Background: Stac3, an uncharacterised gene, was identified by its high expression levels in vertebrate muscle.
Results: Knockdown of stac3 inhibited zebrafish myofibrillar protein assembly and differentiation in C2C12 cells by perturbing Akt signaling and cell cycle exit.
Conclusion: Stac3 is a novel regulator of myogenic differentiation.
Significance: Identifying new genes controlling myogenic differentiation will allow greater understanding of processes leading to muscular diseases.
Keywords: Akt, Insulin-like Growth Factor (IGF), Mouse, mTOR, Myogenesis, Zebrafish
Abstract
Stac3 was identified as a nutritionally regulated gene from an Atlantic salmon subtractive hybridization library with highest expression in skeletal muscle. Salmon Stac3 mRNA was highly correlated with myogenin and myoD1a expression during differentiation of a salmon primary myogenic culture and was regulated by amino acid availability. In zebrafish embryos, stac3 was initially expressed in myotomal adaxial cells and in fast muscle fibers post-segmentation. Morpholino knockdown resulted in defects in myofibrillar protein assembly, particularly in slow muscle fibers, and decreased levels of the hedgehog receptor patched. The function of Stac3 was further characterized in vitro using the mammalian C2C12 myogenic cell line. Stac3 mRNA expression increased during the differentiation of the C2C12 myogenic cell line. Knockdown of Stac3 by RNAi inhibited myotube formation, and microarray analysis revealed that transcripts involved in cell cycle, focal adhesion, cytoskeleton, and the pro-myogenic factors Igfbp-5 and Igf2 were down-regulated. RNAi-treated cells had suppressed Akt signaling and exogenous insulin-like growth factor (Igf) 2 was unable to rescue the phenotype, however, Igf/Akt signaling was not blocked. Overexpression of Stac3, which results in increased levels of Igfbp-5 mRNA, did not lead to increased differentiation. In synchronized cells, Stac3 mRNA was most abundant during the G1 phase of the cell cycle. RNAi-treated cells were smaller, had higher proliferation rates and a decreased proportion of cells in G1 phase when compared with controls, suggesting a role in the G1 phase checkpoint. These results identify Stac3 as a new gene required for myogenic differentiation and myofibrillar protein assembly in vertebrates.
Introduction
Stac are members of a poorly characterized family of putative signal transduction adaptor proteins. Stac1 was cloned as a neurone-specific gene encoding a protein with a cysteine-rich domain of the protein kinase C family and two contiguous SH3 domains (1). Stac family members are encoded by three independent genes (Stac1, -2, and -3) located on three different chromosomes in the mouse. Stac1 and -2 were identified as markers of discrete subsets of neurons (2). Stac1 was mainly expressed in nociceptive peptidergic neurones, whereas Stac2 was localized in three different subpopulations of sensory neurones (2). Protein microarray studies identified STAC family members as 14-3-3-binding proteins (3). The 14-3-3 protein family act as molecular adaptors that interact with signaling molecules involved with cell differentiation, proliferation, and apoptosis to modify their function (4). STAC contains the highly stringent 14-3-3 binding consensus RYYSSP and co-immunoprecipitates with 14-3-3 proteins (3). However, co-immunoprecipitation studies with truncated STAC proteins indicated that the 14-3-3-interacting domain was located in the N-terminal segment and was independent of serine/threonine phosphorylation of the binding domain of STAC (3). A splice variant of STAC was implicated in transcriptional networks controlling senescence in human mammary fibroblasts interacting with nuclear factor-κB and C/EBP transcription factors (5). To date there is no information regarding the function of STAC3.
Through a suppression subtractive hybridization approach, we recently identified stac3 as a gene that was strongly up-regulated in skeletal muscle of Atlantic salmon (Salmo salar L.) following transition from maintenance to fast growth (6). In the present study, stac3 expression was further characterized in Atlantic salmon and its function investigated in vitro using the mammalian C2C12 myogenic cell line and in vivo using the zebrafish model system. We demonstrate that Stac3 is necessary for myogenic differentiation by RNAi knockdown of Stac3 mRNA levels in vitro. We also demonstrate a role for stac3 in myogenic differentiation and myofibrillar protein assembly in vivo, and identify its embryonic expression pattern is restricted to somites. Together, this data identifies Stac3 as a novel regulator of myogenic differentiation in vertebrates, probably interacting with multiple signaling pathways.
EXPERIMENTAL PROCEDURES
Suppression Subtraction Library Production and Salmon Re-feeding and Cell Culture Experiments
Details of the suppression subtraction cDNA library production and re-feeding experiments involving Atlantic salmon (S. salar) have previously been described (6). For the refeeding experiment, fish were fed a reduced ration for 21 days, fasted for 7 days, and then fed to satiation for 21 days. Cell culture experiments were performed using an Atlantic salmon primary cell culture isolated from a freshwater pair ∼60 g (7). For cell culture starvation, cells were grown until day 9 of culture, washed once with amino acid-deprived (starve) media (Earle's balanced salt solution, supplemented with 9 mm NaHCO3, 20 mm HEPES (pH 7.4), 2 g/liter of glucose, supplemented with 1× vitamins, 1× antibiotics), and then grown for 72 h in starve media. For amino acid and insulin-like growth factor (Igf)4 treatments (recombinant salmon protein, GroPep, Adelaide, Australia), cells were starved of amino acids and serum for 72 h and then fed amino acids (DMEM, 9 mm NaHCO3, 20 mm HEPES (pH 7.4), 1× antibiotics), Igf1 (100 ng/ml), amino acids plus 1 μm insulin, or amino acids plus Igf1 (100 ng/ml). Controls were unfed, 72-h starved cells replenished with starve media. RNA was extracted using a RNeasy Plus kit (Qiagen) from the cells at 0, 6, 12, 24, 36, 48, and 72 h following treatment. Cloning of the stac3 mRNA sequence was performed using standard techniques. 5′ RACE was performed using the T-RACE method (8).
C2C12 Cell Culture and Transfection
C2C12 cells (ATCC) were grown and maintained in growth media (GM), Dulbecco's modified Eagle's medium with 20% (v/v) fetal bovine serum (Sigma), and 1× penicillin/streptomycin (Sigma). Reverse transfection of RNAi was performed in 12- or 24-well plates using RNAi directed against Stac3 (Stealth RNAi, Invitrogen) with Lipofectamine RNAiMAX (Invitrogen) as per the manufacturer's recommendations. As control, transfection of a scrambled high GC content RNAi (Stealth RNAi siRNA Negative Control High GC, Invitrogen) with no homology to known mammalian genes was performed. Duplicate wells were reverse transfected to give a seeding density of 30–50% confluence. 8 h after transfection, the media from one of the duplicate wells was removed and put aside and the cells from this well were trypsinized. The media that was put aside (containing the RNAi) was then added to the trypsinized cells, and combined with the duplicate well, which resulted in RNAi-treated cells at ∼90% confluence 24–30 h after the initial RNAi treatment. Upon reaching 90% confluence, cells were washed once with differentiation media (DM) (Dulbecco's modified Eagle's medium with 2% (v/v) horse serum (Sigma)), and myogenic differentiation was induced by growing cells in DM. For rescue experiments, cells were grown in DM containing Igf2 (150 and 300 ng/ml).
Immunofluorescence of Culture Cells
Cells were grown on glass coverslips, fixed with 4% (m/v) paraformaldehyde, and proteins were detected by immunofluoresence as previously described (9). Antibodies specific for myoglobin (Abcam), myosin heavy chain Alexa Fluor 488 (eBioscence), and BrdU (Invitrogen) were used. Nuclei were counterstained with DAPI or 7-aminoactinomycin D (Invitrogen). Cells were imaged using a Leica TCS SP2 confocal microscope.
Microarray
Microarray analysis was performed to comply with MIAME guidelines. Microarray analysis was performed by Cambridge genomics service using an illumina beadchip mouse WG-6 whole genome array (Department of Pathology, Cambridge University). Data were normalized by the percentile method, and mean log transformed values for controls were subtracted from Stac3 RNAi samples. Microarray data were submitted to the NCBI gene expression and hybridization array data repository GEO, (ncbi.nlm.nih.gov/geo) accession number GSE34474.
Cell Proliferation Analysis, Synchronization, and Flow Cytometry
Cell proliferation rates were calculated 24 h after RNAi transfection by culturing cells at 30–50% confluence in the presence of a 1:100 (v/v) dilution of BrdU (Invitrogen) in GM or DM (Invitrogen) for 1 h as previously described (9). Cells were counted from 3 fields of view (0.56 mm2, ×20 magnification) from 3 separate cultures. Cell synchronization was achieved by methionine deprivation as previously described (10). Samples for flow cytometry were grown in DM. Cells were trypsinized and centrifuged at 200 × g for 10 min, washed with PBS, and then resuspended in 300 μl of PBS. 700 μl of ice-cold 100% ethanol was added to the samples, which were then stored for 24 h at −20 °C. Samples were centrifuged again, and washed with PBS before being resuspended in PBS containing 50 μg/ml of propidium iodide (Sigma) and 10 μg/ml of RNase A (Invitrogen). Samples were analyzed on a Beckman Coulter Epics XL Flow cytometer.
RNA Extraction and cDNA Synthesis
RNA was extracted from 4 separate cell cultures. RNA extraction and genomic DNA removal was performed using a RNeasy plus kit (Qiagen Inc.) as per the manufacturer's recommendations and cDNA was synthesized as previously described (9).
Quantitative PCR
The following procedures were performed to comply with the Minimum Information for Publication of Quantitative Real-time PCR experiments MIQE guidelines (11). Quantitative PCR was performed using a Stratagene MX3005P Quantitative PCR system (Stratagene, La Jolla, CA) with Brilliant II SYBR (Stratagene, La Jolla, CA) as described in Ref. 9. The PCR amplification efficiency of each primer pair was calculated using Ling® PCR 2009 (12). Genorm (13) was used to analyze the stability of hypoxanthine phosphoribosyltransferase 1 (Hprt1), peptidylprolyl isomerase A (Ppia) ribosomal protein s29 (Rps29), and ribosomal protein L13 (Rpl13) as reference genes for normalization and revealed that Rps29 and Hprt1 were found to be the most stable genes. Normalization of gene expression was performed using the geometric average of Hprt1 and Rps29, and values are shown as arbitrary units. Statistical analysis was performed using minitab (Minitab Inc). Significant differences in expression between time points were calculated by analysis of variance using Fisher's individual error rate post hoc tests. Primer sequences are listed in supplemental Table S1.
Western Blot Analysis
Total protein was extracted using Cell lysis-M extraction buffer (Sigma) as per the manufacturer's guidelines with the addition of protease inhibitor and phosphatase inhibitors I and II (Sigma). Protein concentration was determined by a Bradford assay (Bio-Rad) using commercially available standards (Bio-Rad) and quantified using a plate reader with a 595-nm filter (VERSAmax, microplate reader, Molecular Devices). Proteins were separated by SDS-PAGE and analyzed by immunoblotting. Antibodies specific for Actin, Akt, phospho-Akt (Ser-473), p38α, mTOR (Ser-2448), phospho-mTOR, phospho-ERK1/2 (Thr-202/Tyr-204), S6k1 (Thr-389), S6k1, phospho-S6 (Ser-235/Ser-236), and S6 (Cell Signaling), phospho-p38α (Thr-180/Tyr-182) (Millipore), phospho-MyoD (Ser-200), and MyoD (Abcam) were used. All antibodies were diluted 1/1000 (v/v) in 0.1% Tween 20, PBS (PBT). Detection was performed using anti-rabbit or anti-mouse secondary antibodies linked to a horseradish peroxidase diluted 1/20,000 (v/v) in PBT. Chemiluminescence was developed using the Amersham Biosciences ECL kit (GE Healthcare). X-ray film (GE Healthcare) was developed in an AFP x-ray film processor. Bands on scanned x-ray film were quantified using TotalLab TL100 software (Nonlinear Dynamics). β-Actin (Sigma) was used as a loading control in all Western blots performed.
Zebrafish Fixation
For analysis of muscle fiber formation, embryos were fixed in 4% paraformaldehyde/PBS overnight at 4 °C at 24 and 48 h postfertilization. For in situ hybridization analysis embryos were fixed at various developmental stages in 4% paraformaldehyde/PBS. In situ hybridization was performed according to Ref. 14. Two cRNA probes were prepared to be specific to zebrafish myoD and stac3 using primers listed in supplemental Table S1 and STAC3 transcripts, respectively. Probes used for morphant analysis for ptc1 and myoD were synthesized from templates as previously described (15).
Morpholinos
The first morpholino (MO-stac3-ATG, 5′-CAGTTGGTCATATTGAGCCATCAGT-3′) was designed to target the translation start site region of the zebrafish stac3 gene. MO-stac3-ATG and the standard control MO (STD control-MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′) were supplied by Gene Tools, LLC (Philomath). Morpholinos were re-suspended in water and injected into embryos at the one-cell stage.
Transgenic Zebrafish Lines
Morpholinos were injected into embryos of the Tg(BAC smyhc1:gfp)i108 transgenic line (16) crossed to tg(bact2-Egfpcaax)pc10 (17).
Whole Mount Imaging (Lateral Views)
Embryos were fixed in 4% paraformaldehyde for 3 h at 4 °C, washed in PBS, and imaged. Slow muscle fibers were analyzed from stage matched whole embryos viewed by confocal microscopy.
Transverse Section Confocal Imaging
Following fixation, embryos were mounted in 2% low melt agarose. Vibratome sections (100-μm transverse sections) were cut immediately caudal to the yolk. Vibratome sections mounted in PBS were imaged using the Leica SPEII confocal microscope (Leica).
Fiber numbers and areas were calculated by Image J info the public domain NIH Image J program. Results are reported as least square mean ± S.E. Results were considered significant when p < 0.05.
RESULTS
Identification and Cloning of Full-length Atlantic Salmon stac3α and -β
Partial stac3 cDNA clones (accession numbers GE618322 and GE622214) were isolated as nutritionally responsive genes from a subtractive hybridization cDNA library comparing fish with zero growth rate to fish growing rapidly as previously described (6). The 2243-nucleotide mRNA of S. salar stac3α (Ss stac3α, accession number JN185194) is comprised of a 266-base 5′ UTR, 1235-bp 3′ UTR, and a 1008-bp coding sequence. The encoded protein of 335 amino acids has a predicted molecular mass of 38.67 kDa and a pI of 5.66. Ss stac3 has 64.7 and 79.2% identity with human and zebrafish Stac3, respectively (Fig. 1). Using T-RACE (8), we identified two alternate transcription start sites at 110 and 75 bp downstream of the transcription start sites of stac3α. Interproscan analysis of motifs identified a protein kinase C domain with phorbal ester diacylglycerol binding domain (PKC domain) at amino acids 62–112, a SRC homology 3 domain (SH3 domain) at amino acids 102–318, which includes a SH3 type 2 domain at amino acids 282–335. The protein is predicted to be phosphorylated at 6 serine, 8 threonine, and 4 tyrosine residues (Fig. 1).
FIGURE 1.
Amino acid alignment Stac3 protein sequences from Atlantic salmon, zebrafish, mouse, and human. The protein kinase C domain and SH3 domains are in bold and boxed, respectively, and the predicted phosphorylation sites in salmon and mouse are underlined.
Ss stac3β mRNA (accession number JN185195) is an alternatively spliced isoform of Ss stac3α, missing nucleotides 227–364 of Ss stac3α, which includes the translational start codon of Ss stac3α. An alternative start codon located 372 nucleotides 3′ of the transcriptional start gives rise to a coding sequence of 774 nucleotides, which encodes a protein of 257 amino acids (Fig. 1), with a predicted molecular mass of 29.44 kDa, and pI of 8.46. This splice variant, which misses the first 79 amino acids of Ss stac3α contains the SH3 domains, but is missing part of the PKC domain based on Interproscan motif searches.
Ss stac3 mRNA Expression Is Nutritionally Regulated and Most Abundant in Muscle
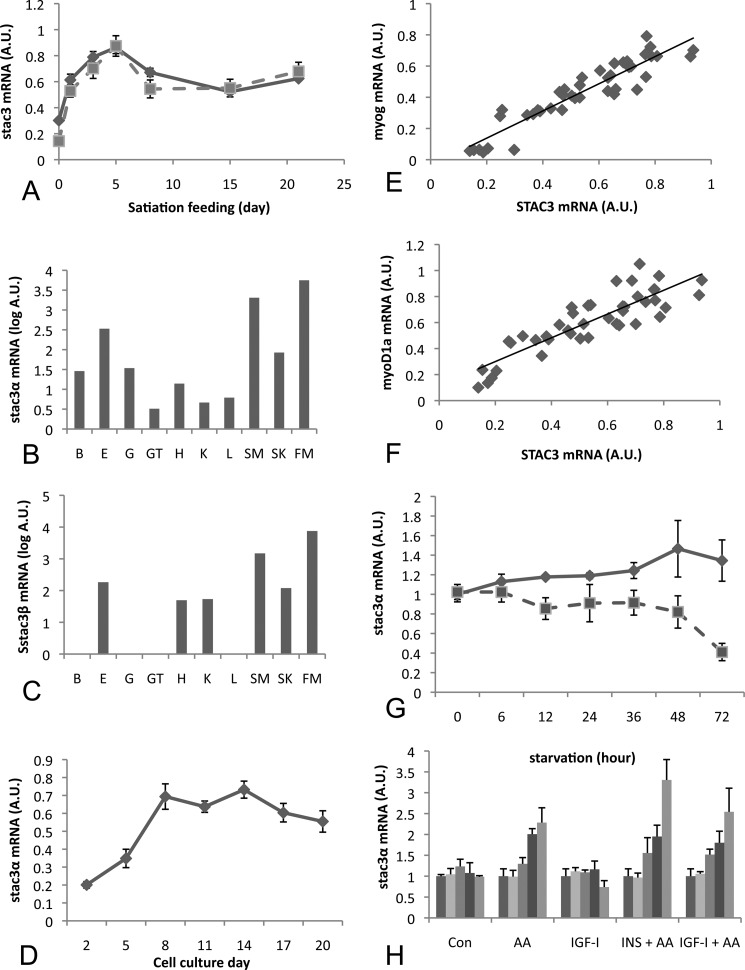
In Atlantic salmon that were fed a reduced ration for 28 days and then fed to satiation for 21 days (as described in Ref. 9), expression of both splice variants increased within 24 h of satiation feeding and remained at levels significantly higher (p < 0.05) than in fasted fish for the remainder of the feeding period, with maximal levels observed 5 days after feeding (Fig. 2A). Ss stac3α was on average 100-fold more abundant than Ss stac3β (data not shown).
FIGURE 2.
Quantitative PCR analysis of stac3 gene expression in Atlantic salmon. A, mRNA expression profile for Ss stac3α and -β in fish fed a reduced ration for 28 days (day 0) followed by refeeding to satiation for 21 days. B, tissue distribution of Ss stac3α mRNA in gill (G), gut (GT), kidney (K), heart (H), liver (L), eye (E), fast muscle (FM), slow muscle (SM), and skin (S). C, tissue distribution for Ss stac3β as described for B. D, stac3α mRNA levels increase during the maturation of an Atlantic salmon primary myogenic cell culture, and its expression is highly correlated with myog (E) and myoD (F). G, Ss stac3α mRNA levels are decreased in cells starved of amino acid and serum for 72 h (dashed line), when compared with controls (solid line). H, in cells starved for 72 h, Ss stac3α mRNA expression increases in response to amino acid stimulation, but not to Igf-I. Vertical bars represent 0, 3, 6, 12, and 24 h poststimulation.
To determine the tissue specificity of Ss stac3, we examined mRNA expression of both splice variants across several tissues in Atlantic salmon. Ss stac3α mRNA was detectable in most tissues by quantitative PCR, although in several, mRNA abundance was very low and at the limit of detection (Fig. 2B). Ss stac3α was most abundant in fast muscle, red muscle, and skin, and was on average, between 200- and 1200-fold higher in fast muscle than in other tissues (other than skin and red muscle). Ss stac3β mRNA had a similar expression profile, but as it is expressed at levels 100-fold less than Ss stac3α, we were unable to detect its expression in some tissues (Fig. 2C).
Ss stac3α Expression Increases during Myogenic Differentiation and Is Highly Correlated with myogenin and myoD1a
To further examine a role for stac3α in muscle growth we examined its expression during the maturation of an Atlantic salmon primary myogenic cell culture. Expression of Ss stac3α was lowest at the start of the culture when mononucleated cells predominated (Fig. 2D). Its expression increased from this time point, reaching peak expression levels at 8 days and remained at this elevated level until 20 days. The increased expression was concomitant with the formation of multinucleated myotubes. To place the expression of Ss stac3α in a myogenic framework, we examined its expression relative to the myogenic regulatory factors myoD1a and myog. Expression of stac3α was highly correlated with that of myog and myoD1a (R = 0.93 p < 0.0001; R = 0.86 p < 0.0001, Fig. 2E). Such high correlations suggest that these genes are on the same signaling pathway or share conserved regulatory elements within their promoters. A bioinformatics search of other stac3 promoters revealed the presence of both MyoD and Myog binding sites within the promoters of Stac3 from various species including teleosts and mammals. Due to the lower level of expression of stac3β we were unable to obtain reliable quantitative PCR data from cell culture experiments for this transcript.
Ss stac3α Expression Is Regulated by Amino Acids
To examine the nutritional regulation of Ss stac3α, we examined its mRNA expression profile in cells that were starved by withdrawal of serum and amino acids for 72 h. mRNA levels in starved cells decreased as soon as 6 h and remained at this level until 36 h, before a further decrease at 72 h (Fig. 2D). To examine the nutritional and hormonal regulation of Ss stac3α, we fed starved cells amino acids, insulin, Igf1, or a combination of amino acids plus the hormonal peptides. In starved cells, expression increased at 12 and 24 h after stimulation by amino acids, and in particular when amino acids were applied with insulin, however, we did not observe a response to Igf1 stimulation alone (Fig. 2E).
Embryonic Expression of stac3 mRNA in Zebrafish
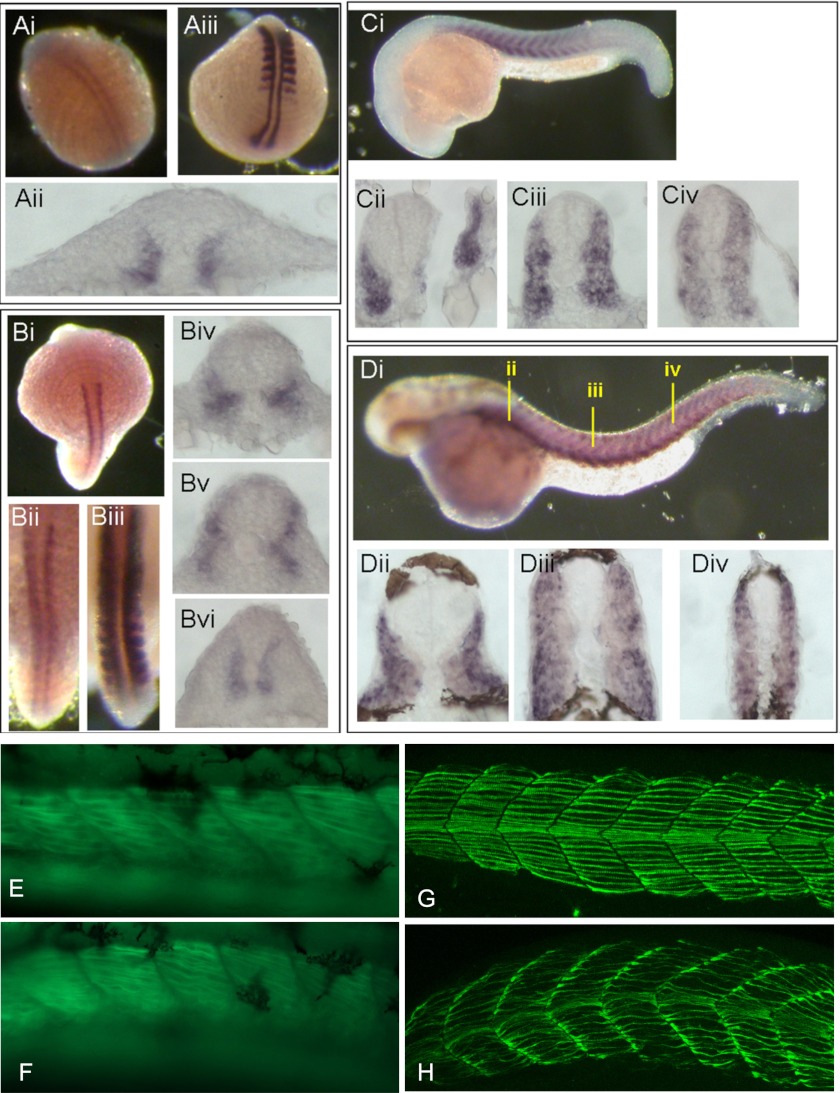
We performed in situ hybridizations on zebrafish embryos with stac3 as a probe and compared the expression pattern with that of myoD. During the early segmentation stage, stac3 transcripts were initially restricted in adaxial cells (Fig. 3A, i and ii) along the anterior-posterior axis of zebrafish embryos, whereas myoD transcripts were expressed in both adaxial cells and posterior somite compartments (Fig. 3A, iii). During the mid-segmentation stage, stac3 transcripts largely remained in adaxial cells (Fig. 3B, i, ii, and iv-vi) and some stac3 transcripts were also expressed in migrating adaxial cells of zebrafish embryos (Fig. 3B, iv and v). At the same developmental stage myoD transcripts were intensively expressed throughout the entire somite (Fig. 3B, iii). During the late segmentation stage, stac3 transcripts started to express within the myotome (Fig. 3C, i–iii) instead of being restricted to the adaxial cells (Fig. 3C, iv). After the completion of segmentation, stac3 transcripts were expressed in the dorsal and ventral extremes of the myotome (Fig. 3D, i) giving rise to a stratified pattern (Fig. 3D, ii–iv). stac3 transcripts were also detectable within the myotome in regions occupied by fast muscle fibers (Fig. 3D, ii–iv).
FIGURE 3.
stac3 and myoD in situ hybridization of zebrafish embryos. stac3 (A, i and ii) and myoD (A, iii) expression during early segmentation. At midsegmentation stac3 (B, i, ii, and iv-vi) is expressed in the adaxial cells, whereas myoD is found throughout the somite (B, iii). stac3 expression is observed in the myotome at late segmentation (C, i–iv), and at the dorsal and ventral extremes of the myotome after the completion of segmentation (D, i–iv). E and F, zebrafish embryos expressing gfp under the control of a fast muscle α actin promoter and in control and stac3 morphant embryos, respectively. Fast muscle fibers in morphant fish (F) have abnormal formation in comparison to controls (E). Antibody specific to slow muscle myosin in stac3 morphant zebrafish embryos showing dysregulated myofibrillar protein assembly in stac3 morphant embryos (H) when compared with controls (G).
stac3 Knockdown Effects Myofibrillar Protein Assembly in Vivo, Perturbs Hedgehog Signaling, and Decreases Slow Muscle Fiber Number
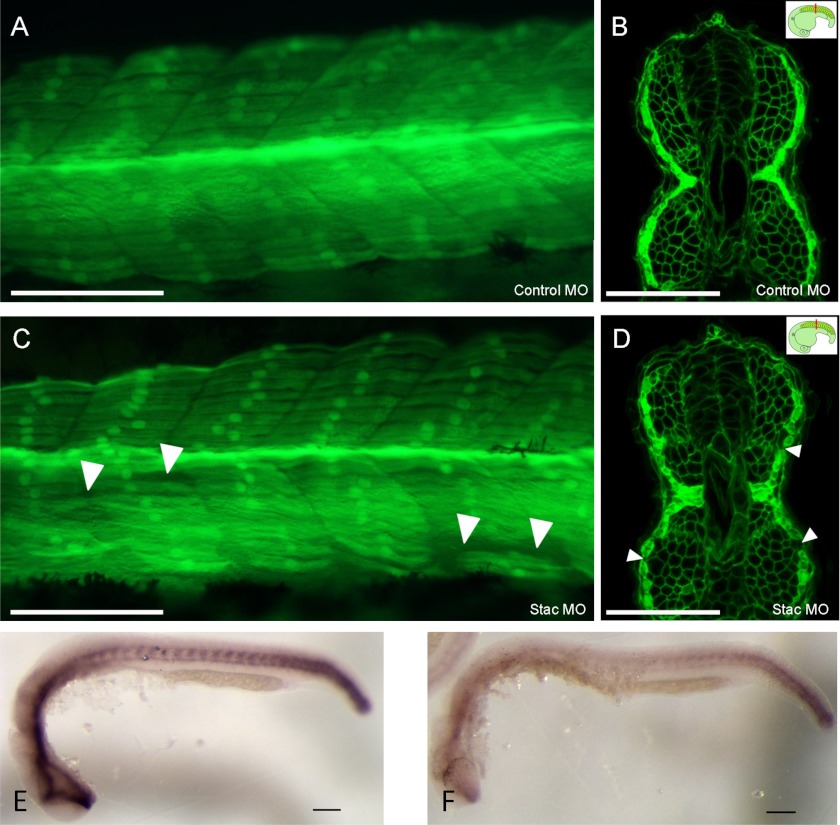
To determine the effect of stac3 expression in vivo, morpholino knockdown of stac3 in zebrafish was performed. In fish expressing GFP under the control of a fast muscle α actin promoter, stac3 morphant fish displayed a phenotype showing abnormal fast muscle fibers when compared with controls (Fig. 3, E and F). When slow muscle myofibrillar protein assembly was examined using an antibody specific to slow muscle myosin, morphant fish displayed impaired myofibrillar protein assembly (Fig. 3, G and H). In transgenic zebrafish expressing GFP under control of the slow myosin heavy chain promoter, and membrane bound GFP under control of the β actin promoter, control embryos displayed normal slow muscle fiber morphology at 24 h postfertilization (Fig. 4, A and B). In contrast, stac3 morphant embryos had abnormal muscle fiber morphology, which included the loss of slow muscle fibers (Fig. 4, C and D). The differential effects of stac3 morpholino on fast and slow muscle myofibrillar assembly are consistent with the observed expression of stac3 in adaxial cells, and the expression in developing fast muscle cells (Fig. 3D, ii–iv). As hedgehog signaling is known to be a critical regulator of slow muscle development, we examined the expression of the hedgehog receptor ptc1. Morphant embryos had reduced levels of ptc1 (19/34) (Fig. 4E) when compared with controls (12/12) by in situ hybridization (Fig. 4F).
FIGURE 4.
Injection of morpholinos in Tg(sMHC-GFP:β-actin-GFPcaax) embryos at 24 h postfertilization. A and B, Mo-control shows normal slow muscle fiber morphology (dose 1.5 ng). A and B, Mo-stac3 shows abnormal slow muscle fiber morphology including loss of slow muscle fibers (white arrowheads) (dose 1.5 ng). Lateral view is shown in A and C. Confocal image of transverse vibratome sections in B and D, insets shows the plane of section cut immediately after the yolk (red line). Expression of ptc1 is reduced in morphant embryos (F) when compared with controls (E). Scale bars represent 100 μm.
The number of slow muscle fibers were counted in somites 6 to 16 in embryos when somite 30 first contained fibers (18 h at 28 °C). There was a significant difference (p < 0.05) in mean number of slow muscle fibers per somite between control and stac3 morpholino-injected groups (p = 0.01; 7.49 ± 0.26 (n = 9) versus 8.48 ± 0.27 (n = 10) such that stac3 morpholino-injected fish had less fibers than control injected fish (mean ± S.E.)).
For fast muscle fibers, at 24 h postfertilization no difference was detected in mean fiber number between treatment groups (p = 0.303; 201 ± 5.8 (n = 10) versus 202 ± 4.14 (n = 10) in stac3 morpholino versus control fish (mean ± S.E.)). There was also no significant difference in fiber areas relative to total muscle area, when fiber area was expressed as a % of total muscle area (p = 0.48, 0.51 ± 0.023 versus 0.52 ± 0.008). At 48 h postfertilization no difference was detected in mean fiber number for fast muscle fibers between treatment groups (p = 0.16; 201 ± 5.8 (n = 8) versus 203 ± 4.14 (n = 7) in stac3 morpholino versus control fish (mean ± S.E.)). Also there was no significant difference in fiber areas relative to total muscle area, when fiber area is expressed as a % of total muscle area (p = 0.68, 0.49 ± 0.022 versus 0.5 ± 0.008).
Stac3 mRNA Is Expressed in C2C12 Myoblasts and Its Expression Increases during Differentiation
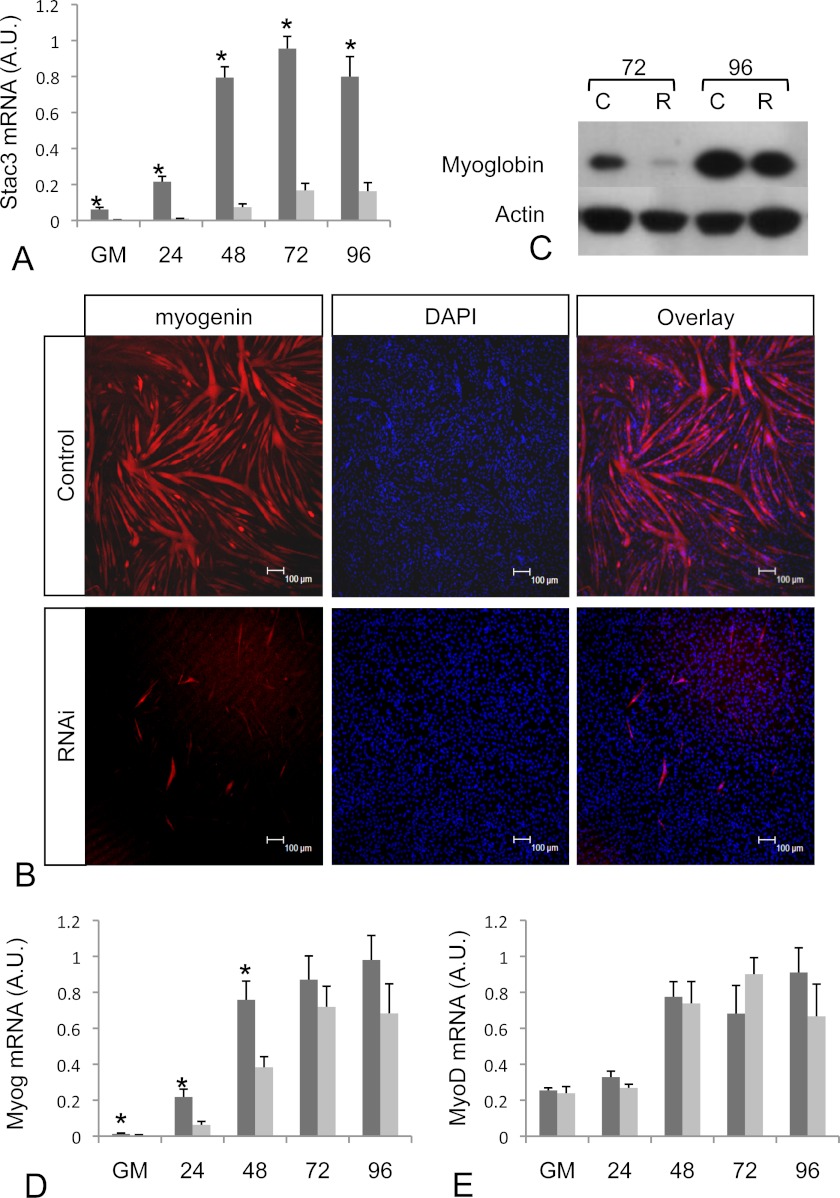
To further characterize Stac3, we examined its role in muscle development in vitro using the mammalian C2C12 myogenic cell line. C2C12 myoblasts are able to undergo terminal differentiation after GM containing 20% (v/v) fetal bovine serum is replaced by DM containing 2% (v/v) horse serum. Stac3 mRNA was detected in C2C12 myoblasts in GM, and after switching to DM, was up-regulated 4-fold within 24 h (Fig. 5A). Stac3 mRNA levels continued to increase reaching peak levels 15-fold higher than cells growing in GM 72 h after switching to DM (Fig. 5A).
FIGURE 5.
stac3 mRNA and protein expression in control and RNAi-treated C2C12 cells. A, stac3 mRNA expression increases during the differentiation of C2C12 cells (dark colored bars), and can be sufficiently knocked down by RNAi (light colored bars). Time points shown are for cells in GM and at 24, 48, 72, and 96 h in DM. B, Myoglobin protein levels (red) in S3RNAi-treated C2C12 cells (lower panels) is decreased in comparison to controls (upper panels) after 72 h in DM. Nuclei were counterstained with DAPI (blue). C, Western blot analysis of Myoglobin protein levels in RNAi-treated (R) and control cells (C) at 72 and 96 h. D, the increased Myog mRNA levels that occur during differentiation of control C2C12 cells (dark bars) is suppressed in S3RNAi-treated cells (light bars). E, there were no differences in myoD mRNA levels between RNAi-treated (light bars) and control cells (dark bars). Scale bars represent 100 μm.
Stac3 Expression Is Required for Myogenic Differentiation
To determine the role that Stac3 plays during differentiation of C2C12 cells, interfering RNAs homologous to Stac3 (S3RNAi) or a scrambled RNAi (Control) were transfected into cells prior to switching to DM. Quantitative real time PCR analysis revealed that S3RNAi-treated cells had on average Stac3 mRNA levels only 5% of those in control cells (Fig. 5A). Upon switching to DM, control cells formed multinucleated myotubes, whereas the S3RNAi-transfected cells failed to fuse, and remained as mononucleated cells. To confirm the lack of differentiation in the S3RNAi-transfected cells, we examined the expression of myoglobin by immunocytochemistry (Fig. 5B) and Western blot (Fig. 5C). Myoglobin expression in both assays revealed reduced myoglobin levels in RNAi-transfected cells. Two other RNAi oligos directed against alternative sites of Stac3 mRNA produced the same phenotype. The lack of differentiation was also confirmed using an antibody against myosin heavy chain (supplemental Fig. S1) with similar results obtained to those described for myoglobin.
Stac3 Knockdown Decreases Myogenin mRNA Levels, but Does Not Alter Those of MyoD
To further examine the role Stac3 plays in differentiation, we examined the mRNA expression of the myogenic regulatory factors MyoD and Myog. Myog mRNA levels were 2.7-fold lower in S3RNAi-treated cells, whereas cells were growing in GM. After switching to DM, Myog levels in control cells increased, whereas levels in S3RNAi-treated cells were 5.4-, 3.1-, and 1.8-fold lower in than controls at 24, 48, and 72 h, with similar levels measured between both groups at 120 h in DM (Fig. 5D) indicating that cells were not exiting the cell cycle when grown in DM. There was no change in the mRNA expression profiles for myoD between treated and control cells (Fig. 5E). As well as having decreased levels of myogenin, further evidence that S3RNAi cells were not leaving the cell cycle was confirmed by the reduced levels of p21 when compared with controls (supplemental Fig. S2A). Additionally, S3RNAi-treated myoblasts were unable to fuse as reflected in reduced levels of Cav3 mRNA (supplemental Fig. S2B).
Identification of Genes Regulated by Stac3
To identify genes that may be regulated by pathways involving Stac3, we performed microarray analysis to compare S3RNAi-transfected cells with control transfected cells. As Stac3 is expressed in myoblasts, and is then up-regulated during differentiation, we analyzed samples at 0 days (24 h after RNAi treatment) and 24 h after switching to DM. Using an adjusted p value cutoff of p < 0.01, 570 genes were identified as being significantly up-regulated and 487 as significantly down-regulated at 0 days. Using the same criteria, 1301 and 1208 genes were up- and down-regulated, respectively, 24 h after switching to DM. Genes identified as up- and down-regulated are listed in supplemental Tables S2 and S3, respectively. Based on the gene ontology term distribution of the differentially regulated genes, the most abundant terms were associated with cell cycle progression, actin cytoskeleton, and focal adhesion (supplemental Tables S2 and S3).
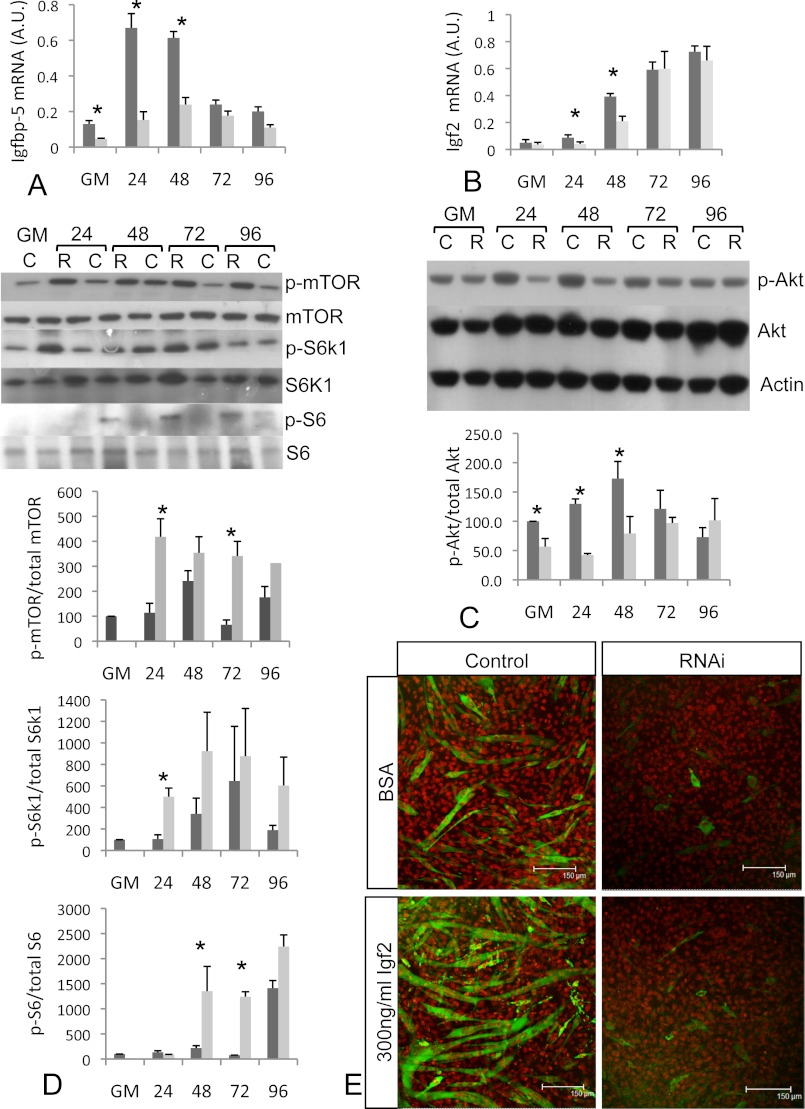
Knockdown of Stac3 Suppresses Igf2 and Igfbp-5 Expression
Within the microarray differentially regulated gene list, Igf2 and Igfbp-5, which are known to be critical for myogenic differentiation (18), were identified as being down-regulated. The down-regulation of both Igf2 and Igfbp-5 was confirmed by quantitative PCR (Fig. 6, A and B). Igfbp-5 mRNA levels were 2.9-fold lower in S3RNAi cells growing in GM (Fig. 6A). During myogenic differentiation, Igfbp-5 mRNA levels in control cells increased 7-fold within 24 h of switching to DM, however, in S3RNAi-transfected cells, the increase in Igfbp-5 mRNA was much lower (Fig. 6A). Igfbp-5 levels were 5.1-, 4.1-, and 1.9-fold lower than in control cells 24, 48, and 72 h after switching to DM (Fig. 6A). Igf2 mRNA levels were 1.4-fold higher in control cells in GM (Fig. 6B). Upon switching to DM, Igf2 mRNA levels were increased in control cells, however, the increase in S3RNAi-treated cells was not as great, with levels 2.1- and 1.9-fold lower at 24 and 48 h, respectively. The reduced mRNA levels for myogenin, Igfbp-5, and Igf2 in S3RNAi-treated cells were restricted mainly to the early stages of differentiation (24 and 48 h in DM). This could be due to degradation of the RNAi at later time points as Stac3 mRNA levels start to increase (Fig. 5A). Additionally, mRNA levels for these genes may increase in nontransfected cells, which are undergoing differentiation due to contact inhibition (19). Importantly, we only observed decreased differentiation when high (>90%) levels of Stac3 mRNA inhibition were obtained at day 0 (cells in GM).
FIGURE 6.
A, Igfbp-5 mRNA levels are lower in S3RNAi-treated cells (light bars) when compared with control cells (dark bars). B, Igf2 mRNA levels are suppressed in S3RNAi-treated cells. C, phosphorylation of Akt (Ser-473) is suppressed in S3RNAi-treated cells (light bars) when compared with controls (dark bars). D, phosphorylation of mTOR (Ser-2448), S6k1 (Thr-389), and S6 ribosomal protein (Ser-235/236) is increased in S3RNAi-treated cells (light bars) when compared with control cells (dark bars). E, exogenously applied Igf2 fails to rescue the S3RNAi-treated cells. Confocal microscopy images showing myosin heavy chain (green) and nuclei (red) of control and RNAi cells stimulated with BSA or 300 ng/ml of Igf2 after growing for 72 h in DM.
Stac3 Knockdown Suppresses Akt Phosphorylation and Increases Phosphorylation of mTOR and Its Downstream Targets S6k1 and S6
Igf2 and Igfbp-5 are known to regulate Igf2 mRNA levels through activating the Igf-1R/PI3K/Akt signaling pathway in C2C12 cells (18). Additionally, knockdown of Igfbp-5 inhibits differentiation of C2C12 cells (18), but does not alter mRNA levels of myoD, similar to what we observed from knockdown of Stac3. This raised the possibility that Igf/Akt signaling might be perturbed in Stac3 RNAi-treated cells. To test this, we measured the levels of phosphorylated Akt to determine whether the reduced expression of Igf2 and Igfbp-5 in Stac3 RNAi-transfected cells was sufficient to effect signaling of this pathway. Phospho-Akt levels in S3RNAi cells were 1.8-fold lower in GM and 3.1 and 2.1 lower at 24 and 48 h after switching to DM (Fig. 6C). Igf/Akt signaling can lead to phosphorylation of downstream targets such as mTOR. To determine whether the effects of Stac3 on Akt signal through this pathway, we examined the phosphorylation of mTOR. Rather than seeing decreased phosphorylation of mTOR, which might have been predicted by the decreased phosphorylation of Akt, we observed increased phosphorylation, with significant increases at 24 and 72 h in DM (Fig. 6D). Additionally, we also observed increased phosphorylation of the mTOR effector S6k1 and its downstream target S6 ribosomal protein with increased phosphorylation at 24 h for S6k1 and 48 and 72 h in DM for S6 ribosomal protein (Fig. 6D).
Exogenous Igf2 Fails to Rescue Stac3 Knockdown Cells
As phosphorylation of Akt was reduced and both Igfbp-5 and Igf2 mRNA levels decreased following RNAi mediated knockdown of Stac3, we hypothesized that the reduced flux through this pathway might be the causal effect inhibiting differentiation. To test this hypothesis and try and “rescue” the Stac3 RNAi-transfected cells, we applied exogenous Igf2 to RNAi-treated cells. Exogenously applied Igf2 (150 and 300 ng/ml in DM) did not recover the phenotype (Fig. 6E).
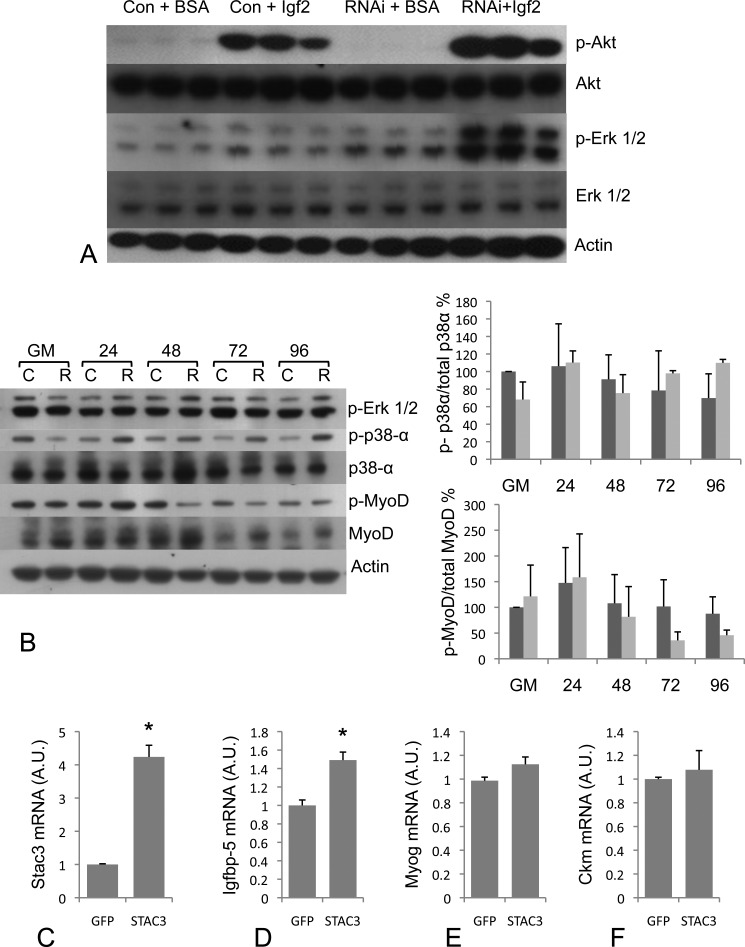
Igf/Akt Signaling Is Not Blocked in S3RNAi Cells
As exogenous Igf2 was unable to rescue the S3RNAi phenotype, this raised the possibility that Igf/Akt signaling was blocked by knockdown of Stac3, which would place Stac3 within the signaling cascade between the Igf1 receptor and Akt. To test this hypothesis, we starved cells of serum for 24 h, and then applied exogenous Igf2, or BSA to control and S3RNAi-treated cells. Phosphorylated Akt was undetectable in control and S3RNAi-starved cells treated with BSA (Fig. 7A), however, both control and S3RNAi cells treated with exogenous Igf2 displayed increased levels of phosphorylated Akt (Fig. 7A), indicating that Igf/Akt signaling is not blocked in S3RNAi cells, thus placing Stac3 external to the Igf/Akt pathway in myogenesis. Interestingly, in starved S3RNAi cells, we observed increased phosphorylation of Erk1/2 by Igf2 stimulation, which was absent from control cells (Fig. 7A). This suggests that during serum starvation, S3RNAi cells do not exit the cell cycle (20).
FIGURE 7.
A, phospho-Akt (Ser-473), total AKT, phospho-Erk1/2 (Thr-202/Tyr-204), and total Erk1/2 protein levels in RNAi-treated and control cells starved of serum for 24 h and then stimulated with Igf2 (150 ng/ml) for 2 h showing that Igf/Akt signaling is not blocked in S3RNAi-treated cells. B, Western blot analysis of phospho-Erk1/2 (Thr-202/Tyr-204), p38α, phospho-p38α (Thr-180/Tyr-182), MyoD, and phospho-MyoD (Ser-200), in RNAi-treated and control cells. There were no differences for these signaling pathways between S3RNAi-treated and control cells. C, mRNA levels for Stac3 in gfp (control) and STAC3 overexpressing cells. D, Igfbp-5 mRNA levels are increased in Stac3 overexpressing cells, but no differences were found for other markers of differentiation Myog (E) and Ckm (F) in cells overexpressing Stac3, or gfp.
Stac3 Knockdown Does Not Affect Erk, p38α, or MyoD Phosphorylation
Activation of several other signaling pathways is required for the differentiation of myogenic cells including the regulation of myoD, Erk1/2, and p38. The Raf-Mek-Erk pathway is known to regulate both p21 and myogenin expression with constitutively active Raf reducing the levels of both genes (21). As we observed increased phosphorylation of Erk1/2 in the Igf2-stimulated serum-starved cells, we next examined the phosphorylation of p42/p44 during C2C12 differentiation to determine whether knockdown of Stac3 affected this pathway, which may explain the reduced expression of myog and p21 in S3RNAi cells. Western blot analysis revealed that knockdown of Stac3 expression does not affect the Raf-Mek-Erk branch of Igf signaling (Fig. 7B).
The p38α pathway has been linked to myogenic differentiation with increased phosphorylation of p38α observed during differentiation of C2C12 cells (22). We were unable to detect any differences in phosphorylation of p38α between control and RNAi-treated cells (Fig. 7B).
During the differentiation of myogenic cells, phosphorylation of MyoD at Ser-200 decreases, leading to increased expression of E-box-dependent genes (10). Similar to p38α, we did not observe any significant differences in levels of phosphorylated myoD between control and RNAi-treated cells (Fig. 7B).
Stac3 Overexpression
We overexpressed Stac3 and examined the expression of several genes that were knocked down in the RNAi-treated cells to see if Stac3 is involved in the transcriptional regulation of these genes and if overexpression can lead to enhanced myogenic differentiation. 48 h after switching to DM, Stac3 overexpressing cells had Stac3 mRNA levels 4.2-fold higher than in controls (Fig. 7C). Cells overexpressing Stac3 had mRNA levels of Igfbp-5, which were 1.5-fold higher than in control cells expressing gfp (Fig. 7D). Although Igfbp-5 levels increased, we did not observe increased expression levels of any other differentiation markers including myog (Fig. 7E) and Ckm (Fig. 7F), demonstrating that increased levels of Stac3 do not lead to increased differentiation. As Stac3 overexpression resulted in increased Igfbp-5 mRNA levels, without increased differentiation, this suggests that Stac3 is involved in multiple pathways leading to myogenic differentiation.
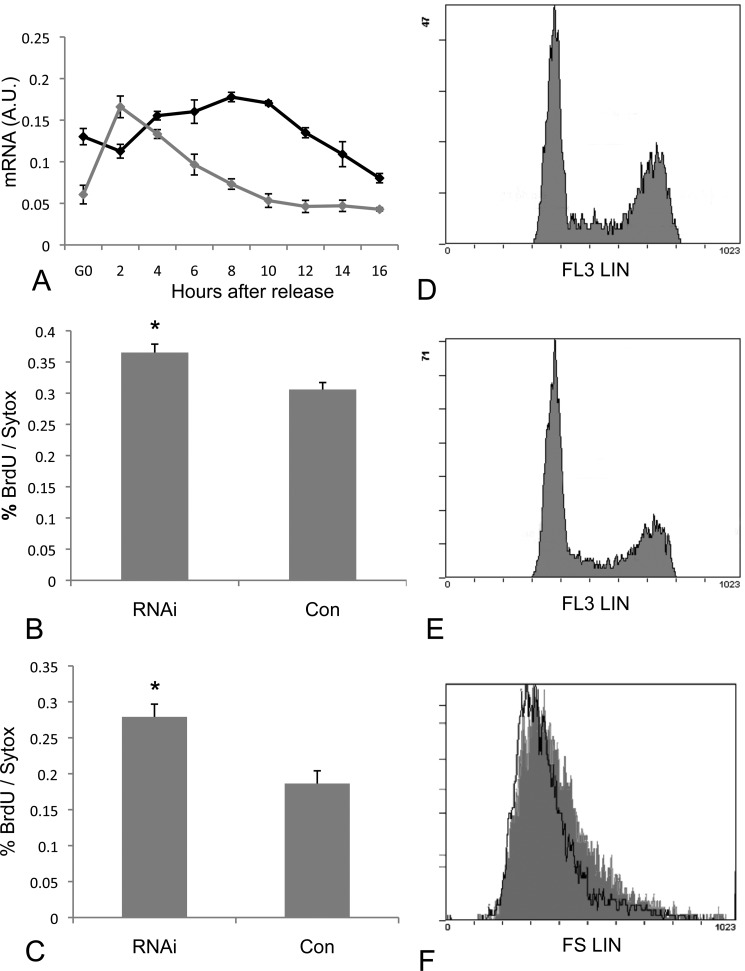
Stac3 Is Expressed during the G1 Phase of the Cell Cycle and RNAi-treated Cells Have Faster Proliferation Rates
To determine at which point within the cell cycle Stac3 is expressed, we synchronized cells in G0 by methionine deprivation. For comparative purposes we measured mRNA levels of MyoD (Fig. 8A), and found an expression profile for MyoD similar to that previously reported in synchronized cells (10) confirming the synchronization of the cells. Stac3 mRNA levels increased from G0 levels 4–10 h after release from methionine deprivation (Fig. 8A). mRNA levels remained elevated through G1, but then declined as cells entered S phase at 12–16 h post release (confirmed by BrdU incorporation). We also measured cell proliferation rates finding higher proliferation in S3RNAi cells in comparison to controls when grown in GM and DM (p < 0.01) (Fig. 8, B and C). Combined, these findings suggest that Stac3 may be involved in regulation of the cell cycle during the G1 phase. This is further supported with data obtained by flow cytometry from cells grown in GM, with S3RNAi-treated cells having a lower proportion of cells in the G1 phase of the cell cycle, and increased proportions in S phase and G2 phase (RNAi G1 = 50.82 ± 2.26%, S = 15.52 ± 0.21%, G2 = 33.65 ± 2.47%; control G1 = 59.65 ± 0.27%, S = 13.79% ± 0.32, G2 = 26.55 ± 0.23% (mean ± S.D.)) with statistically significant differences in the G1, S, and G2 phases of the cell cycle (p = 0.001, 0.0007, and 0.004, respectively) (Fig. 8, D and E). Additionally, S3RNAi cells appeared smaller in culture, and this observation was confirmed by flow cytometry where control cells had a mean forward linear scatter of 407.0 (half-peak coefficient of variation = 2.0), whereas S3RNAi cells had a mean forward linear scatter of 372.6 (half-peak coefficient of variation = 3.3) (Fig. 8F). It is known that during the cell cycle, cellular growth resulting in increased cell size occurs during the G1 phase. Therefore, the difference in size between control and RNAi-treated cells is even more dramatic when it is considered that the controls have a greater proportion of cells within the G1 phase of the cell cycle (Fig. 8E). In comparison, S3RNAi-treated cells have a greater proportion of cells in the S and G2 phases of the cell cycle (Fig. 8D), cells that have already undergone cell growth. These findings add further evidence that Stac3 functions as a regulator of the passage through the G1 phase of the cell cycle.
FIGURE 8.
A, Stac3 (black) and MyoD (gray) mRNA levels in cells synchronized at the G0 phase of the cell cycle and following release of methionine deprivation. Stac3 expression is highest during the G1 phase of the cell cycle and decreases as cells enter S-phase. B, cell proliferation rates in RNAi-treated and control cells 24 h after transfection and growing in GM. C, cell proliferation rates in RNAi-treated and control cells 24 h after transfection and growing in DM. D, flow cytometry analysis showing the proportion of cells in G1, S-phase, and G2 phase of the cell cycle for S3RNAi-treated cells growing in GM (G1 = 50.82 ± 2.26%, S = 15.52 ± 0.21%, G2 = 33.65 ± 2.47%). E, flow cytometry analysis showing the proportion of cells in G1, S-phase, and G2 phase of the cell cycle for control cells growing in GM (G1 = 59.65 ± 0.27%, S = 13.79 ± 0.32%, G2 = 26.55 ± 0.23%). F, flow cytometry analysis of forward linear scatter. S3RNAi-treated cells are smaller (white with black outline, mean = 372.6, half-peak coefficient of variation = 3.3) than control cells (gray, mean = 407.0, half-peak coefficient of variation = 2.0) as indicated by a shift to the left.
DISCUSSION
We have identified Stac3 as a novel regulator of myogenic differentiation and myofibrillar protein assembly, which appears to have conserved roles in teleost fish and mammals. Myogenesis involves the exit of proliferating myoblasts from the cell cycle, cell-cell fusion, elongation events, and sarcomere assembly all of which require extensive modification of the actin cytoskeleton (23). Myogenesis is regulated by the MyoD (MyoD, Myf-5, Myog, and Mrf4) and myocyte enhancer factor-2 families of transcription factors (reviewed in Ref. 24). MyoD and Myf-5 have a role in the specification of cells to the myogenic lineage and are expressed in proliferating myoblasts (25). MyoD is phosphorylated in proliferating myoblasts with dephosphorylation at Ser-200 resulting in exit from the cell cycle and the initiation of the differentiation program, leading to increased expression of E-box-regulated genes (10). In C2C12 cells we did not observe any differences in mRNA levels of MyoD, or in the phosphorylation status of MyoD during the differentiation of S3RNAi or control cells, and microarray analysis did not reveal any difference in transcript levels for Myf5 or Mrf4. In contrast, Myog displayed decreased expression in Stac3 knockdown cells (Fig. 5D). Expression of Myog requires phosphorylation of Akt and inhibition of Erk1/2 phosphorylation (26), and occurs later in the differentiation program than MyoD and Myf5 (27). In S3RNAi cells, phosphorylation of Akt is suppressed, but we observed no difference in Erk1/2 phosphorylation during differentiation. Activation of other signaling pathways including p38 have also been implicated in myogenic differentiation. For example, differentiation required increased phosphorylation of p38α (22), and p38 activation was required for activation of Akt (28). However, we found no difference between S3RNAi and control cells, indicating that Stac3 is not involved in the p38α pathway.
During myogenic differentiation, phosphorylation of Akt also requires signaling through Igf2 and Igfbp-5 (18) and we observed decreased levels of Igf2 and Igfbp-5 mRNA in S3RNAi cells. Despite this, we were unable to rescue S3RNAi-treated cells with exogenous Igf2 (Fig. 6E). By starving cells of serum and then stimulating with Igf2, we were able to show that Akt signaling is not blocked (Fig. 5A). The suppressed phosphorylation of Akt during differentiation (Fig. 6C), without blocking this pathway, combined with the inability to rescue with exogenous Igf2 places Stac3 upstream of Akt, but indicates functionality in more than one of the pathways necessary for myogenic differentiation. In response to Igf signaling, Akt phosphorylates its downstream target mTOR, leading to increased protein synthesis and muscle hypertrophy (29). Although Akt phosphorylation was reduced in S3RNAi-treated cells, we saw an increase in mTOR phosphorylation, its effector S6k1, and downstream target S6 ribosomal protein (Fig. 6D). As mTOR/S6k1 signaling is required for cell cycle progression (30), the increased phospho-mTOR and phospho-S6k1 levels observed, without increased differentiation, support the contention that S3RNAi-treated cells have not exited the cell cycle. Interestingly, in serum-starved cells, Igf2 stimulated phosphorylation of Erk1/2 in S3RNAi cells but not in controls, suggesting that S3RNAi cells had failed to exit the cell cycle during serum starvation (20). Further evidence of a role for Stac3 in the cell cycle regulation was obtained from flow cytometry and proliferation assays. The decreased proportion of cells in the G1 phase of the cell cycle, their smaller size, and increased proliferation rates suggest that S3RNAi-treated cells are traversing the G1 phase of the cell cycle more rapidly than control cells.
Using morpholino knockdown of stac3 in zebrafish, we were able to demonstrate a role for this gene in vivo. Slow and fast muscle fiber types are arranged in discrete layers in teleosts. Early in development, slow muscle progenitors, termed adaxial cells, are found adjacent to the notochord, which, in response to hedgehog signaling derived from the notochord (31), migrate laterally to form a superficial layer of slow muscle (32). The expression pattern we observed for stac3 closely matches that of migrating adaxial cells, indicating a role for stac3 in the development of slow muscle fibers in zebrafish. This is further highlighted in stac3 morphant fish, which have impaired myofibrillar protein assembly within the slow muscle fibers. Stac3 also appears to play a role in fast muscle development as fast muscle fibers in morphant fish had abnormal appearance when compared with controls. This is consistent with the observed expression in developing fast muscle fibers (Fig. 3, C, ii and iii, and D, ii–iv), and the high mRNA expression in Atlantic salmon fast muscle (Fig. 2B). As signals derived from migrating slow muscle cells are responsible for the patterning of fast muscle fibers (33), the observed expression of stac3 in adaxial cells, and impaired development of slow muscle fibers derived from this cell type (Fig. 3, G and H) may also explain the abnormal appearance of fast muscle fibers. The slow muscle phenotype in morphant fish is similar to that described by Wolff et al. (34), when hedgehog signaling was inhibited using cyclopamine, and we observed decreased levels of ptc1 in morphant fish (Fig. 4, E and F). The decreased levels of patched mRNA suggest that hedgehog signaling is perturbed in stac3 morphant fish, however, it should be noted that stac3 expression was observed in the dorsal and ventral extremes of the myotome following segmentation (Fig. 3D, ii–iv), in cells that differentiate to produce slow muscle fibers independent of hedgehog signaling (31, 35).
Thalappilly et al. (36) used the yeast two-hybrid approach to identify proteins that interact with scaffold proteins containing multiple SH3 domains that were expressed in pancreatic ductal adenocarcinoma cells including STAC (Q99469). Proteins that interacted with the STAC bait protein included LMNA, C9orf156, TRIP6, and NCKIPSD. The LMNA genes code for A-type lamins (lamin A/C), a class of intermediate filaments required for disassembly of the nuclear envelop during early mitosis (37). Mutations in human LMNA genes give rise to multiple clinical syndromes called “laminopathies,” including Emery-Dreifuss muscular dystrophy and dilated cardiomyopathy (38). Reduced function of lamin A/C in zebrafish resulted in abnormal muscle development and lipodystrophic phenotypes (39). The deletion of 8 amino acid residues of zebrafish lamin A resulted in embryonic senescence and S-phase accumulation/arrest (39). TRIP6 (thyroid receptor-interacting protein 6), a member of the zyxin family of proteins, functions to relay signals from the cell surface to the nucleus to weaken adherin junctions and promote actin cytoskeletal reorganization and cell migration (40). NCKIPSD encodes NCK interacting protein with the SH3 domain (SPIN90). SPIN90 also plays a role in signal transduction and may function in the maintenance of sarcomeres and in the assembly of myofibrils into sarcomeres (41). These results indicate that STAC family members function as signal adaptor molecules in a variety of tissues through interactions with multiple signaling pathways controlling cell cycle progression, cytoskeletal organization, and cell migration.
CONCLUSIONS
We have identified Stac3 as a novel regulator of myogenic differentiation and myofibrillar assembly, which affects the Akt pathway during differentiation by as yet unknown mechanisms that are independent of p38α, Erk1/2, and MyoD pathways. Based on the affects of Stac3 knockdown on cell differentiation, proliferation, cell size, the increased phosphorylation of Erk1/2 in serum-starved cells and its expression during the G1 phase of the cell cycle, Stac3 may play a role as a regulator of the G1 phase cell cycle checkpoint in myogenic cells.
Acknowledgments
We thank Harry Hodge for help with flow cytometry analysis, Clara Coll for several cell pictures, and Dr. Daniel Macqueen for several primer pairs.
This work was supported in part by the MASTS pooling initiative (The Marine Alliance for Science and Technology for Scotland) supported by Scottish Funding Council Grant HR09011.

This article contains supplemental Tables S1–S3 and Figs. S1 and S2.
- Igf
- insulin-like growth factor
- GM
- growth medium
- DM
- differentiation media
- SH3
- Src homology
- RACE
- rapid amplification of cDNA ends
- mTOR
- mammalian target of rapamycin.
REFERENCES
- 1. Suzuki H., Kawai J., Taga C., Yaoi T., Hara A., Hirose K., Hayashizaki Y., Watanabe S. (1996) Stac, a novel neuron-specific protein with cysteine-rich and SH3 domains. Biochem. Biophys. Res. Commun. 229, 902–909 [DOI] [PubMed] [Google Scholar]
- 2. Legha W., Gaillard S., Gascon E., Malapert P., Hocine M., Alonso S., Moqrich A. (2010) stac1 and stac2 genes define discrete and distinct subsets of dorsal root ganglia neurons. Gene Expr. Patterns 10, 368–375 [DOI] [PubMed] [Google Scholar]
- 3. Satoh J., Nanri Y., Yamamura T. (2006) Rapid identification of 14-3-3 proteins by protein microarray analysis. J. Neurosci. Methods 152, 278–288 [DOI] [PubMed] [Google Scholar]
- 4. Dougherty M. K., Morrison D. K. (2004) Unlocking the code of 14-3-3. J. Cell Sci. 117, 1875–1884 [DOI] [PubMed] [Google Scholar]
- 5. Hardy K., Mansfield L., Mackay A., Benvenuti S., Ismail S., Arora P., O'Hare M. J., Jat P. S. (2005) Transcriptional networks and cellular senescence in human mammary fibroblasts. Mol. Biol. Cell 16, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bower N. I., Johnston I. A. (2010) Discovery and characterization of nutritionally-regulated genes associated with muscle growth in Atlantic salmon. Physiol. Genomics 42, 114–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bower N. I., Johnston I. A. (2010) Paralogues of Atlantic salmon myoblast determination factor genes are distinctly regulated in proliferating and differentiating myogenic cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R1615–R1626 [DOI] [PubMed] [Google Scholar]
- 8. Bower N. I., Johnston I. A. (2010) Targeted rapid amplification of cDNA ends (T-RACE), an improved RACE reaction through degradation of non-target sequences. Nucleic Acids Res. 38, e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bower N. I., Johnston I. A. (2010) Transcriptional regulation of the IGF signaling pathway by amino acids and insulin-like growth factors during myogenesis in Atlantic salmon. PLoS ONE 5, e11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kitzmann M., Vandromme M., Schaeffer V., Carnac G., Labbé J. C., Lamb N., Fernandez A. (1999) cdk1- and cdk2-mediated phosphorylation of MyoD Ser200 in growing C2 myoblasts. Role in modulating MyoD half-life and myogenic activity. Mol. Cell. Biol. 19, 3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., Wittwer C. T. (2009) The MIQE guidelines. Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 [DOI] [PubMed] [Google Scholar]
- 12. Ruijter J. M., Ramakers C., Hoogaars W. M., Karlen Y., Bakker O., van den Hoff M. J., Moorman A. F. (2009) Amplification efficiency. Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH 0034, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cole N. J., Tanaka M., Prescott A., Tickle C. (2003) Expression of limb initiation genes and clues to the morphological diversification of threespine stickleback. Curr. Biol. 13, R951–R952 [DOI] [PubMed] [Google Scholar]
- 15. Lewis K. E., Currie P. D., Roy S., Schauerte H., Haffter P., Ingham P. W. (1999) Control of muscle cell-type specification in the zebrafish embryo by Hedgehog signalling. Dev. Biol. 15, 469–480 [DOI] [PubMed] [Google Scholar]
- 16. Elworthy S., Hargrave M., Knight R., Mebus K., Ingham P. W. (2008) Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development 135, 2115–2126 [DOI] [PubMed] [Google Scholar]
- 17. Williams R. J., Hall T. E., Glattauer V., White J., Pasic P. J., Sorensen A. B., Waddington L., McLean K. M., Currie P. D., Hartley P. G. (2011) The in vivo performance of an enzyme-assisted self-assembled peptide/protein hydrogel. Biomaterials 32, 5304–5310 [DOI] [PubMed] [Google Scholar]
- 18. Ren H., Yin P., Duan C. (2008) IGFBP-5 regulates muscle cell differentiation by binding to IGF-II and switching on the IGF-II auto-regulation loop. J. Cell Biol. 182, 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanaka K., Sato K., Yoshida T., Fukuda T., Hanamura K., Kojima N., Shirao T., Yanagawa T., Watanabe H. (2011) Evidence for cell density affecting C2C12 myogenesis. Possible regulation of myogenesis by cell-cell communication. Muscle Nerve 44, 968–977 [DOI] [PubMed] [Google Scholar]
- 20. Sarbassov D. D., Jones L. G., Peterson C. A. (1997) Extracellular signal regulated kinase-1 and -2 respond differently to mitogenic and differentiative signaling pathways in myoblasts. Mol. Endocrinol. 11, 2038–2047 [DOI] [PubMed] [Google Scholar]
- 21. Rommel C., Clarke B. A., Zimmermann S., Nuñez L., Rossman R., Reid K., Moelling K., Yancopoulos G. D., Glass D. J. (1999) Differentiation stage-specific inhibition of the raf–MEK–ERK pathway by Akt. Science 286, 1738–1741 [DOI] [PubMed] [Google Scholar]
- 22. Perdiguero E., Ruiz-Bonilla V., Gresh L., Hui L., Ballestar E., Sousa-Victor P., Baeza-Raja B., Jardí M., Bosch-Comas A., Esteller M. (2007) Genetic analysis of p38 MAP kinases in myogenesis. Fundamental role of p38α in abrogating myoblast proliferation. EMBO J. 26, 1245–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buckingham M., Bajard L., Chang T., Daubas P., Hadchouel J., Meilhac S., Montarras D., Rocancourt D., Relaix F. (2003) The formation of skeletal muscle. From somite to limb. J. Anat. 202, 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sabourin L. A., Rudnicki M. A. (2000) The molecular regulation of myogenesis. Clin. Genet. 57, 16–25 [DOI] [PubMed] [Google Scholar]
- 25. Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H. H., Jaenisch R. (1993) MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75, 1351–1359 [DOI] [PubMed] [Google Scholar]
- 26. Tiffin N., Adi S., Stokoe D., Wu N. Y., Rosenthal S. M. (2004) Akt phosphorylation is not sufficient for insulin-like growth factor-stimulated myogenin expression but must be accompanied by down-regulation of mitogen-activated protein kinase/extracellular signal-regulated kinase phosphorylation. Endocrinology 145, 4991–4996 [DOI] [PubMed] [Google Scholar]
- 27. Cornelison D. D., Wold B. J. (1997) Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 191, 270–283 [DOI] [PubMed] [Google Scholar]
- 28. Cabane C., Coldefy A. S., Yeow K., Dérijard B. (2004) The p38 pathway regulates Akt both at the protein and transcriptional activation levels during myogenesis. Cell. Signal. 16, 1405–1415 [DOI] [PubMed] [Google Scholar]
- 29. Bodine S. C., Stitt T. N., Gonzalez M., Kline W. O., Stover G. L., Bauerlein R., Zlotchenko E., Scrimgeour A., Lawrence J. C., Glass D. J., Yancopoulos G. D. (2001) Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3, 1014–1019 [DOI] [PubMed] [Google Scholar]
- 30. Fingar D. C., Richardson C. J., Tee A. R., Cheatham L., Tsou C., Blenis J. (2004) mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 24, 200–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barresi M. J., Stickney H. L., Devoto S. H. (2000) The Zebrafish slow-muscle-omitted gene product is necessary for hedgehog signal transduction and the development of slow muscle identity. Development 127, 2189–2199 [DOI] [PubMed] [Google Scholar]
- 32. Devoto S. H., Melançon E., Eisen J. S., Westerfield M. (1996) Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development 122, 3371–3380 [DOI] [PubMed] [Google Scholar]
- 33. Henry C. A., Amacher S. L. (2004) Zebrafish slow muscle cell migration induces a wave of fast muscle morphogenesis. Dev Cell. 7, 917–923 [DOI] [PubMed] [Google Scholar]
- 34. Wolff C., Roy S., Ingham P. W. (2003) Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 13, 1169–1181 [DOI] [PubMed] [Google Scholar]
- 35. Barresi M. J., D'Angelo J. A., Hernández L. P., Devoto S. H. (2001) Distinct mechanisms regulate slow muscle development. Curr. Biol. 11, 1432–1438 [DOI] [PubMed] [Google Scholar]
- 36. Thalappilly S., Suliman M., Gayet O., Soubeyran P., Hermant A., Lecine P., Iovanna J. L., Dusetti N. J. (2008) Identification of multi-SH3 domain-containing protein interactome in pancreatic cancer. A yeast two-hybrid approach. Proteomics 8, 3071–3081 [DOI] [PubMed] [Google Scholar]
- 37. Stuurman N., Heins S., Aebi U. (1998) Nuclear lamins. Their structure, assembly, and interactions. J. Struct. Biol. 122, 42–66 [DOI] [PubMed] [Google Scholar]
- 38. Raharjo W. H., Enarson P., Sullivan T., Stewart C. L., Burke B. (2001) Nuclear envelope defects associated with LMNA mutations cause dilated cardiomyopathy and Emery-Dreifuss muscular dystrophy. J. Cell Sci. 114, 4447–4457 [DOI] [PubMed] [Google Scholar]
- 39. Koshimizu E., Imamura S., Qi J., Toure J., Valdez D. M., Jr., Carr C. E., Hanai J., Kishi S. (2011) Embryonic senescence and laminopathies in a progeroid zebrafish model. PLoS One 6, e17688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yi J. S., Kloeker S., Jensen C. C., Bockholt S., Honda H., Hirai H., Beckerle M. C. (2002) Members of the zyxin family of LIM proteins interact with members of the p130Cas family of signal transducers. J. Biol. Chem. 277, 8580–9588 [DOI] [PubMed] [Google Scholar]
- 41. Lim C. S., Park E. S., Kim D. J., Song Y. H., Eom S. H., Chun J. S., Kim J. H., Kim J. K., Park D., Song W. K. (2001) SPIN90 (SH3 protein interacting with Nck, 90 kDa), an adaptor protein that is developmentally regulated during cardiac myocyte differentiation. J. Biol. Chem. 276, 12871–12878 [DOI] [PubMed] [Google Scholar]