Background: Cyclotides are plant defense peptides that act via membrane interactions.
Results: Cyclotides bind specifically to and progressively insert into phosphoethanolamine-containing lipid bilayers via a predominantly entropy-driven process.
Conclusion: Cyclotides insert deeply into lipid bilayers due to hydrophobic interactions with lipid tails and specific interactions with the phosphoethanolamine headgroup.
Significance: Our study broadens knowledge of how cyclotides interact with membranes.
Keywords: Lipid-binding Protein, Membrane Bilayer, Peptide Conformation, Peptide Interactions, Peptides
Abstract
Cyclotides are a family of plant-derived circular proteins with potential therapeutic applications arising from their remarkable stability, broad sequence diversity, and range of bioactivities. Their membrane-binding activity is believed to be a critical component of their mechanism of action. Using isothermal titration calorimetry, we studied the binding of the prototypical cyclotides kalata B1 and kalata B2 (and various mutants) to dodecylphosphocholine micelles and phosphoethanolamine-containing lipid bilayers. Although binding is predominantly an entropy-driven process, suggesting that hydrophobic forces contribute significantly to cyclotide-lipid complex formation, specific binding to the phosphoethanolamine-lipid headgroup is also required, which is evident from the enthalpic changes in the free energy of binding. In addition, using a combination of dissipative quartz crystal microbalance measurements and neutron reflectometry, we elucidated the process by which cyclotides interact with bilayer membranes. Initially, a small number of cyclotides bind to the membrane surface and then insert first into the outer membrane leaflet followed by penetration through the membrane and pore formation. At higher concentrations of cyclotides, destabilization of membranes occurs. Our results provide significant mechanistic insight into how cyclotides exert their bioactivities.
Introduction
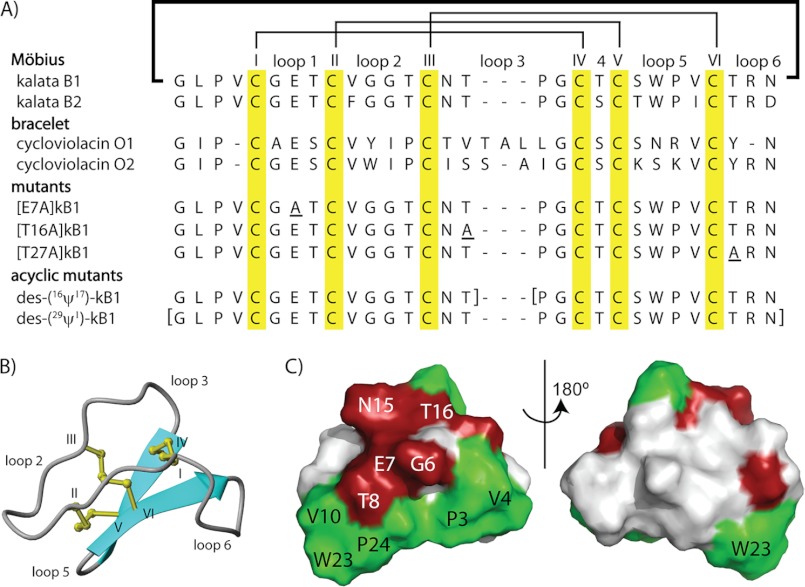
Cyclotides are a large and topologically unique family of plant defense proteins (1). Over 200 cyclotides have been discovered in plants of the Rubiaceae (coffee), Violaceae (violet), and Fabaceae (legume) families (2–9), and several ancestral cyclotide-like precursors have been identified in gramineous crop plants (10). Cyclotides are characterized by a cyclic cystine knot motif, which comprises a circular peptide backbone and a cystine knot core formed from six conserved Cys residues (1). The cyclic cystine knot motif underpins the remarkable stability of cyclotides in harsh thermal, chemical, and enzymatic conditions (11); this stability favors cyclotides as potential frameworks in peptide-based drug design (12). The sequence and compact structure of kalata B1 (kB1),5 the first characterized cyclotide, from Oldenlandia affinis (Rubiaceae) (13), is illustrated in Fig. 1, A and B. Cyclotides have six loops that project away from the cystine knot core and define the surface of cyclotides, which often contains a distinct hydrophobic patch (Fig. 1C).
FIGURE 1.
Sequences, structures, and surface properties of cyclotides. A, an alignment of cyclotide sequences, with the disulfide connectivity and loop nomenclature shown above the alignment. Cyclotides are divided into two main subfamilies, Möbius and bracelet, depending on the presence or absence, respectively, or a cis-Pro peptide bond in loop 5. The alignment also includes the sequences of cyclic and acyclic mutants used in this study. B, a ribbon representation of kalata B1 (Protein Data Bank entry 1NB1) with the loops and Cys residues labeled. C, a surface representation of kalata B1; residues that are important for cyclotide bioactivity are colored in red, and hydrophobic residues are colored in green.
Cyclotides have a diverse set of biological activities, including uterotonic (14), anti-HIV (15), antimicrobial (16), hemolytic (17), cancer cell toxicity (18), neurotensin antagonistic (19), molluscicidal (20), and nematocidal (21, 22) activities. Their native function has been believed to be plant defense-related since it was found that the growth of larvae of the major cotton pests, Helicoverpa punctigera and Helicoverpa armigera, was severely stunted when fed a diet that contained the cyclotide kB1 or kalata B2 (kB2) (23–25).
It is believed that the mechanism of action of cyclotides involves membrane interactions. This is supported by a recent study showing that cyclotides can bind to and disrupt the cell membranes of gut epithelial cells of cyclotide-fed H. armigera larvae (25). Cyclotides can bind directly to phospholipid bilayers, with specificity for phosphoethanolamine (PE)-type lipids (26–28), and binding involves a pore-forming mechanism (29). It is unlikely that cyclotide activity requires recognition by a chiral protein receptor because the all-d-enantiomer of kB1 is bioactive (30). These reports suggest that cyclotides primarily target the lipid components of cell membranes.
A molecular description of how cyclotides interact with lipids has only recently been developed. Spin label experiments using dodecyl phosphocholine (DPC) micelles identified a putative membrane binding surface that is composed mainly of hydrophobic residues and localized to one face of cyclotides (31, 32). NMR was further used to characterize a PE binding region on the surface of kB1 (27). However, despite these findings in DPC membrane mimics, little is known about the structure of cyclotides bound to a lipid bilayer. It has been suggested that this structure may involve cyclotide oligomers following observations that kB1 and kB2 can self-associate in solution (33); however, there is currently no evidence for a mode of action involving oligomerization.
In this study, we were interested in further understanding the molecular mechanism of action of cyclotides by focusing on the two prototypical cyclotides, kB1 and kB2. Using isothermal titration calorimetry (ITC), we defined the thermodynamics of cyclotide binding to DPC micelles and PE-containing phospholipid bilayers. We assessed how thermodynamic parameters of binding are affected by mutations in kB1, PE content of the phospholipid vesicles, temperature, and the presence of metal ions, to identify the factors that mediate cyclotide-lipid binding. Finally, to help define the structure of the cyclotide-containing membranes, we studied the interaction of both kB1 and kB2 with supported lipid bilayers using dissipative quartz crystal microbalance (QCM-D) and neutron reflectometry. The results have helped to elucidate the pathway by which cyclotides insert into and disrupt cell membranes.
EXPERIMENTAL PROCEDURES
Peptide Extraction, Synthesis, and Purification
Native kB1 and kB2 were extracted from O. affinis using a previously described protocol (24). Briefly, this involves an initial separation step where ground plant material is placed in 1:1 dichloromethane/methanol overnight, before repeated rounds of reverse phase C18 HPLC (1%/min gradient of 90% acetonitrile, 10% water, 0.05% trifluoroacetic acid). The purity of fractions was assessed using electron spray ionization-MS and analytical HPLC using the same gradient and solvent as described above. The masses of kB1 and kB2 were verified against the masses provided by CyBase, a database of circular proteins (4, 34).
t-Butoxycarbonyl-based solid phase peptide syntheses of cyclic ([E7A]kB1, [T16A]kB1, and [T27A]kB1) and acyclic (des-(16ψ17)-kB1 and des-(29ψ1)-kB1) variants of kB1 were carried out using a method described previously (35). The cyclic mutants were assembled on resin using a thioester linker that allowed subsequent cyclization by a modified form of native chemical ligation (36). Hydrogen fluoride cleavage was conducted on the deprotected resin using standard protocols (0 °C, 90 min, 90% HF, 8% p-cresol, 2% p-thiocresol). Crude cleavage products were purified by reverse phase C18 HPLC to give linear, reduced, C-terminally thioester capped peptides. The cyclic mutants were cyclized, and both cyclic and acyclic mutants were oxidized overnight in 0.1 m NH4HCO3 at pH 8.5 with 50% 2-propanol and purified as above.
NMR Spectroscopy
NMR was used to check the three-dimensional folds of the mutants. Synthetic mutants of kB1, des-(16ψ17)-kB1 and des-(29ψ1)-kB1, were dissolved in 90% H2O, 10% D2O, and the pH was adjusted using concentrated NaOH and HCl. Bruker ARX 500 or 600 NMR spectrometers were used to record spectra at 303K.
Materials for Membrane Binding Experiments
DPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolimine (POPE), 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine (DMPE), chain-deuterated 1,2-d54-dimyristoyl-sn-glycero-3-phosphoethanolamine (d54-DMPE), and deuterated 1,2 d54-dimyristoyl-sn-glycero-3-phosphocholine (d54-DMPC) were purchased from Avanti Polar Lipids (Alabaster, AL). Sphingomyelin, Tris-HCl, and D2O (>99%) were purchased from Sigma-Aldrich.
Preparation of Vesicles
A defined amount of lipid in chloroform was dried under a nitrogen stream and left under high vacuum overnight to remove traces of solvent. A defined amount of buffer (10 mm Tris-HCl, 100 mm NaCl, pH 7.4) was then added to the lipid, and the dispersion was extensively vortexed. The lipid dispersion was placed in a water bath sonicator for 30 min at 40 °C.
Isothermal Titration Calorimetry
The screening of kB1 and kB2 binding to DPC micelles was performed using a VP ITC instrument (Microcal, Northampton, MA). Samples were degassed in a ThermoVac (Microcal) before use. Typically, 10 injections of 15 μl from a 300-μl syringe containing 15 mm lipid mixtures were injected into a ∼1.4-ml reaction cell loaded with 250 μm cyclotide while stirring at a speed of 300 rpm. Unless noted otherwise, all measurements were made at 30 °C in freshly prepared buffer solutions: 10 mm Tris-HCl (pH 7.4), 100 mm NaCl. Lipid vesicles were suspended in the same buffer as the peptide. As a control, lipid vesicles were injected into pure buffer containing no peptide.
All other ITC binding experiments were conducted using an iTC200 instrument (Microcal), equipped with a 70-μl syringe and a ∼300-μl reaction cell. For these binding experiments, typically 1.9 μl of 3 mm lipid was injected into 100 μm cyclotide solution every 3 min for 20 injections while stirring at 1000 rpm.
The resulting titration data were analyzed and fitted to a one set of sites model using the Origin for ITC software package supplied by Microcal to obtain the number of bound ligand molecules (N), the association constant (Ka), and change in enthalpy (ΔH). Changes in entropy (ΔS) and Gibbs free energy (ΔG) were calculated from the following equation.
Quartz Crystal Microbalance with Dissipation
QCM-D measurements were carried out using a Qsense E4 instrument (Qsense Ltd.) on SiO2-coated quartz crystal sensors at 45 °C. Lipid bilayers were formed by flowing a 0.5 mg/ml solution of 3:1 DMPC/DMPE in 100 mm aqueous NaCl over the sensor surface for 10 min at 100 μl min−1 before a 20–30-min incubation and rinsing off with 100 mm aqueous NaCl at 100 μl min−1 until a stable baseline was reached. The peptide solutions (900 μl) at concentrations of 5–100 μm were introduced at 100 μl min−1, and incubated over the membrane until no more changes in frequency or dissipation were observed. The membrane surface coverage (ng cm−2) and the total mass of peptide, membrane, and associated water were estimated using the Saurbrey calculation. Above 50 μm kB1 and 25 μm kB2, the peptides led to very large dissipation values, indicating a highly hydrated and extended structure beyond the thickness range within which viscoelastic modeling can be used to estimate the membrane thickness and mass.
Supported Membrane Formation for Neutron Reflectometry
The support surfaces (single crystal silicon 111 orientation) were cleaned by a 1:4:5 solution of H2O2/H2SO4/H2O at 80 °C followed by UV ozonolysis for 5–10 min before the experiment. This treatment generates a highly hydrophilic native SiO2 layer of 7–15-Å thickness and 5–10-Å roughness.
Phospholipid bilayers were formed by adsorption of sonicated small unilamellar vesicles of d54-DMPE and d54-DMPC (∼0.5 mg/ml) in 100 mm NaCl to a silicon surface preheated to 45 °C with an incubation period of 20–30 min prior to rinsing off the excess vesicles with 100 mm NaCl. All peptide adsorption experiments were carried out at 45 °C because d54-DMPE and d54-DMPC are fully miscible only above 40 °C (37).
Neutron Reflectometry
Specular neutron reflection was measured on the CRISP reflectometer (38) at the ISIS Neutron and Muon Facility (Rutherford Appleton Laboratory, Didcot, UK), using neutron wavelengths λ of 0.5–6 Å to record reflectivity profiles in the range 0.01 < Q < 0.5 Å−1, where Q = 4πsinθ/λ is the momentum transfer vector of the neutrons in the direction (z) perpendicular to the membrane-water interface. Under these conditions, a typical measurement at three incident angles (0.35°, 0.8°, and 1.8°) to obtain the full reflectivity profile required 75 min. The phospholipid bilayers were formed in the neutron reflectometer sample chamber after measuring the structure of the supporting Si-SiO2 surface in D2O or H2O. Each lipid membrane structure was also measured before exposure to the peptides. Peptide solutions (5 ml) in 10 mm Tris, pH 7.4, 100 mm NaCl at concentrations of 5–100 μm were injected over the lipid membrane consecutively, and changes in membrane structure were recorded. Each contrast (D2O or H2O) was recorded on a separate membrane structure, but the results were fitted simultaneously, because the silicon support and membrane structures were found to be identical.
Details of the analysis of supported lipid membrane structure (39) and interaction with soluble proteins (40) using time-of-flight neutron reflection have been described previously. Briefly, the neutron scattering length density profile ρ(z) of a lipid bilayer can be divided into separate headgroup and acyl chain regions due to their chemical differences and water penetration. Reflectivity analysis is based on modeling the thickness, scattering length density, lipid volume fraction, and roughness of three layers corresponding to the lipid headgroups and the acyl chain region. In membranes containing lipids (L), water (W), and a peptide (P), the scattering length density ρlayer is the sum of the molecular scattering length densities ρi weighted by the volume fractions ϕi of each component and water (W).
Thus, when there are significant differences in the molecular scattering length densities of lipid, peptide, and water, their individual volume fractions can be computed from the fitted scattering length density of the layer. In the present work, all experiments were done in both D2O and H2O subphases, in order to resolve the layer composition.
The Motofit program (41) was used for optical matrix modeling (42) to calculate specular reflectivity from model structures. The scattering length densities of the phospholipids were calculated from volume fractions of the lipid components obtained from molecular dynamics simulations (43), and for the peptides they were calculated from the sequences and amino acid volumes (44) listed in Table 1. The errors in the structural parameters for each sublayer were derived from the maximum acceptable variation in the fitted thickness and lipid volume fraction that allowed a fit to be maintained, subject to a constant molecular area constraint required to maintain a planar bilayer geometry. However, the errors in independent parameters of the model sublayers are coupled in such a way that the overall errors in membrane volume fraction and thickness are ±10% and ±2Å, respectively.
TABLE 1.
Molecular volumes (V) and neutron scattering length densities (ρ) used in fitting data and calculating bilayer structure
| Compound | V | ρ (in D2O)a | ρ (in H2O)a |
|---|---|---|---|
| Å3 | 10−6 Å−2 | 10−6 Å−2 | |
| d54-DMPC | |||
| Headgroups | 326b | 1.8 | 1.8 |
| Chains | 777.5b | 6.8 | 6.8 |
| d54-DMPE | |||
| Headgroups | 243b | 2.8 | 1.8 |
| Chains | 777.5b | 6.8 | 6.8 |
| Kalata B1 | 3313c | 3.7 | 2.2 |
| Kalata B2 | 3313c | 3.7 | 2.2 |
| D2O | 30 | 6.35 | |
| H2O | 30 | −0.56 | |
a Calculated from the molecular component volumes and coherent nuclear scattering lengths.
b From molecular dynamics simulations (43).
RESULTS
Cyclotide Binding to Dodecylphosphocholine Micelles Is Predominantly Entropy-driven
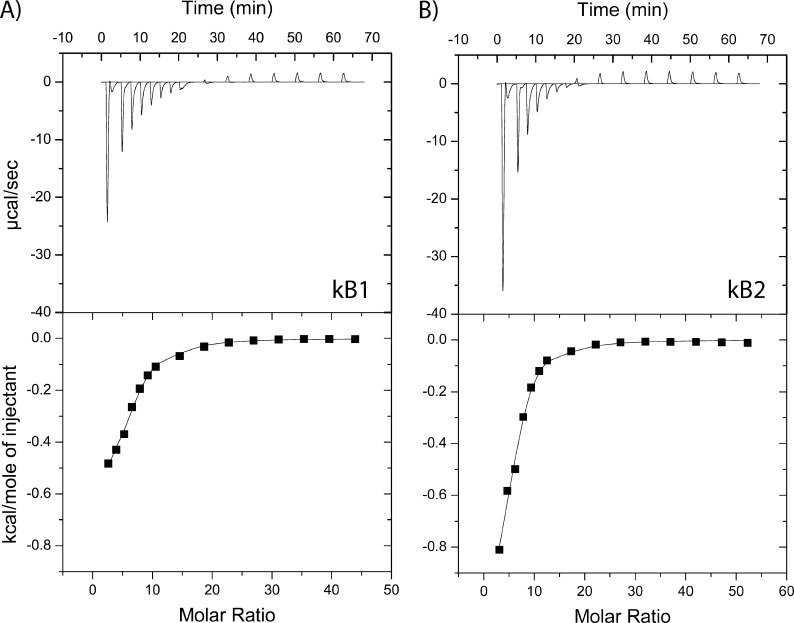
We were interested in characterizing the mechanism of membrane interaction of cyclotides and started by using the simplest membrane-mimic system of DPC micelles, which are monolayer aggregates that have been used to study the membrane binding affinity of cyclotides using NMR (31, 32). Using ITC, a concentrated solution of DPC micelles was titrated into a buffered solution containing either kB1 or kB2. As Fig. 2 shows, both kB1 and kB2 bind to DPC micelles, with saturation of binding after a ∼20:1 molar ratio of DPC/cyclotide, which is in agreement with NMR studies (32). Table 2 shows the results of a thermodynamic analysis of the titration curves, revealing that binding of both kB1 and kB2 to DPC micelles is predominantly entropy-driven. Compared with kB1, the binding affinity of kB2 to DPC micelles is 3–4 times higher, which can be partly attributed to a larger entropy contribution in the binding of kB2 to DPC micelles. We next studied the interaction of cyclotides with lipid bilayers, which are considered to be more physiologically relevant than micelle-based membrane mimics.
FIGURE 2.
Binding of kB1 and kB2 to dodecylphosphocholine (DPC) micelles. Binding isotherms were obtained using isothermal titration calorimetry to measure the titration of DPC micelles (170 mm) into a cell containing 0.42 mm cyclotide at 30 °C (with injections of 1 × 10, 6 × 5, and 8 × 15 μl of titrant). A, isotherm for kB1; B, isotherm for kB2. The integrated data, where the continuous line represents the best curve fit to the experimental data from Microcal Origin, is also shown.
TABLE 2.
Thermodynamic parameters for cyclotide binding to different membrane systems
| System | n | Ka | ΔH | ΔG | ΔS |
|---|---|---|---|---|---|
| 103m−1 | kcal/mol | kcal/mol | cal/mol/K | ||
| DPC | |||||
| kB1 | 5.19 ± 1.03 | 1.70 ± 0.14 | −0.86 ± 0.02 | −6.90 ± 0.05 | 19.93 ± 0.09 |
| kB2 | 6.29 ± 0.82 | 5.85 ± 0.35 | −0.70 ± 0.07 | −7.64 ± 0.04 | 22.91 ± 0.10 |
| PC/PE (5:2) | |||||
| kB1 | 1.94 ± 0.50 | 163.00 ± 37.75 | −1.39 ± 0.16 | −9.63 ± 0.14 | 27.20 ± 0.44 |
| T27A | 1.60 ± 0.13 | 68.90 ± 24.18 | −1.20 ± 0.09 | −9.11 ± 0.22 | 26.11 ± 1.01 |
| kB2 | 2.43 ± 0.33 | 246.50 ± 136.05 | −2.11 ± 0.09 | −9.82 ± 0.33 | 25.47 ± 1.16 |
| PC/PE (6:1) | |||||
| kB2 | 4.23 ± 0.29 | 48.00 ± 8.49 | −1.33 ± 0.11 | −8.90 ± 0.11 | 24.98 ± 0.71 |
| PC/PE (4:3) | |||||
| kB2 | 1.54 ± 0.07 | 416.00 ± 62.23 | −3.11 ± 0.09 | −10.20 ± 0.09 | 23.40 ± 0.01 |
| PC/PE (5:2) at 20 °C | |||||
| kB2 | 2.13 ± 0.23 | 172.00 ± 53.74 | −1.26 ± 0.02 | −9.34 ± 0.18 | 27.59 ± 0.71 |
| PC/PE (5:2) at 40 °C | |||||
| kB2 | 2.15 ± 0.06 | 349.00 ± 12.73 | −2.95 ± 0.02 | −10.44 ± 0.02 | 23.91 ± 0.02 |
Cyclotide Binding to Vesicles Is Entropy-driven and Modulated by PE Content
Studies using surface plasmon resonance have shown that cyclotides bind selectively to PE-containing lipid bilayers (26, 27). Using ITC, we confirmed that both kB1 and kB2 bind strongly to lipid bilayers composed of POPC/POPE (3:1), as shown in Figs. 3A and 4A. In agreement with surface plasmon resonance studies (27), specific binding was not observed when POPC-only vesicles were used (data not shown).
FIGURE 3.
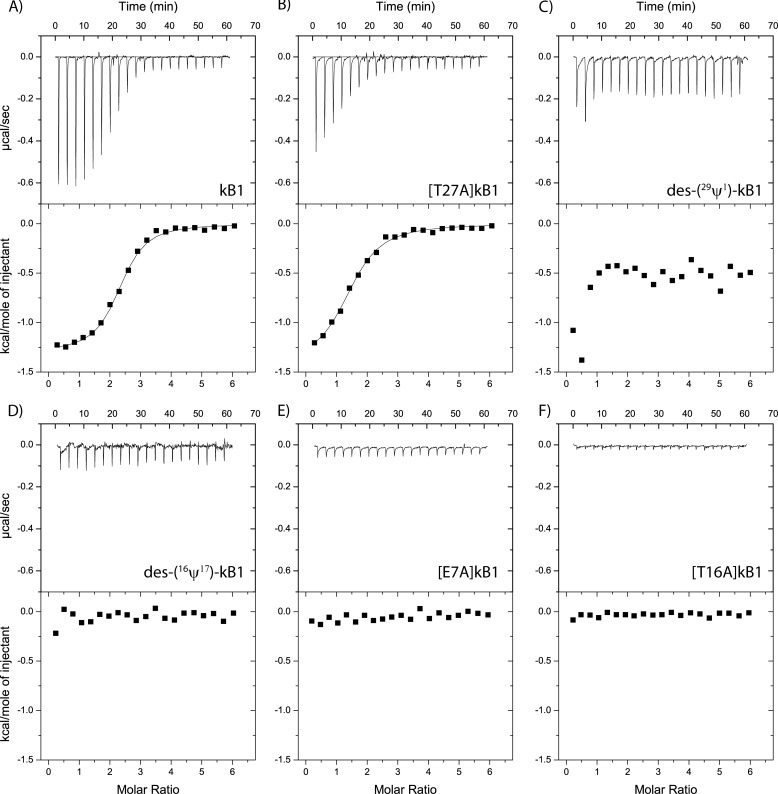
Binding of kB1 mutants to vesicles. Binding isotherms obtained by using ITC to measure the titration of 5:2 POPC/POPE vesicles (3 mm) into a cell containing 0.1 mm cyclotide at 30 °C (with 20 injections of 1.9 μl). Aside from native kB1 (A), several mutants were tested, including [T27A]kB1 (B), kB1-6 (C), kB1-3 (D), [E7A]kB1 (E), and [T16A]kB1 (F).
FIGURE 4.
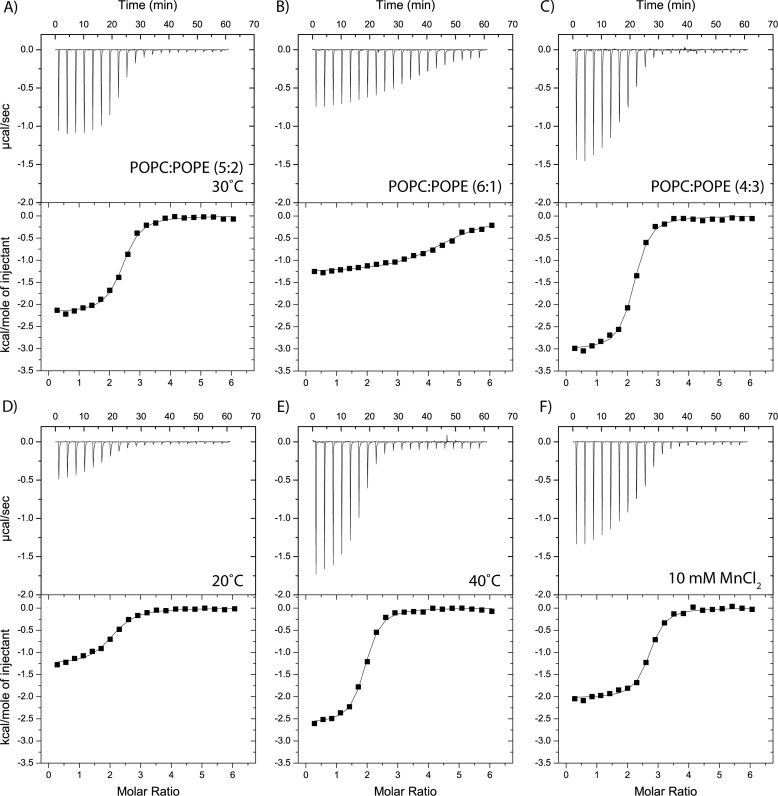
Effects of sample conditions on membrane binding of kalata B2. Binding isotherms obtained by using isothermal titration calorimetry to measure the titration of 3 mm vesicles into a cell containing 0.1 mm (with 20 injections of 1.9 μl). A, standard condition using vesicles composed of 5:2 POPC/POPE at 30 °C. B and C, the effect on membrane binding of variations in POPE content, keeping the temperature constant at 30 °C. D and E, the effect on membrane binding of temperature variation, keeping the membrane composition constant at 5:2 POPC/POPE. F, binding isotherm when 10 mm MnCl2 is included into the initial vesicle and cyclotide solutions.
Using ITC, we further investigated how cyclotides interact with membranes by varying the sample conditions. Because it has been established that cyclotides can bind specifically to PE lipids (28), we were interested in understanding the effect of variations in PE lipid concentration on cyclotide membrane binding. Fig. 4, A–C, and Table 2 show that an increased concentration of PE in the vesicle correlates with stronger binding affinity for kB2.
We next investigated the effect of temperature on cyclotide membrane binding. Fig. 4, A, D, and E, shows that when the temperature was increased, the binding affinity of kB2 became stronger. More specifically, at 20 °C, kB2 bound phospholipid vesicles with a dissociation constant of 5.81 μm, and increasing the temperature to 40 °C gave a dissociation constant of 2.87 μm. From the binding data at different temperatures, the heat capacity (ΔCp) of kB2 was calculated to be −84.5 cal K−1 mol−1.
Based on NMR experiments showing that cyclotides can bind the divalent cation Mn2+, it has been proposed that cyclotides have a metal binding site (45) and that metal binding might facilitate the binding of cyclotides to membranes (32). As Fig. 4F shows, the presence of Mn2+ ions did not influence cyclotide membrane binding affinity, suggesting that although cyclotides may bind metal ions, binding is not important for their bioactivity.
Cyclotide Bioactivity Is Correlated to Membrane Binding
Having quantitatively confirmed that cyclotides can bind directly to lipid membranes, we were interested in determining the biological significance of the membrane binding affinity. To achieve this, a selection of cyclic mutants ([E7A]kB1, [T16A]kB1, and [T27A]kB1) and acyclic mutants (des-(16ψ17)-kB1 and des-(29ψ1)-kB1) of kB1, all of which have previously been tested in various biological assays (46, 47), were synthesized and tested for their membrane binding ability. As Fig. 3 shows, the mutants with no biological activity (i.e. [E7A]kB1, [T16A]kB1, des-(16ψ17)-kB1, and des-(29ψ1)-kB1) demonstrate weak or no affinity for lipid membranes. On the other hand, the biologically active mutant [T27A]kB1 retains the ability to bind lipid vesicles. The clear correlation between the affinity of the cyclotide for lipid vesicles and their observed bioactivity strongly suggests that membrane binding plays a key role in the mechanism of action of cyclotides.
Cyclotide Binding to Lipid Bilayers Leads to the Formation of a Highly Hydrated Bilayer Structure
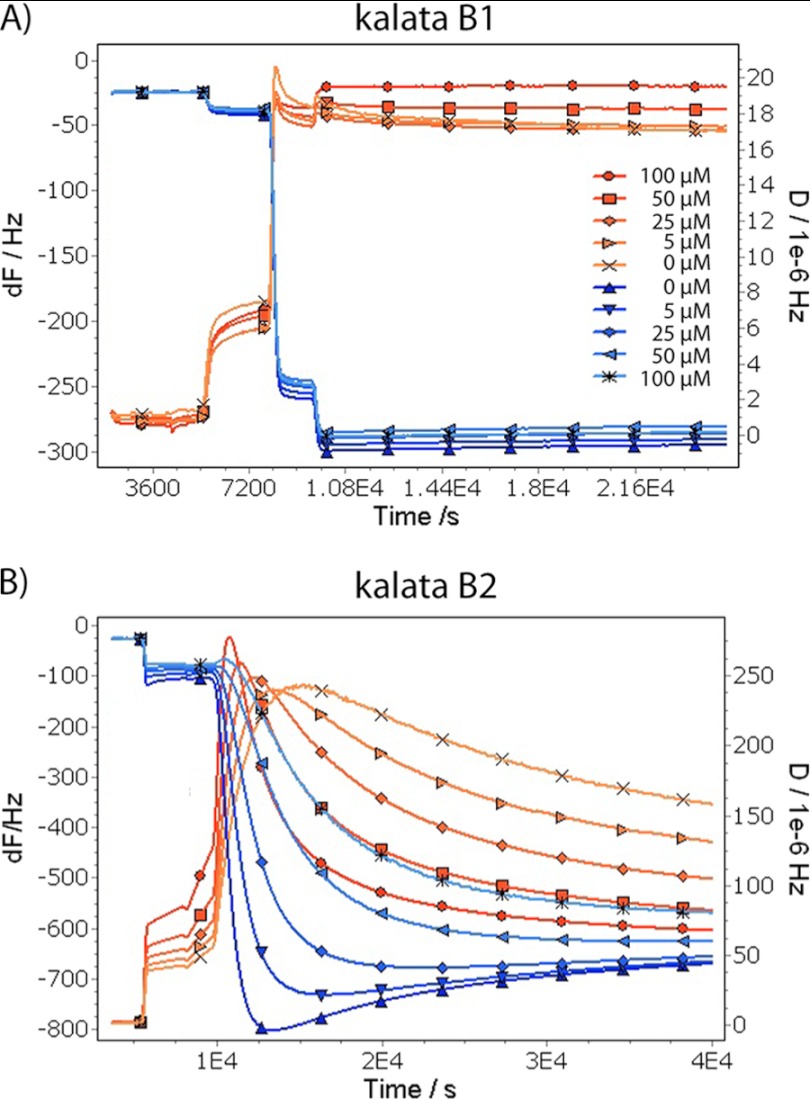
We measured the changes to a lipid bilayer structure upon the addition of cyclotides using a quartz crystal microbalance. The changes in frequency and dissipation (for the 5th to 13th overtones) caused by kB1 and kB2 are shown in Fig. 5, A and B, respectively. The addition of kB1 does not lead to a measurable frequency or dissipation change at 5 μm concentration, but at 25 μm, both a frequency decrease (corresponding to increased membrane mass) and a dissipation increase are observed, showing that the interaction of the peptide leads to formation of a highly hydrated structure, which would be consistent with pore formation. At higher kB1 concentrations and at all concentrations of kB2, the very large frequency and dissipation changes indicate the formation of a very extended structure, which we interpret to correspond to solubilized membrane disks loosely associated with the surface. In either case, it is not possible to calculate the mass of peptide associated or the thickness of the membranes reliably because the dissipation changes indicate that the sample viscoelasticity is too high for modeling.
FIGURE 5.
Quartz crystal microbalance measurements of cyclotide binding to membranes. Quartz crystal microbalance measurements of cyclotide binding to 3:1 DMPC/DMPE supported membranes at 25 °C, pH 6.95: kalata B1 (A) and kalata B2 (B). The frequency (F) and dissipation (D) curves are labeled according to the overtones measured.
The Mechanism of Action of Cyclotides Involves Insertion into the Membrane
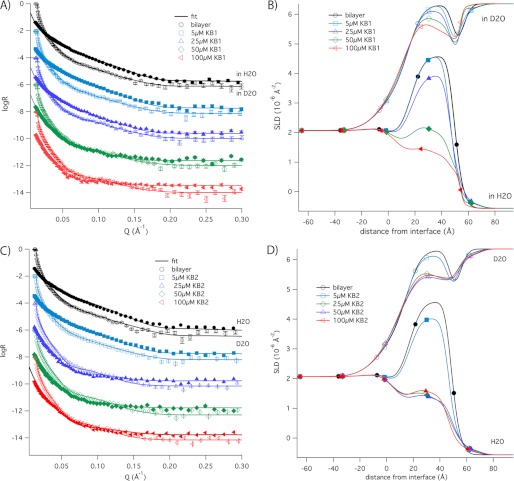
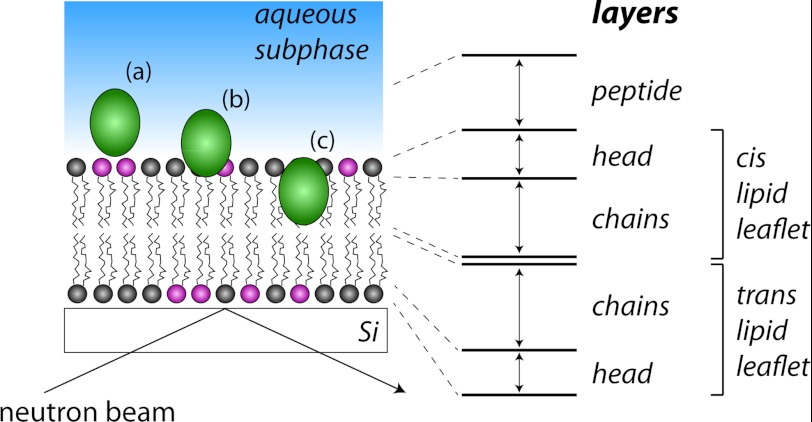
We used neutron reflectometry to investigate the molecular structure of DMPC and DMPE lipid bilayers during interaction with cyclotides and in particular to sense the depth of insertion of cyclotides into membrane bilayers as a function of cyclotide concentration. Reflectivity profiles were measured before lipid injection, after rinsing off excess vesicles, and after consecutive injections of kB1 and kB2 at concentrations of 5, 25, 50, and 100 μm. These titration experiments were initially measured in H2O and then repeated in D2O; although individual samples were used for each contrast, both the silicon oxide and lipid bilayer structures were found to be identical in both contrasts, allowing us to determine peptide and solvent penetration in the bilayer using both data sets. Fig. 6 shows the reflectivity profiles measured after injection of kB1 (A; in D2O and in H2O) and kB2 (C; in D2O and in H2O), with the scattering length density profiles corresponding to the best fits shown in B and D, respectively. The best fit variables (thickness, scattering length density, and lipid volume fraction) as well as the membrane structural parameters calculated from them (average area per lipid molecule A, lipid volume fraction φ, membrane mass per unit area Γ, and volume fraction of peptide) are shown in Tables 3 and 4 for kB1 and kB2, respectively. Fig. 7 provides a schematic illustration of the different layers used to fit the data.
FIGURE 6.
Neutron reflectivity profiles of cyclotide binding to membranes. Neutron reflectivity profiles were recorded for the substrate only, after deposition of the lipid bilayer, and after the addition of 5–100 μm of cyclotide. The reflectivity profiles for kalata B1 are shown in D2O and in H2O (A) with the scattering length density profiles (B). The reflectivity profiles for kalata B2 are shown in D2O and in H2O (C) with the scattering length density profiles (D).
TABLE 3.
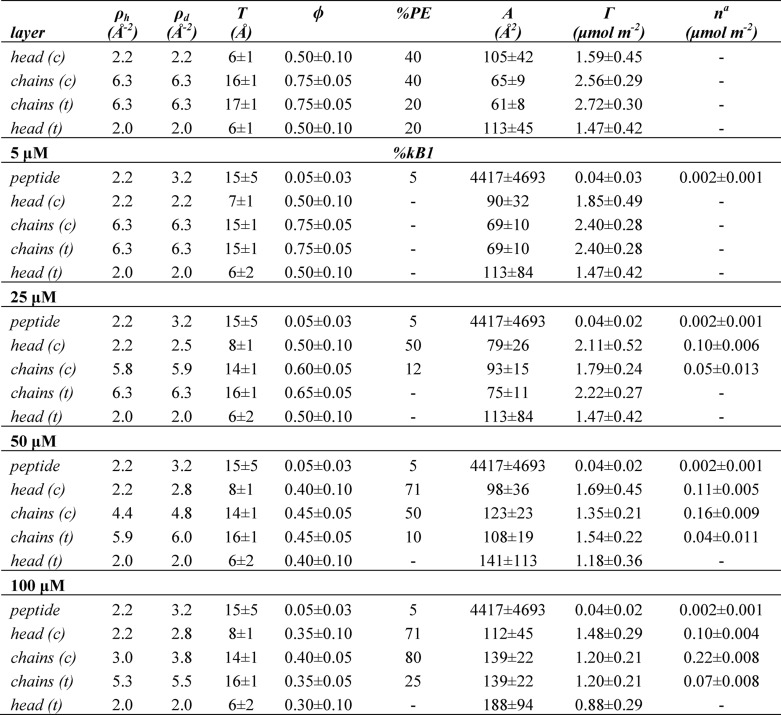
Neuron reflectivity results for kalata B1, including fitted parameters as well as derived structural parameters
T, thickness; ρh, scattering length density in H2O; ρd, scattering length density in D2O; ϕ, volume fraction; %PE, volume percentage of DMPE; %kB1, volume percentage of kB1; A, area available per molecule; Γ, lipid surface coverage; n, number of peptide molecules per unit area; c, cis; t, trans. The errors in A and Γ for each layer were directly propagated from the fitting uncertainty in T and ϕ, whereas the uncertainties in these parameters are coupled, with the overall uncertainty being 10% in each value.
a The total peptide bound is the sum of the n value for all layers (i.e. 0 μm is 0 μmol m−2, 5 μm is 0.002 μmol m−2, 25 μm is 0.152 μmol m−2, 50 μm is 0.312 μmol m−2, and 100 μm is 0.392 μmol m−2).
TABLE 4.
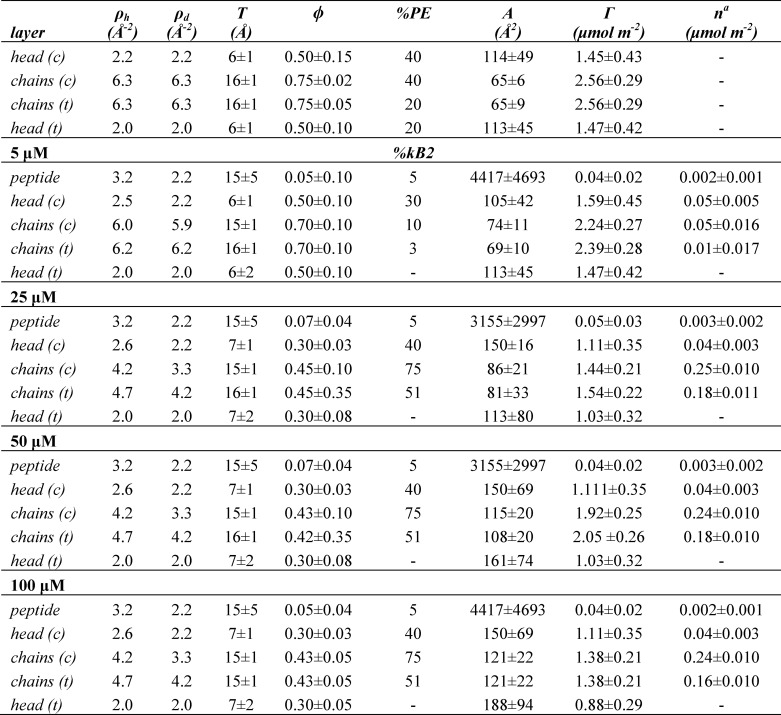
Neutron reflectivity results for kalata B2, including fitted parameters as well as derived structural parameters
T, thickness; ρh, scattering length density in H2O; ρd, scattering length density in D2O; ϕ, volume fraction; %PE, volume percentage of DMPE; %kB2, volume percentage of kB2; A, area available per molecule; Γ, lipid surface coverage; n, number of peptide molecules per unit area; c, cis; t, trans. The errors in A and Γ for each layer were directly propagated from the fitting uncertainty in T and ϕ, whereas the uncertainties in these parameters are coupled, with the overall uncertainty being 10% in each value.
a The total peptide bound is the sum of the n value for all layers (i.e. 0 μm is 0 μmol m−2, 5 μm is 0.112 μmol m−2, 25 μm is 0.473 μmol m−2, 50 μm is 0.463 μmol m−2, and 100 μm is 0.442 μmol m−2).
FIGURE 7.
Schematic illustration of the principle of neutron reflectometry. Targeting of the neutron beam at a shallow angle to the membrane plane allows localization of peptides in the membrane. The definitions of the various layers used in the fitting procedure that generated the fits in Tables 2 and 3 are shown. The silicon support with a thin SiO2 surface layer is represented at the bottom; in the real experiment, the geometry is inverted so that the water subphase is held in a Teflon flow cell below the silicon support. A schematic cyclotide molecule is shown located in the peptide layer (a), the peptide and headgroup layers (b), and the headgroup and chain layers of the upper lipid leaflet (c).
When deposited onto a SiO2 surface, the lipid bilayer had a high surface coverage (φ = 0.75), with a total thickness of 46 ± 2 Å and an area per molecule of 65 ± 8 Å. The scattering length density (ρ) of the lipid chain region (6.3 ± 0.2 × 10−6 Å−2) was somewhat lower than expected for perdeuterated lipid chains and indicated 93% deuteration in the commercial lipids used. The low lipid volume fractions found for the lipid headgroups are an indication of the poorer packing due to the small size of the DMPE headgroup, but the three protons on the PE amine group also exchange with the D2O subphase, giving the layer a higher apparent ρ (the ρ for deuterated PE would be 2.8 × 10−6 Å−2) than for DMPC alone (the ρ of a PC headgroup would be 1.8 × 10−6 Å−2). We observed a slightly higher scattering length density in the headgroup layer that was exposed to the subphase compared with the layer that was in direct contact with the SiO2. This may indicate a higher fraction of d54-DMPE in the inner membrane layer.
After deposition of the bilayer, increasing concentrations of either kB1 or kB2 were injected into the sample cell, and, as shown in Fig. 6, the profiles changed after each injection. Fitting the data using the lipid bilayer structure as a starting point showed that at a concentration of 5 μm, kB1 initially sits on the surface of the membrane but begins to penetrate into the lipid bilayer at higher concentrations, first only into the upper leaflet and then through the bilayer. Eventually, at 100 μm, kB1 solubilized a large fraction of the lipid bilayer, leaving a mixed layer of peptide and lipid with ∼30% surface coverage. kB2 behaved in a similar way, except it interacted with the bilayer much more strongly. Even at a concentration of 5 μm, kB2 had penetrated the lipid bilayer and partially solubilized it.
The total amount of peptide bound at each concentration is shown in Tables 3 and 4. The stronger membrane binding affinity of kB2 is consistent with the ITC results, confirming that kB1 has a weaker affinity for lipids than kB2. Also, the amounts of bound peptide agree well with the QCM-D data, which show that the dissipation change is 10 times higher for kB2 than kB1 at 25 μm.
The neutron reflectivity data report on the average composition of the membrane at different depths but do not provide information on the lateral distribution of lipid, solvent, and peptides. However, because the QCM-D data suggest pore formation at lower peptide concentrations, we can infer that pore formation in the membrane is likely to accompany the peptide penetration and decrease in membrane volume fraction seen in the neutron data.
The average number of peptide molecules per unit area is shown for each membrane layer in Tables 3 and 4. Although more than one cyclotide molecule is required to span the lipid bilayer to form pores, there is no clear correlation between the number of peptide molecules in the different layers. Therefore, we propose that kB1 and kB2 do not form well defined oligomers when inserted into the membrane; however, they may still form oligomers when initially bound to the membrane surface.
DISCUSSION
Cyclotides are a topologically unique family of proteins with exciting therapeutic promise (48–52). It is believed that their mechanism of action involves membrane interaction, and we confirmed in this study, using a selection of kB1 mutants, a correlation between bioactivity and membrane affinity. Given the importance of membrane interactions for cyclotide activity, we were interested in characterizing the molecular basis of the interactions using a combination of ITC, QCM-D, and neutron reflectometry. The results clearly show that cyclotides initially bind to the membrane surface and then progressively insert deeper into the lipid bilayer as their concentration increases. This study of the molecular behavior of cyclotides in a phospholipid bilayer environment provides new insights into the pore formation process by, for the first time, unequivocally demonstrating the ability of cyclotides to deeply penetrate membranes. The approach used here is broadly applicable to a wide range of peptides that interact with membranes, including antimicrobial peptides and pore-forming toxins.
We focused on two prototypical cyclotides, kB1 and kB2, which have been well studied in terms of their structure and bioactivity. These cyclotides have similar structures as well as modes of binding to DPC micelles (31, 32), with the main difference between them being five conservative sequence substitutions (Fig. 1A), which result in slightly different surface profiles. Using ITC, we observed similar binding isotherms to DPC micelles and lipid bilayers for kB1 and kB2, with kB2 displaying stronger affinity than kB1 for these membrane mimics. The results suggest that kB1 and kB2 have similar mechanisms of membrane interaction; differences in their biochemical behavior are thus related to specific amino acid differences rather than to different binding modes.
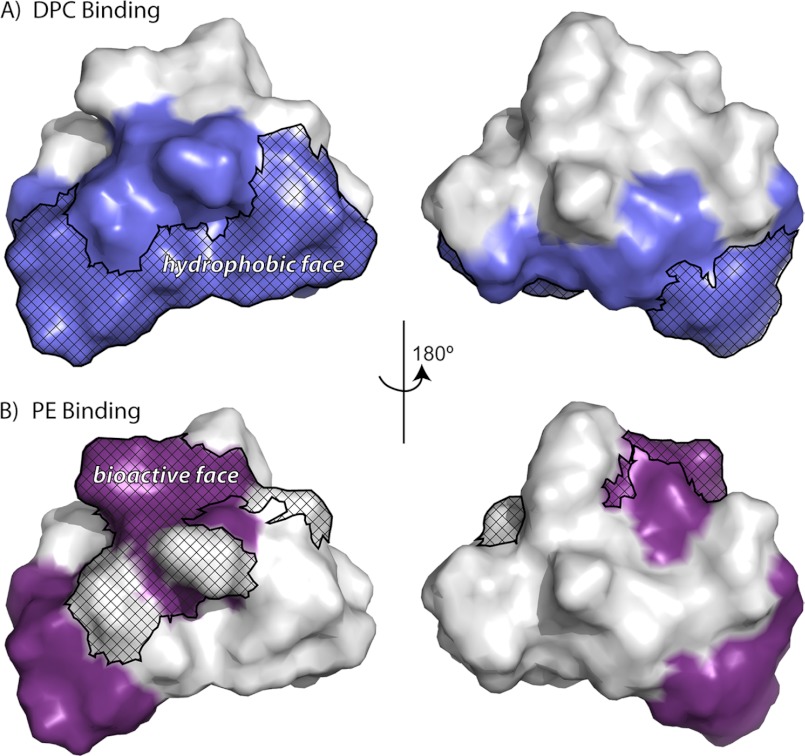
Analysis of the binding isotherms suggests that hydrophobic forces help mediate the interaction between cyclotides and membranes. The thermodynamic parameters obtained show that binding is predominantly an entropy-driven process and is associated with a negative heat capacity, characteristic of the hydrophobic effect (53, 54). In other studies, NMR spin label experiments have shown that kB1 and kB2 interact with DPC micelles via their hydrophobic patch (31, 32), as highlighted in Fig. 8A. Compared with kB1, the hydrophobic patch of kB2 is larger, explaining why kB2 consistently demonstrates stronger affinity toward DPC micelles and phospholipid bilayers. Previous studies have proposed a correlation between cyclotide bioactivity and surface hydrophobicity (55), providing further evidence that hydrophobic forces are important for cyclotide membrane binding. We speculate that these hydrophobic interactions involve the exposed hydrophobic residues of cyclotides and the hydrocarbon tails of phospholipids.
FIGURE 8.
Membrane binding surfaces of kalata B1. A and B, surface representations of kalata B1. Residues that are involved in binding to dodecylphosphocholine micelles are colored in blue (A), and residues that are involved in binding to phosphatidylethanolamine are colored in purple (B). The hydrophobic and bioactive faces identified in previous studies are also shown.
Although hydrophobic interactions are involved in the membrane binding and activity of cyclotides, it is becoming apparent that they are not sufficient. Cyclotides bind specifically and selectively to PE lipids (26–28), which is consistent with the proposed native role of cyclotides in targeting insect cell membranes, which contain a significant proportion of PE-lipids (56). A recent NMR study identified a set of residues in kB1 that form a binding site for PE (27). These residues also form part of the “bioactive face” (Fig. 8B), which is composed of residues that are important for cyclotide bioactivity, suggesting that specific binding to PE-lipids is important for cyclotide activity. This specificity is highlighted by our findings here that variations in the free energy of binding of kB2 to lipid mixtures of varying PE composition is primarily modulated by enthalpic changes, despite entropic effects being the dominant contribution of overall binding to the lipid mixtures. Furthermore, we observed that mutants [E7A]kB1 and [T16A]kB1, which have mutations in the PE binding site, displayed no affinity toward POPC/POPE phospholipid bilayers, whereas the mutant [T27A]kB1, which has a mutation outside of the PE binding site, displayed strong membrane binding affinity, confirming that interactions between the “bioactive face” of cyclotides and PE are necessary for cyclotide membrane binding. The acyclic mutant des-(16ψ17)-kB1, which has a disrupted bioactive face, and des-(29ψ1)-kB1, which has a disrupted hydrophobic face, also showed no binding toward POPC/POPE phospholipid bilayers, further confirming the importance of both PE binding and hydrophobic interactions to the mechanism of membrane interaction.
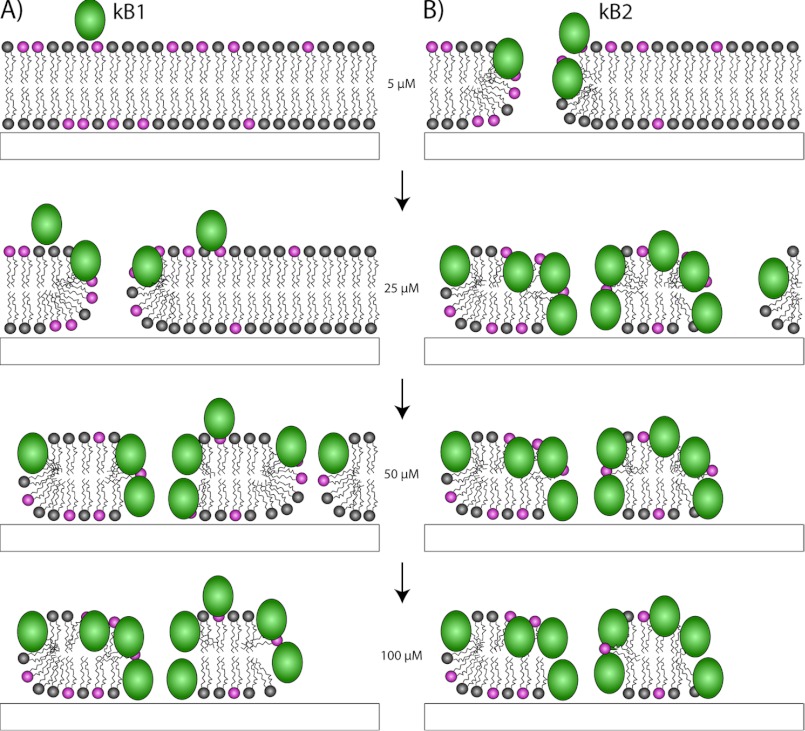
After having established the critical biophysical properties that affect membrane binding, we then defined how cyclotides interact with membranes at a molecular level by using two techniques, QCM-D and neutron reflectometry. The QCM-D data suggested that cyclotides form membrane pores, consistent with a recent electrophysiology study (29). A strength of neutron reflectometry is that it is one of the few biophysical techniques that allow the structural characterization of peptides bound to or inserted in a lipid bilayer under nearly physiological conditions. It provides a measure of the depth of insertion of peptides into bilayers. Here we found that the effective concentration of cyclotides at different depths in a bilayer varies with the applied peptide concentration, with deeper penetration occurring as the peptide concentration increases. As shown in Fig. 9, at low concentrations, kB1 binds to the surface of the membrane and then progressively inserts into the membrane to form pores. At high concentrations, kB1 begins to destabilize the membrane, leaving a sparse layer of peptide attached to the remaining lipids. The interaction of kB2 is similar to kB1, except that it penetrates to form membrane pores and begins to solubilize the bilayer even at 5 μm concentration. The results are consistent with previous studies, which reported that at low concentrations, cyclotides are non-toxic to cells, but at high concentrations, they may cause complete disruption of the cell (57). We propose that there is a threshold concentration; only when the actual concentration exceeds the threshold value do cyclotides insert and form pores in the membrane.
FIGURE 9.
Proposed mechanism for membrane interactions of cyclotides kalata B1 (A) and kalata B2 (B) as function of cyclotide concentration. Kalata B1 interacts with the membrane surface initially, before inserting into the upper membrane leaflet and forming pores. Kalata B2 inserts through the membrane and forms pores at lower concentrations, but both peptides eventually destabilize and solubilize the membrane, leaving small patches of a hydrophobic lipid monolayer with peptide adsorbed on top.
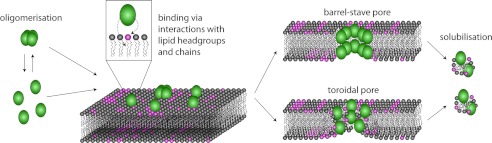
From the results presented in this paper, we can start to build a complete picture of the mechanism of membrane interaction of cyclotides by comparison with known mechanisms of pore-forming anti-microbial peptides. We propose a model as shown in Fig. 10. Cyclotides approach the membrane as either monomers or oligomers (33) and bind to the membrane surface via specific interactions between the cyclotide bioactive face and the PE-lipid headgroup as well as nonspecific hydrophobic interactions between the hydrophobic face and lipid chains. When the localized concentration of cyclotides bound to the membrane surface exceeds a certain threshold value, cyclotides begin to form pores. This pore formation could in principle occur via the barrel-stave or toroidal pore models (58). The barrel-stave model applies to peptides that are not specific for certain membranes, involving direct contact between hydrophobic residues of the peptide and the hydrophobic core of the membrane (58). Although we show here that the hydrophobic residues of cyclotides are important for membrane binding, we also show that cyclotides are specific for PE-lipids. In the toroidal pore model, peptides remain in contact with the lipid headgroup even when penetrating the bilayer (58). Because cyclotides have a specific face for binding to PE, it is likely that cyclotides form tight interactions with the PE-lipid headgroup as they insert into the membrane. Our neutron reflectometry results indicate that the deep penetration into membranes probably does not involve well defined peptide oligomers but rather a progressive insertion by monomers or loosely packed oligomers, possibly providing further support for the toroidal pore model. At high cyclotide concentrations, lipids are extracted from the membrane, leading to membrane solubilization. Magainin is an example of a peptide that forms toroidal pores, which eventually lead to membrane disruption (58). Interestingly, the predicted pore size for magainin of 30–50 Å (59) is similar to that seen for cyclotides (29).
FIGURE 10.
Proposed mechanism of pore formation of cyclotides. Cyclotides reach the membrane as monomers or oligomers, followed by binding to the surface of the membrane by specifically targeting phosphatidylethanolamine lipids. After a threshold concentration of cyclotides has been reached, cyclotides permeate the membrane. This can be achieved via the formation of a “barrel-stave” or “toroidal” pore. Binding of cyclotides at high concentrations also solubilizes the membrane.
In summary, we have shown here that the bioactivities of cyclotides are related to their strong interaction with membranes and elucidated their mechanism of action using a combination of ITC, QCM-D, and neutron reflectivity measurements. Cyclotides can insert deeply into lipid bilayers to form membrane pores, and the membrane penetration is the result of hydrophobic interactions with lipid tails and specific interactions with the PE headgroup. At high peptide concentrations, cyclotides severely destabilize membranes, leading to solubilization. Overall, this work provides a significant advance in our understanding of how cyclotides interact with membranes.
Acknowledgments
We thank Robert Dalgliesh for assistance at ISIS and Sonia Henriques for assistance with neutron reflectometry experiments.
This work was supported in part by Australian Research Council Grant DP0984390. Travel was supported by Australian Nuclear Science and Technology Organisation Grant 04/08-N-40 for neutron reflectometry.
- kB1 and kB2
- kalata B1 and B2, respectively
- PE
- phosphoethanolamine
- DPC
- dodecyl phosphocholine
- ITC
- isothermal titration calorimetry
- QCM-D
- dissipative quartz crystal microbalance
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolimine
- DMPE
- 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine.
REFERENCES
- 1. Craik D. J., Daly N. L., Bond T., Waine C. (1999) Plant cyclotides. A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 2. Gruber C. W., Elliott A. G., Ireland D. C., Delprete P. G., Dessein S., Göransson U., Trabi M., Wang C. K., Kinghorn A. B., Robbrecht E., Craik D. J. (2008) Distribution and evolution of circular miniproteins in flowering plants. Plant Cell 20, 2471–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simonsen S. M., Sando L., Ireland D. C., Colgrave M. L., Bharathi R., Göransson U., Craik D. J. (2005) A continent of plant defense peptide diversity. Cyclotides in Australian Hybanthus (Violaceae). Plant Cell 17, 3176–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang C. K., Kaas Q., Chiche L., Craik D. J. (2008) CyBase. A database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 36, D206–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poth A. G., Colgrave M. L., Lyons R. E., Daly N. L., Craik D. J. (2011) Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proc. Natl. Acad. Sci. U.S.A. 108, 10127–10132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Göransson U., Burman R., Gunasekera S., Strömstedt A. A., Rosengren K. J. (2012) Circular proteins from plants and fungi. J. Biol. Chem. 287, 27001–27006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nguyen G. K., Lim W. H., Nguyen P. Q., Tam J. P. (2012) Novel cyclotides and uncyclotides with highly shortened precursors from Chassalia chartacea and effects of methionine oxidation on bioactivities. J. Biol. Chem. 287, 17598–17607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pinto M. F., Fensterseifer I. C., Migliolo L., Sousa D. A., de Capdville G., Arboleda-Valencia J. W., Colgrave M. L., Craik D. J., Magalhães B. S., Dias S. C., Franco O. L. (2012) Identification and structural characterization of novel cyclotide with activity against an insect pest of sugar cane. J. Biol. Chem. 287, 134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tang J., Wang C. K., Pan X., Yan H., Zeng G., Xu W., He W., Daly N. L., Craik D. J., Tan N. (2010) Isolation and characterization of cytotoxic cyclotides from Viola tricolor. Peptides 31, 1434–1440 [DOI] [PubMed] [Google Scholar]
- 10. Mulvenna J. P., Mylne J. S., Bharathi R., Burton R. A., Shirley N. J., Fincher G. B., Anderson M. A., Craik D. J. (2006) Discovery of cyclotide-like protein sequences in graminaceous crop plants. Ancestral precursors of circular proteins? Plant Cell 18, 2134–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colgrave M. L., Craik D. J. (2004) Thermal, chemical, and enzymatic stability of the cyclotide kalata B1. The importance of the cyclic cystine knot. Biochemistry 43, 5965–5975 [DOI] [PubMed] [Google Scholar]
- 12. Craik D. J., Cĕmažar M., Daly N. L. (2006) The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Dev. 9, 251–260 [PubMed] [Google Scholar]
- 13. Saether O., Craik D. J., Campbell I. D., Sletten K., Juul J., Norman D. G. (1995) Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry 34, 4147–4158 [DOI] [PubMed] [Google Scholar]
- 14. Gran L. (1973) Oxytocic principles of Oldenlandia affinis. Lloydia 36, 174–178 [PubMed] [Google Scholar]
- 15. Gustafson K. R., McKee T. C., Bokesch H. R. (2004) Anti-HIV cyclotides. Curr. Protein Pept. Sci. 5, 331–340 [DOI] [PubMed] [Google Scholar]
- 16. Tam J. P., Lu Y. A., Yang J. L., Chiu K. W. (1999) An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. U.S.A. 96, 8913–8918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schöpke T., Hasan Agha M. I., Kraft R., Otto A., Hiller K. (1993) Hamolytisch aktive Komponenten aus Viola tricolor L. und Viola arvensis Murray. Sci. Pharm. 61, 145–153 [Google Scholar]
- 18. Göransson U., Svangård E., Claeson P., Bohlin L. (2004) Novel strategies for isolation and characterization of cyclotides. The discovery of bioactive macrocyclic plant polypeptides in the Violaceae. Curr. Protein Pept. Sci. 5, 317–329 [DOI] [PubMed] [Google Scholar]
- 19. Witherup K. M., Bogusky M. J., Anderson P. S., Ramjit H., Ransom R. W., Wood T., Sardana M. (1994) Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J. Nat. Prod. 57, 1619–1625 [DOI] [PubMed] [Google Scholar]
- 20. Plan M. R., Saska I., Cagauan A. G., Craik D. J. (2008) Backbone cyclised peptides from plants show molluscicidal activity against the rice pest Pomacea canaliculata (golden apple snail). J. Agric. Food Chem. 56, 5237–5241 [DOI] [PubMed] [Google Scholar]
- 21. Colgrave M. L., Kotze A. C., Huang Y. H., O'Grady J., Simonsen S. M., Craik D. J. (2008) Cyclotides. Natural, circular plant peptides that possess significant activity against gastrointestinal nematode parasites of sheep. Biochemistry 47, 5581–5589 [DOI] [PubMed] [Google Scholar]
- 22. Colgrave M. L., Kotze A. C., Ireland D. C., Wang C. K., Craik D. J. (2008) The anthelmintic activity of the cyclotides. Natural variants with enhanced activity. Chembiochem 9, 1939–1945 [DOI] [PubMed] [Google Scholar]
- 23. Jennings C., West J., Waine C., Craik D., Anderson M. (2001) Biosynthesis and insecticidal properties of plant cyclotides. The cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. U.S.A. 98, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jennings C. V., Rosengren K. J., Daly N. L., Plan M., Stevens J., Scanlon M. J., Waine C., Norman D. G., Anderson M. A., Craik D. J. (2005) Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist. Do Möbius strips exist in nature? Biochemistry 44, 851–860 [DOI] [PubMed] [Google Scholar]
- 25. Barbeta B. L., Marshall A. T., Gillon A. D., Craik D. J., Anderson M. A. (2008) Plant cyclotides disrupt epithelial cells in the midgut of lepidopteran larvae. Proc. Natl. Acad. Sci. U.S.A. 105, 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamimori H., Hall K., Craik D. J., Aguilar M. I. (2005) Studies on the membrane interactions of the cyclotides kalata B1 and kalata B6 on model membrane systems by surface plasmon resonance. Anal. Biochem. 337, 149–153 [DOI] [PubMed] [Google Scholar]
- 27. Henriques S. T., Huang Y. H., Rosengren K. J., Franquelim H. G., Carvalho F. A., Johnson A., Sonza S., Tachedjian G., Castanho M. A., Daly N. L., Craik D. J. (2011) Decoding the membrane activity of the cyclotide kalata B1. The importance of phosphatidylethanolamine phospholipids and lipid organization on hemolytic and anti-HIV activities. J. Biol. Chem. 286, 24231–24241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henriques S. T., Huang Y. H., Castanho M. A., Bagatolli L. A., Sonza S., Tachedjian G., Daly N. L., Craik D. J. (2012) Phosphatidylethanolamine binding is a conserved feature of cyclotide-membrane interactions. J. Biol. Chem. 287, 33629–33643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang Y. H., Colgrave M. L., Daly N. L., Keleshian A., Martinac B., Craik D. J. (2009) The biological activity of the prototypic cyclotide kalata b1 is modulated by the formation of multimeric pores. J. Biol. Chem. 284, 20699–20707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sando L., Henriques S. T., Foley F., Simonsen S. M., Daly N. L., Hall K. N., Gustafson K. R., Aguilar M. I., Craik D. J. (2011) A synthetic mirror image of kalata B1 reveals that cyclotide activity is independent of a protein receptor. Chembiochem 12, 2456–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shenkarev Z. O., Nadezhdin K. D., Lyukmanova E. N., Sobol V. A., Skjeldal L., Arseniev A. S. (2008) Divalent cation coordination and mode of membrane interaction in cyclotides. NMR spatial structure of ternary complex Kalata B7/Mn2+/DPC micelle. J. Inorg. Biochem. 102, 1246–1256 [DOI] [PubMed] [Google Scholar]
- 32. Shenkarev Z. O., Nadezhdin K. D., Sobol V. A., Sobol A. G., Skjeldal L., Arseniev A. S. (2006) Conformation and mode of membrane interaction in cyclotides. Spatial structure of kalata B1 bound to a dodecylphosphocholine micelle. FEBS J. 273, 2658–2672 [DOI] [PubMed] [Google Scholar]
- 33. Nourse A., Trabi M., Daly N. L., Craik D. J. (2004) A comparison of the self-association behavior of the plant cyclotides kalata B1 and kalata B2 via analytical ultracentrifugation. J. Biol. Chem. 279, 562–570 [DOI] [PubMed] [Google Scholar]
- 34. Mulvenna J. P., Wang C., Craik D. J. (2006) CyBase. A database of cyclic protein sequence and structure. Nucleic Acids Res. 34, D192–D194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Daly N. L., Love S., Alewood P. F., Craik D. J. (1999) Chemical synthesis and folding pathways of large cyclic polypeptides. Studies of the cystine knot polypeptide kalata B1. Biochemistry 38, 10606–10614 [DOI] [PubMed] [Google Scholar]
- 36. Tam J. P., Lu Y.-A., Yu Q. (1999) Thia zip reaction for synthesis of large cyclic peptides: mechanisms and applications. J. Am. Chem. Soc. 121, 4316–4324 [Google Scholar]
- 37. Silvius J. R. (1986) Solid- and liquid-phase equilibria in phosphatidylcholine/phosphatidylethanolamine mixtures. A calorimetric study. Biochim. Biophys. Acta 857, 217–228 [DOI] [PubMed] [Google Scholar]
- 38. Penfold J. (1991) Instrumentation for neutron reflectivity. Physica B 173, 1–10 [Google Scholar]
- 39. Vacklin H. P., Tiberg F., Fragneto G., Thomas R. K. (2005) Composition of supported model membranes determined by neutron reflection. Langmuir 21, 2827–2837 [DOI] [PubMed] [Google Scholar]
- 40. Vacklin H. P., Tiberg F., Fragneto G., Thomas R. K. (2005) Phospholipase A2 hydrolysis of supported phospholipid bilayers. A neutron reflectivity and ellipsometry study. Biochemistry 44, 2811–2821 [DOI] [PubMed] [Google Scholar]
- 41. Nelson A. (2006) Co-refinement of multiple-contrast neutron/X-ray reflectivity data using MOTOFIT. J. Appl. Crystallogr. 39, 273–276 [Google Scholar]
- 42. Heavens O. S. (1955) Optical properties of Thin Films, Dover Publications, London [Google Scholar]
- 43. Armen R. S., Uitto O. D., Feller S. E. (1998) Phospholipid component volumes: determination and application to bilayer structure calculations. Biophys. J. 75, 734–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zamyatin A. A. (1972) Protein volume in solution. Prog. Biophys. Mol. Biol. 24, 107–123 [DOI] [PubMed] [Google Scholar]
- 45. Skjeldal L., Gran L., Sletten K., Volkman B. F. (2002) Refined structure and metal binding site of the kalata B1 peptide. Arch. Biochem. Biophys. 399, 142–148 [DOI] [PubMed] [Google Scholar]
- 46. Simonsen S. M., Sando L., Rosengren K. J., Wang C. K., Colgrave M. L., Daly N. L., Craik D. J. (2008) Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J. Biol. Chem. 283, 9805–9813 [DOI] [PubMed] [Google Scholar]
- 47. Simonsen S. M., Daly N. L., Craik D. J. (2004) Capped acyclic permutants of the circular protein kalata B1. FEBS Lett. 577, 399–402 [DOI] [PubMed] [Google Scholar]
- 48. Craik D. J., Simonsen S., Daly N. L. (2002) The cyclotides. Novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Devel. 5, 251–260 [PubMed] [Google Scholar]
- 49. Gould A., Ji Y., Aboye T. L., Camarero J. A. (2011) Cyclotides, a novel ultrastable polypeptide scaffold for drug discovery. Curr. Pharm. Des. 17, 4294–4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thongyoo P., Bonomelli C., Leatherbarrow R. J., Tate E. W. (2009) Potent inhibitors of β-tryptase and human leukocyte elastase based on the MCoTI-II scaffold. J. Med. Chem. 52, 6197–6200 [DOI] [PubMed] [Google Scholar]
- 51. Chan L. Y., Gunasekera S., Henriques S. T., Worth N. F., Le S. J., Clark R. J., Campbell J. H., Craik D. J., Daly N. L. (2011) Engineering pro-angiogenic peptides using stable, disulfide-rich cyclic scaffolds. Blood 118, 6709–6717 [DOI] [PubMed] [Google Scholar]
- 52. Wong C. T., Rowlands D. K., Wong C. H., Lo T. W., Nguyen G. K., Li H. Y., Tam J. P. (2012) Orally active peptidic bradykinin B1 receptor antagonists engineered from a cyclotide scaffold for inflammatory pain treatment. Angew. Chem. Int. Ed. Engl. 51, 5620–5624 [DOI] [PubMed] [Google Scholar]
- 53. Seelig J. (2004) Thermodynamics of lipid-peptide interactions. Biochim. Biophys. Acta 1666, 40–50 [DOI] [PubMed] [Google Scholar]
- 54. Fernández-Vidal M., White S. H., Ladokhin A. S. (2011) Membrane partitioning: “classical” and “nonclassical” hydrophobic effects. J. Membr. Biol. 239, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang C. K., Colgrave M. L., Gustafson K. R., Ireland D. C., Goransson U., Craik D. J. (2008) Anti-HIV cyclotides from the Chinese medicinal herb Viola yedoensis. J. Nat. Prod. 71, 47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parisi M., Li R., Oliver B. (2011) Lipid profiles of female and male Drosophila. BMC Res. Notes 4, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Greenwood K. P., Daly N. L., Brown D. L., Stow J. L., Craik D. J. (2007) The cyclic cystine knot miniprotein MCoTI-II is internalized into cells by macropinocytosis. Int. J. Biochem. Cell Biol. 39, 2252–2264 [DOI] [PubMed] [Google Scholar]
- 58. Brogden K. A. (2005) Antimicrobial peptides. Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250 [DOI] [PubMed] [Google Scholar]
- 59. Ludtke S. J., He K., Heller W. T., Harroun T. A., Yang L., Huang H. W. (1996) Membrane pores induced by magainin. Biochemistry 35, 13723–13728 [DOI] [PubMed] [Google Scholar]