changes in light intensities that occur in nature. In addition, a lack of phenotype in the hox− mutant under many different growth regimes (including fermentation; data not shown) rules out a role for the diaphorase as the sole electron donor to the plastoquinone pool in respiration despite homology to missing Complex I components in cyanobacteria, consistent with previous studies (14, 30, 41).
Conclusions
This extensive study of the Hox hydrogenase in Synechocystis serves to clear up inconsistencies in the literature and reveals new data for subunit and subcomplex interactions that provide a basis for future study of its maturation, assembly, and function. The presence of various complex subforms may represent transient assembly states, suggesting a more complex assembly and maturation scheme than that determined for E. coli [NiFe]-hydrogenases. Our data also imply that this assembly may involve association of the unprocessed, immature hydrogenase large subunit with other subunits of the complex prior to full maturation, a previously uncharacterized phenomenon. Analysis of the various Hox subunit and subcomplex mutants in the same strain background confirms previously reported hydrogenase activity results and reveals a correlation between relative levels of HoxY and processed HoxH and activity. A strain containing a deletion of the entire hox operon is devoid of any outward phenotypes beyond a lack of hydrogenase activity, demonstrating that its hypothesized role as an electron valve may only be very transient and under specific growth conditions.
Acknowledgments
We thank G. Vanzin for construction of hoxH−; T. Ogawa for WT and hox− strains; W. Vermaas (Arizona State University) and R. Burnap (Oklahoma State University) for pPETE and pPSBA2KS plasmids, respectively; J. Golbeck (Penn State) for the PsaD antibody; and D. Vinyard (Princeton University) for assistance with PSII fluorescence data interpretation.
This work was supported by the National Renewable Energy Laboratory's Laboratory Directed Research and Development Program (to P. M., J. Y., C. E., and D. C.), the United States Department of Energy Fuel Cell Technologies Program (contract number DE-AC36-08-GO28308) (to P. M. and J. Y.), the United States Department of Energy Biological and Environmental Research Program (contract KP160103) (to M. B. and A. D.), and Engineering and Physical Sciences Research Council Grant (EP/F002070X/1) (to M. B. and P. N.).

This article contains supplemental Table 1 and Figs. 1–5.
C. Eckert, J. Yu, and P.-C. Maness, unpublished results.
- μE
- microeinsteins
- MV
- methyl viologen
- BN
- blue native
- PSI and PSII
- photosystem I and II, respectively
- ETR
- electron transport rate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Vignais P. M., Billoud B. (2007) Occurrence, classification, and biological function of hydrogenases. An overview. Chem. Rev. 107, 4206–4272 [DOI] [PubMed] [Google Scholar]
- 2. Forzi L., Sawers R. G. (2007) Maturation of [NiFe]-hydrogenases in Escherichia coli. Biometals 20, 565–578 [DOI] [PubMed] [Google Scholar]
- 3. Magalon A., Böck A. (2000) Dissection of the maturation reactions of the [NiFe] hydrogenase 3 from Escherichia coli taking place after nickel incorporation. FEBS Lett. 473, 254–258 [DOI] [PubMed] [Google Scholar]
- 4. Carrieri D., Wawrousek K., Eckert C., Yu J., Maness P. C. (2011) The role of the bidirectional hydrogenase in cyanobacteria. Bioresour. Technol. 102, 8368–8377 [DOI] [PubMed] [Google Scholar]
- 5. Appel J. (2012) The physiology and functional genomics of cyanobacterial hydrogenases and approaches towards biohydrogen production. in Functional Genomics and Evolution of Photosynthetic Systems (Burnap R., Vermaas W., eds) Illustrated Ed., pp. 357–381, Springer; Dordrecht, The Netherlands [Google Scholar]
- 6. Burgdorf T., Lenz O., Buhrke T., van der Linden E., Jones A. K., Albracht S. P., Friedrich B. (2005) [NiFe]-hydrogenases of Ralstonia eutropha H16. Modular enzymes for oxygen-tolerant biological hydrogen oxidation. J. Mol. Microbiol. Biotechnol. 10, 181–196 [DOI] [PubMed] [Google Scholar]
- 7. Schneider K., Cammack R., Schlegel H. G. (1984) Content and localization of FMN, Fe-S clusters and nickel in the NAD-linked hydrogenase of Nocardia opaca 1b. Eur. J. Biochem. 142, 75–84 [DOI] [PubMed] [Google Scholar]
- 8. Schneider K., Schlegel H. G., Jochim K. (1984) Effect of nickel on activity and subunit composition of purified hydrogenase from Nocardia opaca 1b. Eur. J. Biochem. 138, 533–541 [DOI] [PubMed] [Google Scholar]
- 9. Maróti J., Farkas A., Nagy I. K., Maróti G., Kondorosi E., Rákhely G., Kovács K. L. (2010) A second soluble Hox-type NiFe enzyme completes the hydrogenase set in Thiocapsa roseopersicina BBS. Appl. Environ. Microbiol. 76, 5113–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rákhely G., Kovács A. T., Maróti G., Fodor B. D., Csanádi G., Latinovics D., Kovács K. L. (2004) Cyanobacterial-type, heteropentameric, NAD+-reducing NiFe hydrogenase in the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina. Appl. Environ. Microbiol. 70, 722–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rákhely G., Laurinavichene T. V., Tsygankov A. A., Kovács K. L. (2007) The role of Hox hydrogenase in the H2 metabolism of Thiocapsa roseopersicina. Biochim. Biophys. Acta 1767, 671–676 [DOI] [PubMed] [Google Scholar]
- 12. Long M., Liu J., Chen Z., Bleijlevens B., Roseboom W., Albracht S. P. (2007) Characterization of a HoxEFUYH type of [NiFe] hydrogenase from Allochromatium vinosum and some EPR and IR properties of the hydrogenase module. J. Biol. Inorg. Chem. 12, 62–78 [DOI] [PubMed] [Google Scholar]
- 13. Massanz C., Schmidt S., Friedrich B. (1998) Subforms and in vitro reconstitution of the NAD-reducing hydrogenase of Alcaligenes eutrophus. J. Bacteriol. 180, 1023–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boison G., Schmitz O., Schmitz B., Bothe H. (1998) Unusual gene arrangement of the bidirectional hydrogenase and functional analysis of its diaphorase subunit HoxU in respiration of the unicellular cyanobacterium Anacystis nidulans. Curr. Microbiol. 36, 253–258 [DOI] [PubMed] [Google Scholar]
- 15. Antal T. K., Oliveira P., Lindblad P. (2006) The bidirectional hydrogenase in the cyanobacterium Synechocystis sp. strain PCC 6803. Int. J. Hydrogen Energy 31, 1439–1444 [Google Scholar]
- 16. Burgdorf T., van der Linden E., Bernhard M., Yin Q. Y., Back J. W., Hartog A. F., Muijsers A. O., de Koster C. G., Albracht S. P., Friedrich B. (2005) The soluble NAD+-reducing [NiFe]-hydrogenase from Ralstonia eutropha H16 consists of six subunits and can be specifically activated by NADPH. J. Bacteriol. 187, 3122–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Serebriakova L. T., Sheremet'eva M. E. (2006) Characterization of catalytic properties of hydrogenase isolated from the unicellular cyanobacterium Gloeocapsa alpicola CALU 743. Biochemistry 71, 1370–1376 [DOI] [PubMed] [Google Scholar]
- 18. Palágyi-Mészáros L. S., Maróti J., Latinovics D., Balogh T., Klement E., Medzihradszky K. F., Rákhely G., Kovács K. L. (2009) Electron transfer subunits of the NiFe hydrogenases in Thiocapsa roseopersicina BBS. FEBS J. 276, 164–174 [DOI] [PubMed] [Google Scholar]
- 19. Schmitz O., Boison G., Salzmann H., Bothe H., Schütz K., Wang S. H., Happe T. (2002) HoxE. A subunit specific for the pentameric bidirectional hydrogenase complex (HoxEFUYH) of cyanobacteria. Biochim. Biophys. Acta 1554, 66–74 [DOI] [PubMed] [Google Scholar]
- 20. Germer F., Zebger I., Saggu M., Lendzian F., Schulz R., Appel J. (2009) Overexpression, isolation, and spectroscopic characterization of the bidirectional [NiFe] hydrogenase from Synechocystis sp. PCC 6803. J. Biol. Chem. 284, 36462–36472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McIntosh C. L., Germer F., Schulz R., Appel J., Jones A. K. (2011) The [NiFe]-hydrogenase of the cyanobacterium Synechocystis sp. PCC 6803 works bidirectionally with a bias to H2 production. J. Am. Chem. Soc. 133, 11308–11319 [DOI] [PubMed] [Google Scholar]
- 22. Cournac L., Guedeney G., Peltier G., Vignais P. M. (2004) Sustained photoevolution of molecular hydrogen in a mutant of Synechocystis sp. strain PCC 6803 deficient in the type I NADPH-dehydrogenase complex. J. Bacteriol. 186, 1737–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cournac L., Mus F., Bernard L., Guedeney G., Vignais P., Peltier G. (2002) Limiting steps of hydrogen production in Chlamydomonas reinhardtii and Synechocystis PCC 6803 as analysed by light-induced gas exchange transients. Int. J. Hydrogen Energy 27, 1229–1237 [Google Scholar]
- 24. Schütz K., Happe T., Troshina O., Lindblad P., Leitão E., Oliveira P., Tamagnini P. (2004) Cyanobacterial H-2 production. A comparative analysis. Planta 218, 350–359 [DOI] [PubMed] [Google Scholar]
- 25. Appel J., Phunpruch S., Steinmüller K., Schulz R. (2000) The bidirectional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch. Microbiol. 173, 333–338 [DOI] [PubMed] [Google Scholar]
- 26. Gutthann F., Egert M., Marques A., Appel J. (2007) Inhibition of respiration and nitrate assimilation enhances photohydrogen evolution under low oxygen concentrations in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1767, 161–169 [DOI] [PubMed] [Google Scholar]
- 27. Schmitz O., Boison G., Hilscher R., Hundeshagen B., Zimmer W., Lottspeich F., Bothe H. (1995) Molecular biological analysis of a bidirectional hydrogenase from cyanobacteria. Eur. J. Biochem. 233, 266–276 [DOI] [PubMed] [Google Scholar]
- 28. Appel J., Schulz R. (1996) Sequence analysis of an operon of a NAD(P)-reducing nickel hydrogenase from the cyanobacterium Synechocystis sp. PCC 6803 gives additional evidence for direct coupling of the enzyme to NAD(P)H-dehydrogenase (complex I). Biochim. Biophys. Acta Protein Struct. M 1298, 141–147 [DOI] [PubMed] [Google Scholar]
- 29. Boison G., Bothe H., Hansel A., Lindblad P. (1999) Evidence against a common use of the diaphorase subunits by the bidirectional hydrogenase and by the respiratory complex I in cyanobacteria. FEMS Microbiol. Lett. 174, 159–165 [Google Scholar]
- 30. Howitt C. A., Vermaas W. F. J. (1999) Subunits of the NAD(P)-reducing nickel-containing hydrogenase do not act as part of the type-1 NAD(P)H-dehydrogenase in the cyanobacterium Synechocystis sp. PCC 6803. in The phototrophic prokaryotes (Peschek G. A., Löffelhardt W., Schmetterer G., eds), pp. 595–601, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 31. Ikeuchi M., Tabata S. (2001) Synechocystis sp. PCC 6803. A useful tool in the study of the genetics of cyanobacteria. Photosynth. Res. 70, 73–83 [DOI] [PubMed] [Google Scholar]
- 32. Kanesaki Y., Shiwa Y., Tajima N., Suzuki M., Watanabe S., Sato N., Ikeuchi M., Yoshikawa H. (2012) Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res. 19, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuwayama H., Obara S., Morio T., Katoh M., Urushihara H., Tanaka Y. (2002) PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30, E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kommalapati M., Hwang H. J., Wang H. L., Burnap R. L. (2007) Engineered ectopic expression of the psbA gene encoding the photosystem II D1 protein in Synechocystis sp. PCC6803. Photosynth. Res. 92, 315–325 [DOI] [PubMed] [Google Scholar]
- 35. Lagarde D., Beuf L., Vermaas W. (2000) Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 66, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boehm M., Nield J., Zhang P., Aro E. M., Komenda J., Nixon P. J. (2009) Structural and mutational analysis of band 7 proteins in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 191, 6425–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blum H., Beier H., Gross H. J. (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8, 93–99 [Google Scholar]
- 38. Xu Q., Yu L., Chitnis V. P., Chitnis P. R. (1994) Function and organization of photosystem I in a cyanobacterial mutant strain that lacks PsaF and PsaJ subunits. J. Biol. Chem. 269, 3205–3211 [PubMed] [Google Scholar]
- 39. Aubert-Jousset E., Cano M., Guedeney G., Richaud P., Cournac L. (2011) Role of HoxE subunit in Synechocystis PCC6803 hydrogenase. FEBS J. 278, 4035–4043 [DOI] [PubMed] [Google Scholar]
- 40. Pinto F., van Elburg K. A., Pacheco C. C., Lopo M., Noirel J., Montagud A., Urchueguía J. F., Wright P. C., Tamagnini P. (2012) Construction of a chassis for hydrogen production. Physiological and molecular characterization of a Synechocystis sp. PCC 6803 mutant lacking a functional bidirectional hydrogenase. Microbiology 158, 448–464 [DOI] [PubMed] [Google Scholar]
- 41. Schmitz O., Bothe H. (1996) The diaphorase subunit HoxU of the bidirectional hydrogenase as electron transferring protein in cyanobacterial respiration? Naturwissenschaften 83, 525–527 [DOI] [PubMed] [Google Scholar]
- 42. Serebriakova L., Zorin N. A., Lindblad P. (1994) Reversible hydrogenase in Anabaena variabilis ATCC 29413. Presence and localization in non-N-2-fixing cells. Arch. Microbiol. 161, 140–144 [Google Scholar]
- 43. Kentemich T., Bahnweg M., Mayer F., Bothe H. (1989) Localization of the reversible hydrogenase in cyanobacteria. Z. Naturforsch. 44c, 384–391 [Google Scholar]
- 44. Kentemich T., Casper M., Bothe H. (1991) The reversible hydrogenase in Anacystis nidulans is a component of the cytoplasmic membrane. Naturwissenschaften 78, 559–560 [Google Scholar]
- 45. Lutz S., Jacobi A., Schlensog V., Böhm R., Sawers G., Böck A. (1991) Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol. Microbiol. 5, 123–135 [DOI] [PubMed] [Google Scholar]
- 46. Hoffmann D., Gutekunst K., Klissenbauer M., Schulz-Friedrich R., Appel J. (2006) Mutagenesis of hydrogenase accessory genes of Synechocystis sp. PCC 6803. Additional homologues of hypA and hypB are not active in hydrogenase maturation. FEBS J. 273, 4516–4527 [DOI] [PubMed] [Google Scholar]
- 47. Casalot L., Rousset M. (2001) Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9, 228–237 [DOI] [PubMed] [Google Scholar]
- 48. Jones A. K., Lenz O., Strack A., Buhrke T., Friedrich B. (2004) NiFe hydrogenase active site biosynthesis. Identification of Hyp protein complexes in Ralstonia eutropha. Biochemistry 43, 13467–13477 [DOI] [PubMed] [Google Scholar]
- 49. Wünschiers R., Batur M., Lindblad P. (2003) Presence and expression of hydrogenase specific C-terminal endopeptidases in cyanobacteria. BMC Microbiol. 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gutekunst K., Phunpruch S., Schwarz C., Schuchardt S., Schulz-Friedrich R., Appel J. (2005) LexA regulates the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 as a transcription activator. Mol. Microbiol. 58, 810–823 [DOI] [PubMed] [Google Scholar]
- 51. Oliveira P., Lindblad P. (2005) LexA, a transcription regulator binding in the promoter region of the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 251, 59–66 [DOI] [PubMed] [Google Scholar]
- 52. Oliveira P., Lindblad P. (2008) An AbrB-Like protein regulates the expression of the bidirectional hydrogenase in Synechocystis sp. strain PCC 6803. J. Bacteriol. 190, 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kiss E., Kós P. B., Vass I. (2009) Transcriptional regulation of the bidirectional hydrogenase in the cyanobacterium Synechocystis 6803. J. Biotechnol. 142, 31–37 [DOI] [PubMed] [Google Scholar]
- 54. Oliveira P., Lindblad P. (2009) Transcriptional regulation of the cyanobacterial bidirectional Hox-hydrogenase. Dalton Trans. 9990–9996 [DOI] [PubMed] [Google Scholar]
- 55. Dutheil J., Saenkham P., Sakr S., Leplat C., Ortega-Ramos M., Bottin H., Cournac L., Cassier-Chauvat C., Chauvat F. (2012) The AbrB2 autorepressor, expressed from an atypical promoter, represses the hydrogenase operon to regulate hydrogen production in Synechocystis strain PCC6803. J. Bacteriol. 194, 5423–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mitschke J., Georg J., Scholz I., Sharma C. M., Dienst D., Bantscheff J., Voss B., Steglich C., Wilde A., Vogel J., Hess W. R. (2011) An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 108, 2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gruber A. R., Lorenz R., Bernhart S. H., Neuböck R., Hofacker I. L. (2008) The Vienna RNA Websuite. Nucleic Acids Res. 36, W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]