Background: PheVI:09 (6.44) is highly conserved among 7TM receptors.
Results: In the inactive state, PheVI:09 is locked against the backbone of TM-III but slides pass IleIII:16 (3.40) into a tight hydrophobic pocket between TM-III and TM-V during receptor activation.
Conclusion: Mutational analysis and computational chemistry shows that PheVI:09 functions as a microswitch.
Significance: This work clarifies the molecular mechanism of a crucial microswitch in 7TM receptor activation.
Keywords: Cell Signaling, Computational Biology, G Protein Coupled Receptor (GPCR), Receptor Structure-Function, Signal Transduction
Abstract
In seven-transmembrane (7TM), G protein-coupled receptors, highly conserved residues function as microswitches, which alternate between different conformations and interaction partners in an extended allosteric interface between the transmembrane segments performing the large scale conformational changes upon receptor activation. Computational analysis using x-ray structures of the β2-adrenergic receptor demonstrated that PheVI:09 (6.44), which in the inactive state is locked between the backbone and two hydrophobic residues in transmembrane (TM)-III, upon activation slides ∼2 Å toward TM-V into a tight pocket generated by five hydrophobic residues protruding from TM-III and TM-V. Of these, the residue in position III:16 (3.40) (often an Ile or Val) appears to function as a barrier or gate for the transition between inactive and active conformation. Mutational analysis showed that PheVI:09 is essential for the constitutive and/or agonist-induced signaling of the ghrelin receptor, GPR119, the β2-adrenergic receptor, and the neurokinin-1 receptor. Substitution of the residues constituting the hydrophobic pocket between TM-III and TM-V in the ghrelin receptor in four of five positions impaired receptor signaling. In GPR39, representing the 12% of 7TM receptors lacking an aromatic residue at position VI:09, unchanged agonist-induced signaling was observed upon Ala substitution of LeuVI:09 despite reduced cell surface expression of the mutant receptor. It is concluded that PheVI:09 constitutes an aromatic microswitch that stabilizes the active, outward tilted conformation of TM-VI relative to TM-III by sliding into a tight hydrophobic pocket between TM-III and TM-V and that the hydrophobic residue in position III:16 constitutes a gate for this transition.
Introduction
Seven-transmembrane (7TM)3 receptors or G protein-coupled receptors are activated by a diverse range of stimuli. It is, however, generally believed that, despite the very diverse chemical nature of their ligands, the different members of this very large receptor family share a common molecular activation mechanism (1). The structural rearrangement occurring during receptor activation has been studied by biochemical and biophysical means for many years and has recently been further illuminated by a number of high resolution x-ray structures, which in several cases today include both inactive and active receptor structures of the same receptors (2–9). The major component of the global activation mechanism for 7TM receptors comprises a large scale outward movement of the intracellular pole of TM-VI through which a relatively large pocket opens for binding of the C-terminal end of the Gα subunit of the G protein (4–6, 10–12).
A number of highly conserved residues are believed to function either as so-called microswitches, or they participate in the water hydrogen bond network between the intracellular segments of, in particular, TM-I, -II, -VI, and -VII (13). Microswitch residues are found in substantially different conformations and are involved in distinct interactions in the inactive versus the active state of the receptor. An example of this is the ArgIII:26 (3.50)4 microswitch of the DRY motif in TM-III, which in the inactive conformation is locked through a salt bridge to the neighboring AspIII:25 (3.49). In the active state, this residue rotates away to make a hydrogen bond to a conserved TyrV:24 (5.58) in TM-V and possibly interacts directly with the backbone of the Gα subunit. Other examples of microswitch residues are TyrVII:20 (7.53) of the NPXXY motif in TM-VII and TrpVI:13 (6.48) of the CWXP motif in TM-VI (13). Although these microswitch residues are highly conserved, none of them are totally conserved among all rhodopsin-like 7TM receptors, conceivably because they function in parallel and not in a sequential manner in the overall allosteric activation mechanism (1). In other words, the microswitch residues and the hydrogen bond network function together as an extended allosteric interface between the TM segments that perform the large scale, global conformational changes during the activation process.
It was expected that the microswitch residues would be found in a very similar conformation and with very similar interaction partners in the active state of different 7TM receptors. In this respect, the high resolution x-ray structures have displayed surprisingly different pictures. For example, the most highly conserved microswitch residue, ArgIII:26, is found in rather different conformations and interaction patterns in the active form of rhodopsin, opsin, β2-adrenergic receptor (B2AR), and the adenosine A2a receptor (4–7, 9, 12). This is also the case for TyrVII:20 of the NPXXY motif, and TrpVI:13 of the CWXP motif has not been observed to perform the expected rotational change in conformation predicted from previous studies (14, 15).
However, when Rasmussen et al. (5, 12) recently published the crystal structure of the B2AR in complex with both an agonist and an antibody fragment mimicking the G protein, they noticed a rearrangement in the packing between PheVI:09 (6.44) and an isoleucine residue in TM-III, IleIII:16 (3.40), and proposed that this rearrangement could be important in the intramolecular signal transduction events. As shown in Fig. 1, receptor activation not only changes the interaction pattern between PheVI:09 and IleIII:16 but brings PheVI:09 into close proximity with three hydrophobic residues in TM-V: PheV:13 (5.47), LeuV:17 (5.51), and MetV:20 (5.54). This indicates that PheVI:09 could function as a sliding microswitch residue that stabilizes the active conformation of TM-VI relative to not only TM-III but also TM-V. In the present study, the highly conserved (82%) PheVI:09 residue is further analyzed as a potential microswitch through both computational chemistry analysis and receptor mutagenesis. The latter was performed in a series of model receptors displaying different degrees of constitutive activity and further includes mutational analysis of the proposed hydrophobic pocket for the active conformation of PheVI:09 located between TM-III and TM-V.
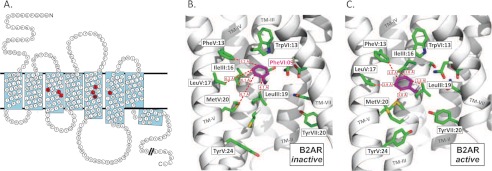
FIGURE 1.
The highly conserved PheVI:09 and its interaction partners in the B2AR. A, serpentine model of the B2AR. The residues indicated in black on red represent the conserved PheVI:09 (Phe6.44) and the residues with which it interacts: SerIII:15, IleIII:16, and LeuIII:19 (in TM-III) and PheV:13, LeuV:17, and MetV:20 (in TM-V). Residues in black on gray indicate the conserved residues AsnI:18 (1.50), AspII:10 (2.50), CysIII:01 (3.25), TrpIV:10 (4.50), ProV:16 (5.50), ProVI:15 (6.50), and ProVII:17 (7.50). B, PheVI:09 (purple) in the inactive conformation of B2AR (PDB entry 2RH1) as viewed from TM-VI (backbone and most residues removed) toward TM-III. C, as in B but in the active conformation of B2AR (PDB entry 3P0G). Distances between PheVI:09 and each of the residues constituting the hydrophobic pocket are indicated by red dotted lines for both receptor conformations.
EXPERIMENTAL PROCEDURES
Molecular Modeling
Knowledge of the inactive and active conformation of 7TM receptors and the basis for conformational changes of microswitches is the key to understanding the activation mechanism of 7TM receptors. In this study, we have used computational methods to investigate the structural and energetic properties of the inactive and active conformations and, in particular, to focus on conformational and energetic properties of the highly conserved residues located in the extended allosteric interface between the TM segments. Our analysis is based on 1) generation of intermediate conformations going from the inactive to the active receptor conformation using a morphed trajectory and 2) rigid and adiabatic energy mapping.
Initially, x-ray structures of the active (PDB entries 3P0G, 3QAK, and 2Y00) and inactive (PDB entries 2RH1, 3EML, and 1GZM) B2AR, adenosine A2a, and rhodopsin receptors were obtained from the Protein Data Bank. The PDB files were manually cleaned to include one receptor domain. Additional domains and molecules, such as T4 lysozyme, cholesterol, lipids, ligands, water, etc., were removed from the coordinate files. The active and inactive structures of the receptors were superimposed with respect to their TM domains using ICM (i.e. extra- and intracellular loops did not contribute to the superposition). The superimposed inactive and active structures were used to create a morphed trajectory including 15 interpolated conformations for each receptor (16, 17). Despite that these are theoretically computed trajectories and consequently lack time scale, kinetic and dynamic information, the intermediates are nevertheless believed to represent fairly realistic structures, which are similar or even indistinguishable from real intermediate conformations. This assumption is based on various structural checks to experimental structures and have previously been used to study known motions associated with, for example, hinged domain or allosteric changes in proteins (16).
Individual conformations in the resulting trajectories, representing the transition from the inactive through intermediate to the active conformation, were subsequently minimized in the simulation package CHARMM (18). Here it was used to analyze and access the role of the highly conserved residues located in an extended allosteric interface between the TM segments associated with large scale conformational changes during activation. Each conformation was minimized in 100 steps using steepest descent followed by 100 steps of the conjugated gradient steps using the CHARMM 22 force field, a 15-Å cut-off radius for the non-bonded interactions, and a distance-dependent dielectric constant of 8. The interaction energy from inactive to active conformations for side chains of interest was computed by constraining the χ1 and χ2 dihedral angles to their initial values using a dihedral harmonic potential with a barrier height of 100 kcal/mol and minimizing the system in 100 steps of steepest descent. All other atoms were free to move, followed by 50 minimization steps of steepest descent with all constraints released.
To evaluate their robustness and reproducibility, the calculations were performed using different minimization protocols (different number of steps and value for distance-dependent dielectric constant). The result of these calculations identifies the same low energy conformations of the studied side chains (data not shown) and gives qualitatively the same results as the protocol described above and presented under “Results.”
Comparative Homology Modeling of the Ghrelin Receptor
The amino acid sequence of the human ghrelin receptor (GHSR_HUMAN accession code Q92847) was obtained from the UniProt Web site. The x-ray structures of the 7TM receptors in the inactive state adenosine A2a (PDB entry 3EML), β1-adrenergic receptor (PDB entry 2VT4), B2AR (PDB entry 2RH1), chemokine CXCR4 (PDB entry 3ODU), dopamine D3 (PDB entry 3PBL), histamine H1 (PDB entry 3RZE), and rhodopsin (PDB entry 1GZM) were obtained from the PDB database. A multiple-sequence alignment between the ghrelin receptor and the above template structures identified dopamine D3 to have slightly higher sequence identity, considering the TM region, compared with the remaining template receptors. A comparative homology model of the ghrelin receptor was constructed based on the template structure of dopamine D3 using the homology modeling package in ICM. Torsion angles of all side chains were simultaneously optimized using the automated Monte Carlo protocol, and best fitting loops were extracted from a database of known loops. The resulting ghrelin model was subsequently minimized for 300 steps (steep descent) using the Merck molecular force field.
Material
The ghrelin and substance P peptides were purchased from Bachem (Bubendorf, Switzerland). MK-677 was purchased from Axon Medchem. Pindolol was purchased from Sigma, and AR-231453 was a generous gift from Arena Pharmaceuticals (San Diego, CA).
Molecular Biology
The B2AR, neurokinin-1 (NK1), GPR39, and GHSR cDNA were cloned into the eukaryotic expression vector pCMV-Tag (2B) (Stratagene, La Jolla, CA). The GPR119 cDNA was cloned into the eukaryotic expression vector pcDNA3.1 (Invitrogen). Mutations were constructed by PCR using the QuikChange method. All PCR experiments were performed using Pfu polymerase (Stratagene) according to the instructions of the manufacturer. All mutations were verified by DNA sequence analysis by MWG (Ebersberg, Germany).
Transfections and Tissue Culture
COS-7 cells were grown in Dulbecco's modified Eagle's medium 1885 supplemented with 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected using 20 μg of DNA/75 cm2 by the calcium phosphate precipitation method with chloroquine addition, as described previously (19).
Competition Binding Assay
Transfected COS-7 cells were plated in poly-d-lysine coated white 96-well plates at a density of 10,000 cells/well aiming at 5–10% binding of the radioactive ligand. The following day, the binding experiments were performed for 3 h at 4 °C using ∼25 pm [3H]MK-677 (Amersham Biosciences). Binding assays were performed in 0.1 ml of a HEPES buffer, pH 7.4, supplemented with 1 mm CaCl2, 5 mm MgCl2, 0.1% (w/v) bovine serum albumin, and 40 μg/ml bacitracin. Nonspecific binding was determined as the binding in the presence of 1 μm unlabeled ghrelin. After two washes in cold buffer, scintillation was added, and the bound radioactivity was counted on Topcount (PerkinElmer Life Sciences).
Phosphatidylinositol Turnover Assay
One day after transfection, COS-7 cells were incubated for 24 h with 5 μCi of myo-[3H]inositol (Amersham Biosciences)/ml of medium supplemented with 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were washed twice in buffer (20 mm HEPES, pH 7.4, supplemented with 140 mm NaCl, 5 mm KCl, 1 mm MgSO4, 1 mm CaCl2, 10 mm glucose, and 0.05% (w/v) fetal bovine serum) and then incubated in 0.5 ml of buffer supplemented with 10 mm LiCl at 37 °C for 30 min. After a 45-min stimulation with various concentrations of ligands at 37 °C, cells were extracted with 10 mm formic acid, followed by incubation on ice for 30 min. The resulting supernatant was purified on Bio-Rad AG 1-X8 anion exchange resin to isolate the negatively charged inositol phosphates. After application of the cell extract to the column, the columns were washed twice with GPI buffer (60 mm sodium formate and 100 mm formic acid) to remove glycerophosphoinositol. Inositol phosphates were eluted by the addition of elution buffer (1 mm ammonium formate, 100 mm formic acid). Determinations were made in duplicates. The columns containing AG 1-X8 anion exchange resin were regenerated by the addition of 3 ml of regeneration buffer (3 m ammonium formate, 100 mm formic acid) and 5 ml water.
For GPR39 transiently transfected in COS-7 cells, on the day after transfection, cells were harvested, and 35,000 cells/well were plated in 96-well plates in Dulbecco's modified Eagle's medium 1885 supplemented with 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 5 μl (20 mCi/mmol) myo-[2-3H]inositol (American Radiolabeled Chemicals)/ml of DMEM. The following day, the cells were washed twice with Hanks' balanced salt solution and incubated for 30 min in Hanks' balanced salt solution with 10 mm LiCl. Following incubation, ligand was added and incubated for an additional 45 min. After removal of the ligand, the cells were placed on ice, and 50 μl of 10 mm cold formic acid was added to each well for 30 min. 20 μl was added to a white 96-well plate, and 80 μl of YSi SPA beads (diluted 1:8 with double-distilled H2O to a final concentration of 1 mg of YSi SPA beads/well) was added to each well. The plate was shaken for 2–4 h and spun down for 5 min. The signal was counted using the Topcount (PerkinElmer Life Sciences).
cAMP Assay
One day after transfection, COS-7 cells were plated into white 96-well plates (20,000 cells/well). The next day, the cAMP assay was performed using the DiscoveRX HitHunterTM cAMPxs+ kit (Freemont, CA) according to the manufacturer's protocol.
Calculations
EC50 values were determined by nonlinear regression using the Prism version 3.0 software (GraphPad Software, San Diego, CA). The basal constitutive activity is expressed as a percentage of the maximal ligand-induced activation of the WT receptor.
Cell Surface Expression (ELISA)
Cells transfected and seeded for cAMP were in parallel seeded for ELISA. The cells were washed twice with phosphate-buffered saline (PBS), fixed for 10 min with formaldehyde, and incubated in blocking solution (PBS added to 3% dry milk) for 30 min at room temperature. Subsequently, the cells were incubated for 1 h at room temperature with anti-FLAG (M2) (Sigma) antibody diluted 1:1000 with PBS plus 3% milk. The cells were washed three times with PBS and incubated for 1 h at room temperature with anti-mouse horseradish peroxidase-conjugated antibody (Sigma) diluted 1:1250 in PBS plus 3% milk. After three additional washing steps with PBS, immunoreactivity was discovered by the addition of horseradish peroxidase, and after 5 min, the reaction was stopped by the addition of H2SO4. The absorbance was read on the Topcount plate reader (PerkinElmer Life Sciences).
RESULTS
Functional Analysis of PheVI:09 in the Ghrelin Receptor
When analyzing activation of efficacy switches, it is particularly appropriate to study effects on constitutive or ligand-independent signaling in order to avoid confusing direct or indirect effects on ligand binding. In this respect, the ghrelin receptor is particularly suited because even in the absence of its endogenous agonist, it displays almost 50% of its maximal signaling capacity, and, importantly, the ghrelin receptor is generally robust with respect to mutational changes (20, 21).
Alanine substitution of PheVI:09 did not affect the surface expression of the ghrelin receptor transiently expressed in COS-7 cells, as measured by ELISA (Fig. 2A (inset) and Table 1). However, the high constitutive signaling through the Gq-mediated pathway observed in the wild-type ghrelin receptor was totally eliminated in the AlaVI:09 mutant form (Fig. 2A and Table 1). Moreover, alanine substitution of PheVI:09 decreased ghrelin-induced signaling to ∼10% with only a minor effect on agonist potency (Fig. 2A and Table 1) and no effect on agonist affinity as determined in competition binding experiments against the radiolabeled non-peptide agonist [3H]MK-677 (Table 2). Interestingly, not even the rather large, flexible, and hydrophobic side chain of a leucine residue could substitute for phenylalanine because the LeuVI:09 mutant form of the ghrelin receptor also displayed a complete lack of constitutive activity (Fig. 2A and Table 1). However, the LeuVI:09 mutant induced an Emax around 35% of that observed for wild-type receptor, again with no effect on ghrelin potency, indicating that PheVI:09 functions as an efficacy switch and is not involved in ligand binding. This is further substantiated by the close to wild type binding properties for ghrelin in these receptor mutants (Table 2). Importantly, substitution of PheVI:09 with a structurally similar, aromatic tyrosine residue decreased the basal activity to 15% and did not affect the ghrelin-induced maximal signaling efficacy. Altogether, the aromatic, rigid hydrophobic properties of the phenyl side chain are required in position VI:09 in particular for constitutive but also for the full ligand-dependent activation of the ghrelin receptor but are apparently not important for agonist binding.
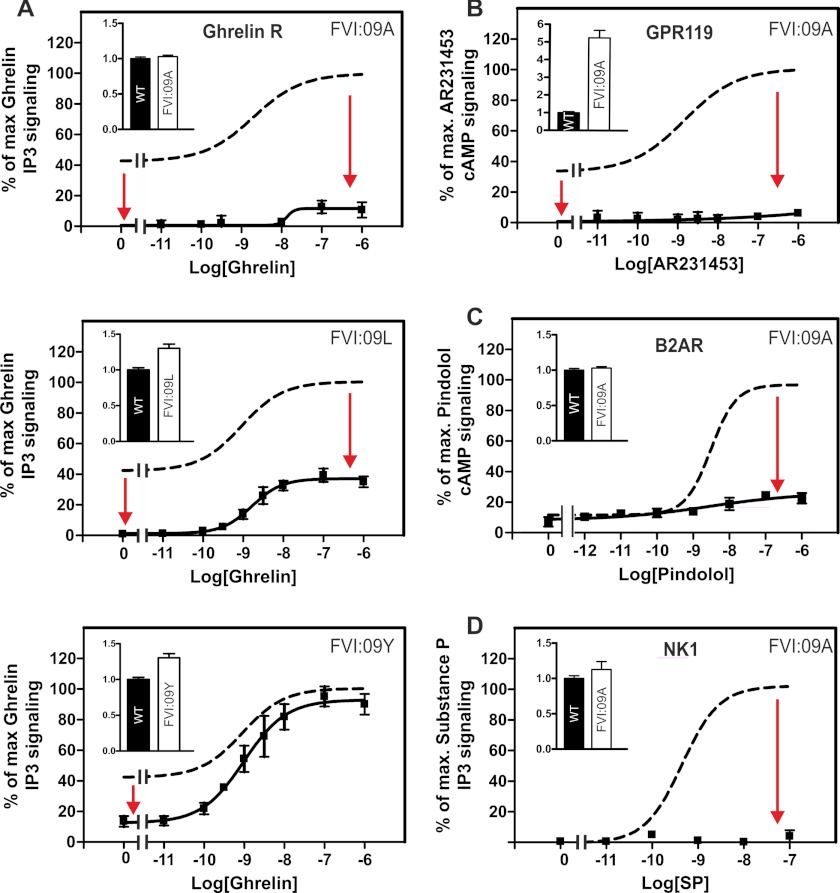
FIGURE 2.
Functional consequence of substituting the PheVI:09 residue. WT (dotted lines) receptor or FVI:09 mutants were all transiently transfected in COS-7 cells. A, basal and agonist (ghrelin)-induced IP3 production of WT ghrelin receptor or mutants of the following: PheVI:09Ala, PheVI:09Leu, and PheVI:09Tyr. B, basal and agonist (AR231453)-induced cAMP production of WT GRP119 and PheVI:09Ala. C, basal and agonist (pindolol)-induced cAMP production of WT B2AR and PheVI:09Ala. D, agonist (SP)-induced IP3 production of WT NK1 or PheVI:09Ala. Cell surface receptor expression, measured by enzyme-linked immunosorbent assay, is shown in the inset column diagrams in each panel. Error bars, S.E.
TABLE 1.
Inositol phosphate signaling of the ghrelin WT receptor and mutant forms with substitutions in positions ValIII:16, IleIII:19, PheV:13, ValV:17, LeuV:20, and PheVI:09
The constructs were expressed in transiently transfected COS-7 cells. The efficacy data on basal activity and maximal response (Emax) are expressed as a percentage of the maximal signaling of the wild-type ghrelin receptor. Values are shown ± S.E.
| Constitutive activity | n | Expression level | n | Ghrelin EC50 | n | Fmut | Ghrelin Emax | n | Fmut | |
|---|---|---|---|---|---|---|---|---|---|---|
| nm | ||||||||||
| WT ghrelin R1a | 45.6 ± 3 | 4 | 1 | 4 | 0.75 ± 0.29 | 4 | 1 | 100 ± 0.7 | 4 | 1 |
| ValIII:16Ala (3.40) | 14.6 ± 3.7 | 4 | 1.4 ± 0.07 | 4 | 1.5 ± 0.43 | 4 | 2 | 100 ± 5.9 | 4 | 1 |
| ValIII:16Ile | 45.5 ± 1.8 | 4 | 1.1 ± 0.04 | 4 | 5.96 ± 2.75 | 4 | 7.9 | 92.7 ± 4.6 | 4 | 0.9 |
| ValIII:16Phe | 25.6 ± 6.1 | 3 | 1.0 ± 0.04 | 3 | 4.55 ± 2.62 | 3 | 6.1 | 88.1 ± 9.9 | 3 | 0.9 |
| IleIII:19Ala (3.43) | 43.5 ± 6.1 | 4 | 0.71 ± 0.05 | 4 | 35.0 ± 17.4 | 4 | 47 | 68.8 ± 11.2 | 4 | 0.7 |
| PheV:13Ala (5.47) | 1.7 ± 1.06 | 5 | 1.07 ± 0.07 | 5 | 1.6 ± 0.33 | 5 | 2 | 70.8 ± 3.7 | 5 | 0.7 |
| ValV:17Ala (5.51) | 52.3 ± 1.3 | 3 | 0.99 ± 0.05 | 4 | 3.34 ± 2.09 | 3 | 4.5 | 101 ± 5.6 | 3 | 1 |
| LeuV:20Ala (5.54) | 11.8 ± 3.7 | 4 | 1.1 ± 0.05 | 4 | 1.31 ± 0.21 | 4 | 1.7 | 118 ± 4.2 | 4 | 1.2 |
| LeuV:20Ile | 51.9 ± 8.8 | 3 | 0.95 ± 0.1 | 4 | 1.17 ± 0.31 | 3 | 1.6 | 114 ± 15.7 | 3 | 1.1 |
| LeuV:20Val | 37.4 ± 6.5 | 3 | 1.0 ± 0.05 | 4 | 2.37 ± 0.74 | 3 | 3.2 | 111 ± 6.5 | 3 | 1.1 |
| LeuV:20Phe | 11.6 ± 3.1 | 3 | 1.0 ± 0.06 | 3 | 1.83 ± 0.62 | 3 | 2.4 | 73.1 ± 6.3 | 3 | 0.7 |
| PheVI:09Ala (6.44) | 1.5 ± 0.8 | 3 | 1.0 ± 0.02 | 4 | 10.7 ± 2.04 | 3 | 14.3 | 13.3 ± 4.0 | 3 | 0.1 |
| PheVI:09Leu | 1.3 ± 1.7 | 6 | 1.3 ± 0.06 | 3 | 1.92 ± 0.99 | 6 | 2.6 | 37.5 ± 4.1 | 6 | 0.4 |
| PheVI:09Tyr | 10.5 ± 3.9 | 5 | 1.3 ± 0.08 | 3 | 0.8 ± 0.27 | 5 | 1.1 | 87.7 ± 7.1 | 5 | 0.9 |
TABLE 2.
Ligand binding properties of substitutions in position ValIII:16, IleIII:16, PheV:13, ValV:17, LeuV:20, and PheVI:09 in the ghrelin receptor using [3H]MK-677 as a radioligand
The constructs were expressed in transiently transfected COS-7 cells. Values are shown ± S.E.
| Bmax | n | MK-677 Kd | n | Ghrelin Ki | n | |
|---|---|---|---|---|---|---|
| fmol/105 cells | nm | nm | ||||
| WT ghrelin R1a | 127 ± 16.7 | 4 | 1.91 ± 0.52 | 4 | 2.47 ± 0.42 | 7 |
| ValIII:16Ala (3.40) | 170 ± 14.9 | 4 | 3.03 ± 0.77 | 4 | 3.41 ± 0.82 | 6 |
| ValIII:16Ile | 165 ± 64.1 | 4 | 2.21 ± 0.4 | 4 | 2.89 ± 0.56 | 7 |
| ValIII:16Phe | 225 ± 6.46 | 4 | 6.3 ± 0.08 | 4 | 1.83 ± 0.01 | 5 |
| IleIII:19Ala (3.43) | 254 ± 55.6 | 3 | 4.79 ± 0.37 | 3 | 5.87 ± 0.14 | 3 |
| PheV:13Ala (5.47) | 208 ± 53.7 | 4 | 4.07 ± 0.96 | 4 | 1.78 ± 0.4 | 5 |
| ValV:17Ala (5.51) | 105 ± 15.7 | 4 | 2.1 ± 0.36 | 4 | 2.07 ± 0.59 | 5 |
| LeuV:20Ala (5.54) | 124 ± 34.1 | 4 | 4.08 ± 0.96 | 4 | 2.5 ± 0.36 | 6 |
| LeuV:20Ile | 164 ± 77.2 | 4 | 2.37 ± 1.22 | 4 | 2.43 ± 0.35 | 6 |
| LeuV:20Val | 238 ± 53.5 | 4 | 4.14 ± 1.08 | 4 | 2.35 ± 0.38 | 6 |
| LeuV:20Phe | 184 ± 25.6 | 4 | 5.31 ± 1.46 | 4 | 4.03 ± 1.21 | 6 |
| PheVI:09Ala (6.44) | 239 ± 52.3 | 4 | 4.43 ± 1.51 | 4 | 1.76 ± 0.39 | 7 |
| PheVI:09Leu | 338 ± 77.0 | 4 | 2.56 ± 0.6 | 4 | 3.66 ± 0.61 | 6 |
| PheVI:09Tyr | 280 ± 66.6 | 4 | 3.37 ± 0.64 | 4 | 2.41 ± 0.17 | 7 |
Mutational Analysis of the PheVI:09 in Other 7TM Receptors
Similar to the ghrelin receptor, GPR119 displays around 50% constitutive activity but through the Gs pathway. As observed in the ghrelin receptor, alanine substitution of PheVI:09 totally abolished the ligand-independent, constitutive activity of GPR119 as determined by cAMP measurements in transiently transfected COS-7 cells (Fig. 2B). Similarly, by use of the non-peptide prototype agonist, AR231453, it was shown that agonist-induced signaling was totally eliminated in the AlaVI:09 mutant form of GPR119 (Fig. 2B). Importantly, these impairments in receptor signaling were observed despite an almost 5-fold increase in receptor cell surface expression for the AlaVI:09 receptor mutant as compared with the wild-type GPR119 receptor.5 The B2AR also signals through Gs. In the present system, we only observed a rather low degree of constitutive activity of the B2AR, which was only slightly affected by alanine substitution of PheVI:09. However, the agonist-induced signaling was almost eliminated in the AlaVI:09 mutant form of the B2AR without any effect on cell surface expression (Fig. 2C).
The NK1 receptor is a very “silent” Gq-coupled receptor with no detectable degree of constitutive activity, as judged by measurement of inositol phosphate accumulation in COS-7 cells. Alanine substitution of PheVI:09 did not affect cell surface expression of the NK1 receptor, but the substance P-induced signaling was abolished for this mutant form of NK1 (Fig. 2D). In several receptors, the results demonstrate the importance of PheVI:09 in receptor activation and show that this residue appears to function as a pure efficacy switch.
GPR39, a Receptor Lacking a Phenylalanine in Position VI:09
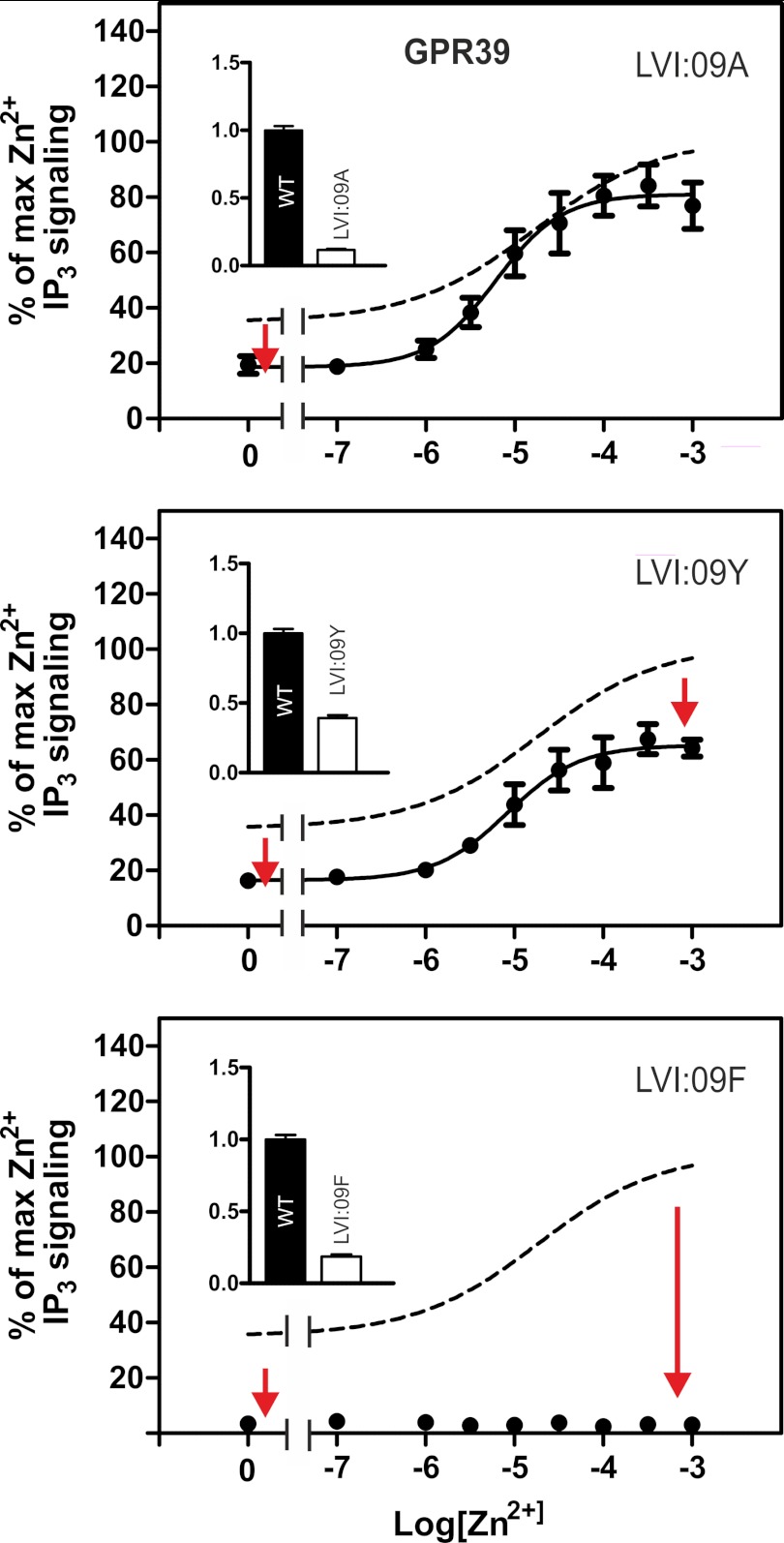
12% of 7TM receptors do not have a phenylalanine or a tyrosine residue in position VI:09. To represent these receptors, we chose GPR39, which is a constitutively active member of the ghrelin receptor family, having a leucine residue in position VI:09. As shown in Fig. 3, substitution of LeuVI:09 with either alanine, tyrosine, or phenylalanine in all three cases reduced the cell surface expression and decreased the apparent constitutive signaling of GPR39. Nevertheless, in contrast to what was observed in the receptors having a phenylalanine in position VI:09 (Fig. 2) and despite the apparently reduced cell surface expression, a normal agonist-induced IP3 response was observed in the GPR39 constructs where LeuVI:09 was substituted with either alanine or tyrosine (Fig. 3). However, introduction of a phenylalanine at position VI:09, as found in the majority of 7TM receptors, was not allowed in GPR39 because it both reduced the expression and completely abolished both the constitutive and the ligand-induced activation of GPR39 (Fig. 3C).
FIGURE 3.
Functional consequence of alanine substitution of the LeuVI:09 residue in GPR39. Shown are basal and agonist (Zn2+)-induced IP3 production in COS-7 cells transiently transfected with either WT GPR39 (dotted line) or the following mutant forms of GPR39: LeuVI:09Ala (A), LeuVI:09Tyr (B), and LeuVI:09Phe (C). Cell surface receptor expression, measured by enzyme-linked immunosorbent assay, is shown in the inset column diagram. Error bars, S.E.
PheVI:09, Analysis of Conformations and Interactions
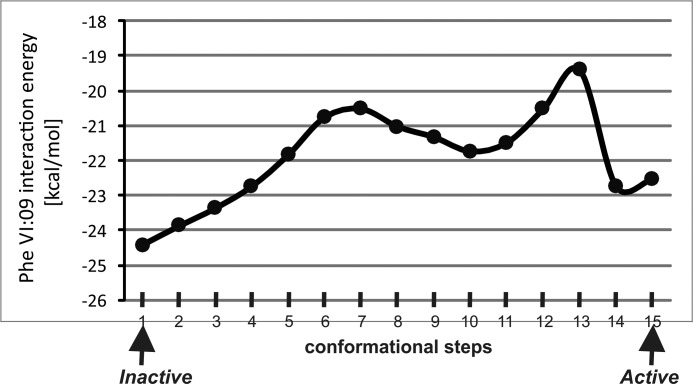
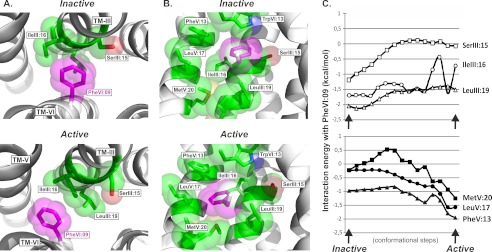
Morphed simulation followed by adiabatic mapping and energy minimization was used to analyze the changes in side chain conformations, interaction pattern, and energy between PheVI:09 and its neighboring residues during the transition from inactive to active conformation of the B2AR based on the high resolution x-ray structures (PDB entries 2RH1 and 3P0G). Initially, the energy between PheVI:09 and the nine closest residues (SerIII:15, IleIII:16, LeuIII:19, PheV:13, ProV:16, ProV:17, MetV:20, MetVI:06, and TrpVI:13) was calculated as a function of the reaction coordinates defined by the morphed trajectory describing the transition from the inactive to the active x-ray structure of the B2AR (Fig. 4). The resulting interaction energies for the transition can by approximations be considered to represent the enthalpic component of the potential of mean force, which indicates two distinct energy minima corresponding to the inactive and the active conformation, respectively, separated by an energy barrier.
FIGURE 4.
Interaction energy between PheVI:09 and the surrounding residues in the B2AR. The interaction energy between PheVI:09 and the surrounding residues in TM-III, TM-V, and TM-VI (SerIII:15, IleIII:16, LeuIII:19, PheV:13, ProV:16, ProV:17, MetV:20, MetVI:06, and TrpVI:13) calculated in 15 conformational steps during “normal mode” simulation of the transition from the inactive (PDB entry 2RH1) to the active structure (PDB entry 3P0G).
In order to understand the interaction mode for PheVI:09 in more detail, we calculated the interaction energy for PheVI:09 with the key residues in TM-III and TM-V. In the inactive conformation of the B2AR (PDB entry 2RH1), the phenyl side chain of PheVI:09 interacts closely with the backbone atoms of SerIII:15 (3.39) and IleIII:16 and the aliphatic side chains of IleIII:16 and LeuIII:19 (Figs. 1B and 5A). This interaction mode is reflected in the favorable interaction energy between the side chain of PheVI:09 and these three residues in TM-III, as shown in Fig. 5C (top). In addition, PheVI:09 makes an edge-to-face aromatic-aromatic interaction with TrpVI:13 located one helical turn above in TM-VI itself.
FIGURE 5.
Interaction mode and energy between PheVI:09 and the hydrophobic residues in TM-III and TM-V in the B2AR. A, the interaction mode of PheVI:09 (purple) with SerIII:15, IleIII:16, and LeuIII:19 (green) in TM-III viewed from the extracellular side of the inactive (PDB entry 2RH1) and active (PDB entry 3P0G) structures of the B2AR shown in the left and right panels, respectively. B, side view (toward TM-III) of the interaction of PheVI:09 (purple) with the surrounding residues TrpVI:13, SerIII:15, IleIII:16, LeuIII:19, PheV:13, LeuV:17, and MetV:20 (green) in the inactive and active structure of B2AR. C, interaction energy landscape between PheVI:09 and the surrounding residues in TM-III (SerIII:15, IleIII:16, and LeuIII:19; top graph) and in TM-V (MetV:20, LeuV:17, and PheV:13; bottom graph), calculated in 15 conformational steps during “normal mode” simulation of the transition from the inactive (PDB entry 2RH1) to the active structure (PDB entry 3P0G).
During receptor activation, the side chain of PheVI:09 moves several Å toward TM-V and hereby totally loses its van der Waal interaction with the backbone of SerIII:15, as evident in the interaction energy, which goes toward zero (Fig. 5C, top). In contrast, in the active conformation of the B2AR (PDB entry 3P0G), PheVI:09 keeps a favorable hydrophobic interaction with LeuIII:19 and IleIII:16, albeit at a slightly higher energy level because it has moved to the other side of IleIII:16 (Fig. 5A). Importantly, the simulation of the conformational changes occurring during activation demonstrated that the phenyl side chain of PheVI:09 has to pass the aliphatic side chain of IleIII:16, which, as expected, constituted an energy barrier and resulted in rather large fluctuations in the interaction energy (Fig. 5C, top). During this, IleIII:16 reorients from a minimum centered around the side chain torsion angle χ1 = 68° in the inactive structure (PDB entry 2RH1) to populate a new rotameric state corresponding to χ1 = −163° found in the active crystal structure (PDB entry 3P0G). These torsion values are in good agreement with the values χ1 = 62° and χ1 = −177°, corresponding to the most populated rotameric states for isoleucine according to the Penultimate Rotamer Library defined in the integrated CCP4 program suite (available at the CCP4 Web site), which is a collection of developed tools for experimental determination and analysis of protein structures.
In the inactive conformation, the distance from the phenyl side chain of PheVI:09 to PheV:13, LeuV:17, and MetV:20 in TM-V is between 5.2 and 6.7 Å (Fig. 1). However, during receptor activation, PheVI:09 moves pass IleIII:16 into a tight hydrophobic pocket generated by these three hydrophobic residues in TM-V with side chain distances narrowing van der Waal contacts between 3.5 and 3.8 Å (Figs. 1 and 5C). This favorable interaction is evident from the energy landscape, which, after a slight increase in the middle of the activation process, reaches a minimum for the calculated interaction energy between PheVI:09 and each of the hydrophobic residues in TM-V in the final active conformation (Fig. 5B, bottom).
It is concluded that PheVI:09 constitutes a microswitch, which in the inactive receptor conformation interacts closely with the backbone and aliphatic hydrophobic residues in the opposing TM-III and which during receptor activation is repositioned to a hydrophobic pocket generated by three hydrophobic residues in TM-V and the two hydrophobic residues in TM-III. IleIII:16 appears to constitute an important gate for this transition of the PheVI:09 side chain.
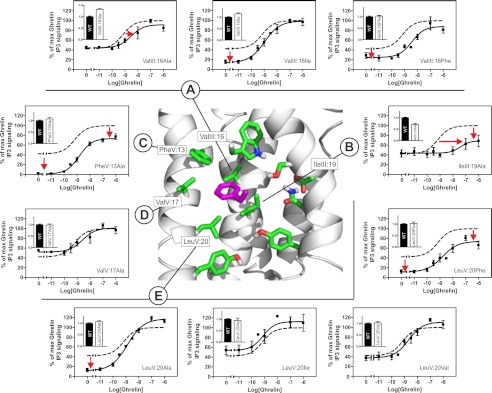
Mutational Analysis of the Hydrophobic Partners for PheVI:09
As described above, in both the inactive and the active conformations, PheVI:09 interacts closely with the hydrophobic residues found in position III:16 and III:19, of which the residue in position III:16 appears to function as a gate between the inactive and active conformation. In the ghrelin receptor, a valine residue is found in position III:16. Alanine substitution of ValIII:16 had a very limited effect on both the constitutive and the agonist-induced signaling (Fig. 6A and Table 1). However, introduction of an isoleucine in position III:16, as found in the B2AR, decreased the constitutive activity, and introduction of a large and relatively rigid phenylalanine residue in this position both decreased the constitutive activity and shifted the agonist dose-response curve ∼10-fold to the right. The binding affinity of the endogenous peptide agonist ghrelin was almost unaffected by this substitution at position III:16 (Table 2).
FIGURE 6.
Functional consequence of substituting the hydrophobic region surrounding the FVI:09 microswitch in the ghrelin receptor. Structure of the hydrophobic region (sticks) (green) surrounding the FVI:09 (purple) microswitch based on homology modeling using the dopamine D3 receptor (PDB entry 3PBL) structure as a template. A–E, basal and agonist (ghrelin)-induced IP3 production in COS-7 cells transiently transfected with either WT (dotted lines) or mutant forms of the following: ValIII:16Ala, ValIII:16Ile, ValIII:16Phe (A); IleIII:19Ala (B); PheV:13Ala (C); ValV:17Ala (D); and LeuV:20Ala, LeuV:20Ile, LeuV:20Val, and LeuV:20Phe (E). Cell surface receptor expression, measured by immunosorbent assay, is shown in the inset column diagrams in each panel. Error bars, S.E.
Alanine substitution of the other hydrophobic interaction partner for PheVI:09 in TM-III, IleIII:19 (leucine in B2AR), shifted the dose-response curve for ghrelin 2 orders of magnitude to the right but without any major change in constitutive activity or in agonist binding affinity or Bmax (Fig. 6B and Table 2).
As previously described and demonstrated here, alanine substitution of PheV:13 eliminated the constitutive activity of the ghrelin receptor and increased the Bmax without affecting agonist potency (Fig. 6C and Tables 1 and 2) (22). In contrast, alanine substitution of the middle hydrophobic interaction partner for PheVI:09 in TM-V, ValV:17, which is furthest away from PheVI:09, had no effect on either constitutive or agonist-induced activity of the ghrelin receptor (Fig. 6D and Table 1). However, alanine substitution of the lower interaction partner in TM-V, LeuV:20 (methionine in B2AR), decreased the constitutive activity without affecting the agonist-induced signaling of the ghrelin receptor (Fig. 6E and Table 1). However, in position V:20, isoleucine or valine substituted well for the naturally occurring leucine, whereas the introduction of a large and more rigid phenylalanine residue decreased both constitutive and agonist-induced signaling but without affecting agonist potency (Fig. 6E and Table 1).
Overall, the mutational analysis of the proposed hydrophobic pocket for PheVI:09 in several positions affected receptor signaling and in particular the ligand-independent signaling. The effect of substituting the proposed interaction partners in TM-III was more complex, in agreement with the fact that these residues appear to interact with PheVI:09 both in the inactive and the active receptor conformation.
DISCUSSION
It has for years been known that PheVI:09 is among the most highly conserved residues in 7TM receptors. However, the structural and functional reason for this was unclear. In the inactive structures of initially rhodopsin and later the B2AR and other ligand-activated 7TM receptors, PheVI:09 did not draw much attention because it basically just pointed toward TM-III. However, the active structure of the B2AR demonstrated a significant rearrangement of the interaction between PheVI:09 and IleIII:16 upon activation (5). The computational chemical analysis and the mutational examination in the present study indicate that PheVI:09 functions as an important microswitch, which alternates between an inactive state, where its phenyl side chain is closely locked against the backbone and between two hydrophobic side chains in TM-III and an active state, where the phenyl side chain is locked in a tight pocket composed of a total of five hydrophobic residues: three protruding from TM-V and two from TM-III (Fig. 5). During the activation process, the side chain of PheVI:09 slides ∼2 Å toward TM-V into the hydrophobic pocket located between TM-III and TM-V. During this process, PheVI:09 has to pass IleIII:16, which needs to change its rotameric state to allow for this passage. Thus, IleIII:16 appears to function as a barrier or gate for the interconversion between the two states of the PheVI:09 microswitch: inactive versus active.
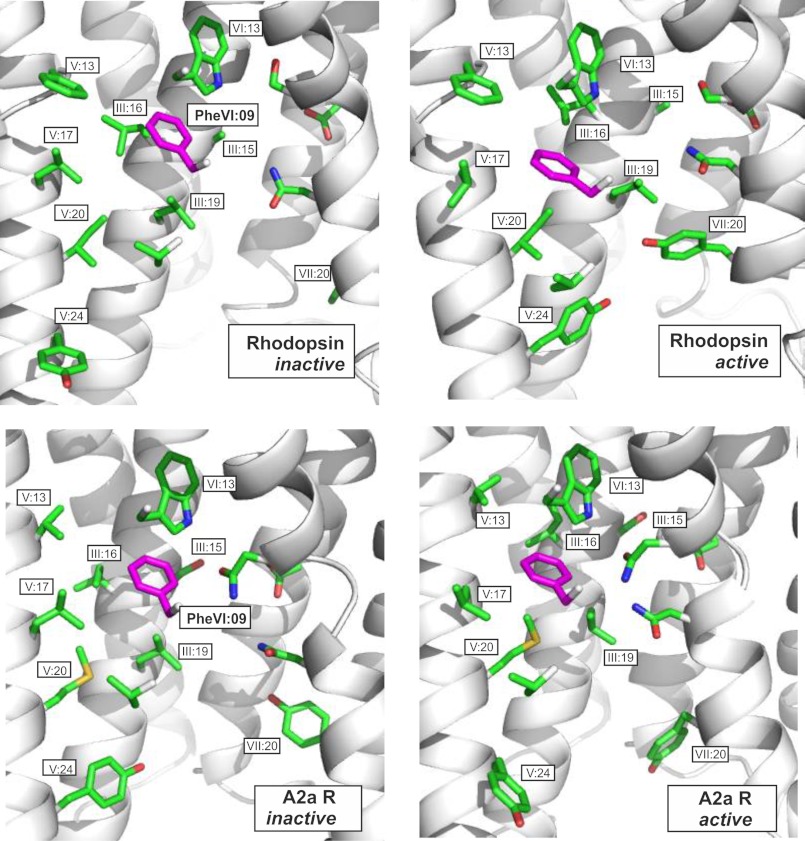
The notion that the PheVI:09 microswitch plays a crucial role in 7TM receptor activation is underlined both by its central location close to the pivot around which TM-VI performs its large scale movement during activation and by the fact that PheVI:09 appears to stabilize the overall active conformation of TM-V and TM-VI relative to TM-III. Notably, whereas other microswitch residues, such as ArgIII:26 of the DRY motif and TyrVII:20 of the NPXXY motif, are found in surprisingly different interaction patterns in the different receptors (3, 4, 6–9, 23), our computational analysis of the available high resolution x-ray structures of pairs of active and inactive conformations of rhodopsin, B2AR, and the adenosine 2a receptor indicates that PheVI:09 is the only microswitch that has almost identical interaction patterns in all three receptor pairs (Fig. 7).
FIGURE 7.
Comparison of PheVI:09 and its interaction partners in the B2AR (top two panels), in rhodopsin (middle two panels), and in the adenosine A2a receptor (bottom two panels). PheVI:09 (highlighted in purple) is shown in the inactive conformation of each of the receptors (PDB entries 2RH1, 1GZM, and 3EML) in the panels to the left and in the active conformations in the panels to the right (PDB entries 3P0G, 2Y00, and 3QAK). In all cases, PheVI:09 and its interaction partners are viewed from TM-VI (backbone and most residues removed) toward TM-III. Distances between PheVI:09 and each of the residues constituting the hydrophobic pocket in B2AR are indicated in Fig. 1, B and C. Note the similar position of the microswitch residue PheVI:09 on each side of the proposed gate or barrier residue III:16 in the inactive versus the active conformation of all three receptors. A couple of other residues are also highlighted (i.e. TrpVI:13 of the CWXP motif in TM-VI, TyrV:24 involved in stabilizing the proposed active conformation of the ArgIII:26 microswitch of the DRY motif in TM-III (not shown), and TyrVII:20 of the NPXXY motif of TM-VII). Note the different positions of TyrV:24 and TyrVII:20 in the three different receptor pairs. TrpVI:13 is positioned very similarly in all three receptor pairs but does not change conformation from inactive to active conformation in any of the receptors.
The mutational analysis of the present study indicates that PheVI:09 is functionally important for the spontaneous and/or agonist-mediated signaling of the ghrelin receptor, GPR119, the B2AR, and the NK1 tachykinin receptor (i.e. different receptors signaling through both Gs and Gq pathways). In these receptors, alanine substitution of PheVI:09 seriously impaired or eliminated signaling. Furthermore, in the ghrelin receptor, the mutational analysis of the residues constituting the hydrophobic pocket for PheVI:09 in several positions affected receptor signaling and in particular the ligand-independent signaling. However, the effects of substituting the proposed interaction partners in TM-III were more complex, which is in agreement with the fact that these residues interact with PheVI:09 both in the inactive and the active receptor conformation and that in particular the residue in position III:16, being valine in the ghrelin receptor and isoleucine in B2AR, apparently constitutes a gate or barrier between the two receptor conformations.
Previously, alanine substitution of PheVI:09 in rhodopsin demonstrated that the mutant receptor reconstituted normally with the 11-cis-retinal chromophore but that it displayed decreased ability to light-dependently activate transducin in analogy with the present observations of functional impairment in a number of other 7TM receptors (24, 25). Similarly, in the cholecystokinin B receptor, alanine substitution of PheVI:09 resulted in a receptor with retained high affinity agonist binding but impaired signaling (26).
However, as often observed with microswitch residues, their functions may vary in different receptors. In the a1B-AR, alanine substitution of PheVI:09 eliminated agonist-induced receptor signaling as described above for a series of other receptors. However, in this case, the mutant receptor showed increased constitutive signaling. Moreover, the a1B-AR leucine substitution of PheVI:09 did not decrease signaling as observed in the ghrelin receptor but instead increased both constitutive signaling and Emax for the agonist (27). Similarly, in the muscarinic M5 receptor, Spalding et al. (28) found that various substitutions of PheVI:09 all introduced constitutive activity and increased agonist-induced maximal signaling. Extensive mutational analysis of the hydrophobic interface between TM-III and TM-VI of the complement C5a receptor demonstrated that, in particular, substitutions of PheVI:09 and its interaction partners in TM-III, IleIII:16 and LeuIII:19, all led to increased constitutive activity (29). Concerning the B2AR, Chen et al. (30) found that alanine substitution of PheVI:09 introduced constitutive signaling and increased the agonist-induced maximal signaling instead of eliminating signaling as observed in the present study. The reason for these differences is unclear. However, studies of B2AR in, for example, COS-7 cells is complicated because these express endogenous receptors that will interfere, and may even interact, with transiently expressed B2AR, especially because these endogenous receptors are also strongly activated by isoproterenol as used by Chen et al. (30), in contrast to pindolol, as deliberately used in the present study. Moreover, we clearly observed no effect of the PheVI:09 mutant on cell surface expression as measured by a specific ELISA for the tagged receptor (Fig. 2C). In contrast, Chen et al. (30) determine receptor expression through adrenergic ligand binding, which may bind to the endogenous receptors also; however, these results were not shown in the article (30). Clearly, more independent studies are required to assess the function of the PheVI:09 microswitch in the B2AR.
It is often expected that highly conserved microswitch residues, such as PheVI:09, have to be essential for 7TM receptor function. However, as previously emphasized, these microswitches do not function as “domino bricks” (i.e. in series, in which case they would all be essential). Instead, the microswitches appear to function in parallel (i.e. as individual, distinct parts of an extended allosteric interface located between the structural elements, the transmembrane helices, which perform the large scale conformational changes) (1, 13). In this Monod, Wymann, and Changeux type of concerted allostery, any of the microswitch residues are in principle dispensable because it is the “strength” of the overall allosteric interface that counts and not any single component. Although PheVI:09 is essential for receptor signaling in a number of receptors, as reported in the present study and discussed above, 12% of 7TM receptors do not have an aromatic residue in position VI:09. In the case of GPR39, the leucine residue found in position VI:09 could be substituted with an alanine residue without affecting agonist-induced signaling despite reduced cell surface expression. This fits very well with the notion that the different microswitches function in parallel and not in series. In this connection, it should be noted that the outward tilted, active conformation of TM-VI and TM-V not only is stabilized by PheVI:09 locked in the hydrophobic pocket between TM-III and TM-V, as shown in Fig. 5, but also is stabilized by another microswitch residue, TyrVII:20, locked in a similar hydrophobic pocket located between TM-V and TM-VI two helical turns further toward the intracellular space (13).
In the glycoprotein hormone family of receptors of which the thyrotropin receptor is a prototype, an aspartic acid residue has been conserved at position VI:09. Interestingly, alanine substitution of AspVI:09 did not impair receptor signaling but instead led to increased constitutive activity of both the thyrotropin and the luteinizing receptors (31, 32).
An interesting aspect of microswitch residues, such as PheVI:09 of the present study and, for example, also ArgIII:26 of the DRY motif in TM-III, is that they apparently are involved in stabilizing both inactive and active states of the receptor. This means that the effect of alanine substitution will vary from receptor to receptor, as discussed above for PheVI:09, where both loss of function and gain of function effects have been observed for receptor signaling in different receptors. The mutational effect is dependent on whether the microswitch residue is most strongly involved in stabilizing the active or the inactive state of the receptor. In the case of ArgIII:26, loss of function is observed in most receptors. However, for example, in the B2AR, no effect is observed upon alanine substitution of ArgIII:26 with respect to Gs stimulation, despite the fact that alanine substitutions of the interaction partners for ArgIII:26 in its active and inactive conformation had major loss of function or gain of function effects, respectively (33, 34).
Acknowledgment
We thank Rob Jones from Arena Pharmaceutical for providing the AR231453 ligand.
The Novo Nordisk Foundation Center for Basic Metabolic Research is supported by an unconditional grant from the Novo Nordisk Foundation to the Faculty of Health Sciences at the University of Copenhagen.
The Schwartz/Baldwin generic numbering system for 7TM receptors, which is based on the actual location of the residues in each transmembrane helix, is used throughout (1).
The large increase in cell surface expression of GPR119 could either be caused indirectly by decreased constitutive activity, because highly constitutively active receptors often are unstable, or it could be caused by an associated effect on the high degree of constitutive internalization observed with GPR119.
- 7TM
- seven-transmembrane
- TM
- transmembrane
- B2AR
- β2-adrenergic receptor
- NK1
- neurokinin-1
- PDB
- Protein Data Bank
- IP3
- inositol 1,4,5-trisphosphate.
REFERENCES
- 1. Schwartz T. W., Frimurer T. M., Holst B., Rosenkilde M. M., Elling C. E. (2006) Molecular mechanism of 7TM receptor activation. A global toggle switch model. Annu. Rev. Pharmacol. Toxicol. 46, 481–519 [DOI] [PubMed] [Google Scholar]
- 2. Altenbach C., Kusnetzow A. K., Ernst O. P., Hofmann K. P., Hubbell W. L. (2008) High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl. Acad. Sci. U.S.A. 105, 7439–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) High resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 5. Rasmussen S. G., Choi H. J., Fung J. J., Pardon E., Casarosa P., Chae P. S., Devree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., Steyaert J., Weis W. I., Kobilka B. K. (2011) Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 7. Choe H. W., Kim Y. J., Park J. H., Morizumi T., Pai E. F., Krauss N., Hofmann K. P., Scheerer P., Ernst O. P. (2011) Crystal structure of metarhodopsin II. Nature 471, 651–655 [DOI] [PubMed] [Google Scholar]
- 8. Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) The 2.6-ångstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu F., Wu H., Katritch V., Han G. W., Jacobson K. A., Gao Z. G., Cherezov V., Stevens R. C. (2011) Structure of an agonist-bound human A2A adenosine receptor. Science 332, 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jensen A. D., Guarnieri F., Rasmussen S. G., Asmar F., Ballesteros J. A., Gether U. (2001) Agonist-induced conformational changes at the cytoplasmic side of transmembrane segment 6 in the β2-adrenergic receptor mapped by site-selective fluorescent labeling. J. Biol. Chem. 276, 9279–9290 [DOI] [PubMed] [Google Scholar]
- 11. Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Kobilka B. K. (2007) GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science 318, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 12. Rasmussen S. G., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., Kobilka B. K. (2011) Crystal structure of the β2-adrenergic receptor-Gs protein complex. Nature 477, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 30, 249–259 [DOI] [PubMed] [Google Scholar]
- 14. Crocker E., Eilers M., Ahuja S., Hornak V., Hirshfeld A., Sheves M., Smith S. O. (2006) Location of Trp265 in metarhodopsin II. Implications for the activation mechanism of the visual receptor rhodopsin. J. Mol. Biol. 357, 163–172 [DOI] [PubMed] [Google Scholar]
- 15. Lin S. W., Sakmar T. P. (1996) Specific tryptophan UV-absorbance changes are probes of the transition of rhodopsin to its active state. Biochemistry 35, 11149–11159 [DOI] [PubMed] [Google Scholar]
- 16. Alexandrov V., Lehnert U., Echols N., Milburn D., Engelman D., Gerstein M. (2005) Normal modes for predicting protein motions. A comprehensive database assessment and associated Web tool. Protein Sci. 14, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Echols N., Milburn D., Gerstein M. (2003) MolMovDB. Analysis and visualization of conformational change and structural flexibility. Nucleic Acids Res. 31, 478–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. (1983) CHARMM. A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 [Google Scholar]
- 19. Holst B., Zoffmann S., Elling C. E., Hjorth S. A., Schwartz T. W. (1998) Steric hindrance mutagenesis versus alanine scan in mapping of ligand binding sites in the tachykinin NK1 receptor. Mol. Pharmacol. 53, 166–175 [DOI] [PubMed] [Google Scholar]
- 20. Holst B., Mokrosinski J., Lang M., Brandt E., Nygaard R., Frimurer T. M., Beck-Sickinger A. G., Schwartz T. W. (2007) Identification of an efficacy switch region in the ghrelin receptor responsible for interchange between agonism and inverse agonism. J. Biol. Chem. 282, 15799–15811 [DOI] [PubMed] [Google Scholar]
- 21. Holst B., Frimurer T. M., Mokrosinski J., Halkjaer T., Cullberg K. B., Underwood C. R., Schwartz T. W. (2009) Overlapping binding site for the endogenous agonist, small-molecule agonists, and ago-allosteric modulators on the ghrelin receptor. Mol. Pharmacol. 75, 44–59 [DOI] [PubMed] [Google Scholar]
- 22. Holst B., Nygaard R., Valentin-Hansen L., Bach A., Engelstoft M. S., Petersen P. S., Frimurer T. M., Schwartz T. W. (2010) A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-transmembrane receptors. J. Biol. Chem. 285, 3973–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Crystal structure of rhodopsin. A G protein-coupled receptor. Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 24. Han M., Lin S. W., Minkova M., Smith S. O., Sakmar T. P. (1996) Functional interaction of transmembrane helices 3 and 6 in rhodopsin. Replacement of phenylalanine 261 by alanine causes reversion of phenotype of a glycine 121 replacement mutant. J. Biol. Chem. 271, 32337–32342 [DOI] [PubMed] [Google Scholar]
- 25. Han M., Lin S. W., Smith S. O., Sakmar T. P. (1996) The effects of amino acid replacements of glycine 121 on transmembrane helix 3 of rhodopsin. J. Biol. Chem. 271, 32330–32336 [DOI] [PubMed] [Google Scholar]
- 26. Jagerschmidt A., Guillaume N., Roques B. P., Noble F. (1998) Binding sites and transduction process of the cholecystokinin B receptor. Involvement of highly conserved aromatic residues of the transmembrane domains evidenced by site-directed mutagenesis. Mol. Pharmacol. 53, 878–885 [PubMed] [Google Scholar]
- 27. Chen S., Lin F., Xu M., Graham R. M. (2002) Phe303 in TMVI of the α1B-adrenergic receptor is a key residue coupling TM helical movements to G-protein activation. Biochemistry 41, 588–596 [DOI] [PubMed] [Google Scholar]
- 28. Spalding T. A., Burstein E. S., Henderson S. C., Ducote K. R., Brann M. R. (1998) Identification of a ligand-dependent switch within a muscarinic receptor. J. Biol. Chem. 273, 21563–21568 [DOI] [PubMed] [Google Scholar]
- 29. Baranski T. J., Herzmark P., Lichtarge O., Gerber B. O., Trueheart J., Meng E. C., Iiri T., Sheikh S. P., Bourne H. R. (1999) C5a receptor activation. Genetic identification of critical residues in four transmembrane helices. J. Biol. Chem. 274, 15757–15765 [DOI] [PubMed] [Google Scholar]
- 30. Chen S., Lin F., Xu M., Riek R. P., Novotny J., Graham R. M. (2002) Mutation of a single TMVI residue, Phe282, in the β2-adrenergic receptor results in structurally distinct activated receptor conformations. Biochemistry 41, 6045–6053 [DOI] [PubMed] [Google Scholar]
- 31. Govaerts C., Lefort A., Costagliola S., Wodak S. J., Ballesteros J. A., Van Sande J., Pardo L., Vassart G. (2001) A conserved Asn in transmembrane helix 7 is an on/off switch in the activation of the thyrotropin receptor. J. Biol. Chem. 276, 22991–22999 [DOI] [PubMed] [Google Scholar]
- 32. Urizar E., Claeysen S., Deupí X., Govaerts C., Costagliola S., Vassart G., Pardo L. (2005) An activation switch in the rhodopsin family of G protein-coupled receptors. The thyrotropin receptor. J. Biol. Chem. 280, 17135–17141 [DOI] [PubMed] [Google Scholar]
- 33. Seibold A., Dagarag M., Birnbaumer M. (1998) Mutations of the DRY motif that preserve β2-adrenoceptor coupling. Receptors Channels 5, 375–385 [PubMed] [Google Scholar]
- 34. Valentin-Hansen L., Groenen M., Nygaard R., Frimurer T. M., Holliday N. D., Schwartz T. W. (2012) The arginine of the DRY motif in transmembrane segment III functions as a balancing micro-switch in the activation of the β2-adrenergic receptor. J. Biol. Chem. 287, 31973–31982 [DOI] [PMC free article] [PubMed] [Google Scholar]