Background: The catabolite control protein A (CcpA) plays an important role in Staphylococcus aureus virulence, biofilm formation, and central metabolism.
Results: Phosphorylation of CcpA negatively affects its DNA binding activity and thus the expression of its target genes.
Conclusion: CcpA activity is modulated by Stk1 phosphorylation.
Significance: This study highlights that phosphorylation of CcpA represents a novel regulatory control mechanism for this major transcriptional factor.
Keywords: Prokaryotic Protein Kinases, Prokaryotic Signal Transduction, Serine/Threonine Protein Kinase, Staphylococcus aureus, Virulence Factors
Abstract
The Staphylococcus aureus serine/threonine protein kinase Stk1 (also known as PknB) affects different key pathways such as cell wall metabolism, antibiotic susceptibility, and regulation of virulence. Here we report that the catabolite control protein A (CcpA), a highly conserved regulator of carbon catabolite repression and virulence in a number of Gram-positive pathogens, was efficiently phosphorylated in vitro and in vivo by Stk1 in S. aureus, whereas the CcpA homologues of Bacillus subtilis and Bacillus anthracis were not affected by the Stk1 orthologue PrkC. Mass spectrometry and mutational analyses identified Thr-18 and Thr-33 as the phosphoacceptors; both are located in the DNA binding domain of this protein. Electrophoretic mobility shift assays demonstrated that the CcpA DNA binding activity was completely abrogated for the phosphorylated CcpA. The physiological relevance of CcpA phosphorylation was assessed by generating CcpA phosphoablative (T18A/T33A) or phosphomimetic (T18D/T33D) mutants. In contrast to the wild-type and phosphoablative ccpA alleles, introduction of the phosphomimetic ccpA allele in a ΔccpA mutant failed to restore the parental biofilm formation profile and the transcription of citZ and hla to levels seen with the wild type. The strong up regulation of ccpA transcripts and CcpA level in the ccpA mutant trans-complemented with the phosphomimetic CcpA variant suggest furthermore that CcpA acts as a negative regulator of its own expression. Together, these findings demonstrate that Stk1-driven phosphorylation of CcpA inhibits its DNA binding activity toward its regulon in S. aureus, representing a novel regulatory mechanism of CcpA activity in addition to the well known regulation via HprKP/Hpr in this clinically important pathogen.
Introduction
Carbon catabolite repression is a widespread, global regulatory phenomenon in bacteria that allows modulation of the expression of genes and operons involved in carbon utilization and metabolization in the presence of the preferred carbon source(s). In carbon catabolite repression, the presence of a preferred carbon source usually induces the repression of genes and operons whose products are involved in the metabolism of alternative, less preferred carbon sources. One of the important and highly conserved regulators of carbon catabolite regulation in low-GC Gram-positive bacteria is the catabolite control protein A (CcpA),3 a member of the LacI/GalR family of bacterial regulatory proteins, which has been intensively studied in Bacillus subtilis (for reviews, see Refs. 1 and 2). In the presence of glucose or other rapidly metabolized carbon sources, CcpA is activated by formation of a complex with its co-regulator Hpr that needs to be phosphorylated on residue Ser-46 by its cognate kinase HprKP. The CcpA·Hpr-Ser(P)-46 complex has an increased affinity for particular cis-acting sequences, termed catabolite-responsive element (cre) sequences, and thereby represses or enhances gene expression depending on the position of the cre in relation to the operator sequence (3, 4). In a number of Gram-positive pathogens such as Bacillus anthracis (5), Clostridium difficile (6), Staphylococcus aureus (7), Staphylococcus epidermidis (8), Streptococcus pneumoniae (9), and Streptococcus pyogenes (10), CcpA is also involved in the regulation of virulence determinants.
In S. aureus, which is one of the leading causes for nosocomial infections and an important human pathogen responsible for a wide variety of diseases, including soft tissue infections, toxic shock syndrome, and necrotizing pneumonia (11), CcpA was shown to affect antibiotic resistance, biofilm formation, expression of toxins such as hla (encoding α-hemolysin) and tst (encoding toxic shock syndrome toxin), and infectivity of this bacterium (7, 12–15). Recent transcriptome and proteome analyses confirmed the broad effects of CcpA on gene expression in S. aureus and suggested this LacI/GalR type of regulator to be active in the presence and absence of glucose (13). However, the regulatory mechanism(s) that modulates the DNA binding properties of the S. aureus CcpA in the absence of glucose has not yet been identified.
Signal transduction through reversible protein phosphorylation is a key regulatory mechanism of both prokaryotes and eukaryotes. To overcome the stressful conditions imposed by a host, pathogens have evolved various protective and offensive responses generally achieved through cascades of phosphorylation reactions. Many of the stimuli encountered are transduced via sensor kinases in the membrane, allowing the pathogen to adapt for survival in hostile environments. In addition to the classical two-component systems, S. aureus contains one eukaryotic-like serine/threonine protein kinase (STPK), Stk1 (16, 17). Recent studies provided the first insights into the regulatory function of Stk1 and the molecular mechanisms of the Ser/Thr phosphorylation system in S. aureus (17). Interestingly, the Ser/Thr kinase Stk1 was identified to influence central metabolic processes in S. aureus (18) and the activity of important regulatory factors such as SarA (19), MgrA (20), and LuxS (21).
Therefore, we investigated the possible regulation of CcpA by Stk1. To this end, we have identified CcpA as a new substrate of Stk1 and identified its phosphorylation sites. This allowed us to address the role and contribution of phosphorylation in regulating CcpA activity. Importantly, with both in vitro and in vivo approaches, we provide for the first time evidence that phosphorylation negatively regulates the activity of CcpA, thus inhibiting biofilm formation and modifying CcpA target gene expression.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37 °C in LB medium supplemented with 100 μg/ml ampicillin or 100 μg/ml spectinomycin when required. S. aureus isolates were plated on tryptic soy agar supplemented with 10 μg/ml chloramphenicol or 50 μg/ml kanamycin when required or grown in filter-sterilized tryptic soy broth (TSB) medium at 37 °C and 150 rpm with a culture to flask volume of 1:10.
TABLE 1.
Strains and plasmids used in this study
| Strains and plasmids | Genotype or descriptiona | Source/Ref. |
|---|---|---|
| E. coli strains | ||
| 10G | E. coli derivative ultracompetent cells used for general cloning; F− mcrA D(mrr-hsdRMS-mcrBC) f80dlacZÄM15 D lacX74 endA1 recA1araD139 D (ara, leu)7697 galU galK rpsL nupG-tonA; | Lucigen |
| BL21(DE3)Star | F2 ompT hsdSB (rB2 mB2) gal dcm (DE3); used to express recombinant proteins in E. coli | Stratagene |
| S. aureus strains | ||
| RN4220 | NCTC 8325-4 r− m+ (restriction-negative, modification-positive) | 26 |
| SA113 | ATCC35556, PIA-dependent biofilm producer, NCTC 8325 derivative, rsbU− | 47 |
| KS66 | SA113 ΔccpA, biofilm-negative SA113 derivative (TetR) | 7 |
| ST1004 | NCTC 8325-4 Δstk1 derivative | 16 |
| E. coli plasmids | ||
| pETPhos | pET15b (Novagen) derivative including the replacement of the thrombin site coding sequence with a tobacco etch virus protease site and Ser to Gly mutagenesis in the N-terminal His tag (AmpR) | 22 |
| pETPhos_ccpA | pETPhos derivative used to express His-tagged fusion of CcpA (AmpR) | This work |
| pETPhos_ccpA_T18A | pETPhos derivative used to express His-tagged fusion of CcpA_T18A (AmpR) | This work |
| pETPhos_ccpA_T33A | pETPhos derivative used to express His-tagged fusion of CcpA_T33A (AmpR) | This work |
| pETPhos_ccpA_T18A/T33A | pETPhos derivative used to express His-tagged fusion of CcpA_T18A/T33A (AmpR) | This work |
| pETPhos_ ccpABs | pETPhos derivative used to express His-tagged fusion of B. subtilis CcpA in E. coli (AmpR) | This work |
| pETPhos_ccpABa | pETPhos derivative used to express His-tagged fusion of B. anthracis CcpA in E. coli (AmpR) | 5 |
| pGEX_prkC | pGEX-4T derivative used to express GST-tagged fusion of B. subtilis PrkC kinase domain in E. coli (AmpR) | This work |
| pCDFDuet-1 | pET vector derivative designed for the co-expression of two proteins under T7lac promoter induction (SpecR) | Novagen |
| pDuet_ccpA | pET vector derivative used for the co-expression of Stk1 and CcpA proteins under T7lac promoter induction (SpecR) | This work |
| S. aureus plasmids | ||
| pMK4_Pprot | pMK4-derived E. coli-S. aureus shuttle vector with a constitutive promoter (AmpR in E. coli/CmR in S. aureus) | 25 |
| pMK4_Pprot_ccpA | pMK4_Pprot derivative used to express His-tagged fusion of CcpA in S. aureus (AmpR in E. coli/CmR in S. aureus) | This work |
| pMK4_Pprot_ccpA_T18A/T33A | pMK4_Pprot derivative used to express His-tagged fusion of CcpA_T18A/T33A in S. aureus (AmpR in E. coli/CmR in S. aureus) | This work |
| pCN34 | E. coli-S. aureus shuttle vector (AmpR in E. coli/KmR in S. aureus) | 27 |
| pCN34_ccpA | pCN34 derivative harboring ccpA and its native promoter (AmpR in E. coli/KmR in S. aureus) | This work |
| pCN34_ccpA_Ala | pCN34 with a ccpA derivative carrying the CcpA_T18A/T33A mutations under the control of the ccpA promoter (AmpR in E. coli/KmR in S. aureus) | This work |
| pCN34_ccpA_Asp | pCN34 with a ccpA derivative carrying the CcpA_T18D/T33D mutations under the control of the ccpA promoter (AmpR in E. coli/KmR in S. aureus) | This work |
a AmpR, ampicillin-resistant; CmR, chloramphenicol-resistant; KmR, kanamycin-resistant; SpecR, spectinomycin-resistant; TetR, tetracycline-resistant; PIA, polysaccharide intercellular adhesin.
Cloning, Expression, and Purification of CcpA and Mutant Proteins from S. aureus
The ccpA gene was amplified by PCR using S. aureus N315 chromosomal DNA as a template with the primers listed in Table 2 containing an NdeI and a BamHI restriction site, respectively. The corresponding amplified product was digested with NdeI and BamHI and ligated into the pETPhos plasmid (22), a variant of pET15b (Novagen) that includes a tobacco etch virus protease site instead of the thrombin site and an N-terminal His tag free of Ser/Thr/Tyr residues, thus generating pETPhos_ccpA. pETPhos_ccpA derivatives harboring threonine to alanine substitutions at Thr-18 and/or Thr-33 of the ccpA open reading frame were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and resulted in the construction of plasmids pETPhos_ccpA_T18A, pETPhos_ccpA_T33A, and pETPhos_ccpA_T18A/T33A, respectively. The duet strategy was used to generate a hyperphosphorylated CcpA protein as described previously (23). The ccpA gene was cloned into the pCDFDuet-1 vector containing the Stk1 kinase domain (21), generating plasmid pDuet_ccpA, which was used to transform E. coli BL21(DE3)Star cells. pDuet_ccpA E. coli cells were used to overexpress and purify His-tagged CcpA as described below. All constructs were verified by DNA sequencing. Recombinant strains harboring the different constructs were used to inoculate 200 ml of LB medium supplemented with ampicillin or spectinomycin, and resulting cultures were incubated at 37 °C with shaking until the optical density of the culture reached an A600 of 0.5. Isopropyl 1-thio-β-d-galactopyranoside (1 mm final) was added to induce the overexpression, and growth was continued for 3 h at 37 °C. Purifications of the His-tagged recombinants were performed as described by the manufacturer (Qiagen).
TABLE 2.
Primers used in this study
Nterm, N-terminal; Cterm, C-terminal; forw, forward; rev, reverse.
| 5′ to 3′ sequencea | |
|---|---|
| Primers | |
| NtermCcpA | TAATAGCTCATATGACAGTACATATATGATGTAGCAAGAGAAGCGCG (NdeI) |
| CtermCcpA | TATGGATCCTTATTTTGTAGTTCCTCGGTATTCAATTCTGTGAGG (BamHI) |
| NtermCcpABs | TAATAGCTGCTAGCATGAGCAATATTACGATCTACGAT (NheI) |
| CtermCcpABs | TATGGATCCCTATGACTTGGTTGA CTTTCTAAGCTC (BamHI) |
| NtermPrkC | TATGGATCCGTGCTAATCGGCAAGCGGATCAGCGGG (BamHI) |
| CtermPrkC | TATAAGCTTCTACCACGGCCACTTTTTTCTTTTGCC (HindIII) |
| pMK4_NtermCcpA | TAATAGCTGGATCCATGACAGTTACTATATATGATGTAGCAAGAGAAGCG (BamHI) |
| pMK4_CtermCcpA | TATCTGCAGTTAGTGATGATGATGATGATGTTTTGTAGTTCCTCGGTATTC (PstI) |
| pCN34_NtermCcpA | TATGGATCCTGTTGAAGTAGAATATTATC (BamHI) |
| pCN34_CtermCcpA | GAAGAATTCTTATTTTGTAGTTCCTCGGTATTCAATTCTGTG (EcoRI) |
| Nterm hla probe | TAATTAATACCCTTTTTCTCTATTTC |
| Cterm hla probe | GTTACTGAGCTGACTATACGTGTTTTCAT |
| Nterm citZ probe | ATTGTAAAATTCATGGATTATTCAC |
| Cterm citZ probe | CTAAACCTCTTTGTAATTCTGCCAT |
| Nterm ald probe | TGCCCATTCCGCTTCATG TGAACC |
| Cterm ald probe | CTCCCTTGGTATACCAATTTTCAT |
| Nterm tst probe | CACTTTGTTTTTTGCTATTTGTAAC |
| Cterm tst probe | CATTAGTAATTTTTTATTCAT |
| Nterm malR probe | TGACTAAACAATCGATTTTAA |
| Cterm malR probe | GCAACATCTTTAATCGTAACCAT |
| Nterm pckA probe | GCTGGGTTTCATTGGGTCCATGTC |
| Cterm pckA probe | CAGTGTATGTGTCTACTGACAT |
| RT-PCR primers | |
| ccpA forw | AAAGGCAATTTGCCAGATGC |
| ccpA rev | AATTGCTTCTTCGTCGCTGATAC |
| citZ forw | CCGTAGGTTCTCTGAAAGGGC |
| citZ rev | AACATCGTCATAACTTGTTCGTTTG |
| gyrB forw | GACTGATGCCGATGTGGA |
| gyrB rev | AACGGTGGCTGTGCAATA |
| hla forw | AACCCGGTATATGGCAATCAACT |
| hla rev | CTGCTGCTTTCATAGAGCCATTT |
| 16 S rRNA forw | CCATAAAGTTGTTCTCAGTT |
| 16 S rRNA rev | CATGTCGATCTACGATTACT |
a Restriction sites are underlined and specified in parentheses.
Mass Spectrometry Analysis
Purified His-tagged hyperphosphorylated CcpA from the E. coli strain carrying pDuet_ccpA and co-expressing the Stk1 kinase was subjected to mass spectrometry without further treatment. Subsequent mass spectrometric analyses were performed as reported previously (24).
Overexpression and Purification of CcpA in S. aureus
The ccpA gene was amplified by PCR using S. aureus N315 chromosomal DNA as a template with the primers listed in Table 2 containing a BamHI and a PstI site, respectively. The PCR product harboring a C-terminal His-tagged fusion was digested with BamHI/PstI, enabling direct cloning into the pMK4-pProt expression vector cut with the same enzymes, thus generating pMK4_ccpA, which allows constitutive expression in S. aureus as described previously (16, 25). The double mutant corresponding to T18A and T33A was generated using the QuikChange site-directed mutagenesis kit (Stratagene), generating the pMK4_ccpA_T18A/T33A construct. The resulting vectors were used to transform S. aureus strain RN4220 (26), which served as donor for transducing the plasmids into the stk1 mutant ST1004 (S. aureus 8325-4 Δstk1) (16). Purification of soluble CcpA proteins was performed on nickel-nitrilotriacetic acid-agarose beads as described by the manufacturer (Qiagen) and used for immunoblotting.
In Vitro Kinase Assay
In vitro phosphorylation was performed with 4 μg of CcpA protein or mutant derivatives in 20 μl of buffer P (25 mm Tris-HCl, pH 7.0, 1 mm dithiothreitol (DTT), 5 mm MgCl2, 1 mm EDTA, 50 μm ATP) with 200 μCi ml−1 (65 nm) [γ-33P]ATP (PerkinElmer Life Sciences; 3000 Ci mmol−1) and 0.5 μg of Stk1 kinase for 30 min at 37 °C. Control experiments were conducted under the same conditions but without kinase. Cloning, expression, and purification of the Stk1 kinase from S. aureus were described previously (21).
Electrophoretic Mobility Shift Assays
The DNA probes for electrophoretic mobility shift assays (EMSAs) were generated by PCR using S. aureus N315 chromosomal DNA as a template, which encompasses the promoter regions of hla, citZ, ald, tst, malR, and pckA, with respective primer pairs listed in Table 2. The 5′-ends of the double-stranded PCR products were labeled using [γ-32P]ATP and T4 polynucleotide kinase. A typical assay mixture contained (in 20 μl) 10 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm EDTA, 1 mm DTT, 0.1 μg of nonspecific competitor (poly(dI-dC)), 5% (v/v) glycerol, radioactive DNA probe (2000 cpm ml−1), and various amounts (0, 15, 65, 130, and 200 nm) of the purified CcpA proteins. After 30 min of incubation at room temperature, 20 μl of this mixture was loaded onto a native 4% (w/v) polyacrylamide Tris borate-EDTA Ready Gel (Bio-Rad) and electrophoresed in 1% Tris borate-EDTA (v/v) buffer for 1 h at 100 V cm−1. Radioactive species were detected by autoradiography using direct exposure to films.
Complementation Studies
For the trans-complementation of the S. aureus SA113 ΔccpA mutant KS66 (12), the following plasmids harboring the wild-type ccpA and 500 bp of the upstream region (pCN34_ccpA), the phosphoablative ccpA variant carrying the T18A/T33A modifications (pCN34_ccpA_Ala), and the phosphomimetic ccpA variant carrying the T18D/T33D modifications (pCN34_ccpA_Asp), respectively, were constructed as follows. The ccpA derivatives were synthesized by Genscript (Aachen, Germany) using the N315 sequence information as master template; amplified with specific primers (Table 2) introducing BamHI and EcoRI sites, respectively; and cloned into the pCN34 vector (27) digested with the same restriction enzymes. The resulting constructs were sequenced and electroporated into S. aureus strain RN4220, which served as donor for transducing the plasmids into KS66.
Antibody Production and Immunoblotting
Polyclonal anti-CcpA antibodies were raised by injecting 500 μg of the His-tagged recombinant CcpA into rabbits (Eurogentec, Liege, Belgium). The resulting crude antisera were purified against the immobilized CcpA antigen. For the determination of CcpA, cytoplasmic protein extracts were isolated from S. aureus cell cultures grown for 5 h in TSB at 37 °C as described previously (28), and protein fractions (10 μg/lane) were separated using SDS-PAGE, blotted onto a nitrocellulose membrane, and subjected to Western blot analysis using the antigen-purified polyclonal anti-CcpA antiserum. Phosphorylated CcpA derivatives were detected by immunoblotting as described (23) using anti-phosphothreonine, anti-phosphoserine, or anti-phosphotyrosine antibodies (Cell Signaling Technology), respectively.
Measurement of Gene Expression by Quantitative Real Time PCR
RNA isolation and real time RT-PCR were carried out as described by Chatterjee et al. (29) using the primer pairs listed in Table 2. The level of mRNA was normalized against the internal controls gyrB and 16 S rRNA. Transcript amounts were expressed as the -fold difference relative to the control (2ΔΔCT where ΔCT represents the difference in threshold cycle between the target and control genes).
Biofilm Assay
Bacterial cultures were grown in TSB supplemented with 0.1% (w/v) glucose in glass culture tubes with aeration (150 rpm) at 37 °C. Growth medium was inoculated with 0.5 McFarland unit. Biofilm formation on the glass surface and clumping of the cells were monitored after 18 h of growth.
Cloning, Expression, and Purification of PrkC and CcpA from Bacillus Species
The ccpA gene from B. subtilis was amplified by PCR using B. subtilis 168 chromosomal DNA as a template with the primers listed in Table 2 containing NheI and BamHI restriction sites, respectively. The corresponding amplified product was then digested with NheI and BamHI and ligated into the pETPhos plasmid, thus generating pETPhos_ccpABa, which was transformed and overexpressed, and the His-tagged CcpA was purified as described above. The B. anthracis ccpA recombinant vector was a kind gift from Marta Perego and purified as described by Chiang et al. (5) to generate CcpABa. The prkC kinase domain from B. subtilis was amplified by PCR using B. subtilis 168 chromosomal DNA as a template with the primers listed in Table 2 containing a BamHI and a HindIII restriction site, respectively. The corresponding amplified product was then digested with BamHI and HindIII and ligated into the pGEX(M) vector, thus generating pGEX_prkC, which was transformed and overexpressed, and the GST-tagged PrkCBs was purified as described previously (30).
Statistical Analysis
The significance of changes was assessed using the Wilcoxon rank sum test. p values <0.05 were considered significant.
RESULTS
Stk1-mediated Phosphorylation of CcpA
The S. aureus genome encodes one Ser/Thr protein kinase named Stk1 or PknB (16, 18). Although Stk1 appears to be involved in different key pathways such as cell wall metabolism, antibiotic susceptibility, and regulation of virulence (17–19, 31–33), only a little is known about the nature of the corresponding substrates. Because of our interests in Ser/Thr kinase regulation in S. aureus and in central metabolic pathways of this pathogen, we wished to investigate whether the CcpA might be regulated by Ser/Thr phosphorylation in this organism.
To establish whether the CcpA protein undergoes post-translational modification by phosphorylation, we first investigated in vitro whether CcpA could be phosphorylated in the presence of the purified Stk1 kinase. Stk1 was expressed as a His-tagged fusion protein and purified from E. coli as described previously (19, 21). The purified kinase was incubated with CcpA and [γ-33P]ATP, the proteins were resolved by SDS-PAGE, and the phosphorylation profile was analyzed by autoradiography (Fig. 1A). The upper bands illustrate the autokinase activity of Stk1, whereas the lower bands represent phosphorylated CcpA (CcpA-P) proteins. The presence of an intense radioactive signal indicated that CcpA was indeed phosphorylated by Stk1. As expected, no radioactive bands were observed in the absence of Stk1 in this assay. These data indicated that CcpA is phosphorylated by Stk1 and suggested that this protein is regulated via this Ser/Thr kinase.
FIGURE 1.
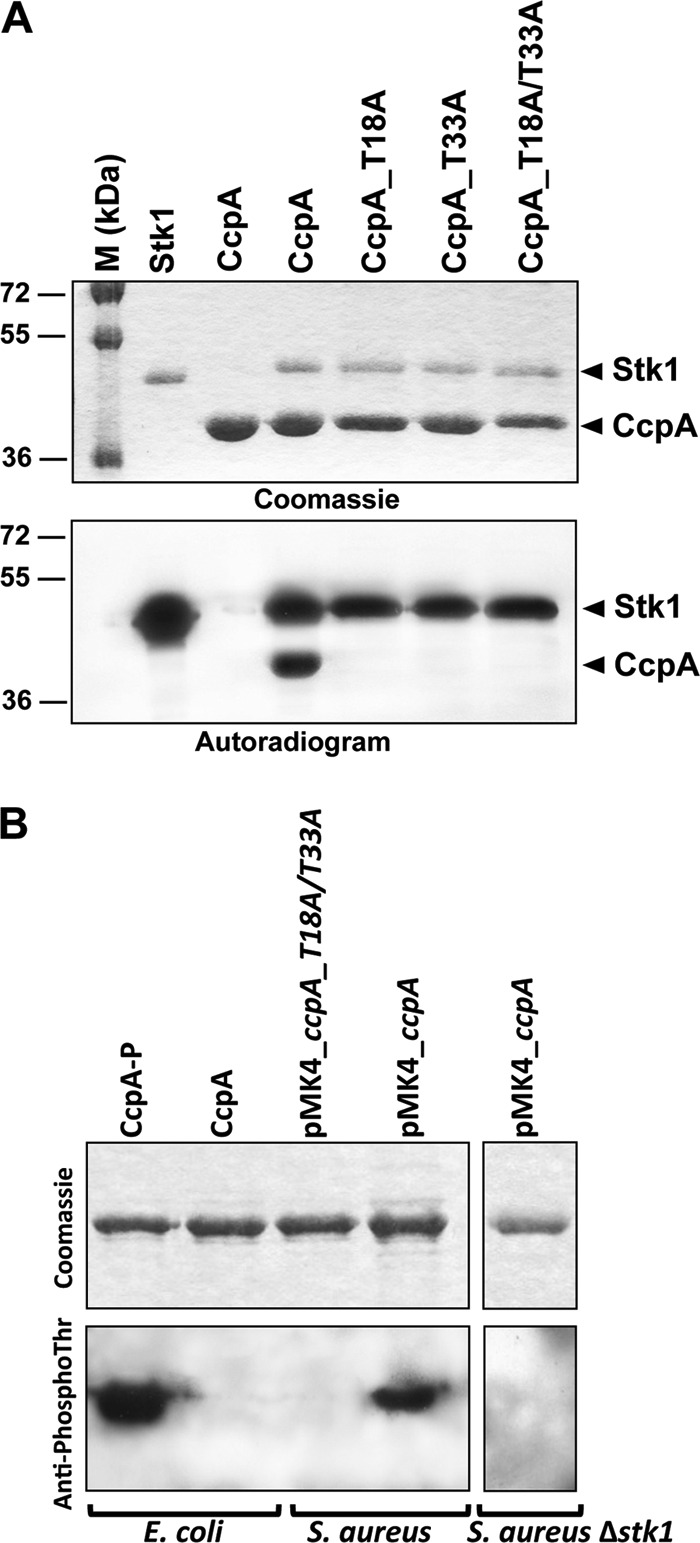
A, in vitro phosphorylation of CcpA and mutant derivatives. The recombinant Stk1 kinase encoded by the S. aureus genome was expressed and purified as a His-tagged fusion in E. coli and incubated in the presence of [γ-33P]ATP with either the purified His-tagged S. aureus CcpA (CcpA_WT) or the mutated variant CcpA_T18A (harboring a Thr to Ala substitution at Thr-18), CcpA_T33A (harboring a Thr to Ala substitution at Thr-33), or CcpA_T18A/T33A (harboring Thr to Ala substitutions at Thr-18 and Thr-33). Samples were separated by SDS-PAGE, stained with Coomassie Blue (upper panel), and visualized by autoradiography (lower panel). The upper bands illustrate the autokinase activity of Stk1, and the lower bands represent phosphorylated CcpA proteins. Standard proteins of known molecular masses were run in parallel (M (kDa) lane). B, in vivo phosphorylation of CcpA. Five micrograms of recombinant CcpA proteins purified from either E. coli (pDuet_ccpA and pETPhos_ccpA, generating a phosphorylated CcpA-P and an unphosphorylated CcpA, respectively), S. aureus (pMK4_ccpA and pMK4_ccpA_T18A/T33A), or S. aureus Δstk1 (pMK4_ccpA) were analyzed by SDS-PAGE after staining with Coomassie Blue (upper panel) and detected on an independent SDS-polyacrylamide gel by immunoblotting using anti-phosphothreonine antibodies (Cell Signaling Technology) (lower panel).
CcpA Is Phosphorylated on Two Threonine Residues, Thr-18 and Thr-33
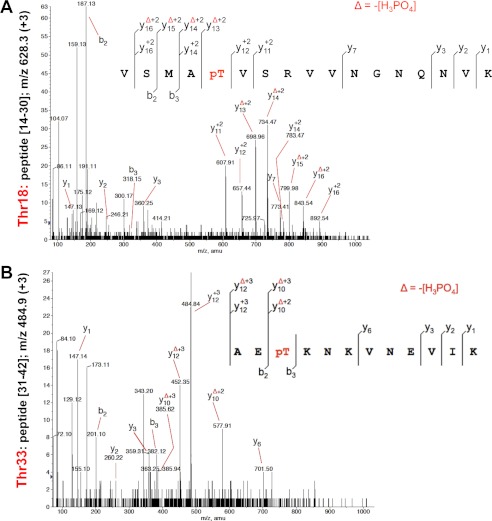
Mass spectrometry (MS) was subsequently used to identify the number and nature of phosphorylation sites on CcpA as reported previously for other STPK substrates (21, 24, 34–38). Phosphorylated CcpA-P was purified from E. coli co-expressing Stk1 and CcpA (pDuet_ccpA) and subjected to mass spectrometry analysis after tryptic digestion. We obtained a sequence coverage of 97%, bearing all possible Ser and Thr residues. The ProteinPilot® database searching software (using the Paragon method with phosphorylation emphasis) was then used to detect and identify the phosphorylated peptides. The MS/MS spectra unambiguously identified the presence of two phosphate groups on peptide(14–30) (Fig. 2A) and peptide(31–42) (Fig. 2B), thus indicating that CcpA is phosphorylated on two threonines, corresponding to Thr-18 and Thr-33.
FIGURE 2.
Identification of the CcpA phosphorylation sites. A, MS/MS spectra at m/z 628.3 (+3) of peptide(14–30) of CcpA. Localization of the phosphate group on Thr-18 was shown by observation of the “y” C-terminal daughter ion series. Starting from the C-terminal residue, all y ions lose phosphoric acid (−98 Da) after the phosphorylated residues. B, MS/MS spectra at m/z 484.9 (+3) of peptide(31–42) of CcpA. Localization of the phosphate group on Thr-33 was shown by observation of the y C-terminal daughter ion series. Starting from the C-terminal residue, all y ions lose phosphoric acid (−98 Da) after the phosphorylated residues.
To confirm the phosphorylation sites identified by MS/MS, we substituted Thr-18 and Thr-33 by alanines either individually or together by site-directed mutagenesis to introduce either single or double mutations (Thr to Ala) in CcpA. The corresponding CcpA_T18A, CcpA_T33A, and CcpA_T18A/T33A proteins were expressed and purified as His-tagged proteins in E. coli BL21(DE3)Star cells harboring either pETPhos_ccpA_T18A, pETPhos_ccpA_T33A, or pETPhos_ccpA_T18A/T33A constructs. Following incubation with Stk1 and [γ-33P]ATP, SDS-PAGE/autoradiogram revealed that all three mutant proteins were no longer phosphorylated by this kinase (Fig. 1A). Surprisingly, single Thr to Ala substitutions were already sufficient to abrogate phosphorylation.
In Vivo Phosphorylation of CcpA
To assess the relevance of our in vitro phosphorylation findings, we also investigated the in vivo phosphorylation of CcpA in a recombinant S. aureus RN4220 strain by Western blot analyses using anti-phosphothreonine, anti-phosphoserine, or anti-phosphotyrosine antibodies, respectively. First, the specificities of the antibodies for the phosphorylated isoforms were determined using the CcpA isoforms purified from either E. coli or E. coli co-expressing Stk1 based on the strategy described by Molle et al. (23). Only the phosphorylated CcpA isoform derived from pDuet_ccpA (CcpA-P) reacted with the anti-phosphothreonine antibodies as expected, whereas the unphosphorylated CcpA protein derived from the pETPhos_ccpA construct (CcpA) failed to give a signal with these antibodies (Fig. 1B). To assess the phosphorylation state of CcpA in S. aureus, we cloned the open reading frames of ccpA and ccpA_T18A/T33A (harboring the Thr to Ala substitutions at Thr-18 and Thr-33) as C-terminal His-tagged fusions into the shuttle vector pMK4-pProt and transformed the resulting plasmids, pMK4_ccpA and pMK4_ccpA_T18A/T33A, respectively, into the S. aureus strain RN4220. Testing the His tag-purified proteins derived from these RN4220 derivatives in a Western blot analysis using anti-phosphothreonine antibodies revealed a clear signal for the wild-type CcpA, whereas no such signal was detectable with the mutated CcpA (Fig. 1B). This demonstrated that CcpA is phosphorylated in S. aureus in vivo as well and that phosphorylation occurs at Thr-18 and Thr-33 both in vitro and in S. aureus. Moreover, no signal could be detected with either the anti-phosphoserine or the anti-phosphotyrosine antibodies, confirming that CcpA is only phosphorylated at threonine residues (data not shown). To confirm the direct phosphorylation of CcpA by Stk1 in vivo, we transduced the pMK4_ccpA vector into the stk1-null mutant strain ST1004 (16) and overexpressed and purified the CcpA protein from this strain devoid of the Stk1 kinase. As shown in Fig. 1B, we could not detect any phosphorylated CcpA isoform when overexpressed in the S. aureus Δstk1 strain, strongly supporting that CcpA is a substrate of Stk1 in vivo.
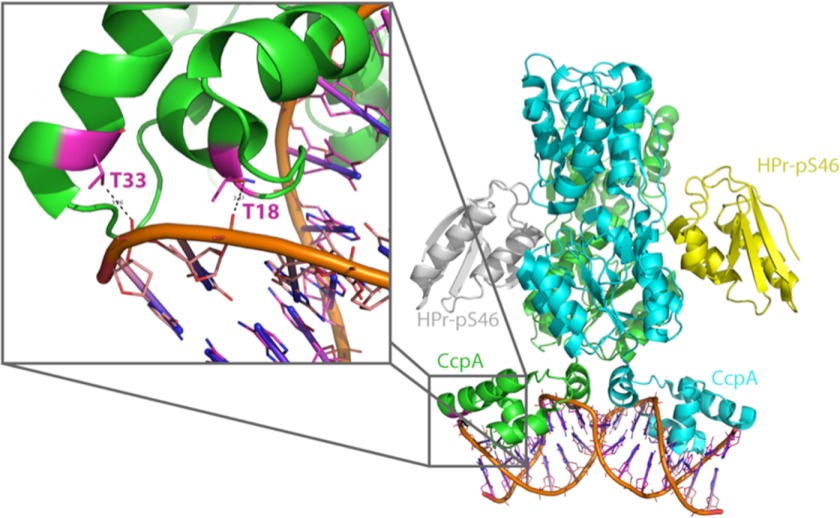
Structural Analysis of CcpA
To get more insight into the possible role of the double phosphorylation on CcpA by Stk1, we looked at various crystal structures of CcpA from different homologues. For our structural analysis depicted in Fig. 3, we used the tertiary complexed form of CcpA, Hpr-Ser(P)-46, and its target DNA from Bacillus megaterium (39). The high degree of conservation between the S. aureus and B. megaterium CcpA amino acid sequences (sequence identity, 57% overall and 82% for the first 60 residues representing the sole DNA binding domain) and their target DNAs allowed us to use this structure for our analysis. This structural complex described the two-component allosteric DNA binding activation mechanism. The two phosphorylated threonine residues are located on helices 2 and 3 of the helix-turn-helix domain responsible for the DNA binding (Fig. 3). Both residues established multiple contacts with the DNA phosphate through the hydroxyl groups of the lateral chain of both threonines. The phosphorylation of one of these threonines will not only generate a steric clash but also cause an electrostatic repulsion between the DNA binding domain and its target DNA.
FIGURE 3.
Mapping of the Thr-18 and Thr-33 phosphorylation sites on CcpA. A schematic representation of the crystal structure of the tertiary complex of CcpA (in blue and green), Hpr-Ser(P)-46 (HPr-pS46; in yellow), and its target DNA (in gray) from B. megaterium (Protein Data Bank code 1RZR) is shown. The close view highlights the interaction of threonines 18 and 31 with the phosphate moieties of the DNA.
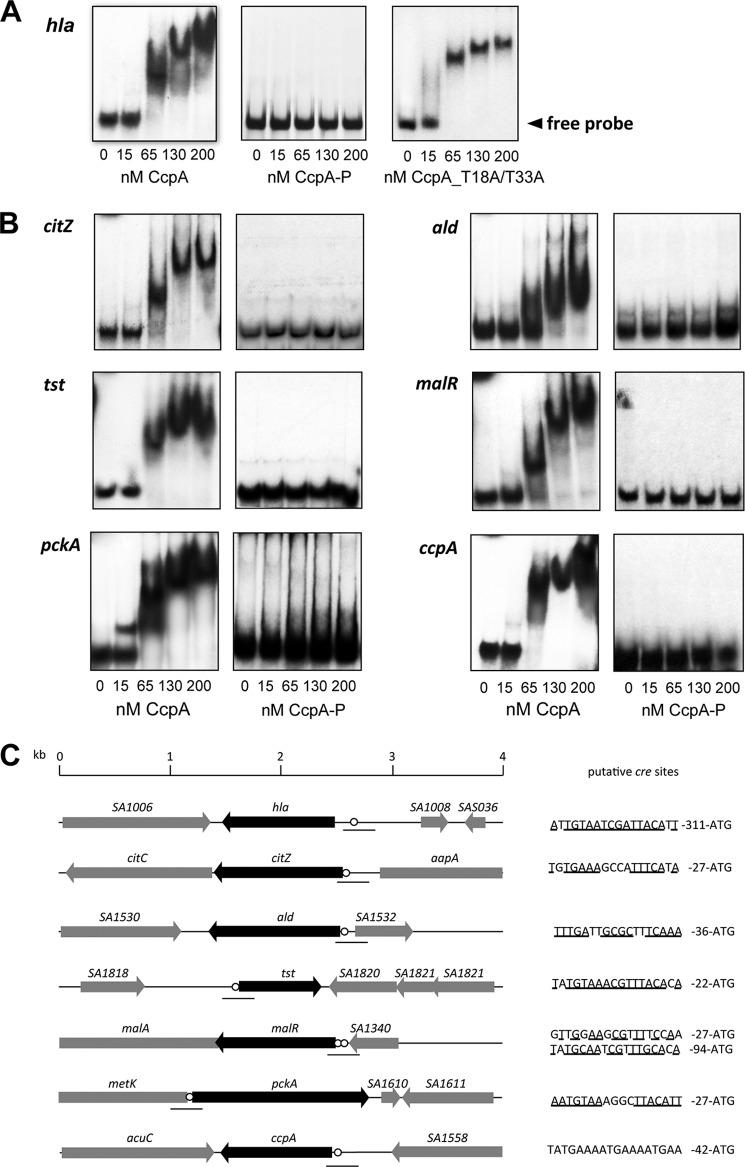
Phosphorylation Negatively Regulates CcpA-DNA Binding Properties
The structural mapping of the phosphorylation sites suggested that the phosphorylation of CcpA might negatively influence the DNA binding activity of this protein. To test this hypothesis, we analyzed the binding capacities of the non-phosphorylated CcpA form (CcpA) and that of the hyperphosphorylated CcpA form (CcpA-P) to a couple of gene promoter elements that were recently shown to be affected by this regulatory protein (Fig. 4C) (7, 12–14). Although the unphosphorylated CcpA interacted with all the double-stranded DNA probes tested in our electrophoretic mobility shift assays (Fig. 4, A and B), the phosphorylated CcpA-P failed to shift any of these probes (Fig. 4A; only the hla probe profile is shown), indicating that CcpA-P had lost its ability to bind to its cre sequences. On the contrary, our EMSAs carried out with the CcpA_T18A/T33A phosphoablative mutant protein showed that this CcpA derivative retained its DNA binding activity (Fig. 4A; only the hla probe profile is shown), confirming the critical role of phosphorylation on the DNA binding capacity of this regulatory protein. It is noteworthy to emphasize that the S. aureus CcpA does not essentially require the association with Hpr being phosphorylated by its cognate HprKP kinase (HPr-P) for efficient binding to its DNA targets (15). This observation was confirmed in our EMSAs for which the DNA binding activity of CcpA was assayed in the absence of Hpr-P, which did not essentially modify the DNA binding properties in this in vitro assay (Fig. 4B).
FIGURE 4.
DNA binding activity of CcpA derivatives. A gel EMSA of CcpA binding to the cre sequences of the hla, citZ, ald, tst, malR, ccpA, and pckA promoters. The cre regions of target genes were amplified by PCR; radioactively labeled; and incubated with 15, 65, 130, or 200 nm purified CcpA. A, binding of the unphosphorylated CcpA, CcpA-P, or the CcpA_T18A/T33A double mutant to the hla promoter region. B, binding of the unphosphorylated CcpA and CcpA-P to the cre sequences of the citZ, ald, tst, malR, ccpA, and pckA promoters. The CcpA_T18A/T33A double mutant is not represented as all the probes tested produced profiles identical to those of CcpA. C, schematic representation of the cre sequences (circles) and open reading frames (arrowed boxes) of the promoter elements studied. The putative cre sequences and the probes (lines) used for the EMSA experiments are indicated. The annotation/numbering of genes are based on the genome sequence of S. aureus strain N315.
Lack of Complementation with the CcpA Phosphomimetic Mutant in ΔccpA Mutant Strain
Acidic residues such as aspartic acid (Asp) qualitatively mimic the phosphorylation effect with regard to functional activity (24, 34, 36, 38, 40, 41). Therefore, phosphoablative (Thr to Ala replacements) and phosphomimetic (Thr to Asp replacements) ccpA alleles were generated and cloned into the E. coli-S. aureus shuttle plasmid pCN34 under the control of the ccpA promoter to trans-complement the S. aureus SA113 ΔccpA mutant KS66 (12). In these constructs, wild-type, phosphoablative (T18A/T33A), and phosphomimetic (T18D/T33D) CcpA isoform expression was monitored by Western blotting using anti-CcpA antibodies. As expected, S. aureus SA113 ΔccpA failed to express CcpA, whereas increased amounts of CcpA were synthesized in the KS66 derivatives complemented with the phosphoablative ccpA variant (pCN34_ccpA_Ala) and the ccpA wild-type allele (pCN34_ccpA), respectively, probably due to a multicopy effect of the introduced plasmids (Fig. 5A). In the KS66 derivative complemented with the phosphomimetic ccpA variant (pCN34_ccpA_Asp), however, a marked increase in CcpA was detected in line with the transcriptional data showing a 63-fold increase in ccpA transcripts in the pCN34_ccpA_Asp-harboring KS66 derivative compared with the wild type (Fig. 5B), suggesting that the expression of this transcriptional regulator is negatively autoregulated in S. aureus.
FIGURE 5.
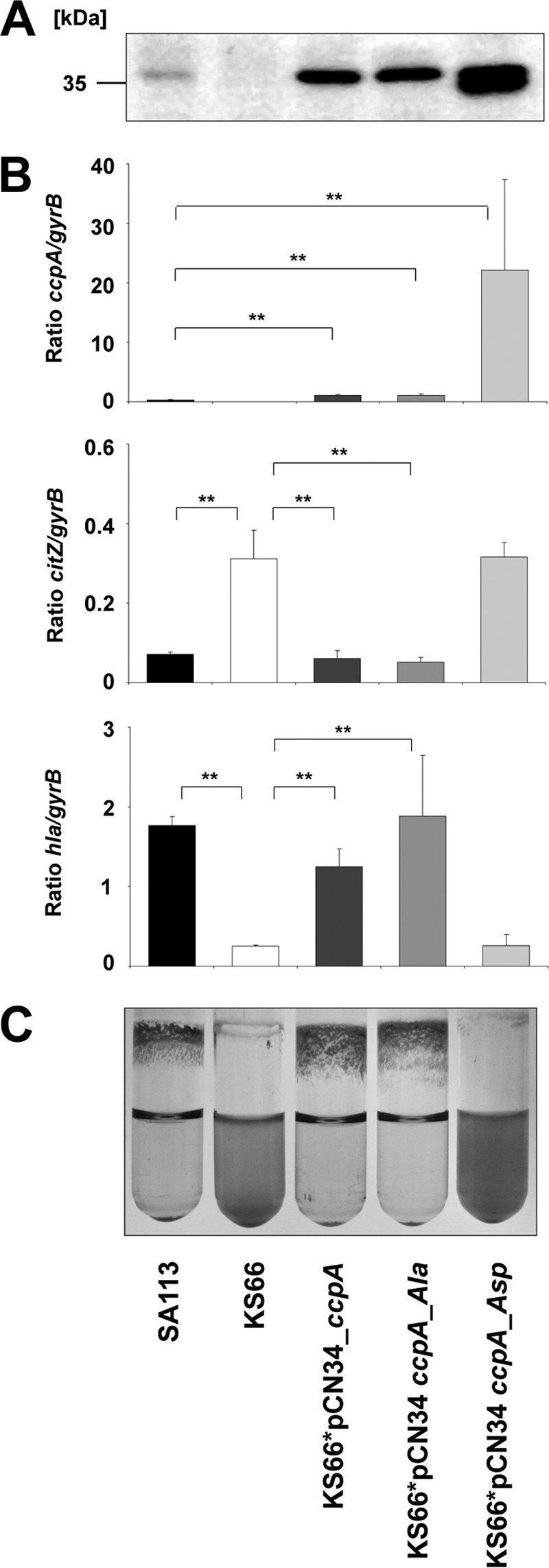
Phenotypic complementation of the ccpA phenotype in S. aureus. A, Western blot analysis of cytosolic protein extracts obtained from 6-h cultures of S. aureus strain SA113, its respective ΔccpA mutant KS66, and the KS66 derivatives complemented with the wild-type allele (KS66*pCN34_ccpA), the CcpA_T18A/T33A variant (KS66*pCN34_ccpA_Ala), and the CcpA_T18D/T33D version (KS66*pCN34_ccpA_Asp) with the polyclonal anti-CcpA antibody SY490. B, quantitative transcript analysis of ccpA, citZ, and hla of S. aureus strain SA113 and its derivatives grown in TSB supplemented with 0.1% (w/v) glucose for 3 (ccpA and citZ) and 8 h (hla) at 37 °C. C, biofilm formation capacities of S. aureus strains SA113 and its derivatives grown in TSB supplemented with 0.1% (w/v) glucose for 18 h at 37 °C. The results are representative of at least two independent experiments (A and C) or are presented as the mean ± S.D. (error bars) of five independent experiments each determined in duplicate (B). *, p < 0.05; **, p < 0.01 for wild type versus ccpA mutant and for trans-complemented derivatives versus ccpA mutant, respectively (Wilcoxon rank sum test).
The impact of constitutive CcpA phosphorylation on gene expression of specific CcpA-regulated target genes was next examined by determining the transcription of citZ encoding the tricarboxylic acid cycle enzyme citrate synthase and hla (encoding α-hemolysin) in SA113 and its derivatives (Fig. 5B). In line with our previous findings (7, 14), we found a significant increase in citZ transcription in the ccpA mutant KS66 in the early exponential growth phase (i.e. after 3 h of growth in TSB supplemented with 0.1% (w/v) of glucose), whereas transcription of hla, which is expressed mainly during the later stages of growth, was markedly down-regulated in KS66 after 8 h of growth compared with the wild type. Trans-complementation of the ccpA mutant with the ccpA wild-type allele (pCN34_ccpA) and the phosphoablative ccpA variant (pCN34_ccpA_Ala) led to the production of citZ and hla transcripts to levels seen in the wild type, whereas the introduction of the T18D/T33D-carrying CcpA version (pCN34_ccpA_Asp) into KS66 did not alter the transcription of both genes (Fig. 5B). This result suggests that the phosphomimetic CcpA variant is unable to modulate the expression of CcpA-regulated target genes.
Whether phosphorylation of CcpA affects biofilm formation as described previously for a ccpA knock-out mutant (12) was investigated by growing the SA113 derivatives in glucose-supplemented TSB under aeration in glass tubes. Under these conditions, SA113 forms a strong biofilm on the glass surface and shows a high degree of cell clumping, whereas cells of the S. aureus SA113 ΔccpA mutant KS66 remain mainly planktonic (12). Here we found that both phenotypes could be restored by complementation with either the wild-type allele or the phosphoablative ccpA variant (Fig. 5C). In contrast, introduction of CcpA_T18D/T33D (ccpA_Asp) failed to restore the parental biofilm profile and clumping, indicating that the phosphomimetic isoform is unable to complement the lack of CcpA activity in biofilm formation presumably as a consequence of its impaired DNA binding activity toward specific regulated genes involved in these processes. It is noteworthy that these effects were not due to a tertiary structure change as confirmed by analysis of the trypsinolysis kinetics of wild-type and mutated CcpA proteins (supplemental Fig. S1) and were consistent with the structural analysis indicating that introduction of Asp at positions 18 and 33 does not seem to induce steric or electrostatic conflicts that could alter the protein fold (Fig. 3). Thus, it can be inferred that CcpA phosphorylation in S. aureus reduces/abolishes its activity, supporting the view that this post-translational modification plays a role in regulating CcpA activity.
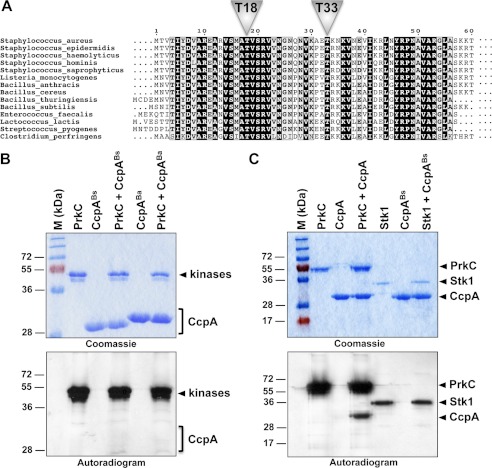
CcpA Orthologue Phosphorylation and Cross-phosphorylation between Species
Given the high degree of similarity between the CcpA proteins of S. aureus and bacilli and based on the finding that the Thr-18 and Thr-33 sites are well conserved among CcpA orthologues (Fig. 6A), we wondered whether the CcpAs from Bacillus species might be phosphorylated as well. Interestingly, CcpA from B. subtilis (CcpABs) could not be phosphorylated by its cognate kinase PrkC (Fig. 6B). This result raised the question whether the phosphoregulation of CcpA might be restricted to pathogens, which led us to test the corresponding CcpA orthologue of B. anthracis (CcpABa). As presented in Fig. 6B, no phosphorylation signal could be detected with CcpABa, indicating that the STPK-driven control of CcpA activity does not seem to be present in bacilli.
FIGURE 6.
A, conservation of the phosphoacceptors in CcpA orthologues within the DNA binding domain. The multiple sequence alignment of CcpA orthologues was performed using ClustalW and ESPript. Numbering of the amino acids corresponds to the CcpA from S. aureus strain COL. Residues conserved in all species are presented in black boxes. The positions of phosphorylated sites of CcpA from S. aureus are indicated by triangles. B, in vitro phosphorylation of CcpA from B. subtilis and B. anthracis orthologues. The recombinant PrkC kinase encoded by the B. subtilis genome was expressed and purified as a GST-tagged fusion in E. coli and incubated in the presence of [γ-33P]ATP with either the purified His-tagged B. subtilis CcpA (CcpABs) or B. anthracis CcpA (CcpABa). The vector allowing CcpABa overexpression and purification was kindly provided by Perego and co-workers (5). Samples were separated by SDS-PAGE, stained with Coomassie Blue (upper panel), and visualized by autoradiography (lower panel). Standard proteins of known molecular masses were run in parallel (M (kDa) lane). C, in vitro cross-phosphorylation. The recombinant PrkC kinase encoded as a GST-tagged fusion was incubated in the presence of [γ-33P]ATP with the purified His-tagged S. aureus CcpA. In parallel, the recombinant His-tagged fusion Stk1 kinase encoded by the S. aureus genome was incubated in the presence of [γ-33P]ATP with the purified His-tagged B. subtilis CcpA (CcpABs). Samples were separated by SDS-PAGE, stained with Coomassie Blue (upper panel), and visualized by autoradiography (lower panel). The upper bands illustrate the autokinase activity of the kinases, and the lower bands represent phosphorylated CcpAs. Standard proteins of known molecular masses were run in parallel (M (kDa) lane).
To determine whether the lack of CcpA phosphorylation observed with the Bacillus orthologues might be due to STPK specificity, we investigated the Ser/Thr cross-phosphorylation between the S. aureus kinase and the B. subtilis CcpA and vice versa. As presented in Fig. 6C, we found that the B. subtilis kinase PrkC was able to cross-phosphorylate CcpA from S. aureus, whereas the CcpA orthologue from B. subtilis remained unphosphorylated in the presence of Stk1. These findings suggest a critical role for the substrate phosphorylation site environment rather than for the kinase specificity at least under in vitro conditions. Interestingly, the in silico comparison of the DNA binding domains of the CcpA orthologues identified a strict conservation around Thr-18 (residues 14–26), whereas the conservation around Thr-33 was much lower (Fig. 6A). Of note, as mentioned above, the conservation of the N-terminal DNA binding domain of the CcpA protein is very high within orthologues (>80%), and the only divergent area is centered around Thr-33. All Bacillus CcpAs contained a proline at position 31, which might explain the absence of phosphorylation in this genus. This proline is likely to N-cap one of the α-helices of the CcpA DNA binding domain, thus stabilizing this structure. Therefore, unfolding of the region critical for Stk1 kinase recognition may be in part altered by the presence of this proline residue in Bacillus species. Moreover, it remains to be investigated whether the CcpA orthologues of other G+ organisms such as Clostridium perfringens, Lactococcus lactis, and S. pneumoniae, which do not possess a proline residue at position 31 (Fig. 6A), might be regulated by Ser/Thr phosphorylation.
DISCUSSION
Recent studies in various S. aureus strains suggest that Ser/Thr phosphorylation by Stk1 plays an important role in central metabolic processes such as cell wall metabolism, glycolysis, the citrate cycle, purine and pyrimidine synthesis, translation, quorum sensing, and virulence (17, 19, 21, 31, 32) presumably due to the regulation of selected global regulators. In line with this assumption, Stk1 was recently shown to affect the activities of regulatory factors such as MgrA, SarA, and LuxS (19–21). With the present study, we extend previous work demonstrating the regulatory role of Stk1-mediated phosphorylation in central metabolism and virulence pathways by providing first evidence of a Ser/Thr kinase phosphorylation-dependent regulation of the major transcriptional regulator CcpA. This mechanism is of special relevance as CcpA contributes to the pathogenicity of S. aureus (7, 8, 12, 14, 15). We demonstrated that phosphorylation of Thr-18 and Thr-33 residues abrogated CcpA DNA binding activity in vitro. Moreover, in the case of growing bacteria, a phosphomimetic mutant displayed a major alteration of the biofilm formation, probably due to the lack of CcpA activity, similar to what was observed for a ΔccpA mutant (12). Likewise, the CcpA phosphomimetic strain exhibited reduced transcription of citZ and increased transcription of hla similar to the ΔccpA mutant profile, confirming that phosphorylation prevents CcpA regulation toward its target genes in vivo. Previous studies identified Stk1 to affect the expression of a variety of proteins, including virulence determinants like hemolysins (32). Of notable interest is the S. aureus stk1 mutant for which transcription of the hla gene was increased (32), thus supporting the view that the increased α toxin expression in the Δstk1 mutant could be linked, at least in part, to the phosphorylation state of CcpA.
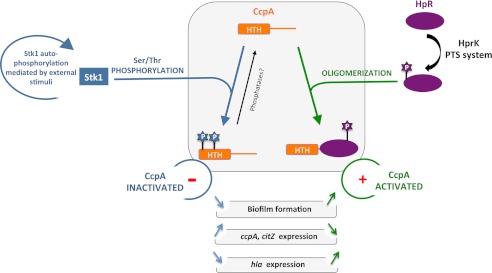
Our data presented here now strongly suggest that the activity of this S. aureus global regulator is not only modulated by metabolic signals that lead to the formation of the coregulator Hpr-Ser(P)-46 but also by Stk1 (Fig. 7). However, the environmental stimuli that activate Stk1 kinase activity have not yet been identified, making it difficult to predict under which conditions CcpA activity might be affected by Stk1 phosphorylation. However, our findings presented here and elsewhere that this regulatory circuit does not seem to be present in the closely related genus Bacillus (Fig. 7) and that ccpA transcription appears to be autoregulated in staphylococci (Fig. 5 and Ref. 42) but not in bacilli (43, 44) highlight that some fundamental differences in the regulation of CcpA activity between staphylococci and bacilli exist. Moreover, in combination with our recent observations that CcpA also seems to regulate the expression of some of its target genes in the absence of glucose (13) and that CcpA can readily bind to its cre sequences in absence of its coregulator Hpr-Ser(P)-46 (Fig. 4 and Ref. 15), it is tempting to speculate that the staphylococcal CcpAs might be fairly active even in the absence of Hpr-Ser(P)-46 and therefore need to be controlled by further transcriptional and post-translational mechanisms, whereas the Bacillus CcpA homologues might not bind efficiently to their cre sequences in the absence of Hpr-Ser(P)-46 and thus do not need to be controlled by these regulatory circuits.
FIGURE 7.
Proposed regulatory role of the different isoforms of CcpA as a molecular switch in carbon catabolite repression and virulence gene expression in S. aureus. The unphosphorylated form of CcpA binds to its target DNA sequences, and this binding is modulated via the HprKP/Hpr system, which is thought to promote the binding of CcpA to its DNA targets if CcpA is complexed with Hpr-Ser(P)-46. The Stk1-mediated phosphorylation of CcpA inactivates the DNA binding activity of this regulatory molecule, thereby preventing the transcriptional control of its target genes. PTS, PhosphoTransferase System; HTH, helix-turn-helix domain.
In summary, this study provides conceptual advances in our understanding of how pathogenic bacteria adapt to their physiology. This work also strengthens the biological importance of CcpA in the physiology and virulence of S. aureus as such Ser/Thr kinase-dependent fine-tuning mechanism is not ubiquitous.
Our results furthermore suggest that displacement of the CcpA/CcpA-P balance in favor of the phosphorylated isoform leads to biofilm inhibition and modification of gene expression. Although very challenging, future studies should now focus on the identification of the extracellular cues that are sensed by the Stk1 kinase and lead to CcpA phosphorylation. However, it remains unclear under which conditions and to what extent Stk1 phosphorylates CcpA in this pathogen, how this phosphorylation affects the expression of the regulon controlled by this LacI/GalR type of regulator, and whether this phosphorylation might be reversible by a serine/threonine phosphatase, such as Stp, that is co-transcribed with Stk1 (45). Moreover, the lack of this regulatory circuit in the closely related genus Bacillus together with the observed low degree of conservation in the amino acid sequence surrounding Thr-33 of the CcpA orthologues analyzed here suggests that this regulatory control mechanism of CcpA might only be present in some of the G+ organisms harboring CcpA and Stk1 orthologues. Investigations are currently ongoing to address these questions.
From an application standpoint, the selective inhibition of CcpA activity through constitutive phosphorylation may strongly impair S. aureus pathogenicity at least in terms of biofilm formation, thus opening new and original perspectives for future drug development. It is indeed noteworthy that small molecules such as bryostatin (46) can activate STPK, which may be of great therapeutic value in inhibiting S. aureus biofilm formation in vivo.
Acknowledgments
We thank I. Zanella-Cléon from the Mass Spectrometry Unit (Centre Commun de Microanalyse des Protéines), UMS3444/US8 Biosciences Gerland-Lyon Sud for excellent expertise and technical assistance, Marta Perego for providing plasmid pET_ccpABa, and Michel Débarbouillé for providing S. aureus Δstk1.
This work was supported by National Research Agency Grant ANR-09-MIEN-004 (to V. M.) and Deutsche Forschungsgemeinschaft Grants BI 1350/1-1 and 1-2 to (to M. B.).

This article contains supplemental Fig. S1.
- CcpA
- catabolite control protein A
- STPK
- Ser/Thr protein kinase
- cre
- catabolite-responsive element
- TSB
- tryptic soy broth
- CcpA-P
- phosphorylated CcpA
- HprKP
- Hpr Kinase Phosphatase
- Hpr-P
- phosphorylated Hpr
- Bs
- B. subtilis
- Ba
- B. anthracis.
REFERENCES
- 1. Fujita Y. (2009) Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73, 245–259 [DOI] [PubMed] [Google Scholar]
- 2. Sonenshein A. L. (2007) Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5, 917–927 [DOI] [PubMed] [Google Scholar]
- 3. Seidel G., Diel M., Fuchsbauer N., Hillen W. (2005) Quantitative interdependence of coeffectors, CcpA and cre in carbon catabolite regulation of Bacillus subtilis. FEBS J. 272, 2566–2577 [DOI] [PubMed] [Google Scholar]
- 4. Singh K. D., Schmalisch M. H., Stülke J., Görke B. (2008) Carbon catabolite repression in Bacillus subtilis: quantitative analysis of repression exerted by different carbon sources. J. Bacteriol. 190, 7275–7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chiang C., Bongiorni C., Perego M. (2011) Glucose-dependent activation of Bacillus anthracis toxin gene expression and virulence requires the carbon catabolite protein CcpA. J. Bacteriol. 193, 52–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Antunes A., Martin-Verstraete I., Dupuy B. (2011) CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 79, 882–899 [DOI] [PubMed] [Google Scholar]
- 7. Seidl K., Bischoff M., Berger-Bächi B. (2008) CcpA mediates the catabolite repression of tst in Staphylococcus aureus. Infect. Immun. 76, 5093–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sadykov M. R., Hartmann T., Mattes T. A., Hiatt M., Jann N. J., Zhu Y., Ledala N., Landmann R., Herrmann M., Rohde H., Bischoff M., Somerville G. A. (2011) CcpA coordinates central metabolism and biofilm formation in Staphylococcus epidermidis. Microbiology 157, 3458–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iyer R., Baliga N. S., Camilli A. (2005) Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187, 8340–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shelburne S. A., 3rd, Keith D., Horstmann N., Sumby P., Davenport M. T., Graviss E. A., Brennan R. G., Musser J. M. (2008) A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. U.S.A. 105, 1698–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lowy F. D. (1998) Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 12. Seidl K., Goerke C., Wolz C., Mack D., Berger-Bächi B., Bischoff M. (2008) Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 76, 2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seidl K., Müller S., François P., Kriebitzsch C., Schrenzel J., Engelmann S., Bischoff M., Berger-Bächi B. (2009) Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 9, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seidl K., Stucki M., Ruegg M., Goerke C., Wolz C., Harris L., Berger-Bächi B., Bischoff M. (2006) Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50, 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li C., Sun F., Cho H., Yelavarthi V., Sohn C., He C., Schneewind O., Bae T. (2010) CcpA mediates proline auxotrophy and is required for Staphylococcus aureus pathogenesis. J. Bacteriol. 192, 3883–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Débarbouillé M., Dramsi S., Dussurget O., Nahori M. A., Vaganay E., Jouvion G., Cozzone A., Msadek T., Duclos B. (2009) Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J. Bacteriol. 191, 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohlsen K., Donat S. (2010) The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int. J. Med. Microbiol. 300, 137–141 [DOI] [PubMed] [Google Scholar]
- 18. Donat S., Streker K., Schirmeister T., Rakette S., Stehle T., Liebeke M., Lalk M., Ohlsen K. (2009) Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus. J. Bacteriol. 191, 4056–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Didier J. P., Cozzone A. J., Duclos B. (2010) Phosphorylation of the virulence regulator SarA modulates its ability to bind DNA in Staphylococcus aureus. FEMS Microbiol. Lett. 306, 30–36 [DOI] [PubMed] [Google Scholar]
- 20. Sun F., Ding Y., Ji Q., Liang Z., Deng X., Wong C. C., Yi C., Zhang L., Xie S., Alvarez S., Hicks L. M., Luo C., Jiang H., Lan L., He C. (2012) Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 109, 15461–15466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cluzel M. E., Zanella-Cléon I., Cozzone A. J., Fütterer K., Duclos B., Molle V. (2010) The Staphylococcus aureus autoinducer-2 synthase LuxS is regulated by Ser/Thr phosphorylation. J. Bacteriol. 192, 6295–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Canova M. J., Kremer L., Molle V. (2008) pETPhos: a customized expression vector designed for further characterization of Ser/Thr/Tyr protein kinases and their substrates. Plasmid 60, 149–153 [DOI] [PubMed] [Google Scholar]
- 23. Molle V., Leiba J., Zanella-Cléon I., Becchi M., Kremer L. (2010) An improved method to unravel phosphoacceptors in Ser/Thr protein kinase-phosphorylated substrates. Proteomics 10, 3910–3915 [DOI] [PubMed] [Google Scholar]
- 24. Slama N., Leiba J., Eynard N., Daffé M., Kremer L., Quémard A., Molle V. (2011) Negative regulation by Ser/Thr phosphorylation of HadAB and HadBC dehydratases from Mycobacterium tuberculosis type II fatty acid synthase system. Biochem. Biophys. Res. Commun. 412, 401–406 [DOI] [PubMed] [Google Scholar]
- 25. Archambaud C., Gouin E., Pizarro-Cerda J., Cossart P., Dussurget O. (2005) Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol. Microbiol. 56, 383–396 [DOI] [PubMed] [Google Scholar]
- 26. Kreiswirth B. N., Löfdahl S., Betley M. J., O'Reilly M., Schlievert P. M., Bergdoll M. S., Novick R. P. (1983) The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712 [DOI] [PubMed] [Google Scholar]
- 27. Charpentier E., Anton A. I., Barry P., Alfonso B., Fang Y., Novick R. P. (2004) Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl. Environ. Microbiol. 70, 6076–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schulthess B., Bloes D. A., François P., Girard M., Schrenzel J., Bischoff M., Berger-Bächi B. (2011) The σB-dependent yabJ-spoVG operon is involved in the regulation of extracellular nuclease, lipase, and protease expression in Staphylococcus aureus. J. Bacteriol. 193, 4954–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chatterjee I., Becker P., Grundmeier M., Bischoff M., Somerville G. A., Peters G., Sinha B., Harraghy N., Proctor R. A., Herrmann M. (2005) Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J. Bacteriol. 187, 4488–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Molle V., Kremer L., Girard-Blanc C., Besra G. S., Cozzone A. J., Prost J. F. (2003) An FHA phosphoprotein recognition domain mediates protein EmbR phosphorylation by PknH, a Ser/Thr protein kinase from Mycobacterium tuberculosis. Biochemistry 42, 15300–15309 [DOI] [PubMed] [Google Scholar]
- 31. Tamber S., Schwartzman J., Cheung A. L. (2010) The role of PknB kinase in antibiotic resistance and virulence in community acquired methicillin resistant Staphylococcus aureus strain USA300. Infect. Immun. 78, 3637–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burnside K., Lembo A., de Los Reyes M., Iliuk A., Binhtran N. T., Connelly J. E., Lin W. J., Schmidt B. Z., Richardson A. R., Fang F. C., Tao W. A., Rajagopal L. (2010) Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS One 5, e11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Truong-Bolduc Q. C., Hooper D. C. (2010) Phosphorylation of MgrA and its effect on expression of the NorA and NorB efflux pumps of Staphylococcus aureus. J. Bacteriol. 192, 2525–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Veyron-Churlet R., Molle V., Taylor R. C., Brown A. K., Besra G. S., Zanella-Cléon I., Fütterer K., Kremer L. (2009) The Mycobacterium tuberculosis β-ketoacyl-acyl carrier protein synthase III activity is inhibited by phosphorylation on a single threonine residue. J. Biol. Chem. 284, 6414–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barthe P., Roumestand C., Canova M. J., Kremer L., Hurard C., Molle V., Cohen-Gonsaud M. (2009) Dynamic and structural characterization of a bacterial FHA protein reveals a new autoinhibition mechanism. Structure 17, 568–578 [DOI] [PubMed] [Google Scholar]
- 36. Veyron-Churlet R., Zanella-Cléon I., Cohen-Gonsaud M., Molle V., Kremer L. (2010) Phosphorylation of the Mycobacterium tuberculosis β-ketoacyl-acyl carrier protein reductase MabA regulates mycolic acid biosynthesis. J. Biol. Chem. 285, 12714–12725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Canova M. J., Kremer L., Molle V. (2009) The Mycobacterium tuberculosis GroEL1 chaperone is a substrate of Ser/Thr protein kinases. J. Bacteriol. 191, 2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Molle V., Gulten G., Vilchèze C., Veyron-Churlet R., Zanella-Cléon I., Sacchettini J. C., Jacobs W. R., Jr., Kremer L. (2010) Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis. Mol. Microbiol. 78, 1591–1605 [DOI] [PubMed] [Google Scholar]
- 39. Schumacher M. A., Allen G. S., Diel M., Seidel G., Hillen W., Brennan R. G. (2004) Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell 118, 731–741 [DOI] [PubMed] [Google Scholar]
- 40. Kang C. M., Nyayapathy S., Lee J. Y., Suh J. W., Husson R. N. (2008) Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology 154, 725–735 [DOI] [PubMed] [Google Scholar]
- 41. Corrales R. M., Molle V., Leiba J., Mourey L., de Chastellier C., Kremer L. (2012) Phosphorylation of mycobacterial PcaA inhibits mycolic acid cyclopropanation: consequences for intracellular survival and for phagosome maturation block. J. Biol. Chem. 287, 26187–26199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Egeter O., Brückner R. (1996) Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol. Microbiol. 21, 739–749 [DOI] [PubMed] [Google Scholar]
- 43. Miwa Y., Saikawa M., Fujita Y. (1994) Possible function and some properties of the CcpA protein of Bacillus subtilis. Microbiology 140, 2567–2575 [DOI] [PubMed] [Google Scholar]
- 44. Hueck C. J., Kraus A., Schmiedel D., Hillen W. (1995) Cloning, expression and functional analyses of the catabolite control protein CcpA from Bacillus megaterium. Mol. Microbiol. 16, 855–864 [DOI] [PubMed] [Google Scholar]
- 45. Beltramini A. M., Mukhopadhyay C. D., Pancholi V. (2009) Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect. Immun. 77, 1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shah I. M., Laaberki M. H., Popham D. L., Dworkin J. (2008) A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iordanescu S., Surdeanu M. (1976) Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96, 277–281 [DOI] [PubMed] [Google Scholar]