Background: YchF is a universally conserved ATPase of unknown function.
Results: YchF inhibits catalase activity, and high YchF levels cause H2O2 hypersensitivity. YchF is negatively regulated by OxyR in response to H2O2.
Conclusion: YchF is a universally conserved negative regulator of the oxidative stress response that acts by a post-translational mechanism.
Significance: This is the first functional description of the YchF protein family.
Keywords: ATPases, Bacterial Metabolism, Cancer Biology, GTPase, Hydrogen Peroxide, Oxidative Stress, YchF, Catalase, hOla1
Abstract
YchF is an evolutionarily conserved ATPase of unknown function. In humans, the YchF homologue hOla1 appears to influence cell proliferation and was found to be up-regulated in many tumors. A possible involvement in regulating the oxidative stress response was also suggested, but details on the underlying mechanism are lacking. For gaining insight into YchF function, we used Escherichia coli as a model organism and found that YchF overexpression resulted in H2O2 hypersensitivity. This was not caused by transcriptional or translational down-regulation of H2O2-scavenging enzymes. Instead, we observed YchF-dependent inhibition of catalase activity and a direct interaction with the major E. coli catalase KatG. KatG inhibition was dependent on the ATPase activity of YchF and was regulated by post-translational modifications, most likely including an H2O2-dependent dephosphorylation. We furthermore showed that YchF expression is repressed by the transcription factor OxyR and further post-translationally modified in response to H2O2. In summary, our data show that YchF functions as a novel negative regulator of the oxidative stress response in E. coli. Considering the available data on hOla1, YchF/Ola1 most likely execute similar functions in bacteria and humans, and their up-regulation inhibits the ability of the cells to scavenge damaging reactive oxygen species.
Introduction
P-loop GTPases execute and regulate essential biological processes and show high sequence conservation, resulting in a set of eight GTPase families that are universally conserved in eukaryotes, bacteria, and archaea (1). These GTPases include YihA and HlfX, which are involved in ribosome biogenesis; initiation factor 2, elongation factor Tu, and elongation factor G, which are required for protein synthesis; and the signal recognition particle and its receptor (FtsY), which are essential for co-translational protein targeting. It also includes YchF, a member of the Obg family of universally conserved GTPases. YchF differs from the other GTPases because it preferentially hydrolyzes ATP over GTP (2), thereby functioning as an ATPase. This altered nucleotide specificity is due to the replacement of a conserved lysine residue within the nucleotide binding motif by valine in the Escherichia coli YchF or by leucine in the human YchF homologue hOla13 (2, 3).
Although the x-ray structures of YchF from Haemophilus influenzae and human Ola1 have been determined (2, 4), the exact function of these ATPases is unknown. Because of its structural characteristics, YchF was proposed to bind to ribosomes as well as to nucleic acids (4) and was classified as a member of the translation factor family of nucleotide-hydrolyzing proteins (TRAFAC family). Consistent with this idea, YchF was shown to bind to ribosomes in the protozoan parasite Trypanosoma cruzi and in the proteobacterium E. coli (3, 5). Additionally, the yeast homologue of YchF, Ybr025c, was suggested to interact with the elongation factor eEF1 (6). These data support a role for YchF during translation, but the physiological significance of YchF binding to ribosomes and possible consequences for translation are currently unknown.
A translation-independent function was suggested for hOla1. Reducing the concentration of hOla1 in HeLa cells increased the cellular resistance to peroxide oxidants and thiol-depleting chemicals, whereas the overexpression of hOla1 increased the sensitivity of HeLa cells toward oxidative stress (7). This could indicate that hOla1 functions as a negative regulator of the oxidative stress response. Whether hOla1 inhibits directly one or more antioxidant enzymes or whether it acts by another mechanism, e.g. by influencing protein degradation, is not known. The latter possibility is supported by data showing an interaction of the yeast homologue Ybr025c with subunits of the proteasome (8).
The possible involvement of hOla1 in regulating the stress response is consistent with data showing its down-regulation in cells treated with DNA-damaging agents or UV light (9). hOla1 overexpression has been observed in many tumor cells (9–11), which could indicate that at high hOla1 concentrations the cellular response to potentially mutagenic substances is impaired. A link between hOla1 and cancer is also deduced from the observation that down-regulation of hOla1 inhibits the motility and invasion of breast cancer cells (12) and the proliferation of neuronal and pancreatic cells (13). Thus, hOla1 appears to play an important role in the cellular stress response, cell proliferation, and tumor development, but details on its mode of action are entirely unknown.
Equally little is known about the bacterial YchF homologues. They are predicted to be located in the cytosol of bacterial cells and show high sequence similarity to hOla1; e.g. the E. coli YchF amino acid sequence is 45% identical and 62% similar to the amino acid sequence of hOla1. YchF is not essential in either E. coli or in Bacillus subtilis, and the only phenotype reported so far for an E. coli ΔychF mutant is weak cold sensitivity (14, 15). A ΔychF mutant of the halophilic Gram-negative pathogen Vibrio vulnificus was shown to exhibit reduced cytotoxic effects on macrophages (16), suggesting an involvement of YchF in inactivating the host defense. Reduced virulence was also observed in a ΔychF mutant of Streptococcus pneumonia (17). In contrast, a ΔychF mutant of the facultative intracellular pathogen Brucella melitensis did not show reduced cytotoxicity but was, like the V. vulnificus ΔychF mutant, impaired in iron metabolism (16, 18). Although these phenotypic reports link bacterial YchF homologues to important biological processes, details on its function are missing.
In the current study, we analyzed the effect of ychF overexpression and ychF deletion on stress tolerance in E. coli. Our data identify YchF as an important regulator of the oxidative stress response that functions by a post-translational mechanism.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
E. coli BW25113 and BL21 were used as wild type E. coli strains and were routinely grown on LB medium at 37 °C. The E. coli strains JW1194 (ΔychF) and JW3933 (ΔoxyR) were provided by the National BioResource Project (National Institute of Genetics, Japan). Both strains were grown on LB medium supplemented with 25 μg/ml kanamycin. Strains carrying pTrc99a-YchF (pYchF) plasmids were supplemented with 50 μg/ml ampicillin, and strains carrying pASK-IBA3C were supplemented with 35 μg/ml chloramphenicol.
Plasmid Construction
ychF was isolated from chromosomal E. coli DNA and cloned via XhoI and NdeI into the plasmid pET-22b. This construct was a gift from S. Angelini, Freiburg, Germany and used for subcloning ychF into plasmid pTrc99a. The primers YchFfw and YchFrev (Table 1) were used to insert XbaI and HindIII restriction sites. pth-ychF was amplified from chromosomal DNA and cloned into pTrc99a as well. The PCR was performed with Phusion High Fidelity PCR Master Mix (Finnzymes, Vantaa, Finland). PCR fragment purification from the agarose gel was done with a QIAquick gel extraction kit (Qiagen, Hilden, Germany), and the purified fragment as well as the plasmid pTrc99a was used for the restriction digest with HindIII and XbaI from New England Biolabs, Ipswich, MA. Vector and insert were ligated with the Quick Ligation kit (New England Biolabs). After ligation, pTrc99a-YchF was used to construct the mutants YchF(S16A), YchF(S16E), and YchF(P11/N12A) via inverse PCR with the primer pairs listed in Table 1. These constructs were transformed into E. coli BW25113 and JW1194. The plasmid pASK-IBA3C-OxyR was constructed by amplifying oxyR from chromosomal E. coli DNA using the primers OxyRFw and OxyRrev. After purification of the PCR product and BsaI digestion, the PCR product was cloned into BsaI-digested pASK-IBA3C vector (IBA, Göttingen, Germany), resulting in OxyR with a C-terminal Strep tag under Tet promoter control. YchF amber stop codon mutants were constructed for in vivo cross-linking by inverse PCR on pTrc99a-YchF using the primers given in Table 1. These constructs were transformed into E. coli BL21 containing pSUP-BpaRS-6TRN (19) encoding the orthogonal aminoacyl-tRNA synthetase/tRNA pair for incorporation of para-benzoyl-l-phenylalanine (pBpa) into the position of the amber codon.
TABLE 1.
Nucleotide primers used in this study
| Name | Sequence |
|---|---|
| YchFfw | 5′-ACAATTCCCCTCTAGAAATAATTTTG-3′ |
| YchFrev | 5′-CAACTCAGCAAGCTTTCGGGCT-3′ |
| Pthfw | 5′-CGCCAGTTATCTAGACACTCAGG-3′ |
| Pth+YchF_rev | 5′-GATTTAGCGAAAGCTTATGAGAC-3′ |
| YchF(S16A)fw | 5′-TCGGGAAAGCAACCCTG-3′ |
| YchF(S16A)rev | 5′-CGTTGGGCAAACCGAC-3′ |
| YchF(S16E)fw | 5′-TCGGGAAAGAGACCCTG-3′ |
| YchF(S16E)rev | 5′-CGTTGGGCAAACCGAC-3′ |
| YchF(P11AN12A)fw | 5′-TTGGCCGCCGTCGGGAAATC-3′ |
| YchF(P11AN12A)rev | 5′-ACCGACGATACCGCATTTGAATCC-3′ |
| OxyRFw | 5′-ATGGTAGGTCTCAAATGAATATTCGTGATCTTGAGTACCTGG-3′ |
| OxyRrev | 5′-ATGGTAGGTCTCAGCGCTAACCGCCTGTTTTAAAACTTATCG-3′ |
| OxyR_RTforward | 5′-TGGCAAGCCAGCAGGGCG-3′ |
| OxyR_RTreversed | 5′-CACAGCCAGCGCTGGCAG-3′ |
| rplB_RTforward | 5′-AGTTGTTAAATGTAAACCGACAT-3′ |
| rplB_RTreversed | 5′-ATTTG-CTACGGCGACGTACGATG-3′ |
| YchF_RTforward | 5′-GCGGTATCGTCGGTTTGCCC-3′ |
| YchF_RTreversed | 5′-AAAAGGAAGTTCATCACATCGCC-3′ |
| YchF-300fw+bio | 5′-CATCGGAATCGGTCA-TACCGG-3′ |
| YchF-300rev | 5′-AAGTTGGCCGC-TTCAATACCGG-3′ |
| YidC-300fw+bio | 5′-ACCTCGATAACCGTGCTCTCTCGG-3′ |
| YidC-300rev | 5′-TGGGCCTGAGGTTGCGGG-3′ |
| YchF(N20pBa)fw | 5′-TCTACCCTGTTCTAGGCGCTGACCAAA-3′ |
Growth Analyses
Cells were grown overnight, diluted 1:100 in liquid LB medium, and further incubated until they reached an A600 of 0.5–0.8. These cells were then used for measuring H2O2 sensitivity by the following methods.
Spot Assay
The culture was adjusted to an A600 of 0.5 and serially diluted. 10 μl of each dilution was spotted onto LB plates supplemented with or without H2O2. After overnight incubation at 37 °C, plates were analyzed.
Cell Viability Assay
Cells were adjusted to an A600 of 0.5 and diluted 1:10 in phosphate-buffered saline (PBS) before treatment with 10 mm H2O2 in PBS for 50 min at 25 °C; control cells were treated with PBS. Subsequently, cells were harvested by centrifugation, washed with PBS, and resuspended in 1 ml of PBS. 100 μl of this cell culture was transferred to a 96-well plate, and 100 μl of the BacTiter-Glo microbial cell viability assay solution (Promega Corp., Mannheim, Germany) was added. The luminescence of H2O2-treated wild type cells was set to 100%.
Inhibition Assay
Cultures were adjusted to an A600 of 0.5, mixed with LB-Top agar, and poured on LB plates. Subsequently, sterile filter discs were soaked in 2 mm H2O2 solution and placed on top of these plates. After approximately 5 h of incubation at 37 °C, the inhibition zones around the discs were quantified.
Isolation of RNA and RT-PCR
RNA isolation was performed by using the Illustra RNAspin minikit from GE Healthcare. RNA was eluted with RNase-free water and tested for quality and purity by agarose gel electrophoresis and absorption measurement. Possible DNA contaminations were detected by running a standard PCR without prior reverse transcriptase treatment. If necessary, an additional DNase treatment was performed. RNA was stored at −80 °C. RT-PCR was performed with the RT-PCR kit from Qiagen according to the enclosed manual in 25-μl reactions. The following primers were used: OxyR_RTforward/OxyR_RT reversed, rplB_RTforward/rplB_RT reversed, and YchF_RTforward/YchF_RT reversed.
Electrophoresis Mobility Shift Assays
DNA binding of OxyR was detected by the GelshiftTM EMSA kit (Active Motif, La Hulpe, Belgium). In brief, the 300-nucleotide upstream region of ychF was amplified using the biotinylated primer YchF-300fw+bio and the primer YchF-300rev. The PCR product was purified using the QIAquick gel extraction kit (Qiagen). As a control, the 300-nucleotide upstream region of yidC was amplified using the biotinylated primer YidC-300fw+bio and the primer YidC-300rev. The DNA (20-fmol final concentration) was incubated with 20 fmol or 1,500 fmol of purified OxyR for 20 min at 23 °C in the provided EMSA buffer. Subsequently, 5 μl of the provided loading dye was added, and the sample was separated on a 5% Tris borate-EDTA acrylamide gel at 100 V and 4 °C. The gel was blotted onto a nylon membrane (Whatman Nytran SPC membrane), and the DNA was UV-cross-linked to the membrane for 1.5 min. Binding of OxyR to biotinylated DNA was detected by chemiluminescence using horseradish peroxidase-coupled streptavidin.
Protein Purification
YchF was purified via its C-terminal His tag. Cells were grown to an A600 of ∼0.8 and then for additional 30 min incubated with or without H2O2. Protein expression was not induced by isopropyl β-d-1-thiogalactopyranoside because the basal expression level from the lac promoter was sufficient. Cells were collected by centrifugation and resuspended in extraction buffer (50 mm HEPES, pH 7.6, 1 m ammonium acetate, 10 mm magnesium acetate) using 1 ml of buffer/1 g of wet cell pellet. Before cells were lysed by French pressing (two passes at 8,000 p.s.i.), 0.5 mm PMSF (Roche Applied Science) and 1× Complete protease inhibitor mixture (Roche Applied Science) were added to prevent protein degradation. Cell debris was removed by 30-min centrifugation at 15,500 rpm (Sorvall SS-34 rotor), and the supernatant was incubated with Talon metal affinity beads (Clontech) for 1 h at 4 °C. After centrifugation at 1,000 rpm for 10 min, the supernatant was removed, and the Talon-bound material was washed four times with 5 mm imidazole in extraction buffer. After washing, the material was placed on a column and eluted with 200 mm imidazole in extraction buffer. This buffer was exchanged with HT buffer (100 mm HEPES, pH 7.6, 200 mm potassium acetate, 20 mm magnesium acetate) via a PD-10 column (GE Healthcare). The quality of the purified protein was controlled via SDS-PAGE and PAGE blue staining, and the purified protein was stored at −20 °C.
OxyR was purified from E. coli BL21 cells carrying pASK-IBA3C-OxyR that were induced at an A600 of 0.3 with 200 μg of anhydrotetracycline chloride, and cell extracts were prepared as described above with the exception that all steps were performed in buffer W (100 mm Tris/HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA). Cell extracts were applied to buffer W-equilibrated streptavidin columns and washed five times with 1 column volume of buffer W. Proteins were eluted with 2.5 mm desthiobiotin in buffer W, and purification was controlled via SDS-PAGE. Fractions containing pure OxyR were pooled and subjected to protease Xa cleavage to remove the Strep tag. The protease digestion was performed in a 50-μl reaction at 23 °C overnight under the following conditions: 50 μg of OxyR, 20 mm Tris/HCl, pH 8.0, 100 mm NaCl, 2 mm CaCl2, 1 μl of Protease Xa (2 units/μl; Qiagen). The cleaved Strep tag was removed via streptavidin columns, and complete cleavage was controlled via Western blot and anti-Strep antibodies (IBA).
In Vivo Formaldehyde Cross-linking
E. coli BW25113 pYchF was grown to an A600 of 1.0 in LB medium and treated for 30 min with 20 mm H2O2. Subsequently para-formaldehyde (PFA) in PFA buffer (136 mm NaCl, 2.7 mm KCl, 1.8 mm KH2PO4, 10 mm Na2HPO4, pH 6.5) was added to a final concentration of 0.1%, and cells were incubated for 30 min at 37 °C before the cells were harvested. For cell fractionation, cells were resuspended in extraction buffer (see above) supplemented with 50 μm PMSF and 100 μg/μl lysozyme. Cells were incubated for 30 min at 37 °C and subjected to repeated freezing and thawing cycles before French pressing. Further processing and YchF purification followed the protocol described above.
In Vivo Site-directed Cross-linking
E. coli BL21 cells carrying plasmids pSUP-BpaRS-6TRN and pYchF or its derivatives were grown overnight and diluted 1:200 in 400 ml of minimal medium (19). 440 μl of 0.8 m pBpa in 1 m NaOH was added to each culture, and the cells were grown at 30 °C for 22 h. The expression of YchF and the YchF amber stop codon mutants was then induced by the addition of 1 mm isopropyl β-d-1-thiogalactopyranoside, and growth was continued for another 4 h. Subsequently, the cells were harvested by centrifugation at 15,800 rpm (Sorvall SLA-3000 rotor) and resuspended in 20 ml of 50 mm triethanolamine acetate, pH 7.5, 30% glycerol for storage at −80 °C.
Cells were thawed and centrifuged for 12 min at 5,000 rpm before the pellet was washed with 1× PBS buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.76 mm KH2PO4, pH 7.8) and centrifuged a second time. Subsequently, the cells were resuspended in 8 ml of PBS buffer and transferred into two 6-well microtiter plates for photocross-linking. One plate was exposed to UV light for 20 min, and the other remained unexposed. The cells were then centrifuged again for 7 min at 5,000 rpm and resuspended in EPTx buffer (50 mm HEPES, 1 m ammonium acetate, 20 mm magnesium acetate, 1% Triton X-100). Prior to French pressing, 10 μl each of 0.5 mm PMSF (Roche Applied Science) and 1× Complete protease inhibitor mixture (Roche Applied Science) was added to each tube. The cells were then subjected three times to a French pressure step and centrifuged at 15,800 rpm (Sorvall SS-34 rotor) for 20 min. YchF was purified by its C-terminal His tag as described above. Protease inhibitors were again added to prevent protein degradation. 100 μl of the eluted material was separated by SDS-PAGE and analyzed by Western blot.
Western Blot Analysis
After SDS-PAGE, proteins were electrotransferred by tank blotting to a nitrocellulose membrane (GE Healthcare). Peptide antibodies were raised in rabbits by GenScript (Piscataway, NJ). The following peptides were used: YchF, VNEDGFENNPYLDQC; OxyR, CRPGSPLRSRYIQLA. The Dps antiserum used in this work was kindly supplied by Regine Hengge, Freie Universität Berlin, Germany. The RplB antibody was a gift from Richard Brimacombe, Max Planck Institute for Molecular Genetics, Berlin, Germany. The YidC antibody was raised in rabbits against the complete protein. KatG antibodies were obtained from Agrisera, Vännas, Sweden, and the LexA antibody was from Active Motif. All the antibodies were gained from rabbit serum except for RplB, which was raised in goat. Horseradish peroxidase-coupled goat anti-rabbit antibodies or sheep anti-goat antibodies from Caltech Laboratories (Burlingame, CA) were used as secondary antibodies, and enhanced chemiluminescence (ECL) reagent (GE Healthcare) was used as detection substrate.
Catalase Activity Assay
Overnight-grown cells were diluted 1:100 in LB medium and grown at 37 °C to an A600 of 0.8 before H2O2 was added, and the cells were incubated for additional 30 min. The cell pellets were resuspended in lysis buffer (100 mm Tris/HCl, pH 8.0, 500 mm NaCl, 1 mm EDTA), and lysozyme was added to a final concentration of 100 μg/ml. After incubation at 37 °C for 30 min, samples were subjected to at least four cycles of freezing in liquid nitrogen and thawing at 37 °C. After 15-min incubation with DNase (final concentration, 50 μg/ml) at 30 °C, samples were centrifuged for 10 min at 10,000 × g, and the supernatant was collected in a fresh tube. The samples were tested for catalase activity using the Catalase Assay kit (Cayman Europe, Tallinn, Estonia).
ATP Hydrolysis Assay
Purified YchF was incubated with γ-33P-labeled ATP to analyze the ATP hydrolysis rate. The radioactive nucleotide was diluted 1:500 with non-labeled ATP, and a 1–100 μm concentration of this solution was incubated with YchF (50 ng if not indicated otherwise) in reaction buffer (50 mm Tris-Cl, pH 7.5, 5 mm MgCl2, 200 mm NaCl) at 37 °C for 10 min. Subsequently, 800 μl of 5% charcoal in 20 mm phosphoric acid was added for absorbing ATP and ADP, and the samples were incubated for 10 min on ice. After a 10-min centrifugation step at 13,000 rpm in a table top centrifuge, 200 μl of the supernatant, which contained the hydrolyzed 33P, was added to 3 ml of scintillation mixture. The samples were measured in a scintillation counter (PerkinElmer Life Sciences), and after preparing a calibration curve, the hydrolyzed ATP was calculated.
Mass Spectrometric Analysis
For the identification of putative interaction partners, YchF was affinity-purified as described above in the presence of phosphatase inhibitors (2 mm sodium orthovanadate, 9.5 mm NaF, 10 mm β-glycerophosphate, 10 mm sodium pyrophosphate), and eluates were subjected to tryptic in-solution digestion (trypsin to protein ratio, 1:50) for 4 h at 42 °C and analyzed by LC/electrospray ionization-MS using an UltiMate 3000 RSLCnano/LTQ-Orbitrap XL system (Thermo Fisher Scientific, Bremen, Germany) essentially as described previously (20). Mass spectrometric data sets were processed using the software suite MaxQuant (version 1.2.7.4) (21, 22). Proteins were identified by searching peak lists against the EcoProt database (v.31.01.2012; 4,275 entries; tagged YchF sequence added) applying a false discovery rate of <1% at both the peptide and protein levels. Criteria for protein identification were as follows: MS and MS/MS mass tolerances of 6 ppm and 0.5 Da, respectively; at least one unique peptide (≥7 amino acids); a maximum of two missed cleavages; methionine oxidation and phosphorylation at serine, threonine, and tyrosine were considered as variable modifications. Quantitative information about proteins was obtained using the label-free protein quantification option in MaxQuant with default settings and the “match between runs” option with a retention time window of 2 min. Only razor and unique peptides were considered.
RESULTS
YchF Overexpression Causes H2O2 Hypersensitivity in E. coli
For analyzing the possible contribution of YchF to the oxidative stress response in E. coli, the H2O2 resistance of wild type (WT) E. coli cells, a YchF deletion strain (ΔychF), and E. coli cells expressing YchF under the control of the lac promoter from the plasmid pTrc99A (pYchF) was determined. On LB agar plates without H2O2, cell growth of all strains was comparable (Fig. 1A), but on plates containing 2 mm H2O2, wild type cells overexpressing YchF showed a significant growth inhibition. The same H2O2 concentration did not significantly impair the growth of WT E. coli cells lacking an additional plasmid copy of ychF. Growth of the ΔychF strain in the presence of H2O2 was slightly better than that of the WT but was also impaired by overexpression of ychF from a plasmid although to a lower degree than WT E. coli.
FIGURE 1.
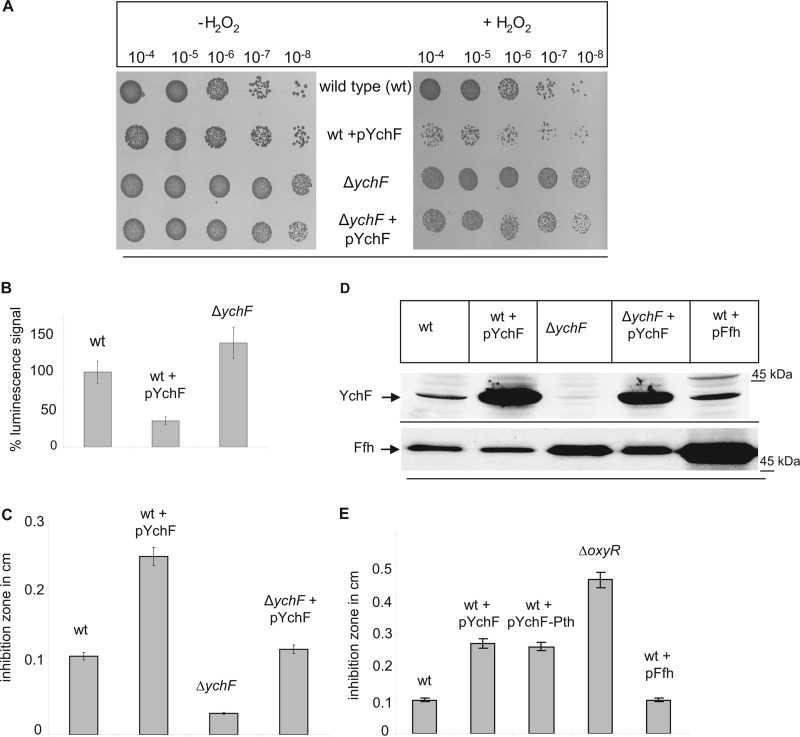
Overexpression of YchF in E. coli leads to H2O2 hypersensitivity. A, the indicated strains were inoculated in LB medium and grown to an A600 of ∼0.5 before stepwise dilution in LB medium. From each dilution, 10-μl cell suspensions were spotted onto LB plates in the presence or absence of 2 mm H2O2. Cell growth was analyzed after overnight incubation at 37 °C. B, cells were adjusted to an A600 of 0.5 and after a 1:10 dilution treated with 10 mm H2O2 in PBS for 50 min at 25 °C. After a wash step, 100 μl of the cell culture was transferred to a 96-well plate, 100 μl of the BacTiter-Glo microbial cell viability assay solution was added, and luminescence was recorded. The luminescence signal of wild type E. coli was set to 100%. Shown are the mean values of at least three independent experiments. The error bars indicate the standard deviation. C, for the agar diffusion assay, E. coli cells were mixed with top agar and poured onto LB agar plates. Sterile discs of filter paper were soaked with 2 mm H2O2 and placed on top of the cell agar mixture. After 4 h of incubation, inhibition zones were measured. The data were obtained in at least three independent experiments, and the mean values are shown. D, Western blot using whole cells (approximately 0.5 × 108 cells) precipitated on ice for 30 min with trichloroacetic acid (TCA). After centrifugation, the pellet was resuspended in SDS loading buffer and separated by SDS-PAGE. After Western transfer, the membrane was probed with α-YchF antibodies. As a control, antibodies against Ffh were used. E, the agar diffusion assay using the indicated strains was performed as described in C.
The H2O2 hypersensitivity of ychF-overexpressing cells was confirmed by two additional quantitative assays. In cell viability assays, the metabolic activity of cells after a 50-min exposure to 10 mm H2O2 was analyzed by measuring their ATP content via luciferase luminescence. The luminescence signal of H2O2-treated wild type cells was set to 100% and taken as reference. Cells overexpressing ychF showed a significantly reduced luminescence signal, indicating that viability of these cells was drastically compromised after H2O2 treatment (Fig. 1B), whereas the signal of the ΔychF strain was slightly stronger than in the wild type (Fig. 1B). These data confirm the results from the spot test (Fig. 1A). An agar disk diffusion test further validated that increased YchF concentrations caused increased H2O2 sensitivity in E. coli. Paper disks were soaked in H2O2 and placed on LB plates overlaid with E. coli cells in top agar. The diameter of the inhibition zone of WT+pYchF approximately doubled in comparison with wild type cells (Fig. 1C). The ΔychF strain showed a smaller inhibition zone, indicating that in the absence of YchF cells are more resistant toward H2O2, which is also visible in the spot assay (Fig. 1A). The H2O2 sensitivity in the ΔychF strain containing pYchF was weaker than in the pYchF-carrying wild type strain, which is probably related to the lower amount of ychF in this strain (Fig. 1D).
For correlating the observed growth defects with the cellular YchF concentrations, we performed Western blotting using peptide antibodies against YchF. The YchF concentration was significantly increased in cells carrying a plasmid-borne copy of ychF (pYchF), but the concentration in ΔychF+pYchF was lower than in WT+pYchF (Fig. 1D), which probably explains why H2O2 sensitivity was less pronounced in ΔychF+pYchF (Fig. 1, A and C). Antibodies against Ffh, the protein component of the bacterial signal recognition particle, were used as a control.
In E. coli and many enterobacteria, ychF is co-transcribed with pth, which encodes an essential peptidyl-tRNA hydrolase (23–25). We therefore tested whether overexpression of ychF-pth caused the same H2O2 sensitivity as overexpression of ychF alone. The H2O2 hypersensitivity of WT and ΔychF strains expressing either ychF alone or ychF-pth was comparable (Fig. 1E), indicating that H2O2 hypersensitivity is correlated with the YchF concentration and that Pth had no significant impact. As additional controls, we analyzed the H2O2 hypersensitivity of a strain lacking OxyR (ΔoxyR), the major transcriptional regulator of the oxidative stress response (26), and a wild type strain overexpressing Ffh (pFfh), which is, like YchF, one of the eight universally conserved NTPases (1). The ΔoxyR strain displayed an enhanced sensitivity toward H2O2, which was more pronounced than the H2O2 sensitivity of ychF-overexpressing strains, suggesting that overexpressing ychF does not completely block the oxidative stress response. Overexpression of Ffh did not significantly influence the H2O2 sensitivity of E. coli, further substantiating that H2O2 hypersensitivity is specifically the result of YchF overexpression. In summary, our data show that high levels of YchF cause hypersensitivity toward oxidative stress in E. coli and thus result in the same phenotype as the overexpression of hOla1 in humans (7).
YchF Does Not Influence the Cellular Concentrations of KatG and OxyR
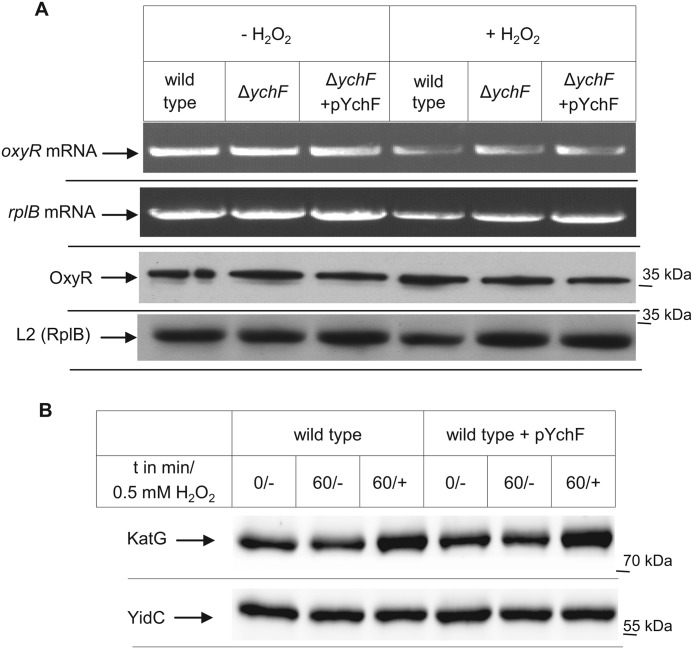
Based on the presence of a putative nucleic acid binding motif within YchF (2, 4) and its ability to bind to E. coli ribosomes (3), it appeared likely that ychF overexpression influenced the expression of oxidative stress response proteins like OxyR and the catalase KatG (26). The expression of these proteins was therefore analyzed by RT-PCR and Western blot analyses. Treating E. coli cells with H2O2 did not significantly influence the amount of the oxyR mRNA (Fig. 2A, upper panels). This is in agreement with previous data showing that H2O2 activates OxyR by specific cysteine oxidation (27) but does not significantly change the cellular OxyR concentration (28). H2O2 stress also did not change the amount of oxyR mRNA in ΔychF cells or in cells overexpressing ychF (Fig. 2A, upper panels). As a control, the mRNA level of the ribosomal protein L2 was analyzed, and this revealed that comparable amounts of total mRNA were used in these assays. Like the mRNA level, we did not notice any difference in the OxyR protein levels (Fig. 2A, lower panels). This demonstrated that the amount of YchF does not significantly influence the transcription or translation of OxyR. Thus, YchF probably functions downstream of OxyR, which would also explain why H2O2 hypersensitivity in the ΔoxyR strain is more pronounced than in the ychF-overexpressing strain (Fig. 1E).
FIGURE 2.
YchF does not influence the steady-state stability of OxyR or KatG. A, RNA was prepared from the indicated strains that were grown at 37 °C up to an A600 of 0.5 and then further incubated for 15 min in the presence or absence of 20 mm H2O2. RT-PCR was performed, and products were analyzed on a 1% agarose gel. 100 ng of RNA was used per 25-μl reverse transcriptase reaction. Primers specific for the ribosomal protein L2 (rplB) served as a control. An aliquot of the same cells (approximately 0.5 × 108 cells) used for RNA preparation was directly TCA-precipitated as described in the legend to Fig. 1, separated by SDS-PAGE, transferred to a nitrocellulose membrane, and subsequently decorated with antibodies against OxyR and L2 (rplB). B, cells were grown to an A600 of 0.5 and incubated for an additional 1 h in the presence or absence of 0.5 mm H2O2. 0.5 × 108 cells were processed as described in the legend to Fig. 1. Samples were taken immediately before H2O2 addition (0/−), 60 min after incubation without H2O2 (60/−), and 60 min after incubation with H2O2 (60/+). As a loading control, the Western blot was also decorated with antibodies against YidC.
KatG is the major catalase during exponential growth of E. coli, and its expression is induced by OxyR in response to H2O2 (26, 27). When cells were treated with H2O2, we observed an increase in the cellular amounts of KatG by Western blotting using α-KatG antibodies (Fig. 2B). However, a similar increase in the cellular KatG concentration was also observed in cells overexpressing ychF (Fig. 2B). The membrane protein YidC served as a loading control. In summary, these data demonstrate that the H2O2 hypersensitivity at high YchF concentrations is not the result of a YchF-dependent down-regulation of major oxidative stress response proteins like KatG and OxyR and not the result of increased KatG degradation.
YchF Suppresses the Oxidative Stress Response by Direct Interaction with KatG
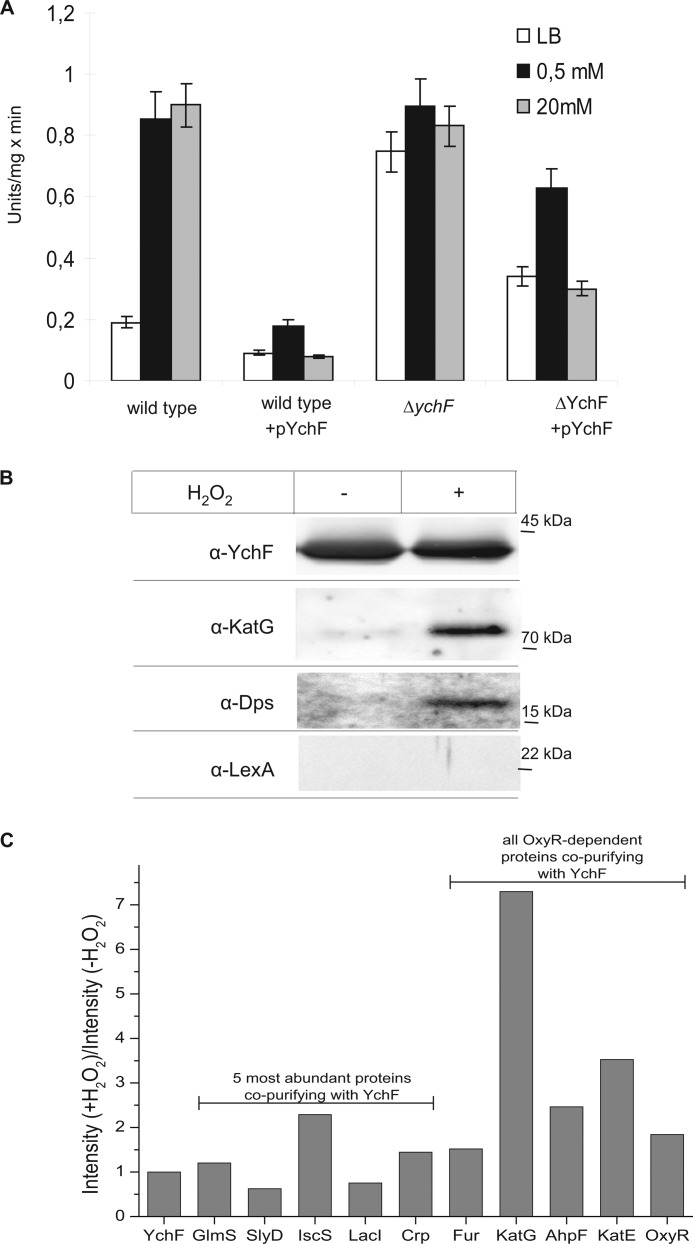
We next analyzed whether H2O2 hypersensitivity was the result of a direct YchF-dependent inhibition of H2O2-detoxifying enzymes like catalases. Catalase activity was measured in cell extracts from H2O2-treated wild type cells, ΔychF cells, and ychF-overexpressing cells. Catalase activity was low in wild type cells without H2O2 treatment (Fig. 3A) but was stimulated approximately 5-fold in extracts from cells treated with 0.5 or 20 mm H2O2 (Fig. 3A). This increased activity is in agreement with the higher KatG concentration in H2O2-treated cells as observed by Western blotting (Fig. 2B). In comparison, the basal catalase activity was reduced by approximately 50% in cell extracts from ychF-overexpressing cells (Fig. 3A). Importantly, we did not observe a strong H2O2-dependent stimulation of catalase activity in these cells, which explains why ychF-overexpressing cells are H2O2-hypersensitive. In ΔychF cell extracts, the basal catalase activity was about 4-fold higher than in wild type extracts and only slightly increased further when cells were treated with 0.5 or 20 mm H2O2. In summary, these data demonstrate that high concentrations of YchF inhibit catalase activity in E. coli.
FIGURE 3.
YchF inhibits catalase activity in E. coli. A, the indicated strains were grown in the presence or absence of H2O2 and subsequently lysed by lysozyme and freezing/thawing cycles. Catalase activity was measured as peroxidatic activity using methanol as a substrate. The values represent the mean values of two independent experiments. Error bars indicate the standard deviation. B, YchF was purified via metal affinity chromatography from cells treated for 30 min with or without 20 mm H2O2. The purified protein (approximately 10 μg) was separated by SDS-PAGE and transferred to nitrocellulose for immune detection using antibodies against YchF, KatG, Dps, and LexA. C, YchF was purified as described in B, and co-purifying proteins were identified by mass spectrometry. Data were normalized to the intensity ratio of YchF from H2O2-treated versus non-treated cells. Shown are the ratios (+H2O2/−H2O2) of the five most abundant proteins detected in the YchF sample as well as the ratios of all identified OxyR-regulated proteins. Note that Dps could not be identified by mass spectrometry.
H2O2 is detoxified in E. coli by three different enzymes. AhpCF functions as an FAD-dependent hydroperoxide reductase that can reduce H2O2 in an NADH/H+-dependent reaction. However, AhpCF is easily saturated when intracellular H2O2 concentrations exceed 20 μm, and subsequently, KatG becomes the primary scavenging enzyme (26). Although AhpCF and KatG expression is regulated by OxyR in an H2O2-dependent manner, the third catalase in E. coli, KatE, is also under the control of σS and is induced during stationary phase (29). Thus, under our experimental conditions, KatG is probably mainly responsible for H2O2 elimination. The catalase assay system used in our study also mainly determines KatG activity because it is based on the peroxidatic activity of catalases and uses methanol as a substrate. This activity is very low in KatE and AhpCF but high in catalase-peroxidases like KatG (30).
For elucidating whether YchF interacts directly with KatG, E. coli cells expressing YchF were treated with H2O2, and YchF was purified via its C-terminal His tag. As a control, YchF was also purified from cells that were not H2O2-treated, and both YchF preparations were analyzed for co-purifying proteins. Western blot analysis revealed that comparable amounts of YchF were purified from both cultures (Fig. 3B). Purified YchF from the control cells (−H2O2) contained only a small amount of KatG (Fig. 3B), but the amount of KatG co-purifying with YchF increased significantly when YchF was purified from H2O2-treated cells (Fig. 3B). Like KatG, the iron-scavenging protein Dps also co-purified with YchF in H2O2-treated cells (Fig. 3B). As a control, we used antibodies against LexA, another stress response protein that is not directly linked to oxidative stress but involved in the SOS DNA damage control pathway. LexA did not co-purify with YchF, supporting a specific interaction of YchF with proteins of the oxidative stress response.
It is important to emphasize that the intracellular levels of KatG and Dps increase upon H2O2 treatment (26), and therefore, their co-purification with YchF could just reflect this increase in concentration. On the other hand, LexA expression is also increased upon H2O2 treatment (31), but it did not co-purify with YchF. Nevertheless, we also followed an unbiased mass spectrometry-based approach for identifying proteins that co-purified with YchF in the presence or absence of H2O2 (Fig. 3C). Data were normalized to the intensity ratio of YchF from H2O2-treated versus non-treated cells. The five most abundant proteins were GlmS, SlyD, and cAMP receptor protein (Crp), which are usual contaminants during His tag purification in E. coli (32); LacI, which controls the expression of the lac promoter and was probably found because ychF was cloned under the control of the lac promoter; and IscS, which is involved in Fe-S cluster biosynthesis. The co-purification of GlmS, SlyD, cAMP receptor protein, and LacI was not significantly influenced by H2O2 treatment, whereas IscS slightly increased upon H2O2 treatment (Fig. 3C). IscS is a cysteine desulfurase and is involved in the biosynthesis of Fe-S clusters, which are a main target of reactive oxygen. A possible direct interaction between YchF and IscS requires further analyses.
In E. coli, approximately 34 proteins are OxyR-controlled, and 22 of them are up-regulated upon oxidative stress (26). In our MS analysis, we found only five OxyR-regulated proteins (supplemental Table S1): all three catalases of E. coli (AhpF, KatG, and KatE); OxyR itself, which is down-regulated by oxidative stress; and Fur, which is up-regulated by OxyR but is also a usual contaminant during His tag purification (32, 33). In particular, KatG showed a strong H2O2-dependent co-purification with YchF (Fig. 3C). Thus, the co-purification of KatG with YchF does not simply reflect OxyR-induced up-regulation of stress response proteins but instead is the result of a selective interaction between both proteins. Surprisingly, we did not detect Dps by MS, although Dps is the protein that is most strongly induced upon oxidative stress (26). Whether this is related to its small size or to its tendency to form homododecamers upon oxidative stress is currently unknown.
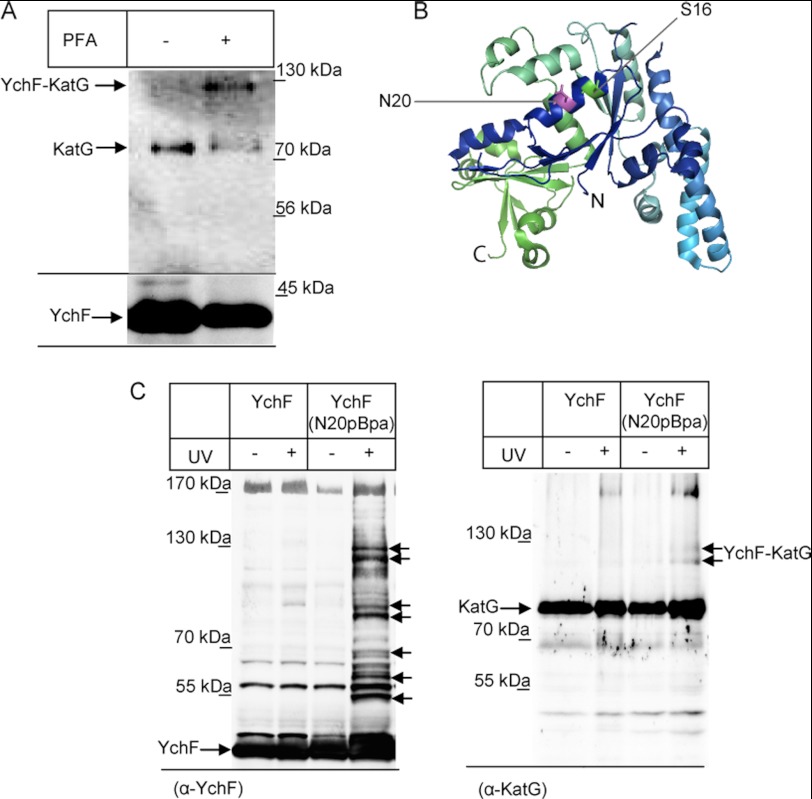
The proposed interaction between YchF and KatG was further verified by two in vivo cross-linking approaches. When E. coli cells expressing YchF were treated with the membrane-permeable homobifunctional cross-linker PFA before YchF purification, we noticed an additional band at approximately 120 kDa that was recognized by α-KatG antibodies and was PFA-dependent (Fig. 4A). The size of the cross-linking product fits with a cross-link between the 40-kDa YchF and the 80-kDa KatG. We also used an in vivo site-directed cross-linking approach using the phenylalanine derivative pBpa. pBpa can be incorporated specifically at amber stop codon positions in the presence of a specific plasmid-borne orthogonal aminoacyl-tRNA synthetase/tRNACUA pair (19). We incorporated pBpa into position 20 of YchF, which is surface-exposed and close to the phosphorylation site Ser-16 of YchF that has been identified in a recent phosphoproteome study of E. coli (34) (Fig. 4B). Whole cells expressing YchF(N20pBpa) or wild type YchF were exposed to UV light to activate pBpa, and YchF and possible cross-linked partner proteins were then purified. For YchF(N20pBpa), we observed several UV-specific cross-linking products that were not present in cells expressing wild type YchF (Fig. 4C). This demonstrates that the N-terminal helix of YchF is a major protein-protein interaction site. When the same material was probed with KatG antibodies, we observed a UV-dependent double band at 120 kDa, demonstrating that KatG is in close proximity to the N-terminal helix of YchF. The reason for the double band is currently unknown, but it is likely that the second band corresponds to an YchF-KatG cross-linking product from which the C-terminal His tag was cleaved. In summary, these data strongly support that YchF interacts directly with KatG and that this interaction is probably responsible for KatG inhibition.
FIGURE 4.
YchF cross-links to KatG in vivo. A, cells expressing YchF were grown as described under “Experimental Procedures” and incubated after H2O2 treatment with the membrane-permeable homobifunctional cross-linker PFA. As a control, cells were treated with PFA buffer. Cells were subsequently fractionated, and YchF and possible cross-linking products were purified and processed as described in Fig. 3. The upper panel was decorated with α-KatG antibodies, and the lower panel was decorated with α-YchF antibodies. B, crystal structure of H. influenzae YchF (Ref. 4; Protein Data Bank code 1JAL). The position Ser-16 (green) and the position Asn-20 (magenta) where pBpa was incorporated for in vivo site-directed cross-linking are indicated. C, cells expressing either YchF or YchF(N20pBpa) were exposed to UV light and fractionated before YchF was purified as described in the legend to Fig. 3. The left panel shows the immune detection using α-YchF antibodies, and putative UV-dependent cross-linking products are indicated by arrows. The right panel shows the immune detection of the same material using α-KatG antibodies.
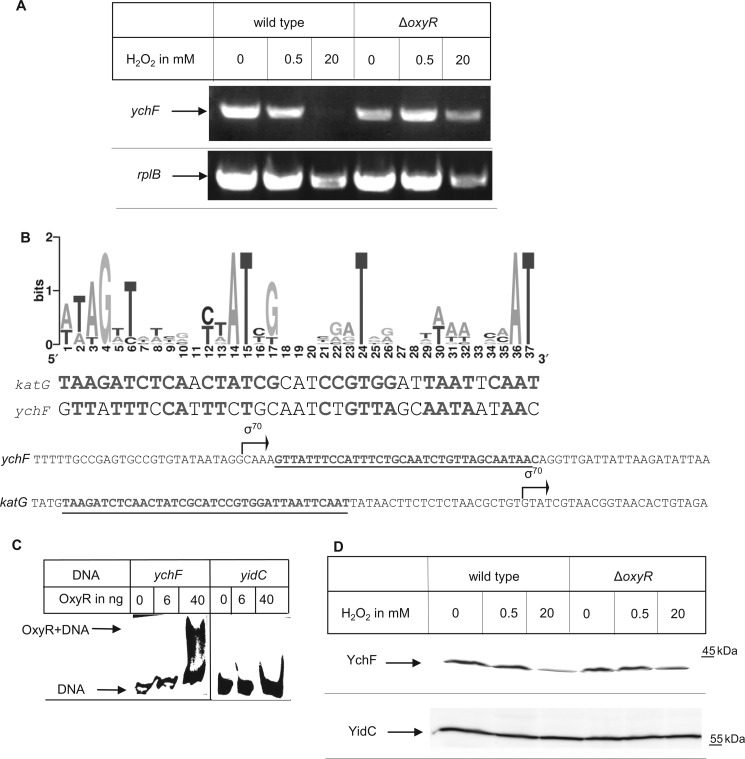
YchF Expression Is Controlled by OxyR in Response to H2O2
The YchF-dependent inhibition of KatG in the presence of H2O2 would be detrimental for the cell, and therefore, we addressed how cells prevented this inhibition under physiological conditions, i.e. when ychF was not overexpressed. First, we determined whether the cellular ychF mRNA concentration was influenced by H2O2. Total mRNA was isolated from either untreated wild type cells or from wild type cells treated with 0.5 or 20 mm H2O2. RT-PCR using a ychF-specific primer demonstrated that the ychF mRNA was slightly reduced in cells treated with 0.5 mm H2O2 and undetectable in cells treated with 20 mm H2O2 (Fig. 5A). As a control, we also analyzed the mRNA for the ribosomal protein L2 (rplB) and noticed a slight decrease in cells treated with 20 mm H2O2, but this reduction was significantly less pronounced than the reduction of ychF mRNA. Thus, the transcription of ychF is down-regulated upon oxidative stress, which would be in line with the possible role of YchF as an inhibitor of the oxidative stress response. Whether down-regulation of ychF was OxyR-dependent was analyzed by determining the ychF mRNA concentration in a ΔoxyR strain. In comparison with wild type cells, the addition of 20 mm H2O2 caused only a slight decrease in ychF mRNA levels in ΔoxyR cells comparable with the decrease of the L2 control. This suggests that ychF expression is regulated by OxyR. The binding sites of OxyR-dependent genes are not completely conserved, but based on known target sites in E. coli, a consensus sequence has been identified (28) (Fig. 5B). A sequence very similar to this consensus sequence is present immediately downstream from the transcription start site of ychF (Fig. 5B). Importantly, its location downstream of the transcription start site is typical for genes that are repressed by OxyR (33), whereas for genes that are activated by OxyR, the consensus motif is located upstream of the start site as in KatG (Fig. 5B).
FIGURE 5.
YchF expression is negatively regulated by the transcription factor OxyR in response to H2O2. A, RNA was prepared from the indicated strains that were grown at 37 °C to an A600 of 0.5 and then further incubated for 15 min with or without 0.5 or 20 mm H2O2. RT-PCR was performed, and products were analyzed on a 1% agarose gel. 100 ng of RNA was used per 25-μl reverse transcriptase reaction. Primers specific for the ribosomal protein L2 served as a control. B, upper panel, sequence logo based on previously identified OxyR binding sites (27, 33, 59). The katG OxyR binding motif and the putative ychF OxyR binding motif are shown below the sequence logo. Lower panel, localization of the OxyR binding sites (underlined) and the transcription start sites (σ70) of katG and ychF. The transcription start sites were taken from the PortEco data bank. C, electrophoresis mobility shift assay using a biotinylated DNA fragment corresponding either to the promoter region of ychF or to the promoter region of yidC. Approximately 20 fmol of DNA was incubated with the indicated amount of purified OxyR and after incubation separated by Tris borate-EDTA PAGE and transferred to a nylon membrane. Binding of OxyR to biotinylated DNA was detected via streptavidin-conjugated horseradish peroxidase. D, immune detection of YchF in cells grown as in A. Approximately 0.5 × 108 cells were processed as described in the legend to Fig. 1. Antibodies against YidC served as a control.
Binding of OxyR to the YchF promoter was verified by electrophoresis mobility shift assays using purified OxyR and a biotinylated DNA fragment comprising the promoter region of ychF. Incubation of the DNA fragment with purified OxyR resulted in a reduced electrophoretic mobility that was detected by horseradish peroxidase coupled to streptavidin after transfer to a nylon membrane (Fig. 5C). As a control, we used a biotinylated DNA fragment comprising the promoter region of the OxyR-independent yidC. Here, no shift was observed. In summary, these data demonstrate that ychF is negatively regulated by OxyR in response to oxidative stress.
The H2O2-dependent decrease of YchF was also detectable by Western blotting, whereas the protein concentration of YidC, an integral membrane protein not involved in stress response, was unchanged (Fig. 5D). However, the effects at the protein level were less pronounced than at the mRNA level, which could indicate that the YchF turnover is rather slow under these conditions.
The Function of YchF Is Regulated by Phosphorylation and by Its ATPase Activity
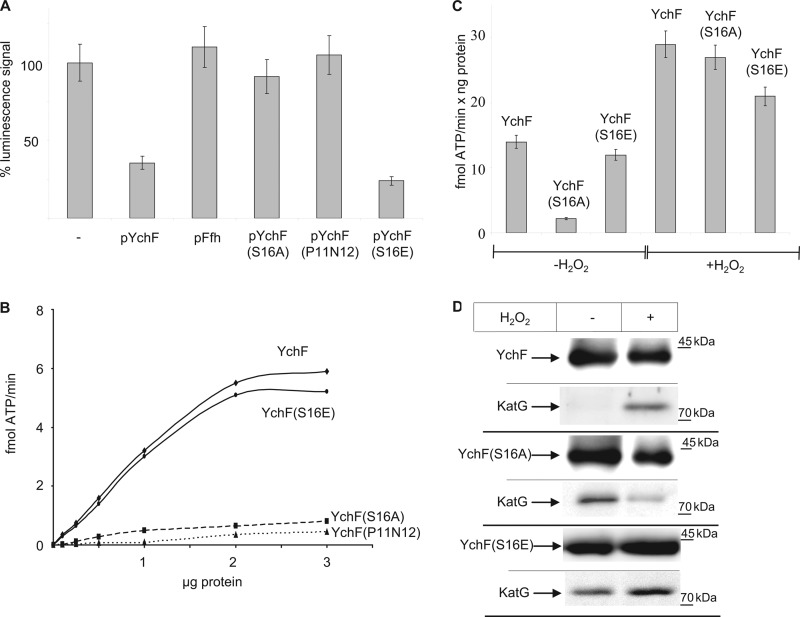
YchF belongs to the Obg family of NTPases and has been shown to hydrolyze ATP (3) and to contain a phosphate group at serine residue 16 (34). This latter analysis was performed on LB-grown cells, and thus a significant portion of YchF exists in the phosphorylated state in the absence of H2O2. For determining the influence of ATP hydrolysis and phosphorylation on H2O2 hypersensitivity, we constructed three plasmid-encoded YchF mutants. In YchF(S16A), serine residue 16 was replaced by alanine, and in YchF(P11AN12A), two residues within the conserved G1 ATP binding motif were mutated to alanine. In addition, we constructed a phosphomimetic YchF(S16E) mutant. All three YchF derivatives were expressed in wild type E. coli cells, and cell viability was tested after H2O2 treatment. Cells expressing only the endogenous chromosomal ychF copy and cells overexpressing either wild type ychF or ffh from a plasmid served as controls. Cells overexpressing the YchF(S16A) or YchF(P11AN12A) derivatives did not show H2O2 hypersensitivity (Fig. 6A). In contrast, cells overexpressing YchF(S16E) were hypersensitive like cells overexpressing YchF (Fig. 6A). This indicates that the phosphorylation of Ser-16 and ATP hydrolysis are important for the inhibitory effect of YchF on the oxidative stress response. Expressing wild type YchF did not impede the H2O2-induced up-regulation of KatG (Fig. 2B), and this was also confirmed for cells expressing the YchF derivatives (data not shown). This excludes that the phosphorylation state of YchF or its ATPase activity influences the expression of KatG and further supports the hypothesis that YchF functions at the post-translational level.
FIGURE 6.
ATP hydrolysis and the phosphorylation state of YchF influence H2O2 hypersensitivity. A, H2O2 hypersensitivity was tested by cell viability assays as described in Fig. 1. In pYchF(S16A), the phosphorylation site Ser-16 in YchF was replaced by alanine, and YchF(S16E) corresponds to a phosphomimetic mutant. In pYchF(P11N12), two residues within the G1 nucleotide binding motif were replaced. pFfh corresponds to plasmid-borne copy of ffh, which encodes the protein component of the bacterial signal recognition particle. Shown are the mean values of two independent experiments and the error bars indicate the standard deviation. B, purified protein was incubated with γ-33P-labeled ATP in reaction buffer for 10 min at 37 °C. ATP and ADP were removed by the addition of 5% charcoal in 20 mm phosphoric acid and a 10-min centrifugation step. The released γ-33P was determined by a scintillation counter. C, ATPase activity was measured as in B in the presence of 50 ng of purified protein. The values represent the mean values of three independent experiments. The error bars indicate the standard deviation. D, wild type YchF, YchF(S16A), and YchF(S16E) were purified as described in Fig. 3 and probed with antibodies against YchF and KatG.
The ATPase activity of YchF was measured directly for the wild type protein and the YchF mutants purified from cells that were not treated with H2O2. The YchF(P11AN12A) mutant displayed only very low activity even when analyzed at high protein concentration (Fig. 6B). On the other hand, the ATPase activity of the phosphomimetic YchF(S16E) derivative was comparable with wild type YchF (Fig. 6B). Importantly, the phosphorylation-deficient YchF(S16A) mutant showed only very low activity (Fig. 6B). This suggested either that the phosphorylation of serine 16 was required for full ATP hydrolysis or that replacing it with alanine interfered with ATP binding because Ser-16 is located within the ATP binding motif. For further analysis, we determined whether ATPase activity responded to H2O2 by purifying YchF and the YchF(S16A)/YchF(S16E) mutants from H2O2-treated and untreated cells. We observed a significant increase in ATPase activity when YchF was purified from H2O2-treated cells, and a significant increase in ATPase activity was also observed for the YchF(S16A) mutant (Fig. 6C). This demonstrates that H2O2 treatment results in increased ATPase activity of YchF. It also shows that YchF(S16A) is able to bind and hydrolyze ATP but has a very low basal activity in the absence of H2O2. The YchF(S16E) mutant was also further stimulated by the presence of H2O2, but the stimulation appeared to be weaker than for wild type YchF (Fig. 6C).
YchF(S16A) probably mimics the dephosphorylated state of YchF, and the increase of ATPase activity upon H2O2 treatment could indicate that the ATPase activity is regulated by an additional H2O2-dependent modification. Like OxyR, YchF contains several cysteine residues, and their oxidation by H2O2 could influence the ATPase activity. However, we did not detect any influence of reducing agents like β-mercaptoethanol on the H2O2-dependent stimulation of ATP hydrolysis (data not shown).
Finally, we tested whether co-purification of KatG and YchF was also observed for YchF(S16A) or YchF(S16E). KatG co-purified with wild type YchF from H2O2-treated cells (Fig. 6D). For YchF(S16A), we observed significant co-purification of KatG in the absence of H2O2, whereas in the presence of H2O2, the amount of KatG co-purifying with YchF(S16A) was significantly reduced. The reduced KatG co-purification with the YchF(S16A) mutant in the presence of H2O2 probably explains why the overexpression of YchF(S16A) did not cause H2O2 hypersensitivity (Fig. 6A). These data further support the hypothesis that YchF function is not only controlled by phosphorylation/dephosphorylation but probably also by additional, so far unknown modifications. KatG co-purified with YchF(S16E) both in the presence and absence of H2O2, but the amount was higher after H2O2 treatment. This explains why cells expressing this phosphomimetic YchF mutant are H2O2-hypersensitive.
DISCUSSION
YchF is a member of the universally conserved Obg-related NTPase family (2), but despite its conservation, the cellular role of YchF and its homologues in different species remained unclear. Our current study has revealed that YchF overexpression causes H2O2 hypersensitivity in E. coli and thus displays the same phenotype as overexpression of its homologue Ola1 in humans. Furthermore, we provide the first mechanistic details on YchF function because we show that YchF does not influence the expression of major oxidative stress response proteins but instead inhibits stress response proteins like KatG by direct interaction. This interaction is regulated by the phosphorylation state of YchF and by ATP hydrolysis. Finally, we show that YchF expression is repressed by the transcription factor OxyR in an H2O2-dependent manner. Based on these observations, we propose that YchF/Ola1 is a universally conserved negative regulator of the oxidative stress response.
YchF Is Part of the OxyR Regulon
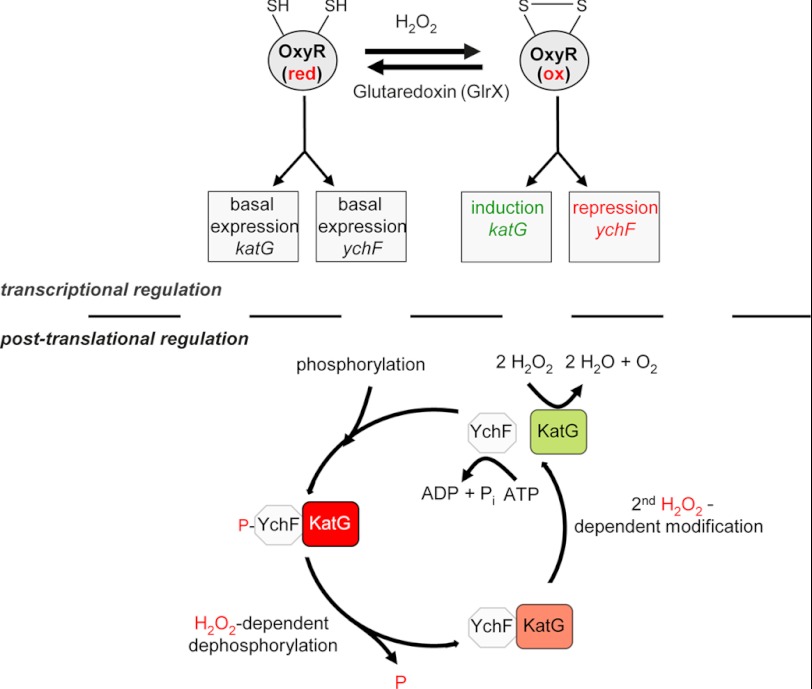
The H2O2 response in most bacteria is coordinated by the LysR-type transcriptional regulator OxyR (32, 35). OxyR interacts in its oxidized state with RNA polymerase and stimulates the expression of at least 30 proteins (35). Additional genes are indirectly controlled by OxyR as it induces the synthesis of OxyS, a small non-coding RNA that controls the expression of additional genes (36). Independently of its oxidation state, OxyR also functions as a repressor. It inhibits its own synthesis (37) and the synthesis of Antigen 43, a self-recognizing surface adhesin found in most E. coli strains (38, 39). We now show that OxyR also inhibits ychF synthesis, but this inhibition is only observed after exposing cells to H2O2. H2O2 concentrations between 0.1 and 1 mm have been shown to oxidize cysteine residues within OxyR (40, 41), and it is therefore likely that only oxidized OxyR is able to repress YchF synthesis. This observation is also in agreement with a global microarray study, which found that ychF is down-regulated after H2O2 treatment (31). The putative OxyR binding site upstream of ychF overlaps with the RNA polymerase binding site, which is typical for genes that are negatively regulated by LysR-type transcriptional regulators. In contrast, activating OxyR binding sites as in katG are located 40–60 nucleotides upstream of the RNA polymerase binding site (32). Thus, upon H2O2 exposure, OxyR activates the transcription of H2O2-scavenging enzymes like KatG and at the same time represses the transcription of the KatG inhibitor YchF (Fig. 7).
FIGURE 7.
Tentative model for YchF function in E. coli. In the absence of H2O2, the transcription factor OxyR is kept in its reduced (red) state by the glutaredoxin GlrX, which allows the basal expression of katG and ychF. Under these conditions, YchF inhibits KatG activity. In the presence of H2O2, OxyR is oxidized (ox) and induces the expression of oxidative stress response genes like catalases. Simultaneously, it represses the expression of the catalase inhibitor YchF. In addition to this transcriptional regulation, the activity of YchF is also regulated by H2O2 at a post-translational level. YchF is phosphorylated at serine 16 in the absence of H2O2 (34), and a phosphorylation-deficient YchF mutant does not cause H2O2 hypersensitivity. Thus, KatG inhibition in the absence of H2O2 is probably caused by phosphorylated YchF. In the presence of H2O2, YchF is dephosphorylated and undergoes a second H2O2-dependent modification, which needs to be identified. These modifications stimulate the ATPase activity of YchF, resulting in the dissociation of KatG. KatG then detoxifies H2O2. At low H2O2 concentrations, repression of YchF by OxyR is relieved, and phosphorylation of YchF increases its affinity for KatG, resulting in its inhibition.
YchF Acts as Negative Regulator of H2O2-scavenging Enzymes
YchF inhibits catalase activity and interacts directly with KatG, leading to H2O2 hypersensitivity when YchF is overexpressed. Although the requirement for an oxidative stress response is apparent, it is less obvious why cells need to inhibit KatG and possibly other proteins of the oxidative stress response in the absence of H2O2. YchF-inhibited KatG could serve as a reservoir that can be rapidly activated when cells encounter H2O2. KatG of E. coli belongs to the catalase-peroxidase family of catalases (30), which can also catalyze a peroxidatic reaction in which ethanol or short-chain aliphatic alcohols are oxidized to potentially toxic aldehydes (42). Catalase-peroxidases also efficiently bind and oxidize NADH, and this NADH oxidase activity is linked to the formation of the reactive superoxide radical (43). Thus, the inhibition of KatG in the absence of oxidative stress might be required for reducing the accumulation of potentially toxic KatG side products and for reducing futile NADH oxidation. Studies in mammalian cells have shown that high levels of catalase activity can have adverse physiological effects (44, 45), which probably also reflects the importance of intracellular H2O2-dependent signaling (46, 47). Whether small amounts of H2O2 are also beneficial for unicellular organism is currently unknown, but it has been shown that bacterial cells that are treated with very low H2O2 concentrations survive a subsequent treatment with high H2O2 concentrations much better than non-adapted cells (48).
We also found that Dps co-purified with YchF by Western blotting (most likely due its small size) but not by MS. Thus, it is currently unknown whether YchF specifically interacts with Dps and if so whether this interferes with Dps activity. Dps is a ferritin-like protein and, like KatG, is under OxyR control (49, 50). Dps in E. coli forms a shell-like dodecamer that may contain up to 400–500 iron atoms (51). Iron sequestration depends on the ferro-oxidase activity of Dps for which H2O2 serves as oxidant (52). Thus, the protective role of Dps during oxidative stress is 2-fold: it reduces the H2O2 concentration and the free iron concentration. As iron availability is a major determinant for bacterial growth, rapid inactivation of Dps by YchF might be important for allowing growth after H2O2 stress ceases. A possible link between iron availability and YchF has already been proposed for V. vulnificus (16) and B. melitensis (18), and it was suggested that YchF is involved in iron metabolism. In these organisms, the absence of YchF has been shown to interfere with growth under iron-limiting conditions. Although this would be in line with increased Dps-dependent iron sequestration in the absence of YchF, further experiments are needed to determine the impact of YchF on Dps function. The in vivo site-directed cross-linking approach (Fig. 4C) revealed that YchF interacts with many more partner proteins, and MS analyses are currently in progress for identifying these additional partner proteins.
Post-translational Regulation of YchF Activity
YchF is phosphorylated in E. coli (34), and phosphorylation of the yeast homologue, probably via the serine/threonine kinases PTK1 (48) and Yck2 (53, 54), has also been observed. E. coli lacks orthologues of both kinases; therefore, the kinase responsible for YchF phosphorylation and the cognate phosphatase need to be identified. The function of YchF is clearly influenced by its phosphorylation state. 1) Overexpression of the phosphorylation-deficient YchF(S16A) derivative did not cause H2O2 hypersensitivity. 2) KatG co-purification with YchF(S16A) was independent of H2O2. 3) YchF(S16A) showed no basal ATPase activity. The first observation suggests that phosphorylation of YchF is required for KatG inhibition, but the second observation implies that the YchF-KatG interaction can occur independently of the Ser-16 phosphorylation. This probably indicates that the inactivation of KatG by YchF is not only regulated by phosphorylation but additionally by a second modification. The presence of a second, so far unknown modification is also deduced from the following observations. 1) YchF(S16A) mimicked the dephosphorylated state but still required H2O2 for ATPase activation. 2) Although YchF(S16E) imitated a permanently phosphorylated state, it still showed H2O2-dependent ATPase activity. We currently favor a model in which the inhibitory effect is executed by phosphorylated YchF and dephosphorylation attenuates this inhibition. This is also in line with the observation that phosphorylation of Ser-16 was found in non-stressed, exponentially growing E. coli cells (34). The stimulated ATPase activity in the presence of H2O2 could induce the dissociation of YchF from KatG, subsequently allowing the detoxification of H2O2 (Fig. 6). A similar mechanism is used for the dissociation of signal recognition particle from its receptor, which is induced by GTP hydrolysis (55). The observation that KatG binds close to the phosphorylation site and ATPase domain of YchF also supports this hypothesis.
In summary, the H2O2 hypersensitivity of YchF-overexpressing cells is the result of (i) a continuously high YchF level that is disconnected from the OxyR regulation and (ii) most likely an inefficient post-translational modification (dephosphorylation of Ser-16 and a so far unknown modification). This results in persistent inhibition of KatG and possibly other proteins of the oxidative stress response. Our data do not exclude that YchF also regulates the translation of some target genes, which would explain the ribosome interaction that has been observed (3, 5).
E. coli YchF as a Model for Human Ola1 Function
The overexpression of YchF in E. coli and the overexpression of hOla1 in humans cause hypersensitivity toward oxidative stress in both organisms, indicating that YchF function has been conserved during evolution. This is further corroborated by our data that experimentally prove that YchF inhibits oxidative stress by a post-translational mechanism, which was also put forward for human Ola1 (7).
Whether hOla1 also functions by inhibiting catalases is so far unknown. Catalases in mammalian cells are found in the cytosol, in peroxisomes, and probably in mitochondria (30, 56, 57). hOla1 is located in the cytoplasm and does not contain a typical peroxisomal or mitochondrial localization signal. Thus, a direct inhibition of cytosolic catalases by hOla1 is possible and needs to be experimentally verified. However, in plants, stress exposure was shown to influence the localization of YchF homologues (58), and it is therefore possible that hOla1 does not exclusively function in the cytosol.
Increased hOla1 expression is observed in many tumor cells, and mutations that lead to enhanced hOla1 expression could be a primary cause for these tumors. High levels of hOla1 would suppress the ability of cells to cope with and inactivate damaging reactive oxygen species, resulting in the accumulation of DNA damage and oncogenic transformation. Thus, our study on E. coli YchF provides an important framework for further exploring the mechanism and the biological and medical significance of the YchF/Ola1 family of ATPases.
Acknowledgments
We gratefully acknowledge Regine Henge, Freie Universität Berlin, Germany for the gift of a-Dps antibodies and Richard Brimacombe, Max-Planck-Institut für Molekulare Genetik, Berlin, Germany for a-L2 antibodies. We also thank Sandra Angelini for constructing plasmid pET22b-YchF and the National BioResource Project E. coli (National Institute of Genetics, Japan) for providing the ΔychF strain, the ΔoxyR strain, and the corresponding wild type strain.
This work was supported in part by Deutsche Forschungsgemeinschaft Grants DFG-FOR 967 and IRTG1478 (to H. G. K.) and FOR1352 (to B. W.) and the Excellence Initiative of the German Federal and State Governments (Grant GSC-4, Spemann Graduate School for Biology and Medicine (to H. G. K.) and Grant EXC 294 BIOSS (to B. W.)).

This article contains supplemental Table S1.
- hOla1
- human Ola1
- Dps
- DNA protection during starvation
- Ffh
- fifty-four homologue
- LB
- lysogeny broth
- pBpa
- para-benzoyl-l-phenylalanine
- PFA
- para-formaldehyde
- Pth
- peptidyl-tRNA hydrolase.
REFERENCES
- 1. Verstraeten N., Fauvart M., W., Michiels J. (2011) The universally conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 75, 507–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koller-Eichhorn R., Marquardt T., Gail R., Wittinghofer A., Kostrewa D., Kutay U., Kambach C. (2007) Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J. Biol. Chem. 282, 19928–19937 [DOI] [PubMed] [Google Scholar]
- 3. Tomar S. K., Kumar P., Prakash B. (2011) Deciphering the catalytic machinery in a universally conserved ribosome binding ATPase YchF. Biochem. Biophys. Res. Commun. 408, 459–464 [DOI] [PubMed] [Google Scholar]
- 4. Teplyakov A., Obmolova G., Chu S. Y., Toedt J., Eisenstein E., Howard A. J., Gilliland G. L. (2003) Crystal structure of the YchF protein reveals binding sites for GTP and nucleic acid. J. Bacteriol. 185, 4031–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gradia D. F., Rau K., Umaki A. C., de Souza F. S., Probst C. M., Correa A., Holetz F. B., Avila A. R., Krieger M. A., Goldenberg S., Fragoso S. P. (2009) Characterization of a novel Obg-like ATPase in the protozoan Trypanosoma cruzi. Int. J. Parasitol. 39, 49–58 [DOI] [PubMed] [Google Scholar]
- 6. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 7. Zhang J., Rubio V., Lieberman M. W., Shi Z. Z. (2009) OLA1, an Obg-like ATPase, suppresses antioxidant response via nontranscriptional mechanisms. Proc. Natl. Acad. Sci. U.S.A. 106, 15356–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guerrero C., Tagwerker C., Kaiser P., Huang L. (2006) An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics 5, 366–378 [DOI] [PubMed] [Google Scholar]
- 9. Sun H., Luo X., Montalbano J., Jin W., Shi J., Sheikh M. S., Huang Y. (2010) DOC45, a novel DNA damage-regulated nucleocytoplasmic ATPase that is overexpressed in multiple human malignancies. Mol. Cancer Res. 8, 57–66 [DOI] [PubMed] [Google Scholar]
- 10. Jones S., Zhang X., Parsons D. W., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., Hong S. M., Fu B., Lin M. T., Calhoun E. S., Kamiyama M., Walter K., Nikolskaya T., Nikolsky Y., Hartigan J., Smith D. R., Hidalgo M., Leach S. D., Klein A. P., Jaffee E. M., Goggins M., Maitra A., Iacobuzio-Donahue C., Eshleman J. R., Kern S. E., Hruban R. H., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V. E., Kinzler K. W. (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nabilsi N. H., Broaddus R. R., Loose D. S. (2009) DNA methylation inhibits p53-mediated survivin repression. Oncogene 28, 2046–2050 [DOI] [PubMed] [Google Scholar]
- 12. Zhang J. W., Rubio V., Zheng S., Shi Z. Z. (2009b) Knockdown of OLA1, a regulator of oxidative stress response, inhibits motility and invasion of breast cancer cells. J. Zhejiang Univ. Sci. B 10, 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kira Y., Nishikawa M. (2008) The identification and characterization of a new GTP-binding protein (Gbp45) involved in cell proliferation and death related to mitochondrial function. Cell. Mol. Biol. Lett. 13, 570–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arigoni F., Talabot F., Peitsch M., Edgerton M. D., Meldrum E., Allet E., Fish R., Jamotte T., Curchod M. L., Loferer H. (1998) A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16, 851–856 [DOI] [PubMed] [Google Scholar]
- 15. Morimoto T., Loh P. C., Hirai T., Asai K., Kobayashi K., Moriya S., Ogasawara N. (2002) Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148, 3539–3552 [DOI] [PubMed] [Google Scholar]
- 16. Chen Y. C., Chung Y. T. (2011) A conserved GTPase YchF of Vibrio vulnificus is involved in macrophage cytotoxicity, iron acquisition, and mouse virulence. Int. J. Med. Microbiol. 301, 469–474 [DOI] [PubMed] [Google Scholar]
- 17. Fernebro J., Blomberg C., Morfeldt E., Wolf-Watz H., Normark S., Normark B. H. (2008) The influence of in vitro fitness defects on pneumococcal ability to colonize and to cause invasive disease. BMC Microbiol. 8, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Danese I., Haine V., Delrue R. M., Tibor A., Lestrate P., Stevaux O., Mertens P., Paquet J. Y., Godfroid J., De Bolle X., Letesson J. J. (2004) The Ton system, an ABC transporter, and a universally conserved GTPase are involved in iron utilization by Brucella melitensis 16M. Infect. Immun. 72, 5783–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ryu Y., Schultz P. G. (2006) Efficient incorporation of unnatural amino acids into proteins in E. coli. Nat. Methods 3, 263–265 [DOI] [PubMed] [Google Scholar]
- 20. Kaller M., Liffers S. T., Oeljeklaus S., Kuhlmann K., Röh S., Hoffmann R., Warscheid B, Hermeking H. (2011) Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol. Cell. Proteomics 10, M111.010462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 22. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 23. Cuzin F., Kretchmer N., Greenberg R. E., Hurwitz R., Chapeville F. (1967) Enzymatic hydrolysis of N-substituted aminoacyl-tRNA. Proc. Natl. Acad. Sci. U.S.A. 58, 2079–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Atherly A. G., Menninger J. R. (1972) Mutant E. coli strain with temperature sensitive peptidyl-transfer RNA hydrolase. Nat. New Biol. 240, 245–246 [DOI] [PubMed] [Google Scholar]
- 25. Das G., Varshney U. (2006) Peptidyl-tRNA hydrolase and its critical role in protein biosynthesis. Microbiology 152, 2191–2195 [DOI] [PubMed] [Google Scholar]
- 26. Imlay J. A. (2008) Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77, 755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng M., Aslund F., Storz G. (1998) Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721 [DOI] [PubMed] [Google Scholar]
- 28. Storz G., Tartaglia L. A., Ames B. N. (1990) Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248, 189–194 [DOI] [PubMed] [Google Scholar]
- 29. Mulvey M. R., Switala J., Borys A., Loewen P. C. (1990) Regulation of transcription of katE and katF in Escherichia coli. J. Bacteriol. 172, 6713–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zamocky M., Furtmüller P. G., Obinger C. (2008) Evolution of catalases from bacteria to humans. Antioxid. Redox Signal. 10, 1527–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., Storz G. (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183, 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robichon C., Luo J., Causey T. B., Benner J. S., Samuelson J. C. (2011) Engineering Escherichia coli BL21 (DE3) derivative strains to minimize E. coli protein contamination after purification by immobilized metal affinity chromatography. Appl. Environ. Microbiol. 77, 4634–4646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maddocks S. E., Oyston P. C. (2008) Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154, 3609–3623 [DOI] [PubMed] [Google Scholar]
- 34. Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 35. Chiang S. M., Schellhorn H. E. (2012) Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525, 161–169 [DOI] [PubMed] [Google Scholar]
- 36. Altuvia S., Weinstein-Fischer D., Zhang A., Postow L., Storz G. (1997) A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90, 43–53 [DOI] [PubMed] [Google Scholar]
- 37. Toledano M. B., Kullik I., Trinh F., Baird P. T., Schneider T. D., Storz G. (1994) Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78, 897–909 [DOI] [PubMed] [Google Scholar]
- 38. Schembri M. A., Hjerrild L., Gjermansen M., Klemm P. (2003) Differential expression of the Escherichia coli autoaggregation factor antigen 43. J. Bacteriol. 185, 2236–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wallecha A., Correnti J., Munster V., van der Woude M. (2003) Phase variation of Ag43 is independent of the oxidation state of OxyR. J. Bacteriol. 185, 2203–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aslund F., Zheng M., Beckwith J., Storz G. (1999) Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. U.S.A. 96, 6161–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carmel-Harel O., Storz G. (2000) Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54, 439–461 [DOI] [PubMed] [Google Scholar]
- 42. Vetrano A. M., Heck D. E., Mariano T. M., Mishin V., Laskin D. L., Laskin J. D. (2005) Characterization of the oxidase activity in mammalian catalase. J. Biol. Chem. 280, 35372–35381 [DOI] [PubMed] [Google Scholar]
- 43. Singh R., Wiseman B., Deemagarn T., Donald L. J., Duckworth H. W., Carpena X., Fita I., Loewen P. C. (2004) Catalase-peroxidases (KatG) exhibit NADH oxidase activity. J. Biol. Chem. 279, 43098–43106 [DOI] [PubMed] [Google Scholar]
- 44. Doonan R., McElwee J. J., Matthijssens F., Walker G. A., Houthoofd K., Back P., Matscheski A., Vanfleteren J. R., Gems D. (2008) Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 22, 3236–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sen C. K., Roy S. (2008) Redox signals in wound healing. Biochim. Biophys. Acta 1780, 1348–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sartoretto J. L., Kalwa H., Pluth M. D., Lippard S. J., Michel T. (2011) Hydrogen peroxide differentially modulates cardiac myocyte nitric oxide synthesis. Proc. Natl. Acad. Sci. U.S.A. 108, 15792–15797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tormos K. V., Anso E., Hamanaka R. B., Eisenbart J., Joseph J., Kalyanaraman B., Chandel N. S. (2011) Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab. 14, 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Christman M. F., Morgan R. W., Jacobson F. S., Ames B. N. (1985) Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41, 753–762 [DOI] [PubMed] [Google Scholar]
- 49. Altuvia S., Almirón M., Huisman G., Kolter R., Storz G. (1994) The dps promoter is activated by OxyR during growth and by IHF and σS in stationary phase. Mol. Microbiol. 13, 265–272 [DOI] [PubMed] [Google Scholar]
- 50. Almirón M., Link A. J., Furlong D., Kolter R. (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6, 2646–2654 [DOI] [PubMed] [Google Scholar]
- 51. Ilari A., Ceci P., Ferrari D., Rossi G. L., Chiancone E. (2002) Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. J. Biol. Chem. 277, 37619–37623 [DOI] [PubMed] [Google Scholar]
- 52. Chiancone E., Ceci P. (2010) The multifaceted capacity of Dps proteins to combat bacterial stress conditions: detoxification of iron and hydrogen peroxide and DNA binding. Biochim. Biophys. Acta 1800, 798–805 [DOI] [PubMed] [Google Scholar]
- 53. Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) The genetic landscape of the cell. Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G, Breitkreutz A., Sopko R., McCartney R. R., Schmidt M. C., Rachidi N., Lee S. J., Mah A. S., Meng L., Stark M. J., Stern D. F., De Virgilio C., Tyers M., Andrews B., Gerstein M., Schweitzer B., Predki P. F., Snyder M. (2005) Global analysis of protein phosphorylation in yeast. Nature 438, 679–684 [DOI] [PubMed] [Google Scholar]
- 55. Koch H. G., Moser M., Müller M. (2003) Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev. Physiol. Biochem. Pharmacol. 146, 55–94 [DOI] [PubMed] [Google Scholar]
- 56. Radi R., Turrens J. F., Chang L. Y., Bush K. M., Crapo J. D., Freeman B. A. (1991) Detection of catalase in rat heart mitochondria. J. Biol. Chem. 266, 22028–22034 [PubMed] [Google Scholar]
- 57. Salvi M., Battaglia V., Brunati A. M., La Rocca N., Tibaldi E., Pietrangeli P., Marcocci L., Mondovì B., Rossi C. A., Toninello A. (2007) Catalase takes part in rat liver mitochondria oxidative stress defense. J. Biol. Chem. 282, 24407–24415 [DOI] [PubMed] [Google Scholar]
- 58. Cheung M. Y., Xue Y., Zhou L., Li M. W., Sun S. S., Lam H. M. (2010) An ancient P-loop GTPase in rice is regulated by a higher plant-specific regulatory protein. J. Biol. Chem. 285, 37359–37369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wei Q., Minh P. N., Dötsch A., Hildebrand F., Panmanee W., Elfarash A., Schulz S., Plaisance S., Charlier D., Hassett D., Häussler S., Cornelis P. (2012) Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 40, 4320–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]