Background: AKAPs tethering the type I regulatory subunit of cAMP-dependent kinase (PKA-RI) have only been marginally described.
Results: Here a novel small AKAP (smAKAP) is identified and characterized as a PKA-RI-specific AKAP.
Conclusion: smAKAP is a novel AKAP that localizes PKA-RI specifically to the plasma membrane.
Significance: PKA-RI is specifically localized through a novel AKAP.
Keywords: AKAP, Cyclic AMP (cAMP), Plasma Membrane, Protein Kinase A (PKA), Proteomics, Chemical Proteomics
Abstract
Protein kinase A-anchoring proteins (AKAPs) provide spatio-temporal specificity for the omnipotent cAMP-dependent protein kinase (PKA) via high affinity interactions with PKA regulatory subunits (PKA-RI, RII). Many PKA-RII-AKAP complexes are heavily tethered to cellular substructures, whereas PKA-RI-AKAP complexes have remained largely undiscovered. Here, using a cAMP affinity-based chemical proteomics strategy in human heart and platelets, we uncovered a novel, ubiquitously expressed AKAP, termed small membrane (sm)AKAP due to its specific localization at the plasma membrane via potential myristoylation/palmitoylation anchors. In vitro binding studies revealed specificity of smAKAP for PKA-RI (Kd = 7 nm) over PKA-RII (Kd = 53 nm) subunits, co-expression of smAKAP with the four PKA R subunits revealed an even more exclusive specificity of smAKAP for PKA-RIα/β in the cellular context. Applying the singlet oxygen-generating electron microscopy probe miniSOG indicated that smAKAP is tethered to the plasma membrane and is particularly dense at cell-cell junctions and within filopodia. Our preliminary functional characterization of smAKAP provides evidence that, like PKA-RII, PKA-RI can be tightly tethered by a novel repertoire of AKAPs, providing a new perspective on spatio-temporal control of cAMP signaling.
Introduction
In the cellular context signaling regulated by the small second messenger, cAMP, involves a plethora of signaling proteins. A key player is cAMP-dependent protein kinase (PKA). In the resting state the catalytic subunits (PKA-C)3 are tightly bound and inhibited by regulatory subunits (PKA-R). This inactive holoenzyme is organized as a heterotetramer ((PKA-R)2(PKA-C)2) and becomes activated by cAMP binding (1). The dimerization and docking (D/D) domain of the PKA-R isoforms allows binding to the so-called protein kinase A-anchoring proteins (AKAPs), the key regulator scaffolds of cAMP intracellular specificity (2–4). The hallmark signature motif of the AKAPs is a small sequence of 17–20 amino acids that forms an amphipathic helix that associates tightly with the PKA-R D/D domain (5). Besides targeting PKA activity to a specific location within the cell, most AKAPs function as scaffolds for a variety of other signaling proteins such as phosphatases, phosphodiesterases, and other kinases. These macromolecular protein complexes create efficient signaling hubs that allow for a localized cAMP regulatory mechanism (4). For example, AKAP18δ localizes to the heart sarcoplasmic reticulum and creates a signaling complex comprising AKAP18δ, phospholamban, PKA, and sarcoplasmic reticulum Ca2+-ATPase to regulate intracellular Ca2+ levels upon β-adrenergic stimulation (6).
In 1984 Lohmann et al. (7) reported on the first AKAP, namely the microtubule-associated protein 2 (MAP2), when purifying PKA-RIIα. Since then >70 different AKAPs originating from approximately 30 different genes have been identified (3, 4, 8). Initially, PKA-R overlay assays (7) and yeast two-hybrid screens were used for the discovery of new AKAPs. For instance, Rab32, a member of the Ras superfamily of small G proteins, was shown by yeast two-hybrid screens to interact with PKA-RII and co-localizes to mitochondria (8). More recently, mass spectrometry-based chemical proteomics experiments with immobilized cAMP have proven their value in capturing, identifying, and characterizing novel AKAPs (3, 9). This specific PKA-AKAP enrichment approach led, for instance, to the discovery of novel AKAPs such as sphingosine kinase type 1-interaction protein (SPHKAP) and the PALM2-AKAP2 fusion protein (10, 11).
In mammalian systems there are four genetically distinct and functionally nonredundant isoforms of PKA regulatory subunits: RIα, RIβ, RIIα, and RIIβ, which all contain a D/D domain capable of AKAP binding. Subtle differences, especially between the RI and RII isoforms, seem to induce selectivity in binding to a subset of AKAPs. Most reported AKAPs have a preferred specificity for PKA-RII, whereas three AKAPs: d-AKAP1 (12), d-AKAP2 (13), and the recently discovered Opa1 (14), are dual-specific AKAPs. We identified SPHKAP as the first PKA-RI-specific AKAP (10, 15). Detailed structural analysis of the RI and RII D/D domain in complex with the earlier mentioned amphipathic helix protein kinase A binding (AKB) domain of the dual-specific d-AKAP2 revealed that interaction with PKA-RI requires a larger interaction surface which leads to a larger set of unique constraints to bind PKA-RI (5, 16). These restrictions also seem to drive the specificity of the very small subset of the AKAPs known to interact tightly with PKA-RI. Array-based synthetic peptide screening led to detailed knowledge on the importance of particular amino acid combinations at key locations in the AKB domain that contribute to RI over RII specificity (17, 18).
Here we report on the identification and preliminary functional characterization of a novel, small (i.e. 11 kDa) PKA-RI-specific protein kinase A-anchoring protein, which we refer to as the small-membrane AKAP (smAKAP). smAKAP is tethered to the plasma membrane most likely through a dual acylation of its N-terminal Met-Gly-Cys- motif (myristoylation and palmitoylation, respectively). Through biochemical experiments in vitro and in vivo, we show that smAKAP has the capacity to target specifically PKA-RI isoforms to the plasma membrane; similar to both AKAP250 (gravin) and AKAP18α, which perform this task for PKA-RIIα (19, 20). Using a novel singlet oxygen-generating electron microscopy probe (miniSOG) (25) we confirm smAKAP localization to plasma membranes, its enrichment at cell-cell junctions, and its association with filopodia.
EXPERIMENTAL PROCEDURES
Human left ventricular heart tissue and human platelets were collected from individuals without any diagnosed cardiovascular disease. All samples were obtained with the appropriate consent according to the Helsinki convention. The lysate preparation and cAMP pulldowns were performed in earlier work in human heart (21) and platelets (22).
RT-PCR
Total RNAs of spleen, liver, kidney, brain, lung, uterus, stomach, intestine, ovaries, skeletal muscle, heart, and colon were isolated from a female mouse using TRIzol reagent (Invitrogen). Subsequently the RNAs were treated by DNase I followed by addition of oligo(dT)12-VN (Promega) and Superscript II (Invitrogen). Finally, the PCR was completed using Taq polymerase (Invitrogen) and the appropriate primers (Eurogentec) for mouse smAKAP and mouse GAPDH.
Creating Plasmids
The human smAKAP gene was isolated from mouse heart cDNA, and by applying the enzyme free cloning method, the amplified cDNA was cloned into pLICHIS vector (23). Human smAKAP was subcloned from pLICHIS into the BamHI-HindIII site of a mCherry vector (24), into the BamHI-HindIII site of the miniSOG vector (25), and into the EcoRI-BlpI site of the mGFP vector (26). The single site mutations (G2A, C3A) and double site mutation (G2A/C3A) of the human smAKAP sequence in the mGFP vector were made by QuikChange mutagenesis (Agilent Technologies). PKA-RIα tagged with mCherry and RI-specific d-AKAP2 were described previously (27, 28). The mKO2-tagged PKA regulatory (R) subunits were generated by fusing respective R subunits with mKO2 to the C terminus with PCR and inserted between EcoRI and NotI sites in pcDNA3 (Invitrogen). There is a SalI site as linker in RIα, RIβ, and RIIβ constructs and a BamHI site as linker in the RIIα construct.
Binding Affinity
Full-length PKA-RIα/β and PKA-RIIα/β dimers were purified as described previously (29, 30). The 5-TAMRA N terminus tagged TVILEYAHRLSQDILCDALQQWAC peptide was synthesized at the peptide facility of The Netherlands Cancer Institute (Amsterdam, Netherlands). Flat bottom black 96-well plates (Thermo) were used for the fluorescence polarization readings of each regulatory subunit dimer with the 5-TAMRA-tagged peptide, which was excited at 535 nm (5–10-nm bandpass) and emission monitored at 580 nm (5–10-nm bandpass), by the Tecan Genios Pro 96/384 Multifunction Microplate Reader (Tecan). The binding experiments were each performed four times after which they were put into the nonlinear regression model of one-site saturated binding in GraphPad Prism 5.0.
Cell Culture
HeLa and HEK293 cells were acquired from the American Type Culture Collection. Before and after the transfections, the cells were sustained in Dulbecco's modified Eagle's medium (DMEM, BioWhittaker) containing 10% heat-inactivated fetal bovine serum (Omega Scientific Inc., San Diego, CA) and 2 mm Glutamax (Invitrogen) in 35-mm glass-bottom dishes (MatTek). The dishes for HEK293 cells were coated with poly-d-lysine. Cells were grown to 75% confluence and transfected using PolyFect according to the manufacturer's protocol for the specific cell type (Qiagen).
Fluorescence Imaging
Cells were fixed by ice-cold 4% formaldehyde in PBS and subsequently washed twice with PBS. The confocal images were acquired with a Leica TCS SPE II confocal system with a 63× water objective lens.
Electron Microscopy
smAKAP-miniSOG was co-transfected with either RIα-mCherry or RIα-mKO2 to help identify transfected cells. 14 h after transfection, cells were fixed in 2.0% glutaraldehyde (Electron Microscopy Sciences) in 0.1 m sodium cacodylate buffer (pH 7.4) for 5 min at room temperature and then moved to ice for a duration of 45 min for optimal cell ultrastructural preservation. The cells were washed five times in ice-cold 0.1 m sodium cacodylate buffer (pH 7.4), each taking 2 min, to remove excess aldehydes. Cells were treated for 15 min in blocking buffer (50 mm glycine, 10 mm KCN, and 10 mm aminotriazole) to reduce nonspecific background reaction of diaminobenzidine (DAB). Images of cells were acquired using a Leica TCS SPE II confocal system using both 488-nm and 568-nm laser excitation and a 63× water objective immersion lens. Cells expressing a high level of smAKAP-miniSOG were photooxidized. Afterward, cells were washed with 0.1 m sodium cacodylate buffer, postfixed with 1% osmium tetroxide (Electron Microscopy Sciences) in 0.1 m sodium cacodylate buffer, dehydrated in an ethanol series, embedded in Durcupan AMC Fluka epoxy resin (Sigma), sectioned at 70–80 nm by a Leica ultracut UCT ultramicrotome, poststained in 2% aqueous uranyl acetate (Electron Microscopy Sciences) for 10 min, and imaged with a JEOL JEM1200EX transmission electron microscope at either 60 or 80 kV according to Ref. 25. The stereo images were acquired at 10° intervals.
Statistics and Quantification
The two fluorescent profiles were compared by ImageJ 1.43 by plotting a line through the visible cell and then creating a plot profile. The profiles were compared, and an R2 correlation coefficient was computed using MS Excel.
RESULTS
Discovery of smAKAP by Chemical Proteomics and Bioinformatics
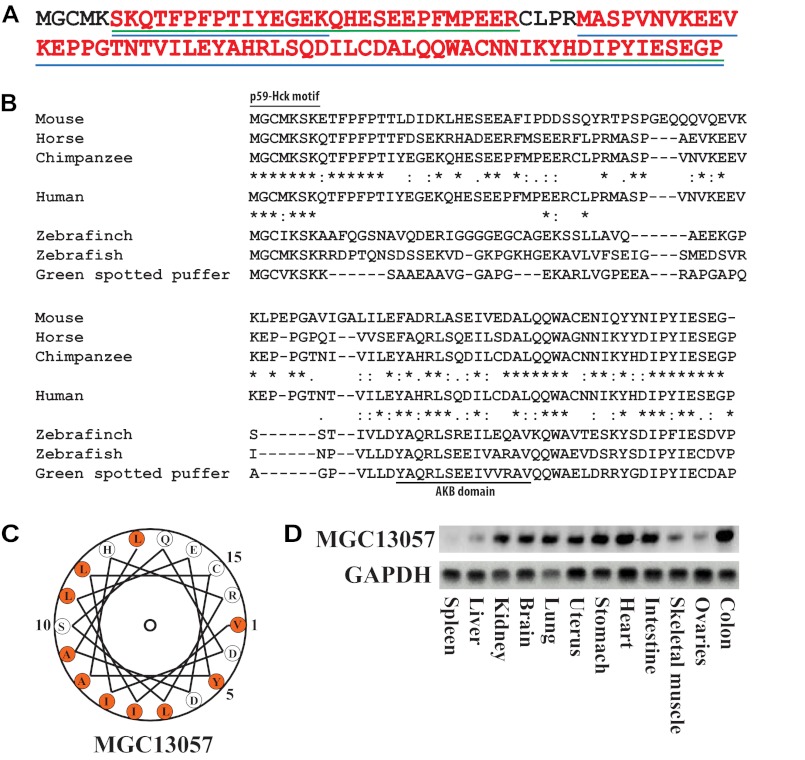
Following a chemical proteomics-based strategy described earlier (9, 31) an unknown protein, with the connotation MGC13057, belonging to the UPF0733 family (Q9BSF0, C2orf88), was observed as selective binder in cAMP-based affinity pulldown experiments (21, 22), in both human heart tissue and human platelets. LC-MS/MS analyses allowed the mapping of 92% of the sequence of MGC13057 affinity-captured from both these tissues, except the ultimate N-terminal residues (Fig. 1A). Applying and aligning our previously established Hidden Markov model (9) with the sequence of MGC13057 suggested that the YAHRLSQDILCDAL (amino acids 61–74) sequence could be a potential PKA anchoring motif making MGC13057 a putative novel AKAP.
FIGURE 1.
Discovery of MGC13057 as a putative AKAP. A, a chemical proteomics analysis by means of cAMP-bound resin performed upon human heart (left ventricle) and platelets leading to the detection of the hypothetical protein MGC13057. Sequence coverage (red text) of this small protein was nearly complete with peptides identified in heart (blue lines) and platelets (green lines). B, sequence alignment of human MGC13057 with various orthologues in other species (from top to bottom: mouse (Q9CPS8), horse (XP_001502030.1), chimpanzee (XP_003309413.1), human (Q9BSF0), zebrafinch (XM_002191648.1), zebrafish (P0C8S0), and green spotted puffer (Q4RTJ5)). Identity (*) and similarity (:) in several regions, including the AKB domain and p59Hck motif are annotated. C, helical wheel alignment of the putative AKB domain revealing an amphipathic helix with a hydrophobic surface on one side (orange). D, mRNA expression of the MGC13057 gene in several mouse tissues by RT-PCR.
BLAST analysis and alignment of the retrieved homologues of MGC13057 revealed a good conservation across mammals, but also to other vertebrate species such as bird (Taeniopygia guttata) and fish (Danio rerio). Although in the latter species the degree of identical amino acids diminished, some regions, including the putative AKB motif and the N terminus, still showed strong similarity/identity (Fig. 1B). A helical wheel alignment revealed the presence of an amphipathic helix, a hallmark of AKAPs, with one side of the helix consisting primarily of hydrophobic residues and the other side of charged/hydrophilic residues, further suggesting that this uncharacterized protein may be a novel AKAP (Fig. 1C).
MGC13057 expression across various mouse tissues was tested by means of mRNA RT-PCR. The mRNA expression level appeared to be ubiquitous although it was much less in spleen and liver (Fig. 1D). As expected, mRNA levels were relatively high in murine heart, because we originally detected MGC13057 in human heart tissue.
MGC13057 Displays PKA-RI Selectivity
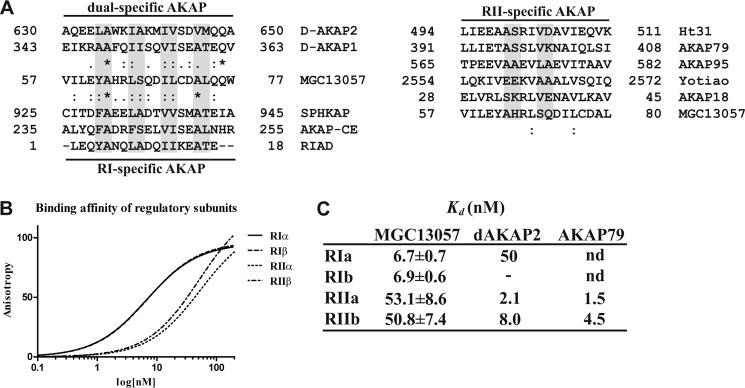
We first further investigated the sequence of the well conserved proposed AKB domain of MGC13057 possibly to define its PKA-R subunit specificity. The AKB domain of MGC13057 was therefore aligned with AKB domains of PKA-RII-specific AKAPs, dual specific AKAPs, and RI-specific AKAPs (Fig. 2A). Similarity between the MGC13057 AKB domain and those of PKA-RII was poor (Fig. 2A); however, the alignments with PKA-RI-binding AKAPs proved very strong. For instance, in MGC13057 there are four helical turns with hydrophobic domains. In the first domain the PKA-RI-specific alanine is conserved, as well as the bulky tyrosine residue (phenylalanine in SPHKAP (10)) that fits well in the deeper grooved D/D domains of the PKA-RI isoforms, but less in the PKA-RII isoforms (16). Similarly, in MGC13057 there are two large polar residues present after the final hydrophobic domain which merely arrest PKA-RII binding and are also found in d-AKAP1, d-AKAP2, AKAP-CE, and partially in SPHKAP and RIAD (17). This suggests that MGC13057 is, like SPHKAP, potentially a PKA-RI-specific AKAP. To verify the isoform specificity, in vitro binding assays were performed with fluorescence anisotropy. Binding between all four full-length PKA-R subunits and a 24-amino acid long synthetic peptide TVILEYAHRLSQDILCDALQQWAC that mimics the AKB domain of MGC13057 (AA56–79) was probed. For read-out of binding, the peptide was labeled with a 5-TAMRA tag. This in vitro assay clearly indicated an almost 10-fold preference in binding affinity between the PKA-RI isoforms (Kd ∼7 nm) and the PKA-RII proteins (Kd ∼ 53 nm) (Fig. 2B), verifying the in vitro MGC13057 preference for PKA-RI. These are the highest affinities reported so far for PKA-RI subunits binding to AKAPs (Fig. 2C).
FIGURE 2.
MGC13057 is a PKA-RI-specific AKAP. A, alignment of the MGC13057 AKB domain (57–77) with the AKB domains of dual (left, top) and PKA-RI (left, bottom)-specific AKAPs. The same was done with the AKB domains of several established PKA-RII-specific AKAPs (right). Identity (*) and similarity (:) are annotated. B, fluorescence anisotropy measurements to determine the binding affinity of the MGC13057 AKB domain peptide, tagged with 5-TAMRA (excitation at 535 nm and emission at 580 nm) with the full-length regulatory subunit dimers: PKA-RIα (solid line), PKA-RIβ (dash), PKA-RIIα (alternating dash), and PKA-RIIβ (short dash). C, Kd values and S.D. (n = 4).
Localization and Specificity in Vivo
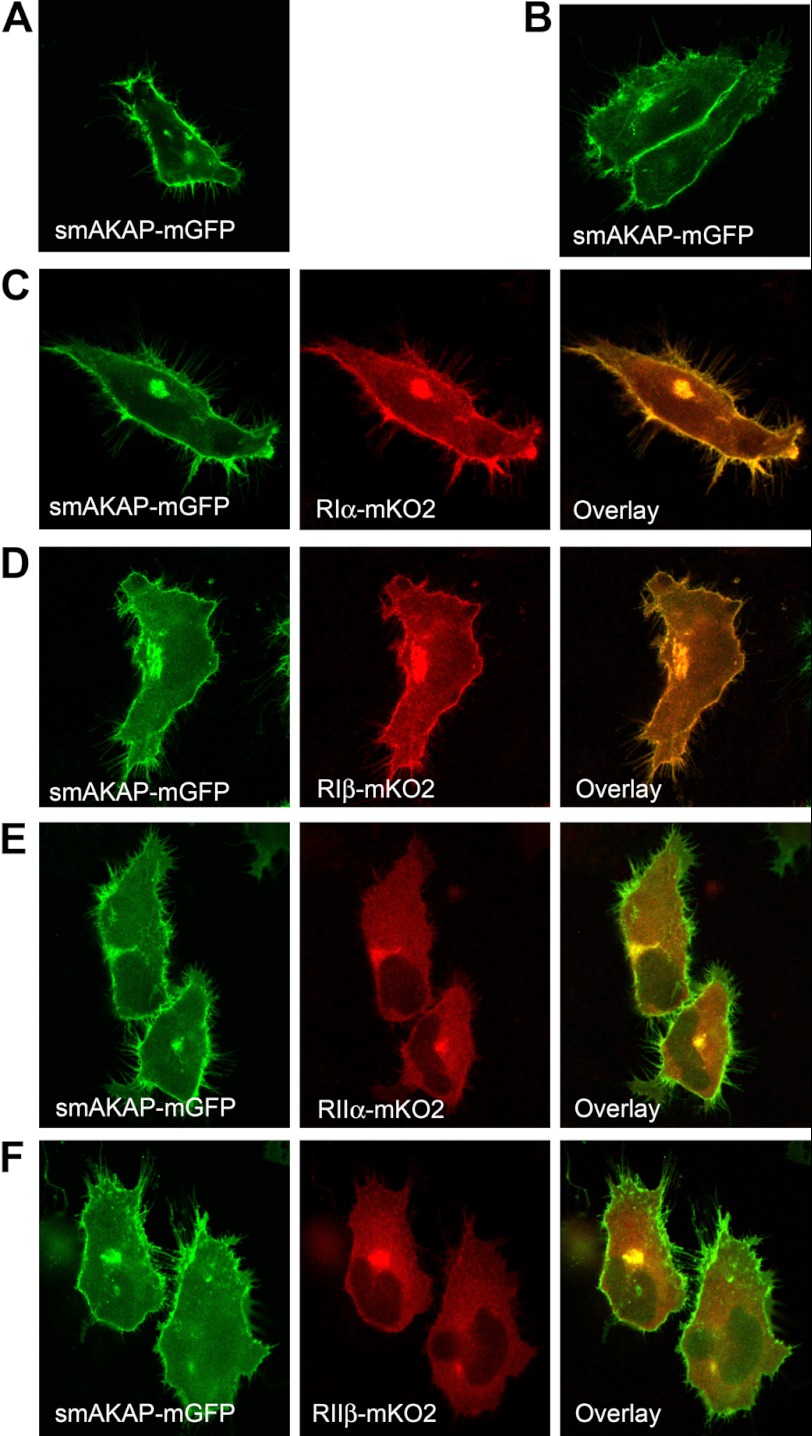
As the previous experiments indicated that MGC13057 is a putative PKA-RI-specific AKAP, we set out to confirm this in a cellular context. The human MGC13057 gene was subcloned and fused to the green fluorescent protein (GFP). Both HEK293 and HeLa cells were transiently transfected with this construct. Fluorescence microscopy showed strong localization of MGC13057 at the plasma membrane and its particular association with filopodia (Fig. 3A, supplemental Videos A and B). Based on its size and membrane localization we subsequently refer to it as smAKAP. Using confocal imaging and scanning through the cell we confirmed the localization to the plasma membrane and its strong presence at cell-cell junctions when smAKAP was expressed in HeLa cells (Fig. 3B and supplemental Video C).
FIGURE 3.
smAKAP assertively relocates PKA-RI to the plasma membrane. A, transfection of HeLa with smAKAP-mGFP shows smAKAP present at the plasma membrane and filopodia. B, a stronger presence of smAKAP at cell-cell junction is displayed by smAKAP-mGFP. C, PKA-RIα-mKO2 and smAKAP-mGFP strongly co-localize. D, smAKAP-mGFP and PKA-RIβ-mKO2 also co-localize. E, in contrast, PKA-RIIα-mKO2 and smAKAP-mGFP display no co-localization. F, likewise, smAKAP-mGFP and PKA-RIIβ-mKO2 do not co-localize. The protein expression levels are presented in supplemental Fig. 2.
Next, fluorescent imaging co-localization studies of smAKAP with the four PKA-R isoforms were performed to further investigate smAKAP specificity and PKA-R localization in the cellular context. For these studies we engineered, in addition to the smAKAP-GFP construct, the four R subunit-mKO2 constructs. When we carried out a dual transfection of smAKAP-GFP and RIα-mKO2 in HeLa cells, smAKAP and PKA-RIα strongly co-localize at the plasma membrane, specifically to filopodia, suggesting that they interact tightly in the cellular context (Fig. 3C). Strong co-localization at the plasma membrane was also observed when PKA-RIβ-mKO2 was co-transfected with smAKAP-GFP (Fig. 3D). In contrast, when we co-expressed smAKAP with either PKA-RIIα-mKO2 or PKA-RIIβ-mKO2, we observed no co-localization with smAKAP-GFP at the plasma membrane (Fig. 3, E and F). Whereas smAKAP-GFP was enriched to the plasma membrane, PKA-RIIα and PKA-RIIβ were localized to the cytoplasm.
To further establish the strong binding affinity of smAKAP to PKA-RI in a cellular context a competition assay was performed by means of a triple transfection in HEK293 cells. smAKAP-GFP and PKA-RIα-mCherry containing HEK293 cells were co-transfected with a d-AKAP2 AKB domain peptide that was engineered toward RI specificity by dedicated mutations in its AKB domain (Q631F, V643W, and M647F, Kd = 5.2 ± 0.5 nm) (27). This peptide poses as a strong competitive antagonist of PKA-RI smAKAP binding. Despite the presence of the competing PKA-RI tethering d-AKAP2, which localizes to the cytosol, PKA-RIα is not released from smAKAP at its location at the plasma membrane (supplemental Fig. 1). These data clearly indicate that smAKAP specifically binds to PKA-RI in cells and demonstrate that these isoforms are not always cytosolic as initially hypothesized (32, 33). This is further supported by the large differences in r2 between the fluorescence intensity profiles of each co-localization (supplemental Fig. 2).
Posttranslational Modifications in the N Terminus of smAKAP Mediate Membrane Interaction
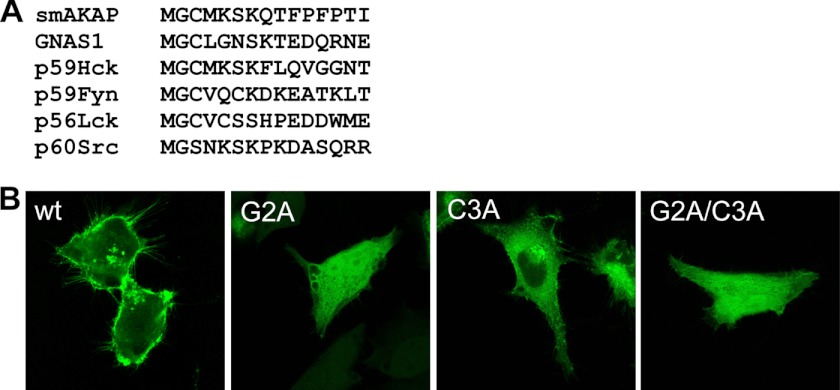
Having established that smAKAP localizes to the plasma membrane we next explored the requirements for this localization. In addition to the AKB domain, another well conserved region of smAKAP (Fig. 1B) is at the N terminus, which contains both a Gly (Gly-2) and a Cys (Cys-3) residue. This “two-signal mode” for protein acylation and membrane anchoring is highlighted by proteins belonging to specific members of the Src family of kinases, most of which are modified at their N termini with both myristate and palmitate (34). Interestingly, the 7-amino acid N-terminal stretch of smAKAP is identical to that of the oncogenic tyrosine kinase p59-Hck, which is known to be localized at the cell membrane (and at caveolae), and membrane localization of Hck requires both palmitoylation at Cys-3 and myristoylation at Gly-2 (Fig. 4A) (35). The guanine nucleotide-binding protein Gs subunit α isoform, which activates adenylyl cyclase (a transmembrane protein) and is thus part of the PKA signaling pathway, can also be both myristoylated and palmitoylated, in turn allowing it to be near smAKAP in regulating PKA activity (36). Myristoylation usually occurs co-translationally, whereas palmitoylation is a posttranslational modification (37, 38). The N terminus of smAKAP thus provides a well known motif that has the ability to undergo palmitoylation and myristoylation, likely mediating a lipid-induced localization of smAKAP to the plasma membrane (36, 39). Unfortunately, we were unable to detect peptides of the N terminus of smAKAP in our proteomics experiments, but with hindsight this may be partly caused by its extensive lipidation. To establish the importance of these acylation sites for membrane localization, we mutated smAKAP at Gly-2 and Cys-3 with two single alanine mutations (G2A, C3A) and a double mutation (G2A/C3A) in the full-length human smAKAP-GFP construct. Fig. 4B clearly shows that plasma membrane localization is abolished when either site is mutated, and thus as well with the double mutation, indicating that both of these residues are essential. This is a strong indirect indication of acylation through both palmitoylation and myristoylation and that they are both essential and likely driving smAKAP localization to the plasma membrane, as described for p59-Hck (35).
FIGURE 4.
Modification by lipids of the N terminus of smAKAP is crucial for localization. A, sequence alignment of the first seven N-terminal amino acids of smAKAP with various proteins, known to carry myristoylated and/or palmitoylated glycines/cysteines. B, transfection of mutated forms of smAKAP-GFP, G2A (left), blocking myristoylation, results in loss of plasma membrane localization. To a lesser degree, the single mutation, C3A (middle), which blocks palmitoylation of smAKAP-mGFP, similarly hampers membrane localization (middle). The double mutation, G2A/C3A, fully negates binding of smAKAP-mGFP to the membrane (right).
Electron Microscopy Imaging Further Defines the Localization of smAKAP
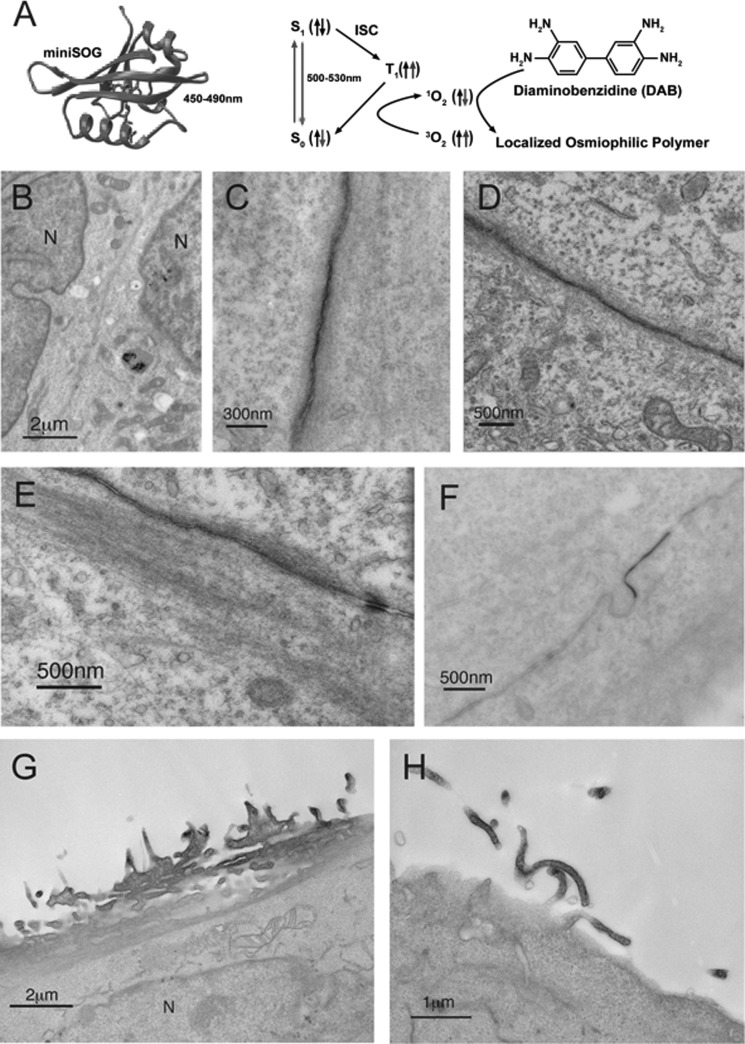
To more precisely map the localization of smAKAP we used electron microscopy and the recently introduced miniSOG (25). MiniSOG is a genetically modified flavoprotein, which produces singlet oxygen upon excitation with green light (Fig. 5A). DAB will then be oxidized to form an insoluble osmium-philic polymer which gives contrast to the ultrastructure near the tagged fusion protein. The smAKAP cDNA was fused to miniSOG and transiently transfected into HEK293 or HeLa cells for photooxidation. RIα-mCherry was co-transfected to help deduce the location of smAKAP-miniSOG via red fluorescence. Via this enhanced imaging method a much higher resolution was obtained allowing us to observe that smAKAP was localized at the plasma membrane (Fig. 5, C–E). In these images one can also very clearly see how the smAKAP is enhanced at cell-cell contact sites (Fig. 5, C–F) and furthermore the localization of smAKAP to filopodia (Fig. 5, G and H). In addition to its localization at the plasma membrane, we could observe some localization to internalized vesicles as expected by both the suspected myristoylation and palmitoylation (35). It is shown how a protrusion of the membrane, containing smAKAP, is formed on the plasma membrane junction between two cells, utilizing both plasma membranes. Potentially, this is then released into the adjacent cell, possibly indicating cell-to-cell signaling (Fig. 5F).
FIGURE 5.
Electron microscopy imaging reveals localization of smAKAP. A, miniSOG is a flavoprotein whose excitation is ∼500–530 nm and leads to the production of singlet oxygen. Upon the addition of DAB to a dish, the DAB located around the excited miniSOG will polymerize and create an osmiophilic polymer. (Adapted from Ref. 25.) B, the control, which was not exposed to oxygen, had no DAB polymerization at the plasma membrane as seen between two cells. N stands for nuclei. C, electron microscopy of the miniSOG-tagged smAKAP reveals plasma membrane localization in HEK293 cells after photooxidation (black areas), and smAKAP is on the intracellular side of the membrane. D and E, in certain regions, actin fibers are visible on the intracellular side of the membrane and smAKAP concentrated at a junction between two cells. F, one area of the plasma membrane of adjoining cells is highly stained with DAB with a bulge occurring. G, smAKAP in an isolated HeLa cell is also located on the plasma membrane and especially enriched in filopodia. H, a higher magnification image of filopodia showing the enrichment of the smAKAP.
DISCUSSION
In cellular biology the ability to organize enzymes into multiprotein complexes allows for a high degree of fidelity, efficiency, and spatial precision in signaling responses. In cAMP-regulated signaling, localization of signaling complexes around AKAP scaffolds is a key element in controlling and regulating intracellular communication. Here, using chemical proteomics, we discovered a novel AKAP, termed smAKAP, which specifically and tightly targets PKA-RI to the inside of the plasma membrane. The tethering of PKA-RI to the plasma membrane by smAKAP contradicts many previous reports that regard PKA-RI to be “freely” diffused in the cytosol (32, 33), although this general view has been questioned more recently (10, 12–14). For instance, a study involving FRET-based reporters of cAMP suggested not only specific PKA-RII but also PKA-RI anchoring sites to be present in cardiomyocytes (40). PKA-RI tethering was initially suggested by the discovery of AKAP-CE in Caenorhabditis elegans, which bound PKA-RI (41). Moreover, in human erythrocytes it was established, via isolating various organelles followed by immunoblotting, that PKA-RI is tightly bound to the plasma membrane (42–44). Although RI subunits were typically found to be more diffuse in the cytoplasm than RII subunits, they could also be recruited in a more dynamic way as evidenced by the migration of RI to the cap site in activated lymphocytes (45) and following treatment of cardiac myocytes with H2O2 (46). Furthermore, it was shown that overexpressed RIα and C subunits were still associated with AKAP220 even though they were diffuse in the cytoplasm; however, targeting of the RIα subunit to multivesicular bodies only occurred upon activation of the complex (47).
The recently identified dual specific AKAP, Opa1, binds PKA-RIα strongly with a Kd = 12.5 ± 2.8 nm, and this is the first dual specific AKAP to bind PKA-RI with higher affinity than PKA-RII (14). For comparison, the first PKA-RIα-specific AKAP, SPHKAP, had a Kd = 73 ± 9 nm for RIα (15), and the dual specific AKAP d-AKAP2 had a Kd of 48 nm for RIα (27). Opa1 was the strongest binding PKA-RI AKAP before we reported here our smAKAP with its apparent Kd = 6.7 ± 0.7 nm (based on in vitro AKB peptide binding, Fig. 2, B and C).
Recently, it was noted that AKAPs bind to lipid rafts via posttranslational modifications such as palmitoylation (48) and bind to multivesicular bodies (47) allowing the signaling scaffolds to be moved throughout the cell. In line with these findings, the EM imaging in Fig. 5 hints at how a double plasma membrane vesicle can form at the cell-cell contact sites and then migrate into the adjacent cell. This possibly suggests a mechanism for how cytosolic content, including the smAKAP complex, can be carried from one cell to the other, but further investigations need to follow to deduce smAKAP physiological functions.
It is also noteworthy that the smAKAP is localized to the filopodia and overexpression of smAKAP appeared to increase the number of filopodia (supplemental Fig. 3). RIα is thought to be associated with cell migration mediated by the α4 integrin, and this localization to the filopodium would be consistent with this function (49).
Acknowledgments
We thank Thijs van Holten and Mark Roest for the platelets; Toon van Veen, Siddarth Soni, and Marc Vos for the human cardiac tissue; and Marien Houtman for technical assistance.
This work was supported, in whole or in part, by the PRIME-XS Project, Grant Agreement 262067, funded by the European Union Seventh Framework Program and by National Institutes of Health Grant DK54441 (to S. S. T.). This work was also supported by The Netherlands Proteomics Centre, embedded in The Netherlands Genomics Initiative (to A. S. and A. J. R. H.) and the Centre for Biomedical Genetics (to A. J. R. H.).

This article contains supplemental Videos A–C and Figs. 1–3.
- PKA-C
- protein kinase A catalytic subunit
- AKAP
- A kinase-anchoring protein
- AKAP250
- gravin
- AKB
- A-kinase binding domain
- DAB
- diaminobenzidine
- D/D
- dimerization and docking domain
- miniSOG
- mini singlet oxygen-generating electron microscopy probe
- PKA-R
- PKA regulatory subunit
- RI/RII
- regulatory subunit I/II
- smAKAP
- small membrane AKAP
- SPHKAP
- sphingosine kinase type 1-interacting protein
- 5-TAMRA
- 5-carboxytetramethylrhodamine.
REFERENCES
- 1. Krebs E. G., Beavo J. A. (1979) Phosphorylation-dephosphorylation of enzymes. Annu. Rev. Biochem. 48, 923–959 [DOI] [PubMed] [Google Scholar]
- 2. Burns-Hamuro L. L., Hamuro Y., Kim J. S., Sigala P., Fayos R., Stranz D. D., Jennings P. A., Taylor S. S., Woods V. L., Jr. (2005) Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 14, 2982–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scholten A., Aye T. T., Heck A. J. (2008) A multi-angular mass spectrometric view at cyclic nucleotide dependent protein kinases: in vivo characterization and structure/function relationships. Mass Spectrom. Rev. 27, 331–353 [DOI] [PubMed] [Google Scholar]
- 4. Wong W., Scott J. D. (2004) AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 5. Carr D. W., Stofko-Hahn R. E., Fraser I. D., Bishop S. M., Acott T. S., Brennan R. G., Scott J. D. (1991) Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 266, 14188–14192 [PubMed] [Google Scholar]
- 6. Lygren B., Carlson C. R., Santamaria K., Lissandron V., McSorley T., Litzenberg J., Lorenz D., Wiesner B., Rosenthal W., Zaccolo M., Taskén K., Klussmann E. (2007) AKAP complex regulates Ca2+ re-uptake into heart sarcoplasmic reticulum. EMBO Rep. 8, 1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lohmann S. M., DeCamilli P., Einig I., Walter U. (1984) High-affinity binding of the regulatory subunit (RII) of cAMP-dependent protein kinase to microtubule-associated and other cellular proteins. Proc. Natl. Acad. Sci. U.S.A. 81, 6723–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alto N. M., Soderling J., Scott J. D. (2002) Rab32 is an A-kinase anchoring protein and participates in mitochondrial dynamics. J. Cell Biol. 158, 659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scholten A., Poh M. K., van Veen T. A., van Breukelen B., Vos M. A., Heck A. J. (2006) Analysis of the cGMP/cAMP interactome using a chemical proteomics approach in mammalian heart tissue validates sphingosine kinase type 1-interacting protein as a genuine and highly abundant AKAP. J. Proteome Res. 5, 1435–1447 [DOI] [PubMed] [Google Scholar]
- 10. Kovanich D., van der Heyden M. A., Aye T. T., van Veen T. A., Heck A. J., Scholten A. (2010) Sphingosine kinase interacting protein is an A-kinase anchoring protein specific for type I cAMP-dependent protein kinase. ChemBiochem 11, 963–971 [DOI] [PubMed] [Google Scholar]
- 11. Scholten A., van Veen T. A., Vos M. A., Heck A. J. (2007) Diversity of cAMP-dependent protein kinase isoforms and their anchoring proteins in mouse ventricular tissue. J. Proteome Res. 6, 1705–1717 [DOI] [PubMed] [Google Scholar]
- 12. Huang L. J., Durick K., Weiner J. A., Chun J., Taylor S. S. (1997) Identification of a novel protein kinase A anchoring protein that binds both type I and type II regulatory subunits. J. Biol. Chem. 272, 8057–8064 [DOI] [PubMed] [Google Scholar]
- 13. Huang L. J., Durick K., Weiner J. A., Chun J., Taylor S. S. (1997) d-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc. Natl. Acad. Sci. U.S.A. 94, 11184–11189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pidoux G., Witczak O., Jarnæss E., Myrvold L., Urlaub H., Stokka A. J., Küntziger T., Taskén K. (2011) Optic atrophy 1 is an A-kinase anchoring protein on lipid droplets that mediates adrenergic control of lipolysis. EMBO J. 30, 4371–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Means C. K., Lygren B., Langeberg L. K., Jain A., Dixon R. E., Vega A. L., Gold M. G., Petrosyan S., Taylor S. S., Murphy A. N., Ha T., Santana L. F., Tasken K., Scott J. D. (2011) An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc. Natl. Acad. Sci. U.S.A. 108, E1227–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sarma G. N., Kinderman F. S., Kim C., von Daake S., Chen L., Wang B. C., Taylor S. S. (2010) Structure of d-AKAP2:PKA RI complex: insights into AKAP specificity and selectivity. Structure 18, 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carlson C. R., Lygren B., Berge T., Hoshi N., Wong W., Taskén K., Scott J. D. (2006) Delineation of type I protein kinase A-selective signaling events using an RI anchoring disruptor. J. Biol. Chem. 281, 21535–21545 [DOI] [PubMed] [Google Scholar]
- 18. Gold M. G., Lygren B., Dokurno P., Hoshi N., McConnachie G., Taskén K., Carlson C. R., Scott J. D., Barford D. (2006) Molecular basis of AKAP specificity for PKA regulatory subunits. Mol. Cell 24, 383–395 [DOI] [PubMed] [Google Scholar]
- 19. Malbon C. C., Tao J., Wang H. Y. (2004) AKAPs (A-kinase anchoring proteins) and molecules that compose their G-protein-coupled receptor signalling complexes. Biochem. J. 379, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trotter K. W., Fraser I. D., Scott G. K., Stutts M. J., Scott J. D., Milgram S. L. (1999) Alternative splicing regulates the subcellular localization of A-kinase anchoring protein 18 isoforms. J. Cell Biol. 147, 1481–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aye T. T., Soni S., van Veen T. A., van der Heyden M. A., Cappadona S., Varro A., de Weger R. A., de Jonge N., Vos M. A., Heck A. J., Scholten A. (2012) Reorganized PKA-AKAP associations in the failing human heart. J. Mol. Cell. Cardiol. 52, 511–518 [DOI] [PubMed] [Google Scholar]
- 22. Margarucci L., Roest M., Preisinger C., Bleijerveld O. B., van Holten T. C., Heck A. J., Scholten A. (2011) Collagen stimulation of platelets induces a rapid spatial response of cAMP and cGMP signaling scaffolds. Mol. bioSyst. 7, 2311–2319 [DOI] [PubMed] [Google Scholar]
- 23. de Jong R. N., Daniëls M. A., Kaptein R., Folkers G. E. (2006) Enzyme free cloning for high throughput gene cloning and expression. J. Struct. Funct. Genomics 7, 109–118 [DOI] [PubMed] [Google Scholar]
- 24. Shu X., Shaner N. C., Yarbrough C. A., Tsien R. Y., Remington S. J. (2006) Novel chromophores and buried charges control color in mFruits. Biochemistry 45, 9639–9647 [DOI] [PubMed] [Google Scholar]
- 25. Shu X., Lev-Ram V., Deerinck T. J., Qi Y., Ramko E. B., Davidson M. W., Jin Y., Ellisman M. H., Tsien R. Y. (2011) A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9, e1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ormö M., Cubitt A. B., Kallio K., Gross L. A., Tsien R. Y., Remington S. J. (1996) Crystal structure of the Aequorea victoria green fluorescent protein. Science 273, 1392–1395 [DOI] [PubMed] [Google Scholar]
- 27. Burns-Hamuro L. L., Ma Y., Kammerer S., Reineke U., Self C., Cook C., Olson G. L., Cantor C. R., Braun A., Taylor S. S. (2003) Designing isoform-specific peptide disruptors of protein kinase A localization. Proc. Natl. Acad. Sci. U.S.A. 100, 4072–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martin B. R., Deerinck T. J., Ellisman M. H., Taylor S. S., Tsien R. Y. (2007) Isoform-specific PKA dynamics revealed by dye-triggered aggregation and DAKAP1α-mediated localization in living cells. Chem. Biol. 14, 1031–1042 [DOI] [PubMed] [Google Scholar]
- 29. Brown S. H., Wu J., Kim C., Alberto K., Taylor S. S. (2009) Novel isoform-specific interfaces revealed by PKA RIIβ holoenzyme structures. J. Mol. Biol. 393, 1070–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim C., Cheng C. Y., Saldanha S. A., Taylor S. S. (2007) PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell 130, 1032–1043 [DOI] [PubMed] [Google Scholar]
- 31. Kovanich D., Aye T. T., Heck A. J., Scholten A. (2012) Probing the specificity of protein-protein interactions by quantitative chemical proteomics. Methods Mol. Biol. 803, 167–181 [DOI] [PubMed] [Google Scholar]
- 32. Brunton L. L., Hayes J. S., Mayer S. E. (1981) Functional compartmentation of cyclic AMP and protein kinase in heart. Adv. Cyclic Nucleotide Res. 14, 391–397 [PubMed] [Google Scholar]
- 33. Corbin J. D., Sugden P. H., Lincoln T. M., Keely S. L. (1977) Compartmentalization of adenosine 3′:5′-monophosphate and adenosine 3′:5′-monophosphate-dependent protein kinase in heart tissue. J. Biol. Chem. 252, 3854–3861 [PubMed] [Google Scholar]
- 34. Resh M. D. (2006) Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE 2006, re14. [DOI] [PubMed] [Google Scholar]
- 35. Robbins S. M., Quintrell N. A., Bishop J. M. (1995) Myristoylation and differential palmitoylation of the HCK protein-tyrosine kinases govern their attachment to membranes and association with caveolae. Mol. Cell. Biol. 15, 3507–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forrester M. T., Hess D. T., Thompson J. W., Hultman R., Moseley M. A., Stamler J. S., Casey P. J. (2011) Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 52, 393–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Resh M. D. (2006) Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2, 584–590 [DOI] [PubMed] [Google Scholar]
- 38. Wright M. H., Heal W. P., Mann D. J., Tate E. W. (2010) Protein myristoylation in health and disease. J. Chem. Biol. 3, 19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smotrys J. E., Linder M. E. (2004) Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73, 559–587 [DOI] [PubMed] [Google Scholar]
- 40. Di Benedetto G., Zoccarato A., Lissandron V., Terrin A., Li X., Houslay M. D., Baillie G. S., Zaccolo M. (2008) Protein kinase A type I and type II define distinct intracellular signaling compartments. Circ. Res. 103, 836–844 [DOI] [PubMed] [Google Scholar]
- 41. Angelo R., Rubin C. S. (1998) Molecular characterization of an anchor protein (AKAPCE) that binds the RI subunit (RCE) of type I protein kinase A from Caenorhabditis elegans. J. Biol. Chem. 273, 14633–14643 [DOI] [PubMed] [Google Scholar]
- 42. Corbin J. D., Sugden P. H., West L., Flockhart D. A., Lincoln T. M., McCarthy D. (1978) Studies on the properties and mode of action of the purified regulatory subunit of bovine heart adenosine 3′:5′-monophosphate-dependent protein kinase. J. Biol. Chem. 253, 3997–4003 [PubMed] [Google Scholar]
- 43. Rubin C. S., Erlichman J., Rosen O. M. (1972) Cyclic adenosine 3′,5′-monophosphate-dependent protein kinase of human erythrocyte membranes. J. Biol. Chem. 247, 6135–6139 [PubMed] [Google Scholar]
- 44. Montoliu C., Piedrafita B., Serra M. A., del Olmo J. A., Rodrigo J. M., Felipo V. (2007) A single transient episode of hyperammonemia induces long-lasting alterations in protein kinase A. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G305–314 [DOI] [PubMed] [Google Scholar]
- 45. Mosenden R., Singh P., Cornez I., Heglind M., Ruppelt A., Moutschen M., Enerbäck S., Rahmouni S., Taskén K. (2011) Mice with disrupted type I protein kinase A anchoring in T cells resist retrovirus-induced immunodeficiency. J. Immunol. 186, 5119–5130 [DOI] [PubMed] [Google Scholar]
- 46. Brennan J. P., Bardswell S. C., Burgoyne J. R., Fuller W., Schröder E., Wait R., Begum S., Kentish J. C., Eaton P. (2006) Oxidant-induced activation of type I protein kinase A is mediated by RI subunit interprotein disulfide bond formation. J. Biol. Chem. 281, 21827–21836 [DOI] [PubMed] [Google Scholar]
- 47. Day M. E., Gaietta G. M., Sastri M., Koller A., Mackey M. R., Scott J. D., Perkins G. A., Ellisman M. H., Taylor S. S. (2011) Isoform-specific targeting of PKA to multivesicular bodies. J. Cell Biol. 193, 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Delint-Ramirez I., Willoughby D., Hammond G. V., Ayling L. J., Cooper D. M. (2011) Palmitoylation targets AKAP79 protein to lipid rafts and promotes its regulation of calcium-sensitive adenylyl cyclase type 8. J. Biol. Chem. 286, 32962–32975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lim C. J., Han J., Yousefi N., Ma Y., Amieux P. S., McKnight G. S., Taylor S. S., Ginsberg M. H. (2007) α4 integrins are type I cAMP-dependent protein kinase-anchoring proteins. Nat. Cell Biol. 9, 415–421 [DOI] [PubMed] [Google Scholar]