Background: Sec12 is a GEF responsible for the initiation of COPII vesicle budding.
Results: The 1.36 Å crystal structure of yeast Sec12 is presented, together with in vitro and in vivo analysis of structure-guided mutants.
Conclusion: A potassium-stabilized “K loop” plays an unprecedented and critical role in GEF activity.
Significance: All of the key proteins for COPII vesicle budding have now been structurally characterized.
Keywords: Endoplasmic Reticulum (ER), Guanine Nucleotide Exchange Factor (GEF), Metals, Vesicles, X-ray Crystallography
Abstract
Coat protein II (COPII)-coated vesicles transport proteins and lipids from the endoplasmic reticulum to the Golgi. Crucial for the initiation of COPII coat assembly is Sec12, a guanine nucleotide exchange factor responsible for activating the small G protein Sar1. Once activated, Sar1/GTP binds to endoplasmic reticulum membranes and recruits COPII coat components (Sec23/24 and Sec13/31). Here, we report the 1.36 Å resolution crystal structure of the catalytically active, 38-kDa cytoplasmic portion of Saccharomyces cerevisiae Sec12. Sec12 adopts a β propeller fold. Conserved residues cluster around a loop we term the “K loop,” which extends from the N-terminal propeller blade. Structure-guided site-directed mutagenesis, in conjunction with in vitro and in vivo functional studies, reveals that this region of Sec12 is catalytically essential, presumably because it makes direct contact with Sar1. Strikingly, the crystal structure also reveals that a single potassium ion stabilizes the K loop; bound potassium is, moreover, essential for optimum guanine nucleotide exchange activity in vitro. Thus, our results reveal a novel role for a potassium-stabilized loop in catalyzing guanine nucleotide exchange.
Introduction
The generation of COPII2-coated transport vesicles, required within the secretory pathway for endoplasmic reticulum (ER)-to-Golgi trafficking, provides a well characterized model system for investigating vesicle budding (1, 2). The assembly of COPII coat components onto the ER membrane is initiated by Sec12, a transmembrane guanine nucleotide exchange factor (GEF) (3, 4). Sec12 catalyzes the exchange of GDP for GTP on the small GTPase Sar1. Nucleotide exchange produces a conformational change in Sar1 that leads to the exposure of its amphipathic N-terminal α-helix and an increase in its affinity for membranes (5). Thus activated, Sar1 recruits Sec23/24 heterodimers, forming the inner layer of the COPII coat (6–9). Sar1 and Sec23/24 then recruit Sec13/31 heterotetramers, which polymerize to form the outer “cage” layer of the coat (7–11).
Insertion of the Sar1 N terminus into the ER membrane contributes to the membrane curvature required for COPII-coated vesicle budding and fission of vesicles from ER membranes, whereas Sec23/24 and Sec13/31 participate in shaping the vesicle through an intrinsic ability to deform membranes (12). In addition to initiating budding, Sec12 has also been proposed to stabilize the COPII coat during the budding process by renewing GTP on Sar1 at the boundary of the budding vesicle (13).
X-ray structures have been determined for all of these COPII vesicle budding machinery components except Sec12 (5, 14, 15). The 38-kDa cytosolic portion of Sec12, which is responsible for its GEF activity (13), was previously predicted to adopt a β propeller fold based on alignment of divergent homologs (16). Experimental validation and high resolution structural information have, however, been lacking. Furthermore, only one structure of a β propeller GEF, RCC1 (regulator of chromosome condensation 1), has previously been reported (17). We have now determined the high resolution crystal structure of the cytosolic portion of Saccharomyces cerevisiae Sec12, revealing the expected β propeller decorated by a novel surface loop critical for GEF function. This “K loop” binds a potassium ion, which in turn enhances GEF activity, providing what is, to our knowledge, the first example of a monovalent metal ion playing a role in GEF function. Asn-40, previously shown to be essential for Sec12 activity (13), sits at the base of the K loop, where it acts as an anchor to stabilize the loop and surrounding region through an elaborate network of hydrogen bonds.
EXPERIMENTAL PROCEDURES
Purification and Expression of Sec12 and Sar1
The cytoplasmic portion of Sec12 from S. cerevisiae (residues 1–354, previously termed Sec12ΔCp (13) but hereafter called simply Sec12), fused at its C terminus to a hexahistidine tag, was produced in Escherichia coli (Rosetta) using the expression plasmid pET21b (Novagen). Cells were grown in either Luria-Bertani medium or, for production of selenomethionine-substituted protein, in M9 minimal medium supplemented with 40 mg/liter selenomethionine. Cells were pelleted and resuspended in lysis buffer (20 mm Tris, pH 7.6, 200–250 mm NaCl, 5 mm imidazole, 1 mm dithiothreitol (DTT)) before being lysed using an Emulsiflex-C5 homogenizer (Avestin). The lysate was supplemented with 2.5 mm phenylmethylsulfonyl fluoride and 1 μl/ml Protease Inhibitor Mixture (Sigma), clarified by centrifugation, and then fractionated using His60 Ni Superflow Resin (Clontech) or Ni2+-charged IMAC Sepharose 6 Fast Flow Resin (GE Healthcare). Sec12 to be used for crystallization was further purified by ion exchange chromatography on a Source Q 10/10 column (GE Healthcare), where it eluted in the flow-through, and size exclusion chromatography (Superdex 200 10/300 GL, GE Healthcare). Sec12 to be used for biochemical experiments was further purified by size exclusion chromatography (Superdex 200 16/60 or Superdex 13/300). Although three Sec12 peaks were generally observed during size exclusion chromatography, only the peak corresponding to the monomer molecular weight was retained. Small aliquots of Sec12 in 20 mm Tris, pH 7.6, 1 mm DTT, 100 (or 200) mm NaCl were flash frozen in liquid N2 and stored at −80 °C.
S. cerevisiae Sar1 was produced as a C-terminally hexahistidine-tagged fusion protein using the expression plasmid pET21b and purified by Ni2+ affinity and size exclusion chromatographies as described above. Purified Sar1p was frozen and stored at −80 °C in 20 mm Tris, pH 7.6, 200 mm NaCl, 1 mm DTT.
Crystallization and Structural Determination of Sec12
Crystals of selenomethionine-labeled Sec12 were grown by sitting drop vapor diffusion from a 1:1 (v/v) mixture of protein stock (3.8 mg/ml) containing 20 mm Tris, pH 7.6, 100 mm NaCl, 1 mm DTT and precipitant solution containing 91 mm Tris, pH 8.0, 181 mm potassium citrate, pH 8.3, 22% (w/v) PEG 3350. Crystals grew with cell dimensions of a = b = 189.5 Å, c = 53.4 Å in space group P64 containing two molecules per asymmetric unit. Multiwavelength anomalous diffraction data were collected at the selenium K edge at beamline X29 of the National Synchrotron Light Source at Brookhaven National Laboratory. Data were processed using the HKL suite (18) (Table 1). Ten selenium atoms were located using the program SHELXD (19), and phases were calculated using the program SHARP (20). The structure was built into the experimentally phased map using the program COOT (21) and refined with PHENIX (22) using non-crystallographic symmetry restraints.
TABLE 1.
Data collection, phasing, and refinement statistics
|
P64 MADa |
P1 | |||
|---|---|---|---|---|
| Peak | Inflection | Remote | ||
| Data collection | ||||
| Wavelength (Å) | 0.9792 | 0.9794 | 0.9640 | 1.075 |
| Resolution (Å) | 100–2.8 (2.9–2.8)b | 100–2.8 (2.9–2.8) | 100–2.6 (2.69–2.60) | 50–1.35 (1.37–1.35) |
| Unique reflections | 27,210 | 27,520 | 34,159 | 132,685 |
| Completeness (%) | 99.5 (99.1) | 99.9 (100.0) | 99.9 (99.8) | 93.6 (79.8) |
| Redundancy | 6.0 (5.3) | 8.2 (7.2) | 8.3 (7.4) | 3.8 (3.5) |
| R-sym | 0.122 (0.423) | 0.147 (0.621) | 0.105 (0.426) | 0.063 (0.610) |
| 〈I/σI〉 | 9.5 (2.7) | 10.1 (2.3) | 11.7 (3.4) | 16.2 (2.2) |
| Phasing | ||||
| Figure of merit | 0.46 | |||
| Refinement | ||||
| Resolution (Å) | 50–2.60 (2.64–2.60) | 50–1.36 (1.38–1.36) | ||
| Reflections F ≥ 0 σF (free) | 34,138 (1,726) | 132,033 (6,617) | ||
| Completeness (%) | 99.9 (99) | 94.5 (82) | ||
| R-factor (%) | 17.87 (21.8) | 17.86 (25.3) | ||
| R-free (%) | 21.64 (27.6) | 19.71 (28.9) | ||
| No. of atoms | ||||
| Protein | 5,261 | 5,249 | ||
| Water | 86 | 428 | ||
| Potassium | 6 | 4 | ||
| RMSDc bond lengths (Å) | 0.011 | 0.011 | ||
| RMSD bond angles (degrees) | 1.189 | 1.248 | ||
| Wilson B-factor (Å2) | 28.6 | 12.8 | ||
| Average B-factors (Å2) | ||||
| All | 34.2 | 15.0 | ||
| Main chain | 32.8 | 12.1 | ||
| Side chain | 35.8 | 16.8 | ||
| Solvent/ligand | 33.9 | 21.7 | ||
| Ramachandran plot | ||||
| Favored (%) | 97.0 | 98.2 | ||
| Allowed (%) | 2.8 | 1.8 | ||
| Outliers (%) | 0.2 | 0.0 | ||
| PDBd code | 4H5J | 4H5I | ||
a Multiwavelength anomalous diffraction.
b Values in parentheses are for the highest resolution shell.
c RMSD, root mean square deviation.
d Protein Data Bank.
During refinement, one metal-binding site per monomer was observed in the electron density maps and was provisionally identified as either Ca2+ or K+ (which have similar x-ray scattering properties). K+ was present at approximately physiological concentration in the crystallization conditions. In addition, the apparent occupancy of the metal site was unaffected by the addition of 10 mm calcium chloride or the chelating agent EGTA (30 mm) or EDTA (10 mm) to the precipitant solution. Soaking crystals in mother liquor plus the Ca2+ homolog Ho3+ (as 25 mm holmium(III) chloride hexahydrate) was similarly without effect. On the other hand, soaking crystals in mother liquor plus the K+ homolog Rb+ (as 50 or 200 mm rubidium chloride) caused an increase in the electron density consistent with substitution of the K loop K+ with Rb+. Together, these observations led us to assign the metal site as K+ rather than Ca2+. As described under “Results,” other alternatives, such as Na+ or Mg2+, were inconsistent with the refined metal occupancy (∼1.0 for K+) and metal-ligand distances.
Subsequently, we found alternative selenomethionine Sec12 crystallization conditions by using 100 mm Tris, pH 8, 400 mm KCl, 22% (w/v) PEG 3350 as the precipitant solution. These crystals grew in a second, higher resolution, crystal form in space group P1 with a = 53.4 Å, b = 53.5 Å, c = 62.0 Å, α = 81.7°, β = 73.4°, γ = 87.58° and two molecules in the asymmetric unit (Table 1). Data were collected to 1.35 Å at National Synchrotron Light Source beamline X29. The structure was phased by molecular replacement, using the program PHASER (23) with the P64 crystal structure as a model and refined using PHENIX. As with the P64 crystal form, the scattering at the metal binding site and the ligand-metal distances were consistent with one bound K+ per K loop in each Sec12 monomer.
Transient State Kinetics
The GEF activity of purified Sec12 proteins was measured by tryptophan fluorescence essentially as described (13). Briefly, reactions were performed in HKM buffer (20 mm HEPES, pH 7.0, 160 mm potassium acetate (KOAc), 3.5 mm magnesium chloride) containing liposomes, prepared as described previously (24), at a final concentration of 0.32 mm. GTP was added to a final concentration of 4 μm, and the reactions were performed with 0.4 μm Sar1 and Sec12 (at the noted concentrations) at 27 °C. Sec12 concentration was determined by UV absorbance following denaturation as described (25). We note that this method gives concentration measurements that are 2.4-fold higher than and presumably more accurate than those obtained using the Bradford assay calibrated to bovine serum albumin. We found that measurements of specific activity were about 10-fold higher than previously reported (13), possibly reflecting improvements in the purification protocol. For example, only those fractions whose elution from a size exclusion column was consistent with monomeric Sec12 were retained for use in crystallographic and biochemical experiments.
To measure the GEF activity displayed by a particular Sec12 construct, Sar1 activation experiments were conducted using a range of Sec12 concentrations. For each concentration, the apparent rate constant of Sar1 activation (kactivation) was determined using single-exponential fitting of the fluorescence time course. kactivation values were then plotted as a function of Sec12 concentration. The rate increases linearly with concentration according to the equation kactivation = kspontaneous + kexchange·[Sec12], where kspontaneous and kexchange, both estimated by least squares fitting, are the rate constant for spontaneous nucleotide exchange and the GEF activity for a given Sec12 construct, respectively (13).
To test the impact of different monovalent cations on exchange activity, GEF assays were performed in which the KOAc was replaced with either 150 mm KCl or 150 mm NaCl. The pH of the HEPES buffer was adjusted with either KOH or NaOH, as appropriate. We note that for reactions performed in KCl, a small amount (∼2 mm) of Na+ was also present, because Sec12 and Sar1 stocks both contained NaCl.
Yeast Methods
To set the stage for plasmid shuffling experiments, an S. cerevisiae heterozygous deletion SEC12/Δsec12 strain (strain background BY4743, Thermo Fisher Scientific) was transformed with the CEN-URA plasmid pRS416-SEC12. pRS416-SEC12 contains SEC12, together with 460 bp of upstream and 455 bp of downstream flanking DNA, inserted into the XhoI and HindIII sites of pRS416 (26). Transformants were sporulated and dissected to obtain haploid sec12Δ cells, whose chromosomal deletion was complemented by pRS416-SEC12. pRS415 CEN-LEU wild-type and mutant SEC12 plasmids were constructed in the same manner and were transformed into the pRS416-SEC12-complemented haploid strains for plasmid shuffling. Individual transformants were streaked onto 5-fluoroorotic acid plates to select against pRS416-SEC12; the plates were then incubated at 22, 30, or 37 °C.
RESULTS
X-ray Structure of the Sec12 Cytoplasmic Domain
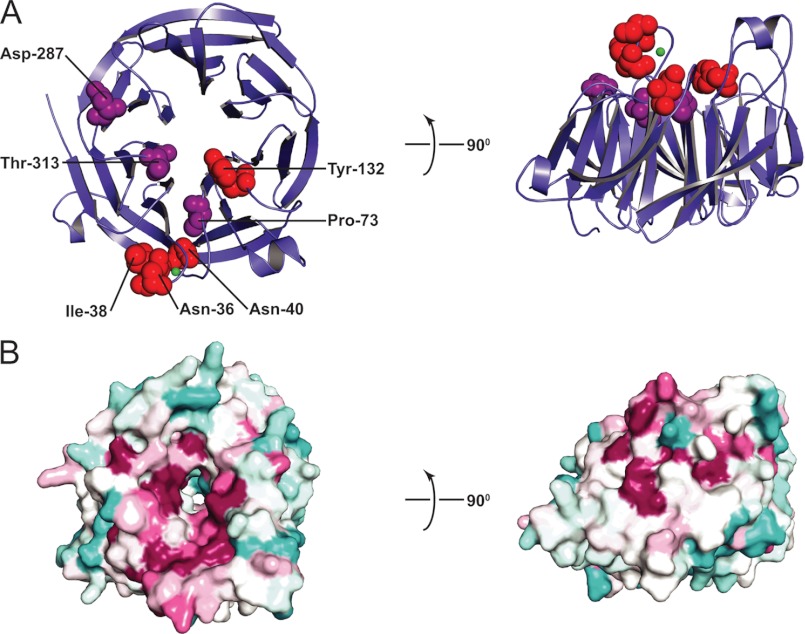
COPII coat assembly is initiated by Sec12, a single-pass transmembrane protein (3, 4). The ER luminal domain of Sec12 is highly variable in length and sequence and thus presumably lacks a conserved function, whereas the cytosolic domain is somewhat more conserved and functions as a Sar1 GEF (13, 27). To develop a detailed understanding of the initiation of COPII-coated vesicle budding, we determined the 1.36 Å resolution crystal structure of the cytosolic portion (residues 1–354) of S. cerevisiae Sec12 (Table 1; see “Experimental Procedures”). The structure confirms a previous proposal, based on sequence analysis (16), that Sec12 adopts a seven-bladed β propeller fold with a “1 + 3 velcro ring closure”; i.e. the seventh propeller blade comprises one β-strand from the N terminus in combination with three β-strands from the C terminus (28) (Fig. 1A).
FIGURE 1.
X-ray structure of Sec12. A, ribbon representation of the cytosolic domain of Sec12 (residues 1–354). Residues mutated in this work are shown in a space-filling representation and are colored to reflect the impact of the mutation on GEF activity in vitro, with red signifying <25% of wild-type specific exchange activity and purple signifying milder defects. The impact of N40A, P73L, and T313A mutations on GEF activity were described previously by Futai et al. (13). B, surface representation of the Sec12 structure, with sequence conservation represented in colors ranging from burgundy (most highly conserved) to turquoise (least highly conserved). These images were based on ConSurf analysis (36). The sequence alignment used to generate conservation scores contained primarily yeast Sec12 sequences, obtained by selecting only those sequences sharing at least 15% sequence identity with S. cerevisiae Sec12, residues 1–354.
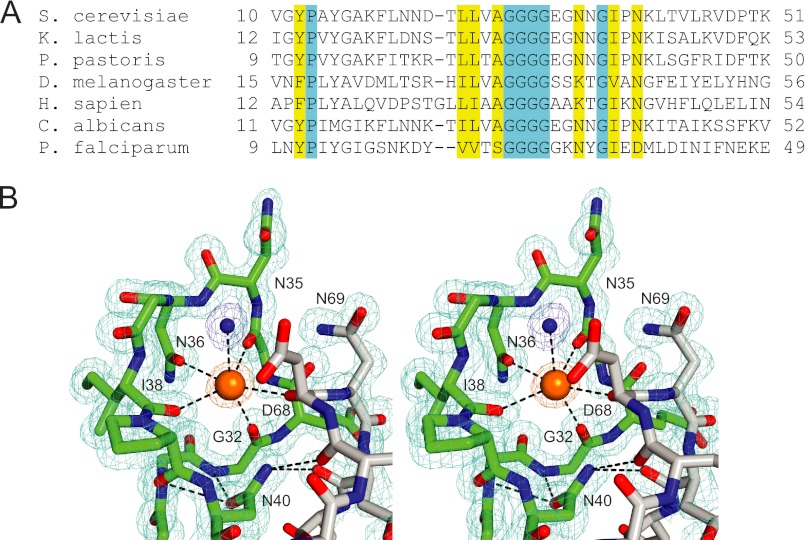
One contiguous region of the Sec12 surface contains most of the highly conserved solvent-accessible residues (Fig. 1B). In the center of this region is an extended loop (the K loop) that projects upward from the first propeller blade. Despite the low overall sequence conservation among Sec12 proteins from distantly related species, the K loop is formed by a universally conserved sequence motif, GGGGXXXXGΦXN (where Φ represents a hydrophobic amino acid; rarely, the fifth glycine is instead serine or the final asparagine is instead aspartate; Fig. 2A). This conserved sequence motif, corresponding to residues 29–40, connects the central two β-strands of the first propeller blade (16). In all four monomers (two distinct monomers per asymmetric unit in each of two different crystal forms), the K loop adopts the same conformation: a vertical loop, reminiscent of a roller coaster inversion, which surrounds and is evidently stabilized by a metal ion (Fig. 2B).
FIGURE 2.
The K loop region of Sec12. A, sequence alignment. Much less homology is observed outside the K loop region of the cytoplasmic domain, and virtually no homology is observed in the luminal domain. B, stereo view of the K loop (green), adjacent loop (gray), and corresponding electron density (2Fo − Fc, contoured to 1σ). The bound K+ and liganding water molecule are shown as a large orange sphere and a small blue sphere, respectively. The residues that contribute the remaining five oxygen ligands (except for Gly-34) are labeled. Also displayed is the hydrogen-bonding network centered on the functionally critical side chain of Asn-40.
As discussed below, all available evidence suggests that the K loop metal is a K+ ion, coordinated in an octahedral geometry by the oxygens of five protein groups and a water molecule (Fig. 2B). Four of the five protein ligands are provided by the K loop itself (green in Fig. 2B); these include the side chain oxygen of Asn-36 and the backbone carbonyl oxygens of Gly-32, Gly-34, and Ile-38. The fifth protein ligand is the backbone carbonyl oxygen of Asp-68, a residue within a neighboring loop (residues 65–73; gray in Fig. 2B). The metal ion coordination is completed by a single water molecule. The relatively high B-factor for this water molecule hints that it could be displaced to allow the bound K+ to interact directly with Sar1. Thus, the water-filled ligand position on the K+ ion might contribute directly to substrate binding, as observed for other enzymes (29) but not previously for nucleotide exchange factors.
Asn-40, previously proposed to act as a catalytic residue because of its strict conservation (16) and critical importance for GEF activity (13), instead plays a structural role, stabilizing both the K loop and the neighboring loop that provides the fifth protein ligand (Fig. 2B). Asn-40 fulfills this structural role by forming a network of hydrogen bonds with the backbone nitrogens of the strictly conserved residues Gly-31 and Gly-32 in the K loop and with the backbone carbonyls of Glu-67 and Asp-70 (flanking the liganding residue Asp-68) in the adjacent loop.
The Sec12 structure explains the strict conservation of the four glycines at the beginning of the K loop. Gly-32, whose carbonyl oxygen is one of the K+ ligands, has a backbone conformation that lies in a region of the Ramachandran space excluded for non-glycine amino acids. The remaining glycines inhabit structural contexts where there is simply no room for a side chain larger than hydrogen. For example, any residue other than glycine at position 30 would collide with the absolutely conserved Asn-40, whereas any residue other than glycine at position 31 would collide with the conserved hydrophobic residue, Ile-38.
Conserved Surface Residues in and around the K Loop Are Critical for GEF Activity
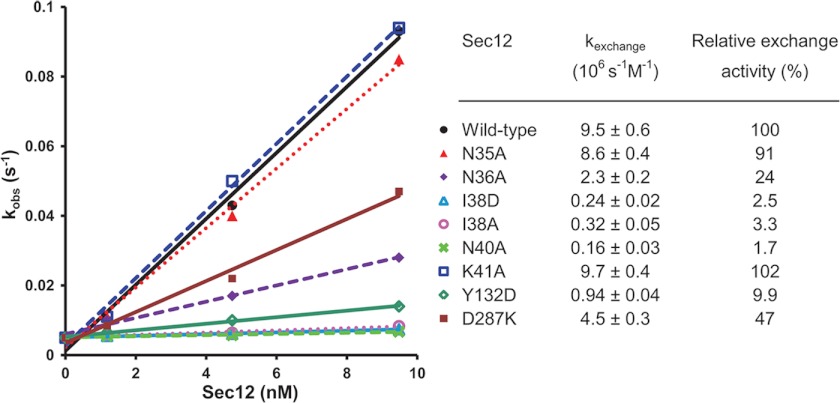
To test the functional significance of the K loop (residues 29–40), we replaced several residues with Ala. We did not test substitutions for Gly (residues 29–32, 34, and 37) or Pro (residue 39) because of the concern that these would perturb the structure. Similarly, Glu-33, whose side chain forms two hydrogen bonds with Ser-91, was not altered. The four remaining K loop residues (Asn-35, Asn-36, Ile-38, and Asn-40), as well as the flanking residue Lys-41, were each replaced by Ala. To determine the activities of these mutants, we used an established in vitro GEF assay that relies on the change in Sar1 fluorescence upon replacement of GDP with GTP (13). As shown in Fig. 3, Ala substitution for Asn-36 (N36A), Ile-38 (I38A), and Asn-40 (N40A) reduced activity 75–98%; the other two substitutions had little or no impact on activity. Asn-36 is especially notable in that its side chain interacts directly with the bound metal (the other liganding groups are backbone carbonyls and a water molecule). Moreover, although Asn-36 is not one of the conserved residues within the K loop motif, multiple-sequence alignment indicates that this position is invariably occupied by a residue capable of interacting with a bound metal (i.e. either Asn, Ser, Thr, Tyr, or His). Nonetheless, although N36A retains only 25% of wild-type activity, it is not completely inactive. This finding may indicate that a bound metal is not absolutely required for activity; alternatively, it may indicate that a water molecule can take the place of the missing side chain ligand. Unfortunately, our inability to crystallize the N36A mutant prevented us from resolving this question definitively.
FIGURE 3.
Exchange activity of Sec12 mutants. Tryptophan fluorescence nucleotide exchange assays measuring the GEF activity of Sec12 (wild type and single substitution mutants) were performed at various Sec12 concentrations. kactivation values from each reaction (derived from exchange time courses, such as those shown in Fig. 4A) were plotted; the slope of these values, as determined by least squares fitting, is reported as the specific exchange activity (kexchange; see “Experimental Procedures” for further details).
Ile-38 is strictly conserved as a hydrophobic residue (Φ) in the K loop. Its carbonyl oxygen interacts directly with the metal ion, whereas its side chain is surface exposed on the “back” side of the loop (as viewed in Fig. 2B). I38A retained only 3.3% of wild-type activity; a second mutant, I38D (substituting Ile-38 with Asp) was similarly inactive (2.6% of wild-type activity). Ile-38 thus appears to play a key role in GEF activity, perhaps interacting directly with Sar1. Finally, consistent with a previous report (13) and with the key structural role of Asn-40 as described above, we find that N40A has very little (<2%) residual activity.
In earlier work, several additional conserved residues proposed, based on the predicted β propeller structure, to lie on the top face of the propeller were investigated using site-directed mutagenesis (13). Two single substitutions, Pro-73 by Leu (P73L) and Thr-313 by Ala (T313A), displayed significantly (>50%) reduced GEF activity. However, although both residues map to the conserved regions of the x-ray structure (Fig. 1), both of their side chains are largely buried, and thus indirect structural effects cannot be excluded. We therefore tested the impact of changing the remaining conserved surface-exposed residues, Tyr-132 and Asp-287. There was a strong reduction in GEF activity, to 9.9% of wild type, when Tyr 132 was changed to aspartate (Y132D). Changing Asp-287, located on the outer edge of the conserved region, to Lys (D287K) had a milder impact (47% of wild-type activity).
The K Loop Binds Potassium, Enhancing GEF Activity
As described above, a single metal ion stabilizing the K loop was evident in electron density maps (Fig. 2). Multiple lines of evidence indicate that this metal ion is K+. First, K+ was present at near (180 mm) or above (400 mm) physiological concentrations in all of our successful crystallization experiments; no diffraction quality crystals could be grown in its absence. Second, the crystallographic occupancy for a single K+ modeled in the K loop binding site refined to 0.98 and 0.86 for the two monomers within each asymmetric unit. This finding rules out the lighter elements Na+ or Mg2+ but not the slightly heavier element Ca2+. Third, the presence of the Ca2+ chelator EGTA or the multivalent cation chelator EDTA had no effect on crystallization or on the electron density observed at the metal position. Fourth, as predicted for a K+ binding site, the electron density increased when Sec12 crystals were soaked in the K+ analog Rb+. Fifth, the observed metal-oxygen distances in our high resolution structure average 2.8 Å, which is much closer to the calculated optimum (assuming a six-ligand symmetrical coordination shell) for K+ (2.77 Å) than Na+ (2.46 Å) (29). Taken together, our results suggest strongly that the physiologically relevant K-loop-stabilizing metal ion is indeed K+.
To determine whether K+ modulates Sec12 function, GEF activity assays were performed in which the standard conditions were modified to exclude Na+ or K+, respectively. Consistent with an important role for K+, the specific activity decreased 80% when K+ was replaced with Na+ (Fig. 4). To determine whether the stimulatory effect of K+ is specifically attributable to K loop stabilization, we repeated the experiment using the N36A mutant. As described above, the K loop residue Asn-36 contains the only side chain that interacts directly with the bound K+; the remaining ligands are contributed by main chain carbonyls and an ordered water molecule. Strikingly, the N36A mutant displayed the same specific activity in the presence and absence of K+ (Fig. 4). These results support the conclusion that the binding of K+ at physiological concentrations to the K loop results in a substantial increase in Sec12 GEF activity.
FIGURE 4.
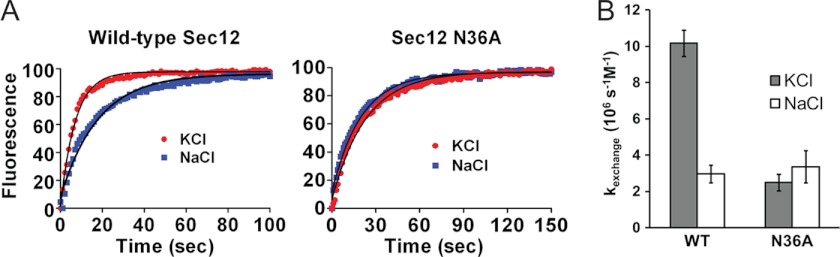
K+ enhances Sec12 GEF activity by binding to the K loop. A, exchange assays with wild-type and N36A Sec12 were performed in the presence of Sec12 (9.5 nm). Conditions were as in Fig. 3 but with KCl or NaCl (each 150 mm) replacing potassium acetate. Fluorescence is shown in arbitrary units; representative time courses are shown together with fits to a single exponential. B, the corresponding kexchange values. Error bars, S.D. (n = 3).
The K Loop Is Critical for Normal Growth of S. cerevisiae
To directly assess the physiological significance of the active site region of the Sec12 β propeller, we used plasmid shuffling to replace wild-type SEC12, which is essential for viability, with a battery of site-directed mutants. To ensure approximately wild-type expression levels, wild-type and mutant SEC12 genes were carried on single-copy CEN plasmids together with ∼500 nucleotides of flanking sequence on each side. We used single residue substitution mutants located within the K loop (Asn-35, Asn-36, Ile-38, or Asn-40) in the region surrounding the K loop (Tyr-15, Lys-41, Ser-72, or Tyr-132) or at the periphery of the conserved region (Glu-175 or Asp-287). The I38D and N40A mutations, which had the most severe effects of any mutants tested on GEF activity measured in vitro (Fig. 3), were lethal in vivo (Table 2). A second Ile-38 mutant, I38A, was not lethal but conferred a severe growth defect. These results demonstrate the essentiality of the K loop for S. cerevisiae viability. Our results also demonstrate that mutant Sec12 proteins retaining as little as 10% in vitro GEF activity were nonetheless capable of supporting apparently normal yeast growth, at least when expressed from a CEN plasmid in rich medium (compare Fig. 3 and Table 1). Thus, yeast containing the mutants N36A and Y132D, with 25 and 9.9% of wild-type GEF activity, respectively, as their sole source of Sec12 grew similarly to wild-type cells.
TABLE 2.
Growth of wild-type and sec12 mutant yeast
| SEC12a | Growthb |
|---|---|
| Wild-type | + |
| pRS415c | − |
| N40A | − |
| I38D | − |
| I38A | +/− |
| Y15K | + |
| N35A | + |
| N36A | + |
| N36Q | + |
| K41A | + |
| S72A | + |
| Y132D | + |
| E175K | + |
| D287K | + |
a Haploid SEC12Δ S. cerevisiae carrying pRS416-SEC12 was transformed with pRS415-SEC12 plasmids and then plated on 5-fluoroorotic acid to select against pRS416-SEC12.
b +, growth similar to wild-type; −, no growth (at 30 °C). I38A (+/−) displayed a significant growth defect at 30 °C, which was markedly exacerbated at 37 °C.
c pRS415 plasmid lacking SEC12.
DISCUSSION
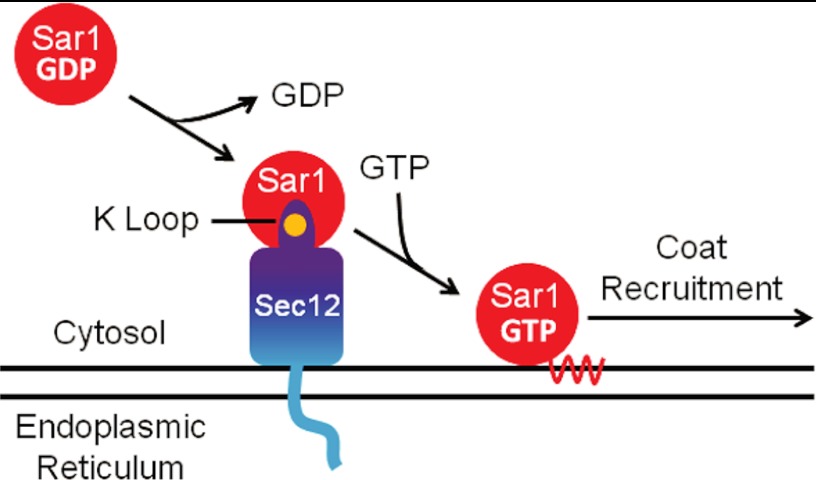
It has long been clear that GEF proteins have a wide variety of three-dimensional structures and use an array of mechanisms to destabilize the interaction between small GTPases and bound nucleotides (30). Our structure of Sec12 adds a second example of a β propeller GEF, similar in overall structure to the Ran GEF RCC1 (31). In both Sec12 and RCC1, small structural elements protruding from one propeller blade (a conserved, potassium-stabilized K loop in the case of Sec12, a “β wedge” in the case of RCC1 (17)) are crucial for GEF activity. A Ran-RCC1 co-crystal structure revealed that the β wedge pushes aside the P loop as a rigid body; a sulfate anion is found in the site normally occupied by the β phosphate of GDP/GTP and presumably serves to stabilize the conformation of the P loop in the absence of bound nucleotide (17). A Sar1-Sec12 co-crystal structure has proved elusive, presumably because of the very low affinity of Sec12 for Sar1 (13); similarly, given the large structural arrangements that GEFs generally provoke in GTPases (30), we have not attempted to dock the known Sar1 structure (5) with our Sec12 structure. Nonetheless, a working hypothesis is that the K loop interacts with the Sar1 P loop in a manner roughly analogous to that observed for the β wedge of RCC1 and Ran (Fig. 5). The rigidity of the β wedge of RCC1 appears to be important because its conformation remains essentially unchanged after binding to Ran (17, 31). K+ may play a similar rigidifying role by stabilizing the K loop. In addition, the K+ ion may participate directly in Sec12-Sar1/GDP binding. One attractive possibility is that the sixth K+ ligand, a relatively weakly bound water molecule, is displaced in the Sec12-Sar1 complex by an oxygen ligand contributed by Sar1/GDP.
FIGURE 5.
Model of the interaction between Sec12 and Sar1. One face of Sec12 is populated by conserved residues (purple), which interact with Sar1-GDP. The K loop helps to dislodge GDP from Sar1, perhaps by clashing with its P loop; this activity is enhanced by bound potassium (orange), which stabilizes the K loop. Following activation, Sar1 binds stably to the ER membrane and initiates COPII vesicle budding.
The catalytic importance of the K loop is clearly established by the strong impact, in vitro and in vivo, of mutations in conserved K loop residues. These include Ile-38, a surface-exposed K loop residue that probably interacts directly with Sar1, and Asn-40, which sits at the base of the K loop and stabilizes its conformation through an elaborate hydrogen bond network (Fig. 2B). Mutations in either residue cause catastrophic drops in catalytic activity in vitro (Fig. 3) and, as demonstrated by plasmid shuffling, are lethal in vivo (Table 2). The finding that all of the mutations that compromise function in vivo also display strongly reduced GEF activity in vitro is consistent with the expectation that GEF activity is, in fact, the physiologically essential function of Sec12.
Although the strict conservation of the GGGGXXXXGΦXN motif that forms the K loop had been previously noted (16), its ability to coordinate K+ was entirely unexpected. Two lines of evidence suggest that bound K+ causes a marked increase in Sec12 GEF activity. First, mutation of Asn-36, whose side chain provides a liganding oxygen atom to the bound K+, reduces in vitro GEF activity 4-fold. Second, substitution of K+ by Na+ in the in vitro assay reduces GEF activity about 5-fold. We note that each of these experiments might, in principle, underestimate the role of a bound metal. In the first case, it is possible that K+ is still able to bind the N36A mutant, perhaps with water providing the missing ligand. In the second case, it remains possible that Na+ can substitute, albeit imperfectly, for K+. However, combining Na+ substitution and N36A replacement did not diminish GEF activity further (Fig. 4). In any case, the level of rate enhancement observed in the presence of K+ is in line with that observed for other enzymes activated by K+ (or Na+) (29).
Our work presents what is, to our knowledge, the first biochemical evidence supporting a role for a metal cation in stimulating GEF activity. Although several members of the RasGRP family of Ras GEFs contain potential Ca2+-binding motifs (outside their catalytic domains) that have been implicated in Ca2+ regulation of GEF activity in vivo (32, 33), these conclusions remain controversial (34) and have not been validated in vitro. Mss4 was initially thought to function as a Zn2+-stabilized GEF for the Rab protein Sec4, but its low GEF activity and the discovery that Sec2 is a much more efficient Sec4 GEF have led to its reclassification as a Rab chaperone (35). Thus, our results establish Sec12 as the first GEF whose catalytic domain is activated by metal ion binding.
Acknowledgments
We gratefully acknowledge the staff of National Synchrotron Light Source beamline X29 for assistance with data collection. We also thank Mark Rose and his group for helpful advice and reagents and especially Abigail Trarbach for assistance with yeast tetrad dissection.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM071574 (to F. M. H.).
The atomic coordinates and structure factors (codes 4H5I and 4H5J) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- COPII
- coat protein II
- ER
- endoplasmic reticulum
- GEF
- guanine nucleotide exchange factor.
REFERENCES
- 1. Lee M. C., Miller E. A. (2007) Molecular mechanisms of COPII vesicle formation. Semin. Cell Dev. Biol. 18, 424–434 [DOI] [PubMed] [Google Scholar]
- 2. Miller E. A., Barlowe C. (2010) Regulation of coat assembly. Sorting things out at the ER. Curr. Opin. Cell Biol. 22, 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barlowe C., Schekman R. (1993) SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365, 347–349 [DOI] [PubMed] [Google Scholar]
- 4. Nakano A., Brada D., Schekman R. (1988) A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 107, 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang M., Weissman J. T., Beraud-Dufour S., Luan P., Wang C., Chen W., Aridor M., Wilson I. A., Balch W. E. (2001) Crystal structure of Sar1-GDP at 1.7 Å resolution and the role of the NH2 terminus in ER export. J. Cell Biol. 155, 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aridor M., Weissman J., Bannykh S., Nuoffer C., Balch W. E. (1998) Cargo selection by the COPII budding machinery during export from the ER. J. Cell Biol. 141, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matsuoka K., Orci L., Amherdt M., Bednarek S. Y., Hamamoto S., Schekman R., Yeung T. (1998) COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell 93, 263–275 [DOI] [PubMed] [Google Scholar]
- 8. Matsuoka K., Schekman R., Orci L., Heuser J. E. (2001) Surface structure of the COPII-coated vesicle. Proc. Natl. Acad. Sci. U.S.A. 98, 13705–13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stagg S. M., LaPointe P., Razvi A., Gürkan C., Potter C. S., Carragher B., Balch W. E. (2008) Structural basis for cargo regulation of COPII coat assembly. Cell 134, 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bi X., Mancias J. D., Goldberg J. (2007) Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31. Dev. Cell 13, 635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shaywitz D. A., Espenshade P. J., Gimeno R. E., Kaiser C. A. (1997) COPII subunit interactions in the assembly of the vesicle coat. J. Biol. Chem. 272, 25413–25416 [DOI] [PubMed] [Google Scholar]
- 12. Lee M. C., Orci L., Hamamoto S., Futai E., Ravazzola M., Schekman R. (2005) Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 122, 605–617 [DOI] [PubMed] [Google Scholar]
- 13. Futai E., Hamamoto S., Orci L., Schekman R. (2004) GTP/GDP exchange by Sec12p enables COPII vesicle bud formation on synthetic liposomes. EMBO J. 23, 4146–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bi X., Corpina R. A., Goldberg J. (2002) Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419, 271–277 [DOI] [PubMed] [Google Scholar]
- 15. Fath S., Mancias J. D., Bi X., Goldberg J. (2007) Structure and organization of coat proteins in the COPII cage. Cell 129, 1325–1336 [DOI] [PubMed] [Google Scholar]
- 16. Chardin P., Callebaut I. (2002) The yeast Sar exchange factor Sec12, and its higher organism orthologs, fold as β-propellers. FEBS Lett. 525, 171–173 [DOI] [PubMed] [Google Scholar]
- 17. Renault L., Kuhlmann J., Henkel A., Wittinghofer A. (2001) Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell 105, 245–255 [DOI] [PubMed] [Google Scholar]
- 18. Otwinowski Z., Minor W. (1998) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 20. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Generation, representation and flow of phase information in structure determination. Recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 21. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX. Building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 23. Storoni L. C., McCoy A. J., Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 24. Antonny B., Madden D., Hamamoto S., Orci L., Schekman R. (2001) Dynamics of the COPII coat with GTP and stable analogues. Nat. Cell Biol. 3, 531–537 [DOI] [PubMed] [Google Scholar]
- 25. Edelhoch H. (1967) Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 26. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weissman J. T., Plutner H., Balch W. E. (2001) The mammalian guanine nucleotide exchange factor mSec12 is essential for activation of the Sar1 GTPase directing endoplasmic reticulum export. Traffic 2, 465–475 [DOI] [PubMed] [Google Scholar]
- 28. Fülöp V., Jones D. T. (1999) β propellers. Structural rigidity and functional diversity. Curr. Opin. Struct. Biol. 9, 715–721 [DOI] [PubMed] [Google Scholar]
- 29. Page M. J., Di Cera E. (2006) Role of Na+ and K+ in enzyme function. Physiol. Rev. 86, 1049–1092 [DOI] [PubMed] [Google Scholar]
- 30. Bos J. L., Rehmann H., Wittinghofer A. (2007) GEFs and GAPs. Critical elements in the control of small G proteins. Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 31. Renault L., Nassar N., Vetter I., Becker J., Klebe C., Roth M., Wittinghofer A. (1998) The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 392, 97–101 [DOI] [PubMed] [Google Scholar]
- 32. Ebinu J. O., Bottorff D. A., Chan E. Y., Stang S. L., Dunn R. J., Stone J. C. (1998) RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science 280, 1082–1086 [DOI] [PubMed] [Google Scholar]
- 33. Kawasaki H., Springett G. M., Toki S., Canales J. J., Harlan P., Blumenstiel J. P., Chen E. J., Bany I. A., Mochizuki N., Ashbacher A., Matsuda M., Housman D. E., Graybiel A. M. (1998) A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc. Natl. Acad. Sci. U.S.A. 95, 13278–13283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stone J. C. (2011) Regulation and function of the RasGRP family of Ras activators in blood cells. Genes Cancer 2, 320–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Itzen A., Rak A., Goody R. S. (2007) Sec2 is a highly efficient exchange factor for the Rab protein Sec4. J. Mol. Biol. 365, 1359–1367 [DOI] [PubMed] [Google Scholar]
- 36. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010. Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]