Background: The 3D molecular structure of the L-type channel is poorly understood.
Results: The 3D locations of the α and β subunits and the II-III loop are identified.
Conclusion: Key membrane targeting and signal transduction elements are placed in the context of the channel and cell membrane.
Significance: This work provides novel insight in the L-type channel structure-function relationships.
Keywords: Calcium Channels, Cell Signaling, Electron Microscopy (EM), Excitation-Contraction Coupling, Protein Expression, 3D Reconstruction, Cav1.1, Dihydropyridine Receptor, L-type Channel, Protein Structure
Abstract
The L-type Ca2+ channel (dihydropyridine receptor (DHPR) in skeletal muscle acts as the voltage sensor for excitation-contraction coupling. To better resolve the spatial organization of the DHPR subunits (α1s or CaV1.1, α2, β1a, δ1, and γ), we created transgenic mice expressing a recombinant β1a subunit with YFP and a biotin acceptor domain attached to its N- and C- termini, respectively. DHPR complexes were purified from skeletal muscle, negatively stained, imaged by electron microscopy, and subjected to single-particle image analysis. The resulting 19.1-Å resolution, three-dimensional reconstruction shows a main body of 17 × 11 × 8 nm with five corners along its perimeter. Two protrusions emerge from either face of the main body: the larger one attributed to the α2-δ1 subunit that forms a flexible hook-shaped feature and a smaller protrusion on the opposite side that corresponds to the II-III loop of CaV1.1 as revealed by antibody labeling. Novel features discernible in the electron density accommodate the atomic coordinates of a voltage-gated sodium channel and of the β subunit in a single docking possibility that defines the α1-β interaction. The β subunit appears more closely associated to the membrane than expected, which may better account for both its role in localizing the α1s subunit to the membrane and its suggested role in excitation-contraction coupling.
Introduction
Voltage-gated Ca2+ channels are key transducers in excitation-contraction (EC)3 coupling, secretion, regulation of gene expression, integration of synaptic input, and synaptic transmission (1). The skeletal muscle L-type Ca2+ channel (dihydropyridine receptor (DHPR)) is a heteropentameric membrane protein complex formed by a Ca2+ channel pore-forming subunit (CaV1.1) and four auxiliary subunits (α2-δ1,β1a, γ). The DHPR senses depolarization in the sarcolemma and induces RyR1 to open and release Ca2+ stored in the sarcoplasmic reticulum into the cytoplasm for muscle contraction (2, 3). Because in skeletal muscle the activation of RyR1 by the L-type Ca2+ channel is independent of extracellular Ca2+ entry, it is widely held that signaling between these two channels involves “conformational coupling” (reviewed in Ref. 4). This conformational EC coupling is mediated by a highly organized macromolecular complex whereby the large cytoplasmic domain of RyR1 acts as a hub for the assembly of up to four DHPR units (5).
The α1S subunit of DHPR, or CaV1.1, is the largest subunit (176 kDa), mainly composed of a transmembrane domain that constitutes both the Ca2+ channel pore and the voltage sensor. It belongs to the voltage-gated ion channel superfamily, consisting of four repeats with six transmembrane helices each (S1-S6) (6, 7), with each repeat connected to the next by a cytoplasmic loop. The I-II loop contains a highly conserved α interaction domain (AID) that has been proposed as the binding site of the β subunit (8). The largest cytosolic loop (the II-III loop, with 138 residues) has a major role in EC coupling (9–12). In solution, the II-III loop displays an intrinsically disordered structure (13–15). The III-IV loop has also been suggested to interact with RyR1 (16, 17), although without a direct role in EC coupling (18). The α2-δ subunit is the disulfide-linked cleavage product of a single gene (19): α2 (147 kDa), extracellular and highly glycosylated, and δ (24 kDa), which anchors α2 to the membrane through its single transmembrane domain. The β1a subunit (56 kDa) is a soluble cytoplasmic protein unless bound to the AID of the CaV1.1 subunit. In addition to facilitating the trafficking of CaV1.1 to the plasma membrane, β1a plays a key role in skeletal EC coupling (9, 20–23). The structure of its core region has been solved to atomic resolution for several isoforms (24–26). The γ subunit (34 kDa) has four transmembrane domains with cytosolic C- and N-termini and interacts with CaV1.1 (27), acting as a Ca2+ channel antagonist (28).
The structure of the DHPR complex has been mainly studied by transmission electron microscopy (TEM) and single-particle analysis. Although two initial 3D reconstructions differed in shape (29, 30), a consensus structure consisting of a globular structure with an appendage is clearly emerging for L- and T- type channels (31, 32), with the α2 subunit assigned to the large appendage (31, 33) and the β subunit assigned to a region within the globular structure (31). Further discernment of subunits has been difficult because of inherent flexibility of the macromolecule, potential loss of subunits during the purification procedure, relatively small size for TEM, and the presence of detergent surrounding the membrane-embedded region. In addition, isolation of a sufficient amount of material for single-particle image processing of this channel complex has been challenging.
Here we devised a comprehensive approach to simplify the purification of an intact macromolecular complex by creating a transgenic mouse that was used to express a functional recombinant DHPR complex with YFP and biotin bound to its β1a subunit (34). Using a negative stain, TEM, and single-particle analysis methodology, we obtained a 19.1-Å 3D reconstruction of the recombinant DHPR complex. The 3D structure consists of a flat irregular main body with five pronounced corners, a flexible extension emerging from one of the faces, and a smaller protuberance extending from the opposite face, and its high definition enabled simultaneous docking of the atomic coordinates of a voltage-gated cation channel, a β subunit, and YFP. Decoration of the DHPR with an anti- II-III loop antibody allowed the mapping of this functional domain within the 3D structure. Our results significantly refine the structure of the DHPR complex and demonstrate the advantage of the use of animal transgenesis for the production and isolation of native heteromeric transmembrane protein complexes for structural analysis.
EXPERIMENTAL PROCEDURES
Construction of the Recombinant cDNA
The YFP-β1a-BAD cDNA construct was made as reported previously (34). Targeted expression to skeletal muscle was achieved by cloning YFP-β1a-BAD cDNA under control of the human skeletal actin (HSA) promoter (35). For this, a 2882-bp fragment from the pHSAvpA vector (a gift from Dr. J. Chamberlain) containing 2409 bp of the HSA promoter, 293 bp of the 5′ end of HSA intron 1, and 473 bp of the splice acceptor sequence from SV40 (VP1 intron) was cloned upstream of the YFP-β1a-BAD coding sequence.
Transgenic Animals
The complete 5648-bp YFP-β1a-BAD expression construct, including a skeletal actin promoter and a SV40 poly(A) region, was excised and microinjected into fertilized mouse eggs (FVB/N) by the Massachusetts General Hospital transgenic facility, Charlestown, MA, with approval of the Harvard University Institutional Animal Care and Use Committee. Genotyping of transgenic mice was performed by PCR analysis of genomic tail DNA using the forward primer 5′-GAAGGGTTGCAAATATCATTTGGGC-3′, corresponding to bp 2624–2648 of the HSA promoter sequence and the reverse primer 5′-GTCGTGCTGCTTCATGTGGTCGGGG-3′, corresponding to bp 224–249 of the YFP cDNA sequence. Transgenic mice expressing HSA-YFP-β1a-BAD were then crossbred with heterozygous β1-null mice (F1) and subsequently backcrossed (F2) to select for homozygous β1-null animals expressing only the transgenic YFP-β1a-BAD subunit.
Gold-Streptavidin Labeling and Confocal Microscopy
Flexor digitorum brevis muscle fibers were isolated and bathed in an “internal solution” of 140 mm Cs-Aspartate, 10 mm Cs2EGTA, 5 mm MgCl2, and 10 mm HEPES (pH 7.3) with CsOH. The cells were permeabilized by exposure for 30 s to saponin (Sigma, 12 mg/ml in internal solution). Following a wash in internal solution, the cells were incubated in Alexa Fluor 594-nanogold streptavidin (Invitrogen, 1:500 in internal solution) for 30 min, washed for 30 min with internal solution, and then viewed with a Zeiss LSM 510 confocal microscope. YFP was visualized with 488-nm excitation and 505–530-nm band-pass emission. Alexa Fluor 594 was visualized with 543-nm excitation and 560-nm long-pass emission. Images were acquired as the average of four to eight line scans per pixel and digitized at 8 bits.
DHPR Purification
DHPR was purified from the skeletal muscle of mice expressing DHPR containing β1a subunit with YFP and a BAD domain attached to its N- and C- termini, respectively. Purification was performed under the presence of protease inhibitors at 4 °C. Skeletal muscle was excised from the animal and homogenized, and whole membrane vesicles were isolated by differential centrifugation. The vesicle fraction (16–75 mg of total protein) was solubilized with 1% (w/v) digitonin (Sigma) in equilibration buffer (50 mm Tris-HCl (pH 7.4), 0.5 m NaCl, 1 mm iodoacetamide) and a protein/detergent (w/w) ratio of 5–7 for 40 min followed by centrifugation at 100,000 × g for 70 min (31, 36, 37). Glycosylated proteins, including the α2-δ1-containing DHPR complex, were then enriched using a wheat-germ agglutinin (WGA) chromatography column. For this the supernantant was loaded to a WGA-Sepharose affinity column (Sigma) (37), and the bound proteins were eluted with 0.5 m N-acetyl-glucosamine. Peak fractions were pooled and loaded onto a HiTrap streptavidin column (GE). Binding occurred by the affinity between the streptavidin resin and the metabolically biotinylated BAD domain of the β1a subunit (34). Bound proteins were eluted with 5 mm D-desthiobiotin, and peak fractions were concentrated ∼10-fold with a centrifugal filter (MWCO, 100 kDa).
Antibody Labeling of the DHPR II-III Loop
Western blot analyses were performed with monoclonal antibodies against CaV1.1 (mA3–920 Thermo) (38), α2 (Abcam), β (Developmental Studies Hybridoma Bank, University of Iowa), and GFP (Clonetech). The secondary antibody (goat anti-mouse antibody, Pierce) was detected via chemiluminescence. For TEM, purified DHPR was incubated with a 10-fold molecular excess of mA3–920 antibody against the II-III loop of CaV1.1for 12 h at 4 °C.
Electron Microscopy
Samples were imaged by negative-stain TEM. The sample (5 μl) was applied to the carbon-coated side of a glow-discharged grid and let to adsorb for 70 s. The grid was washed twice with Milli Q water and then negatively stained with 0.75% uranyl formate as described previously (39). Images were collected on a FEI Tecnai T12 TEM operated at 120 kV and a magnification of ×67,000 at 0°, 45°, or 60° tilt angles under low-dose conditions (< 20 e−/Å2). Images were recorded on image plates. The antibody-labeled DHPR specimen was imaged on a Phillips CM10 TEM at a magnification ×39,000 under low-dose conditions, and images were recorded on a 1k × 1 k Gatan charge-coupled device (CCD) camera.
Image Processing
Particles were selected from the micrographs using the Boxer software (40). A total of 11,929 particles were collected for the DHPR dataset and a total of 625 particles for the antibody-labeled DHPR. 2D and 3D image processing was performed using the SPIDER software suite (41) for the unlabeled and antibody-labeled datasets. Images were binned down to a final pixel size 4.48 Å, and particles were windowed with final window sizes of 1302 pixels for the DHPR complex and 1252 pixels for the DHPR-antibody dataset. The windowed particles underwent alignment, classification, and multi-reference alignment to produce class averages (42). 3D reconstruction of the DHPR complex was produced by projection matching using a 3D reference reconstruction (31) filtered to 40-Å resolution, followed by 3D refinement. Particles with low cross-correlation were discarded, with 2,693 particles in the final 3D reconstruction. Resolution was calculated according to the Fourier shell correlation curve between two half datasets, providing a value of 19.1 Å at the 0.5 cutoff criterion. The 3D reconstruction was filtered to 19.1 Å using a Gaussian low-pass filter. Image rendering was performed with Chimera (43).
RESULTS
Expression of a Recombinant β1a Subunit in a β1a-null Mouse
We constructed a transgenic mouse expressing a recombinant β1a subunit under control of the HSA promoter for specific expression in skeletal muscle (44). The recombinant β1a subunit was engineered as a fusion protein (YFP-β-BAD), with YFP fused to the N terminus and a BAD fused to the C terminus. Transgenic mice overexpressing the recombinant β1a subunit were crossed with heterozygous β1a-null mice and bred to homozygosity for both the transgene and the β1a-null allele. Although homozygous β1a null mice have a birth-lethal phenotype (22), the mice homozygous both for the β1a-null allele and for the YFP-β-BAD transgene were viable and appeared healthy, indicating that DHPR complexes containing the recombinant β1a subunit were functional for EC coupling.
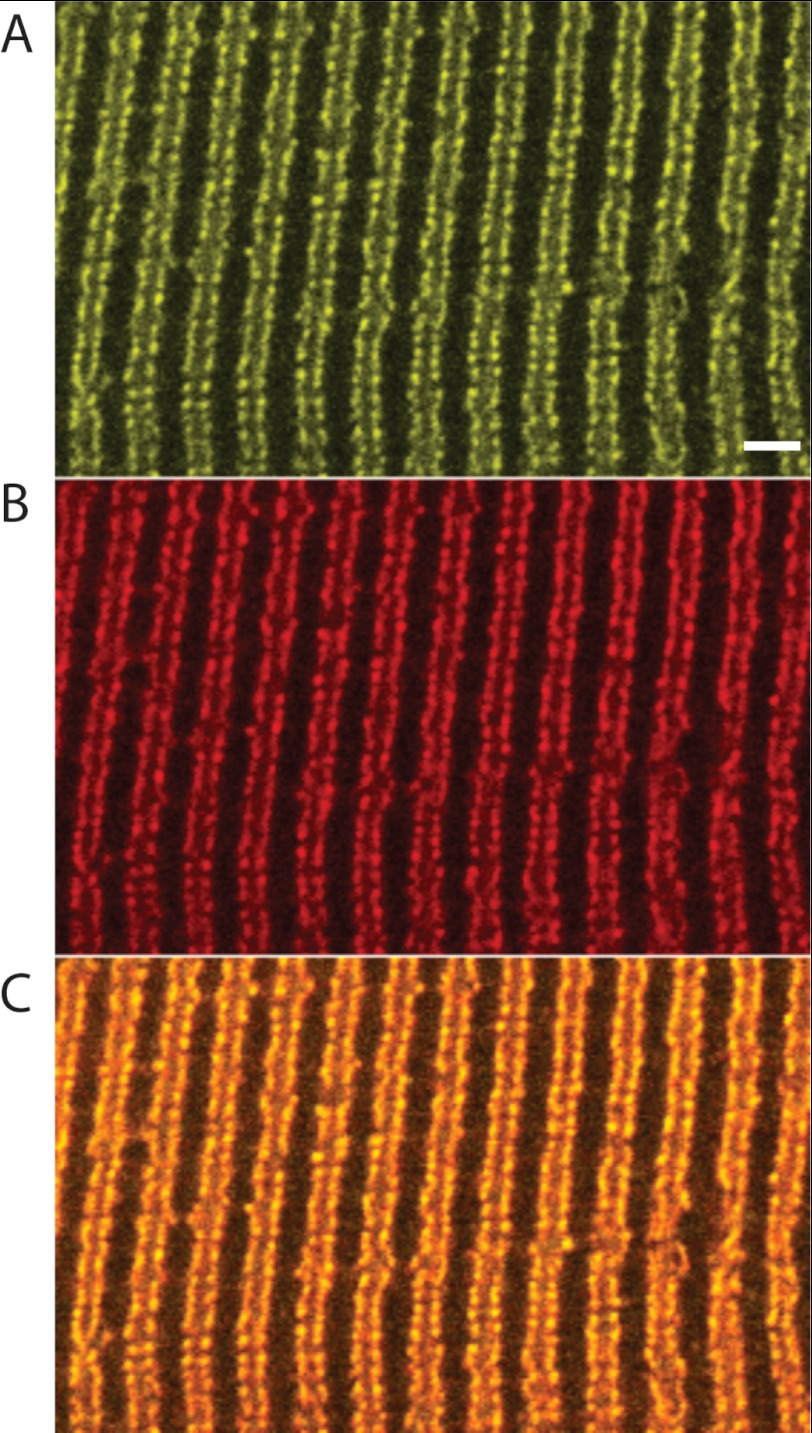
Fig. 1 illustrates the subcellular distribution of the YFP-β-BAD transgene in a flexor digitorum brevis fiber that had been permeabilized with saponin and then exposed to 1 nm fluoro-nanogold streptavidin. The yellow fluorescence arising from the YFP tag was arrayed in double rows having a center-to-center spacing of ∼2 μm, consistent with a T-tubular localization. Furthermore, the red fluorescence arising from the bound fluoro-gold streptavidin colocalized with the yellow fluorescence, indicating that there is effective biotinylation of the BAD attached to the β1a C terminus and that its disposition in triad junctions makes it accessible to a moderately sized (1 nm) probe, consistent with previous work on myotubes (34, 45, 46). This previous work also showed that EC coupling was not impaired when streptavidin bound to YFP-β-BAD in myotubes (45).
FIGURE 1.
Confocal microscopy of skeletal muscle tissue expressing a transgenic DHPR β1asubunit, YFP-β-BAD. A, YFP fluorescence displaying a T-tubular localization. B, fluoro-nanogold streptavidin fluorescence of the same region. C, the overlay of the YFP and fluoro-nanogold fluorescence images indicates full colocalization of the two fluorophores, and the binding of streptavidin demonstrates that the BAD protein is biotinylated. Scale bar = 2 μm.
Protein Purification
As expected for a muscle-specific promoter, the YFP-β-BAD construct was expressed primarily in skeletal muscle, although a modest expression was also observed in liver and heart (Fig. 2A). The DHPR was purified from skeletal muscle, as indicated under “Experimental Procedures.” Silver staining of SDS gels indicated the purity of the preparation, and the presence of the larger subunits identified on the basis of their mass (Fig. 2B). The identity of the α1, α2, β1a subunits and of YFP was further confirmed by Western blot analysis (Fig. 2C). Note that the molecular mass of the β1a subunit was higher than that of WT β1a (96 kDa instead of 56 kDa) because of the presence of the YPF and BAD moieties. Altogether, these results indicate that our methodology provides highly pure DHPR complexes from a relatively small amount of starting material (between 10 and 20 g).
FIGURE 2.
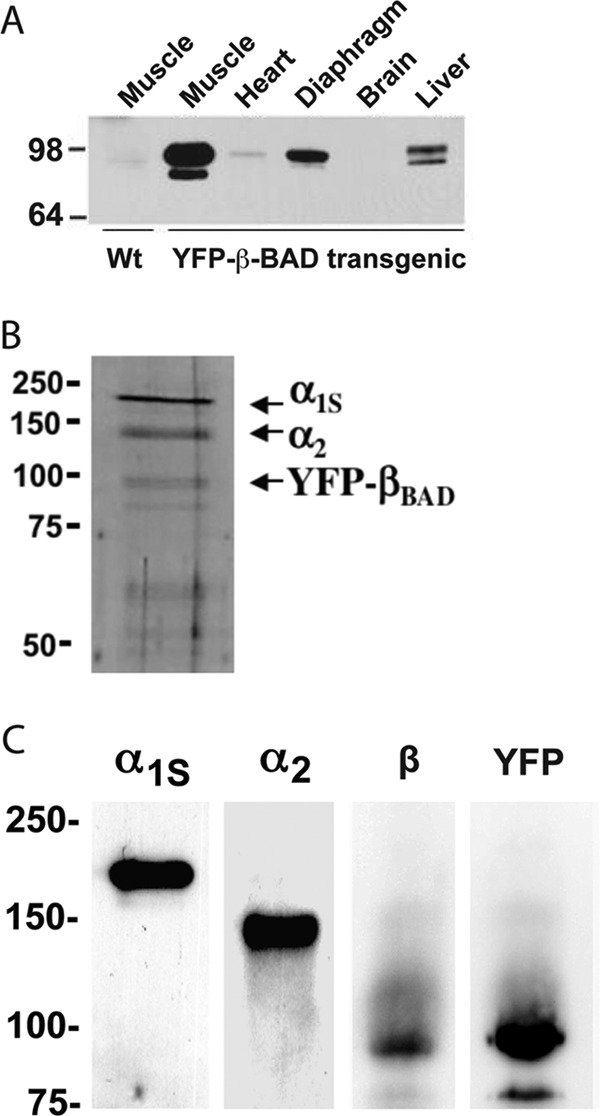
Biochemical characterization of the purified DHPR complex. A, Western blot analysis using anti-GFP antibody showing the expression of YFP-β1a-BAD in different tissues. B, SDS-PAGE silver-stained gel (10% separating gel under reducing conditions) confirms the presence of the high molecular weight components of the purified DHPR complex: α1 (176 kDa), α2 (147 kDa), and YFP-β1a-BAD (96 kDa) subunits. C, Western blot analyses of the skeletal muscle vesicle fraction incubated with antibodies against the α1s-II-III loop, α2, β, and YFP. Both the anti-β and anti-GFP antibodies label the same band corresponding to the YFP-β1a-BAD construct, with a molecular mass of 96 kDa.
3D Reconstruction of the DHPR
TEM images of negatively stained DHPRs show individual particles of homogeneous size with a main body and a bulky appendage attached to it (Fig. 3, A and B). In some views, a smaller protuberance is visible on the opposite side. A total of 11,929 individual DHPRs was analyzed using single-particle image processing (47). Two-dimensional alignment and classification (42) produced stable 2D class averages with defined features and orientational information (Fig. 3C). At least 7 of our 12 2D averages (1, 5, 7, 9–12) strongly resemble most of the DHPR 2D averages published previously (31), and averages 8 and 14, formed by a thick line and a small appendage, strongly resemble one of three 2D averages published by another group (Fig. 4B of (33)). For 3D reconstruction, we used a reference-based approach. Given that all the 2D averages presented in Wolf et al. (31) were represented in our dataset, we performed a reference alignment using their 23-Å resolution 3D reconstruction filtered to 40-Å resolution as a starting reference. This produced a first 3D reconstruction with novel prominent features such as pronounced corners (two of them square corners), a flatter shape of the main globular domain, and an irregular pentagon shape of 17 × 11 × 8 nm. Further refinement of the 3D reconstruction yielded a resolution of 19.1 Å, according to the Fourier shell correlation curve criterion, with a cutoff of 0.5 (Fig. 3D), indicating a net increase in detail from 40 Å, and that our dataset had genuine signal that was absent in the reference. The angular distribution plot (Fig. 3E) indicates a full coverage of the angular range and, consequently, the absence of the missing cone artifact (48).
FIGURE 3.
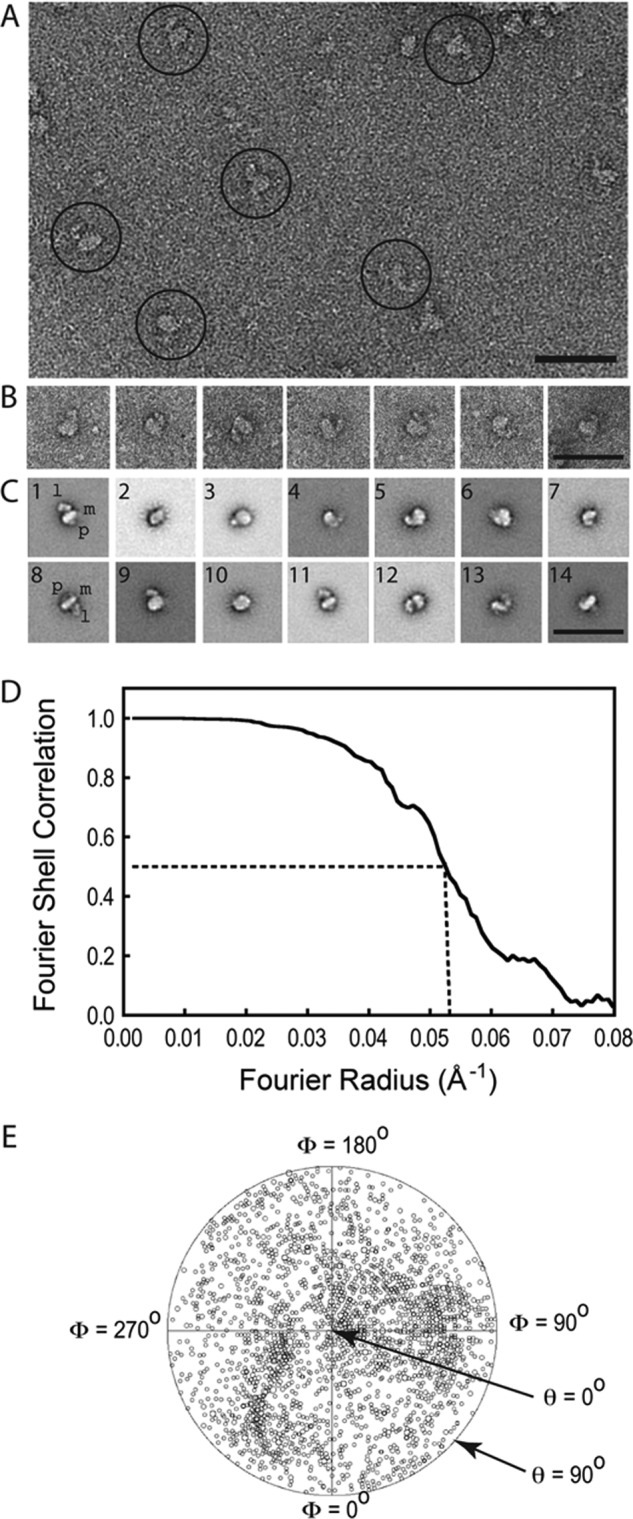
TEM and image processing of the purified DHPR complexes. A, raw electron micrograph displaying a homogeneous distribution of L-type channels. The more representative images are highlighted with circles. B, windowed DHPR particles showing a main body with an appendage. C, gallery of the 2D averages of DHPR after several rounds of classification and multireference alignment. The main body (m), hook- or leg- shaped feature (l), and the small protuberance (p) are indicated in averages 1 and 8. Scale bars for A–C = 50 nm. D, Fourier shell correlation curve indicating a resolution of 19.10 Å according to the 0.5 cutoff criterion. E, plot of the angular distribution of the particles used in the final 3D reconstruction showing a uniform distribution of Euler angular orientations of the DHPR. The size of the small circles represents the number of particles in that orientation, and the center of the circular plot corresponds to a θ angle of 0°.
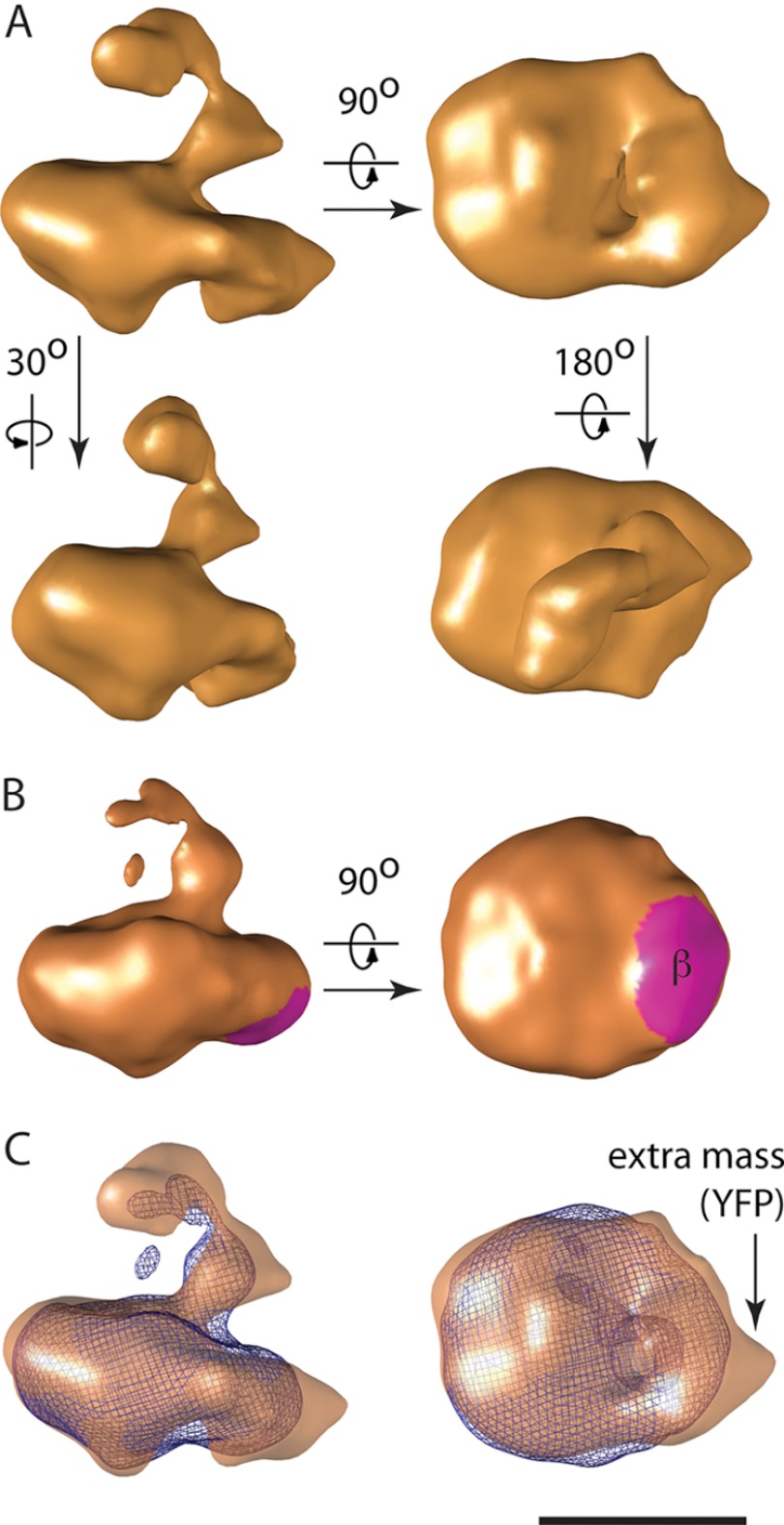
FIGURE 4.
New 3D reconstruction of the DHPR complex and comparison with a previous 3D reconstruction and β1a subunit localization. A, 3D reconstruction of the recombinant DHPR in different orientations. B, 3D reconstruction of DHPR with the region labeled by antibodies against the β1a subunit colored in magenta from Ref. 31. C, superimposition of both 3D reconstructions, A in semitransparent orange and B in blue mesh. An extra mass of the recombinant (YFP-β1a-BAD)DHPR, presumably the β1a-adjoining YFP, emerges from the location labeled by anti-β1a antibodies in the previous study. The blue mesh DHPR reconstruction has been rotated 10° with respect to the plasma membrane to match the new DHPR reconstruction in the new proposed orientation. The left panels show a side view (perpendicular to the plane of the membrane). Scale bar = 10 nm.
The refined 3D reconstruction reveals that the main body is formed by two distinct regions, one forming a square and one forming a triangle, separated by a mass deficiency between them (Fig. 4A). The long appendage attaches to one edge of the square portion (Fig. 4A). The surface contour level was adjusted to the midpoint between the background density level and the highest density level at which the volume presents no discontinuities. Under these conditions, the surface encloses a mass that is 23% larger than the expected molecular weight (49). Nevertheless, it is important to point out that, in addition to the protein moiety, there is also detergent present. Although the change of contour level between these limits had little effect on the appearance of the main body, the hook-shaped feature changed in shape, which is an indicator of flexibility. At the contour level used, the hook-shaped structure forms an L shape with a thickness between 2 and 4 nm that extends 7 nm from the main body.
Antibody Labeling of the II-III Loop
An antibody specific for the II-III loop (38) recognizes the CaV1.1 subunit of the DHPR complex by Western blot analysis (Fig. 2C). Negative staining TEM of the DHPR complex incubated previously with the anti II-III loop antibody shows DHPR particles with antibody bound (Fig. 5A). The raw images of the DHPR were matched to the more similar 2D projection of the DHPR 3D reconstruction, which yielded the orientation of the DHPR with respect to its support and, thus, the localization of the epitope in the 3D model of the DHPR (Fig. 5, B and C). All individual determinations converge on the small protrusion of the main body, indicating that this protrusion corresponds to the II-III loop.
FIGURE 5.
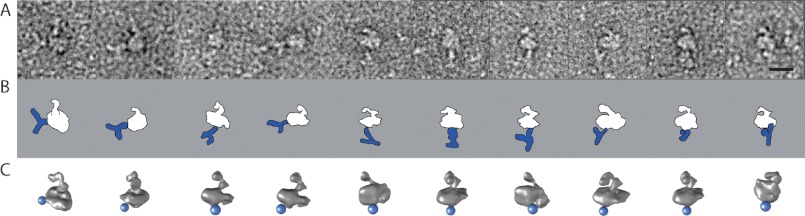
Electron microscopy of the purified DHPR complex bound to anti II-III loop antibody. A, raw images of the L-type channel labeled with anti-α1s-II-III loop antibody. B, schematic representation of the antibody (blue) labeling the DHPR complex (white) in the orientation found in A. C, 3D reconstruction of the DHPR in the corresponding orientation. The blue sphere represents the location found for the antibody. Scale bar = 10 nm.
3D Location and Docking of the YFP Atomic Structure
By employing affinity chromatography using the biotinylated BAD tag (see “Experimental Procedures”), our purification protocol ensured that all purified DHPR complexes used for analysis contained the recombinant YFP-β1a-BAD subunit. The presence of this YFP can account for an extra mass that was present in the 3D reconstruction of the recombinant DHPR but not in the 3D reconstruction of WT DHPR (31) (Fig. 4C). This protrusion, situated along the main globular part of the complex, presents a size and shape compatible with a YFP molecule (a β barrel 45 Å long and 25 Å in diameter) and enabled a straightforward docking of the YFP atomic structure (Fig. 6). YFP can be seen as a peripheral addition to the DHPR macromolecule with a thickness compatible with the diameter of YFP. Therefore, at the current resolution, it appears that the YFP tag is accommodated along the β1a subunit without noticeable changes in its native orientation. Such a location is probably the most benign in the context of the interaction between DHPR and RyR1 in the triad junction and is consistent with the functional viability of the YFP recombinant β1a subunit.
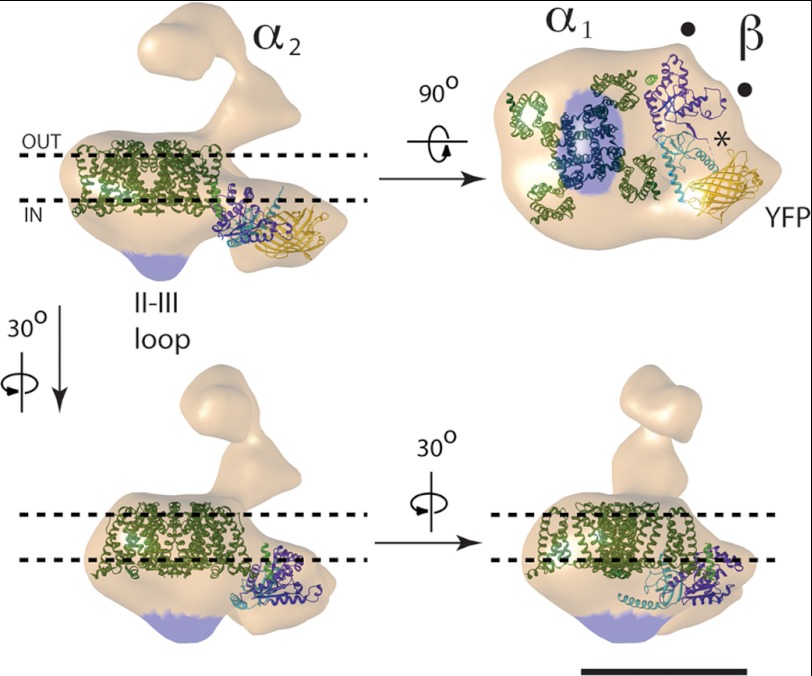
FIGURE 6.
Interpretation of the 3D reconstruction of DHPR. The dashed black lines indicate the approximate boundary of the plasma membrane. The region corresponding to the II-III loop is indicated in blue shading. The atomic structures of an Na+ channel (dark green), the β subunit (SH3 in light blue and GK in purple) bound to the AID (light green), and YFP (yellow) are docked into the DHPR structure. The ● are two features of the density map that match with the atomic structure of the β subunit, and the asterisk shows an empty density that could allocate the N-terminus, C-terminus, and HOOK regions of the β subunit and the γ subunit. YFP has been omitted in the bottom panels for clarity. Scale bar = 10 nm.
Docking of the Atomic Structure of a Voltage Gated Ion Channel
Consistent with the considerable conservation of membrane topology among voltage-gated channels, with four subunits or repeats, each with six transmembrane domains (7), the main body of our 3D reconstruction can be closely fitted with existing atomic structures of voltage-gated cation channels such as the full-length potassium channel KV1.2 (50) or the bacterial Na+ channel, NaVAb (51), by placing the channel with the selectivity filter toward the extracellular side and by aligning two of the corners of the channel to two square corners of the DHPR 3D structure. Because the fitting of these two atomic structures is equivalent at our resolution, we only present the fitting with the Na+ channel (Fig. 6).
3D Location of the β Subunit
CaV β subunits have hypervariable N- and C-termini that flank a core comprised of SH3, HOOK, and GK domains (52–55). Several atomic structures for the core of different β subunit isoforms obtained by x-ray crystallography (24–26) present high structural similarity among them, with RMSD below 1 Å for all the pairs. Because no region of β1a has been crystallized, we first aligned the sequence of β1a to the sequences of the solved structures. From this multiple alignment, the crystallized structure with the highest coverage of β1a was a rabbit β2a subunit (24). Thus, for the docking shown in Fig. 6, we used this structure (24), which was solved alone and bound to the AID of Cav1.1. The atomic structure of rabbit β2a accounts for 62% of the β1a sequence with 84% identity, with full coverage of the SH3 and GK domains with 79 and 91% identity, respectively, and lower coverage of the N-, C-, and HOOK domains, thus leaving 64, 81, and 55 residues of β1a of 524 residues not accounted for in the crystal structure.
After docking the structures of the Na+ channel (representing CaV1.1) and YFP into the main body, there were two unoccupied densities in the DHPR volume: the intracellular protuberance directly “below” CaV1.1 and the region in the irregular triangular region not occupied by the YFP. Both regions were considered as possible locations for the β subunit. The protuberance directly below CaV1.1, however, was too small to contain the atomic model of the β subunit, and furthermore, because our antibody labeling studies indicated that this protuberance includes the II-III loop of CaV1.1, there was even less space available for the β subunit. The atomic model of the β subunit fits in the empty region between CaV1.1 and YFP (Fig. 6), which is consistent with β1a and YFP being continuous in sequence. This area has two characteristic protrusions pointing toward the periphery, separated by 4 nm (● in Fig. 6), and another protrusion pointing toward the cytoplasm. From three possible orientations within this area, only one of them satisfied two essential criteria: that the AID is close to Cav1.1 and that the SH3 domain, more N-terminal than the GK domain, is proximal to the N-terminal YFP. Furthermore, in this orientation, the two prominent lobes of the GK domain point toward the two peripheral spikes, thus providing a very close fit as seen from orthogonal viewing angles (Fig. 6).
DISCUSSION
Use of a Transgenic Animal to Obtain Recombinant Membrane Proteins for Structural Determination
The DHPR presents a formidable challenge for its structural determination. On one hand, being a membrane protein, it requires the use of detergent and a more complex purification process than soluble proteins. In addition, it has a hetero-oligomeric composition, and the presence of detergent throughout the purification could promote subunit dissociation. The use of animal transgenesis as a method to produce recombinant proteins made it possible to introduce several features that facilitated the protein purification process and increased the quantity, integrity, and preservation of this hetero-oligomeric membrane protein complex. First, the introduction of metabolic biotinylation as a purification tag (56) enabled a straightforward 1-day purification from a relatively small quantity of starting material, and the use of two consecutive affinity chromatographies ensured the presence of two key subunits in the complex. The shorter and faster purification protocol very likely promoted a better preservation of the macromolecule. Second, it was possible to insert a decoration tag (YFP) in one subunit for its recognition in the 3D reconstruction. Third, the use of an animal organ (skeletal muscle) where all of the subunits are normally expressed made it possible to obtain sufficient material for TEM. This rapid method of purification contributed to a 3D reconstruction of the DHPR that has more definition, enabling the docking of relevant atomic structures into the main body of the structure.
Novel Features in the 3D Reconstruction of the skeletal muscle l-type Channel Complex
The general outline of DHPR complex with a main body and a large appendage revealed by our studies is compatible with some earlier DHPR 3D reconstructions (31–33) but not with others (29, 30). Below we summarize and interpret the new features revealed in our 3D reconstruction for each subunit.
CaV1.1 Subunit
Our studies localize CaV1.1 to the 11 × 11 × 8 nm square prism in the main body, with the voltage-sensing modules (S1-S4) aligned with two outer corners of our 3D reconstruction and the II-III loop of Cav1.1 to the protuberance on the opposite side of the appendage. Because the II-III loop is downstream of S6 and thus adjacent to the intracellular helical bundle formed by the four S6 helices, our antibody labeling provides an additional unambiguous assignment of the orientation of the 3D reconstruction in the cell membrane, with the selectivity filter oriented toward the extracellular space as defined by the irregular appendage (Fig. 6) and the S6 helical bundle toward the intracellular space. The four CaV1.1 repeats are positioned clockwise as seen from the extracellular side (57). Finally, the assignment of the II-III loop to the cytosolic protuberance is consistent with the demonstrably important role of the II-III loop in EC coupling (11, 16, 58–64) and would allow the loop to extend toward the RyR1 in triad junctions of skeletal muscle.
α2-δ1 Subunit
Previous studies indicate that α2, on the extracellular side of the channel, corresponds to the hook-shaped structure (31). Our localization of the intracellular II-III loop on the opposite face of the main body further substantiates this assignment. We find that changes in the threshold for surface representation affect the shape of α2 more dramatically than that of the rest of the 3D structure, suggesting not only that α2 is a flexible domain, but also that its size is larger than what is represented by our 3D reconstruction. Indeed, although α2 represents 30% of the mass of DHPR, the hook-shaped structure represents only 13% of the mass of the DHPR 3D reconstruction. The δ1 subunit, disulfide-bonded to α2 (19), must cross the membrane near the point of attachment of the hook-shaped structure.
β1a Subunit
The β1a subunit has a known role in trafficking the α1S subunit to the membrane, is intracellular, and is important in EC coupling (9, 20). The core domain (8) has been crystallized for several β subunit isoforms, resulting in very similar atomic maps, which strongly suggests that this domain has a conserved and rigid structure. The core domain has also been cocrystallized bound to AID, its α helical target in Cav1.1, 22 residues after the inner helix (S6) of repeat I (8, 25). Because these intervening residues have a predicted α helix structure, the prevalent model has a physical separation between CaV1.1 and β1a (∼3 nm), situating the β subunit below CaV1.1 (24, 25). However, our 3D reconstruction is incompatible with this model. First, our insertion of YFP, which, along with GFP, constitutes a commonly used method for 3D sequence localization (65, 66), localizes the β subunit beside Cav1.1. Furthermore, the core of the β subunit fits very well when it is docked adjacent to CaV1.1 (Fig. 6), which is compatible with an earlier TEM antibody labeling study with intact DHPR (31) (Fig. 4, B and C) and with a DHPR α1-β subcomplex (33). The adjacent positioning keeps the β subunit close to the cytoplasmic domain of RyR1, consistent with its direct role in skeletal muscle EC coupling (9, 20–23). However, in this adjacent position, the AID is situated 5 nm away from the center of the pore of CaV1.1, which is difficult to conceive if the intervening peptide between S6 and the AID forms an α helix. This position is possible if the peptide is flexible at least in some section, or if in vivo, the interaction with the closely adjoining RyR1 could reorient the cytoplasmic loops of Cav1.1. It is also possible that with the membrane context being lost after its solubilization with detergent, the β1a subunit could relocate slightly to a position that was unavailable in vivo, although the clarity of the β subunit boundaries, characteristic of a non-mobile region, argue against this point. In this localization, the β subunit appears partially embedded in the membrane, a localization that was also implicit in a previous electron microscopy study (31), which is something unexpected, given the cytosolic location suggested for β1a (21, 24, 67–69). Given the recognizable Cav1.1 subunit, α2 subunit, and II-III loop in our 3D reconstruction, we have rotated the main body of the structure by 10° with respect to that previous 3D reconstruction, which we believe provides a more reasonable orientation and results in a more cytosolic β subunit (see Figs. 4 and 6). Although this represents a new interesting possibility, further testing with the full DHPR in the intact membrane is necessary to ascertain the exact location of the β subunit in vivo.
γ Subunit
The γ subunit has four transmembrane domains with intracellular C- and N termini, and interacts both with CaV1.1 (27) and the β subunit (70). After accommodating the atomic structure of a voltage-gated cation channel, the YFP and the core of the β subunit, there remains extra volume in the interphase of the CaV1.1/β subunits that could accommodate the γ subunit (asterisk in Fig. 6, top right panel).
In conclusion, to better ascertain the molecular organization of DHPR, we have expressed a DHPR with a recombinant β1a subunit using a transgenic animal both for an improved purification strategy as well as to provide a 3D decoration tag. Using TEM and 3D image reconstruction, we obtained a 19.1 Å-resolution 3D map of the full DHPR complex, which represents the highest resolution obtained for this complex to date. This reconstruction shows that the six transmembrane voltage-gated ion channel outline is recognizable in the full DHPR complex, and antibody labeling shows that the protuberance on the cytosolic side of CaV1.1 corresponds to the II-III loop. The atomic coordinates of the β subunit core structure can be docked in a location that indicates a closer association to the membrane than reported previously, thus defining a new relative positioning of the CaV1.1-β subunits.
Acknowledgments
We thank Dr. T. Walz for generous sharing of the TEM facility at the Department of Cell Biology at Harvard Medical School.
This work was supported, in whole or in part, by National Institutes of Health Grant 5K01AR054818 (to C. F. P.), 1R01GM081819 (to P. D. A. and M. S.), and 2R01AR05510 (to K. G. B. and P. D. A.). This work was also supported by the Brigham and Women's Hospital Biomedical Research Institute Fund to sustain Research Excellence (to M. S.) and by Virginia Commonwealth University startup funds (to M. S.).
- EC
- excitation-contraction
- DHPR
- dihydropyridine receptor
- AID
- α interaction domain
- TEM
- transmission electron microscopy
- 3D
- three-dimensional
- HSA
- human skeletal actin
- BAD
- biotin acceptor domain
- 2D
- two-dimensional
- GK
- guanylate kinase.
REFERENCES
- 1. Catterall W. A. (2011) Voltage-gated calcium channels. Cold Spring Harb. Perspect. Biol. 3, a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tanabe T., Beam K. G., Powell J. A., Numa S. (1988) Restoration of excitation-contraction coupling and slow calcium current in dysgenic muscle by dihydropyridine receptor complementary DNA. Nature 336, 134–139 [DOI] [PubMed] [Google Scholar]
- 3. Nakai J., Ogura T., Protasi F., Franzini-Armstrong C., Allen P. D., Beam K. G. (1997) Functional nonequality of the cardiac and skeletal ryanodine receptors. Proc. Natl. Acad. Sci. U.S.A. 94, 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Franzini-Armstrong C., Protasi F. (1997) Ryanodine receptors of striated muscles. A complex channel capable of multiple interactions. Physiol. Rev. 77, 699–729 [DOI] [PubMed] [Google Scholar]
- 5. Paolini C., Protasi F., Franzini-Armstrong C. (2004) The relative position of RyR feet and DHPR tetrads in skeletal muscle. J. Mol. Biol. 342, 145–153 [DOI] [PubMed] [Google Scholar]
- 6. Yu F. H., Yarov-Yarovoy V., Gutman G. A., Catterall W. A. (2005) Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacol. Rev. 57, 387–395 [DOI] [PubMed] [Google Scholar]
- 7. Catterall W. A., Perez-Reyes E., Snutch T. P., Striessnig J. (2005) International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol. Rev. 57, 411–425 [DOI] [PubMed] [Google Scholar]
- 8. Pragnell M., De Waard M., Mori Y., Tanabe T., Snutch T. P., Campbell K. P. (1994) Calcium channel β-subunit binds to a conserved motif in the I-II cytoplasmic linker of the α 1-subunit. Nature 368, 67–70 [DOI] [PubMed] [Google Scholar]
- 9. Cheng W., Altafaj X., Ronjat M., Coronado R. (2005) Interaction between the dihydropyridine receptor Ca2+ channel β-subunit and ryanodine receptor type 1 strengthens excitation-contraction coupling. Proc. Natl. Acad. Sci. U.S.A. 102, 19225–19230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Proenza C., O'Brien J., Nakai J., Mukherjee S., Allen P. D., Beam K. G. (2002) Identification of a region of RyR1 that participates in allosteric coupling with the α(1S) (Ca(V)1.1) II-III loop. J. Biol. Chem. 277, 6530–6535 [DOI] [PubMed] [Google Scholar]
- 11. Grabner M., Dirksen R. T., Suda N., Beam K. G. (1999) The II-III loop of the skeletal muscle dihydropyridine receptor is responsible for the bi-directional coupling with the ryanodine receptor. J. Biol. Chem. 274, 21913–21919 [DOI] [PubMed] [Google Scholar]
- 12. Leong P., MacLennan D. H. (1998) A 37-amino acid sequence in the skeletal muscle ryanodine receptor interacts with the cytoplasmic loop between domains II and III in the skeletal muscle dihydropyridine receptor. J. Biol. Chem. 273, 7791–7794 [DOI] [PubMed] [Google Scholar]
- 13. Casarotto M. G., Cui Y., Karunasekara Y., Harvey P. J., Norris N., Board P. G., Dulhunty A. F. (2006) Structural and functional characterization of interactions between the dihydropyridine receptor II-III loop and the ryanodine receptor. Clin. Exp. Pharmacol. Physiol. 33, 1114–1117 [DOI] [PubMed] [Google Scholar]
- 14. Dulhunty A. F. (2006) Excitation-contraction coupling from the 1950s into the new millennium. Clin. Exp. Pharmacol. Physiol. 33, 763–772 [DOI] [PubMed] [Google Scholar]
- 15. Tae H. S., Cui Y., Karunasekara Y., Board P. G., Dulhunty A. F., Casarotto M. G. (2011) Cyclization of the intrinsically disordered α1S dihydropyridine receptor II-III loop enhances secondary structure and in vitro function. J. Biol. Chem. 286, 22589–22599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leong P., MacLennan D. H. (1998) The cytoplasmic loops between domains II and III and domains III and IV in the skeletal muscle dihydropyridine receptor bind to a contiguous site in the skeletal muscle ryanodine receptor. J. Biol. Chem. 273, 29958–29964 [DOI] [PubMed] [Google Scholar]
- 17. Carbonneau L., Bhattacharya D., Sheridan D. C., Coronado R. (2005) Multiple loops of the dihydropyridine receptor pore subunit are required for full-scale excitation-contraction coupling in skeletal muscle. Biophys. J. 89, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bannister R. A., Grabner M., Beam K. G. (2008) The alpha(1S) III-IV loop influences 1,4-dihydropyridine receptor gating but is not directly involved in excitation-contraction coupling interactions with the type 1 ryanodine receptor. J. Biol. Chem. 283, 23217–23223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Jongh K. S., Warner C., Catterall W. A. (1990) Subunits of purified calcium channels. α 2 and Δ are encoded by the same gene. J. Biol. Chem. 265, 14738–14741 [PubMed] [Google Scholar]
- 20. Schredelseker J., Dayal A., Schwerte T., Franzini-Armstrong C., Grabner M. (2009) Proper restoration of excitation-contraction coupling in the dihydropyridine receptor β1-null zebrafish relaxed is an exclusive function of the β1a subunit. J. Biol. Chem. 284, 1242–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beurg M., Sukhareva M., Ahern C. A., Conklin M. W., Perez-Reyes E., Powers P. A., Gregg R. G., Coronado R. (1999) Differential regulation of skeletal muscle L-type Ca2+ current and excitation-contraction coupling by the dihydropyridine receptor β subunit. Biophys. J. 76, 1744–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gregg R. G., Messing A., Strube C., Beurg M., Moss R., Behan M., Sukhareva M., Haynes S., Powell J. A., Coronado R., Powers P. A. (1996) Absence of the β subunit (cchb1) of the skeletal muscle dihydropyridine receptor alters expression of the α 1 subunit and eliminates excitation-contraction coupling. Proc. Natl. Acad. Sci. U.S.A. 93, 13961–13966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sheridan D. C., Cheng W., Ahern C. A., Mortenson L., Alsammarae D., Vallejo P., Coronado R. (2003) Truncation of the carboxyl terminus of the dihydropyridine receptor β1a subunit promotes Ca2+ dependent excitation-contraction coupling in skeletal myotubes. Biophys. J. 84, 220–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Opatowsky Y., Chen C. C., Campbell K. P., Hirsch J. A. (2004) Structural analysis of the voltage-dependent calcium channel β subunit functional core and its complex with the α 1 interaction domain. Neuron 42, 387–399 [DOI] [PubMed] [Google Scholar]
- 25. Van Petegem F., Clark K. A., Chatelain F. C., Minor D. L., Jr. (2004) Structure of a complex between a voltage-gated calcium channel β-subunit and an α-subunit domain. Nature 429, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y. H., Li M. H., Zhang Y., He L. L., Yamada Y., Fitzmaurice A., Shen Y., Zhang H., Tong L., Yang J. (2004) Structural basis of the α1-β subunit interaction of voltage-gated Ca2+ channels. Nature 429, 675–680 [DOI] [PubMed] [Google Scholar]
- 27. Arikkath J., Chen C. C., Ahern C., Allamand V., Flanagan J. D., Coronado R., Gregg R. G., Campbell K. P. (2003) γ 1 Subunit interactions within the skeletal muscle L-type voltage-gated calcium channels. J. Biol. Chem. 278, 1212–1219 [DOI] [PubMed] [Google Scholar]
- 28. Andronache Z., Ursu D., Lehnert S., Freichel M., Flockerzi V., Melzer W. (2007) The auxiliary subunit γ 1 of the skeletal muscle L-type Ca2+ channel is an endogenous Ca2+ antagonist. Proc. Natl. Acad. Sci. U.S.A. 104, 17885–17890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Serysheva I. I., Ludtke S. J., Baker M. R., Chiu W., Hamilton S. L. (2002) Structure of the voltage-gated L-type Ca2+ channel by electron cryomicroscopy. Proc. Natl. Acad. Sci. U.S.A. 99, 10370–10375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang M. C., Velarde G., Ford R. C., Berrow N. S., Dolphin A. C., Kitmitto A. (2002) 3D structure of the skeletal muscle dihydropyridine receptor. J. Mol. Biol. 323, 85–98 [DOI] [PubMed] [Google Scholar]
- 31. Wolf M., Eberhart A., Glossmann H., Striessnig J., Grigorieff N. (2003) Visualization of the domain structure of an L-type Ca2+ channel using electron cryo-microscopy. J. Mol. Biol. 332, 171–182 [DOI] [PubMed] [Google Scholar]
- 32. Walsh C. P., Davies A., Butcher A. J., Dolphin A. C., Kitmitto A. (2009) Three-dimensional structure of CaV3.1. Comparison with the cardiac L-type voltage-gated calcium channel monomer architecture. J. Biol. Chem. 284, 22310–22321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murata K., Nishimura S., Kuniyasu A., Nakayama H. (2010) Three-dimensional structure of the α1-β complex in the skeletal muscle dihydropyridine receptor by single-particle electron microscopy. J. Electron. Microsc. (Tokyo) 59, 215–226 [DOI] [PubMed] [Google Scholar]
- 34. Lorenzon N. M., Haarmann C. S., Norris E. E., Papadopoulos S., Beam K. G. (2004) Metabolic biotinylation as a probe of supramolecular structure of the triad junction in skeletal muscle. J. Biol. Chem. 279, 44057–44064 [DOI] [PubMed] [Google Scholar]
- 35. Muscat G. E., Kedes L. (1987) Multiple 5′-flanking regions of the human α-skeletal actin gene synergistically modulate muscle-specific expression. Mol. Cell. Biol. 7, 4089–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haase H., Striessnig J., Holtzhauer M., Vetter R., Glossmann H. (1991) A rapid procedure for the purification of cardiac 1,4-dihydropyridine receptors from porcine heart. Eur. J. Pharmacol. 207, 51–59 [DOI] [PubMed] [Google Scholar]
- 37. Florio V., Striessnig J., Catterall W. A. (1992) Purification and reconstitution of skeletal muscle calcium channels. Methods Enzymol. 207, 529–546 [DOI] [PubMed] [Google Scholar]
- 38. Kugler G., Weiss R. G., Flucher B. E., Grabner M. (2004) Structural requirements of the dihydropyridine receptor α1S II-III loop for skeletal-type excitation-contraction coupling. J. Biol. Chem. 279, 4721–4728 [DOI] [PubMed] [Google Scholar]
- 39. Ohi M., Li Y., Cheng Y., Walz T. (2004) Negative staining and image classification. Powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ludtke S. J. (2010) 3-D structures of macromolecules using single-particle analysis in EMAN. Methods Mol. Biol. 673, 157–173 [DOI] [PubMed] [Google Scholar]
- 41. Shaikh T. R., Gao H., Baxter W. T., Asturias F. J., Boisset N., Leith A., Frank J. (2008) SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Samsó M., Radermacher M., Frank J., Koonce M. P. (1998) Structural characterization of a dynein motor domain. J. Mol. Biol. 276, 927–937 [DOI] [PubMed] [Google Scholar]
- 43. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 44. Brennan K. J., Hardeman E. C. (1993) Quantitative analysis of the human α-skeletal actin gene in transgenic mice. J. Biol. Chem. 268, 719–725 [PubMed] [Google Scholar]
- 45. Lorenzon N. M., Beam K. G. (2007) Accessibility of targeted DHPR sites to streptavidin and functional effects of binding on EC coupling. J. Gen. Physiol. 130, 379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sheridan D. C., Moua O., Lorenzon N. M., Beam K. G. (2012) Bimolecular fluorescence complementation and targeted biotinylation provide insight into the topology of the skeletal muscle Ca2+ channel β1a subunit. Channels 6, 26–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frank J. (2009) Single-particle reconstruction of biological macromolecules in electron microscopy. 30 years. Q. Rev. Biophys. 42, 139–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Glaeser R. M., Tong L., Kim S. H. (1989) Three-dimensional reconstructions from incomplete data. Interpretability of density maps at “atomic” resolution. Ultramicroscopy 27, 307–318 [DOI] [PubMed] [Google Scholar]
- 49. Quillin M. L., Matthews B. W. (2000) Accurate calculation of the density of proteins. Acta Crystallogr. D Biol. Crystallogr. 56, 791–794 [DOI] [PubMed] [Google Scholar]
- 50. Long S. B., Campbell E. B., Mackinnon R. (2005) Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 [DOI] [PubMed] [Google Scholar]
- 51. Payandeh J., Scheuer T., Zheng N., Catterall W. A. (2011) The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hanlon M. R., Berrow N. S., Dolphin A. C., Wallace B. A. (1999) Modelling of a voltage-dependent Ca2+ channel β subunit as a basis for understanding its functional properties. FEBS Lett. 445, 366–370 [DOI] [PubMed] [Google Scholar]
- 53. Opatowsky Y., Chomsky-Hecht O., Kang M. G., Campbell K. P., Hirsch J. A. (2003) The voltage-dependent calcium channel β subunit contains two stable interacting domains. J. Biol. Chem. 278, 52323–52332 [DOI] [PubMed] [Google Scholar]
- 54. He L. L., Zhang Y., Chen Y. H., Yamada Y., Yang J. (2007) Functional modularity of the β-subunit of voltage-gated Ca2+ channels. Biophys. J. 93, 834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Richards M. W., Leroy J., Pratt W. S., Dolphin A. C. (2007) The HOOK-domain between the SH3 and the GK domains of Cavβ subunits contains key determinants controlling calcium channel inactivation. Channels 1, 92–101 [DOI] [PubMed] [Google Scholar]
- 56. Consler T. G., Persson B. L., Jung H., Zen K. H., Jung K., Privé G. G., Verner G. E., Kaback H. R. (1993) Properties and purification of an active biotinylated lactose permease from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 90, 6934–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li R. A., Ennis I. L., French R. J., Dudley S. C., Jr., Tomaselli G. F., Marbán E. (2001) Clockwise domain arrangement of the sodium channel revealed by (mu)-conotoxin (GIIIA) docking orientation. J. Biol. Chem. 276, 11072–11077 [DOI] [PubMed] [Google Scholar]
- 58. Tanabe T., Beam K. G., Adams B. A., Niidome T., Numa S. (1990) Regions of the skeletal muscle dihydropyridine receptor critical for excitation-contraction coupling. Nature 346, 567–569 [DOI] [PubMed] [Google Scholar]
- 59. Altafaj X., Cheng W., Estève E., Urbani J., Grunwald D., Sabatier J. M., Coronado R., De Waard M., Ronjat M. (2005) Maurocalcine and domain A of the II-III loop of the dihydropyridine receptor Cav 1.1 subunit share common binding sites on the skeletal ryanodine receptor. J. Biol. Chem. 280, 4013–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lu X., Xu L., Meissner G. (1994) Activation of the skeletal muscle calcium release channel by a cytoplasmic loop of the dihydropyridine receptor. J. Biol. Chem. 269, 6511–6516 [PubMed] [Google Scholar]
- 61. el-Hayek R., Antoniu B., Wang J., Hamilton S. L., Ikemoto N. (1995) Identification of calcium release-triggering and blocking regions of the II-III loop of the skeletal muscle dihydropyridine receptor. J. Biol. Chem. 270, 22116–22118 [DOI] [PubMed] [Google Scholar]
- 62. Lu X., Xu L., Meissner G. (1995) Phosphorylation of dihydropyridine receptor II-III loop peptide regulates skeletal muscle calcium release channel function. Evidence for an essential role of the β-OH group of Ser-687. J. Biol. Chem. 270, 18459–18464 [DOI] [PubMed] [Google Scholar]
- 63. Nakai J., Tanabe T., Konno T., Adams B., Beam K. G. (1998) Localization in the II-III loop of the dihydropyridine receptor of a sequence critical for excitation-contraction coupling. J. Biol. Chem. 273, 24983–24986 [DOI] [PubMed] [Google Scholar]
- 64. Dulhunty A. F., Laver D. R., Gallant E. M., Casarotto M. G., Pace S. M., Curtis S. (1999) Activation and inhibition of skeletal RyR channels by a part of the skeletal DHPR II-III loop. Effects of DHPR Ser-687 and FKBP12. Biophys. J. 77, 189–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Meng X., Xiao B., Cai S., Huang X., Li F., Bolstad J., Trujillo R., Airey J., Chen S. R., Wagenknecht T., Liu Z. (2007) Three-dimensional localization of serine 2808, a phosphorylation site in cardiac ryanodine receptor. J. Biol. Chem. 282, 25929–25939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Conway J. F., Cockrell S. K., Copeland A. M., Newcomb W. W., Brown J. C., Homa F. L. (2010) Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J. Mol. Biol. 397, 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kobrinsky E., Kepplinger K. J., Yu A., Harry J. B., Kahr H., Romanin C., Abernethy D. R., Soldatov N. M. (2004) Voltage-gated rearrangements associated with differential β-subunit modulation of the L-type Ca(2+) channel inactivation. Biophys. J. 87, 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vendel A. C., Rithner C. D., Lyons B. A., Horne W. A. (2006) Solution structure of the N-terminal A domain of the human voltage-gated Ca2+channel β4a subunit. Protein Sci. 15, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dolphin A. C. (2003) β Subunits of voltage-gated calcium channels. J. Bioenerg. Biomembr. 35, 599–620 [DOI] [PubMed] [Google Scholar]
- 70. Yang L., Katchman A., Morrow J. P., Doshi D., Marx S. O. (2011) Cardiac L-type calcium channel (Cav1.2) associates with γ subunits. FASEB J. 25, 928–936 [DOI] [PMC free article] [PubMed] [Google Scholar]