Abstract
p53 mutations are present in up to 70% of lung cancer. Cancer cells with p53 mutations, in general, grow more aggressively than those with wild-type p53 or no p53. Expression of tumor-derived mutant p53 in cells leads to up-regulated expression of genes that may affect cell growth and oncogenesis. In our study of this aggressive phenotype, we have investigated the receptor protein tyrosine kinase Axl, which is up-regulated by p53 mutants at both RNA and protein levels in H1299 lung cancer cells expressing mutants p53-R175H, -R273H, and -D281G. Knockdown of endogenous mutant p53 levels in human lung cancer cells H1048 (p53-R273C) and H1437 (p53-R267P) led to a reduction in the level of Axl as well. This effect on Axl expression is refractory to the mutations at positions 22 and 23 of p53, suggesting that p53’s transactivation domain may not play a critical role in the up-regulation of Axl gene expression. Chromatin immunoprecipitation (ChIP) assays carried out with acetylated histone antibodies demonstrated induced histone acetylation on the Axl promoter region by mutant p53. Direct mutant p53 nucleation on the Axl promoter was demonstrated by ChIP assays using antibodies against p53. The Axl promoter has a p53/p63 binding site, which however is not required for mutant p53–mediated transactivation. Knockdown of Axl by Axl-specific RNAi caused a reduction of gain-of-function (GOF) activities, reducing the cell growth rate and motility rate in lung cancer cells expressing mutant p53. This indicates that for lung cancer cell lines with mutant p53, GOF activities are mediated in part through Axl.
Keywords: mutant, p53, Axl, lung
Introduction
p53 mutations are common in lung cancer and range from 33% in adenocarcinomas to 70% in small cell lung cancers. In general, tumors with p53 mutations have poor prognosis.1-6 The majority of clinical studies suggest that lung cancers with p53 alterations carry a worse prognosis and may be relatively more resistant to chemotherapy and radiation.7 More than 50% of the p53 mutations found in cancer are missense mutations, and in most human cancers, only the mutant protein is expressed.8 We and others have identified several genes important for their involvement in growth and oncogenesis that are regulated at the level of expression by gain-of-function (GOF) mutant p53.9-15
Important phenotypes ascribed to GOF activity of mutant p53 are increased tumorigenicity,16,17 increased growth rate and motility,8,18 increased metastasis and invasiveness,19 increased growth in soft agar,20 and decreased sensitivity to chemotherapeutic drugs.9,21,22 It appears that its mutational status determines the efficacy of many of the chemotherapeutic drugs.22-26 The GOF phenotypes can be partially explained by a dominant-negative effect of mutant p53 on p73 and p63.27-31
Receptor tyrosine kinases (RTKs) play an important role in the growth and differentiation of normal cells and represent a major class of proto-oncogenes that are involved in the progression and metastasis of cancer.32 Axl/Mer/Sky represents a comparatively recent class of the RTK family that induces extracellular signals inside cells.33 Axl is a RTK with transforming activity34,35 and may be used as a target for therapy.36
The antiapoptotic, cell adhesion, and chemotactic activities of Axl have been ascribed to an increased expression of Axl and its increased interaction with Gas6.37 The invasiveness and metastasis in various cancer cell types are associated with an elevated level of Axl expression, which is also correlated with a poor prognosis of patients with various cancers such as myeloid leukemia,34,38 metastatic lung cancer,39,40 breast cancer,41 and gastric cancer.42 Use of RNAi techniques and dominant-negative receptor mutants of Axl have resulted in the growth suppression of cancer cells in a xenograft model.43,44
Here, we show that the receptor protein tyrosine kinase Axl is up-regulated by p53 mutants in H1299 lung cancer cells expressing mutant p53-R175H, -R273H, and -D281G. Knockdown of Axl by Axl-specific RNAi caused a reduction of GOF activities in lung cancer cells expressing endogenous mutant p53, suggesting that mutant p53 may induce Axl as one of the target genes to execute its GOF activities.
Results
The transactivation-deficient mutant p53-D281G (L22Q/W23S) is effective in up-regulating many mutant p53 target genes
We focused our attention on potential mechanisms of GOF mutant p53 by studying the role of p53’s transactivation domain in gene regulation. We determined whether the triple amino acid substitution mutant p53-D281G (L22Q/W23S) has retained the ability to up-regulate the expression of mutant p53 target genes identified by our expression analysis.9 We compared gene expression profiles of 3 H1299 cell lines stably transfected with a vector alone (HC5), stably expressing mutant p53-D281G, and the transactivation domain mutant p53-D281G (L22Q/W23S) using Affymetrix GeneChip arrays (Santa Clara, CA). The data shown in Table 1 show a list of example genes induced by p53-D281G and not affected by the 22Q/23S mutations (P = 3 × 10–24). This indicates that a substantial part of the mutant p53 gene expression signature is independent of the transactivation domain at codons 22 and 23.
Table 1.
H1299 Cells Expressing Transactivation-Deficient Mutant p53-D281G (L22Q/W23S) Are Efficient in Up-Regulating Mutant p53 Targets
| Symbol | Fold over the control | P value | Gene |
|---|---|---|---|
| ACTN4 | 5.47 | 0.001 | Actinin, α 4 |
| AXL | 7.04 | 0.0007 | AXL receptor tyrosine kinase |
| BTG1 | 0.49 | 0.0009 | B-cell translocation gene 1, antiproliferative |
| CDC25B | 0.51 | 0.001 | Cell division cycle 25B |
| EEF1A2 | 0.28 | 0.0008 | Eukaryotic translation elongation factor 1 α 2 |
| ERCC1 | 5.23 | 1.04E-05 | Excision repair cross-complementing rodent repair deficiency, complementation group 1 |
| ERCC2 | 2.85 | 0.003 | Excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| ERF | 3.72 | 1.40E-05 | Ets2 repressor factor |
| FN1 | 0.27 | 0.002 | Fibronectin 1 |
| GARS | 5.26 | 0.002 | Glycyl-tRNA synthetase |
| GDI2 | 0.29 | 0.001 | GDP dissociation inhibitor 2 |
| GPR126 | 0.43 | 0.0001 | G protein–coupled receptor 126 |
| IGFBP6 | 17.25 | 0.0004 | Insulin-like growth factor binding protein 6 |
| PRAME | 0.28 | 0.0002 | Preferentially expressed antigen in melanoma |
| PRKAR1A | 0.19 | 8.78E-05 | Protein kinase, cAMP-dependent, regulatory, type I, α (tissue-specific extinguisher 1) |
| PSMB6 | 0.31 | 0.0002 | Proteasome (prosome, macropain) subunit, β type, 6 |
| RAB31 | 0.22 | 0.001 | RAB31, RAS oncogene family member |
| RELB | 4.77 | 3.82E-05 | v-Rel |
| SRPX | 0.37 | 0.0009 | Sushi repeat–containing protein, X-linked |
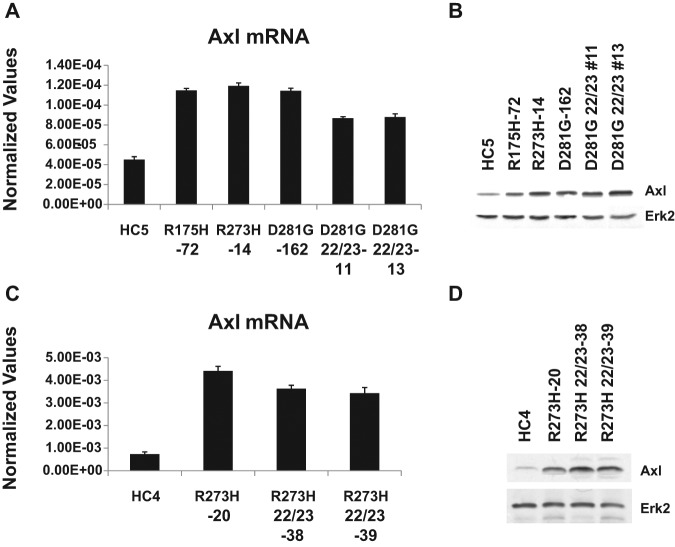
Interaction between mutant p53 and p63 has been demonstrated and shown to be independent of the transactivation domain mutants.45 The mutant p53–induced genes that we identified that are not dependent on the transactivation domain might involve a mutant p53-p63 interaction. We analyzed mutant p53–induced genes for the overrepresentation of genes that contain putative p63 binding sites. The Axl gene was selected for further study based on the presence of a p63 binding site, its known role in oncogenesis,34,40,43,46,47 and its potential role in mutant p53 GOF activities. We first verified the microarray data by performing RT-qPCR analysis of the Axl mRNA, confirming induction by mutant p53. Data presented in Figure 1A show the presence of more Axl RNA in the H1299 cells expressing mutant p53 compared to vector-transfected cells (compare lanes HC5 with R175H, R273H, D281G, and D281G [L22Q/W23S]). We also tested the Axl expression at the protein level by performing immunoblot analysis as described in Materials and Methods. Protein expression analysis shown in Figure 1B demonstrates that cell lines expressing mutant p53 have higher levels of the Axl protein compared to the vector-transfected cells. This induction of Axl protein expression by mutant p53 remains when the transactivation domain mutations at codons 22 and 23 are present, confirming our RNA expression analysis. Similar data were also observed when we compared Axl levels in H1299 cells expressing p53-R273H and -R273H with additional mutations at codons 22 and 23 (Fig. 1C and 1D).
Figure 1.
H1299 cells expressing gain-of-function mutant p53 up-regulate protein receptor tyrosine kinase Axl. (A, C) RT-qPCR analysis of Axl levels in H1299 cells stably transfected with a vector or mutant p53 expression plasmids. RT-qPCR was performed for Axl with cDNA from indicated cell lines. qPCR was performed for GAPDH and was used to normalize with Axl values. (B, D) Western blot analysis of Axl levels in H1299 cells stably transfected with a vector or mutant p53 expression plasmids. PAGE followed by Western blot analysis was performed with extracts made from cells stably transfected with a vector or mutant p53 expression plasmids. Immunoblots were developed for Axl (using an antibody against Axl from Abnova) and Erk2. Data show mutant p53–induced Axl expression both at RNA and protein levels in H1299 cells, and this induction is not disturbed by mutations at amino acids 22 and 23. Experiments were performed in technical triplicates. Experiments were performed multiple times with similar trends. Error bars showing standard deviations are indicated.
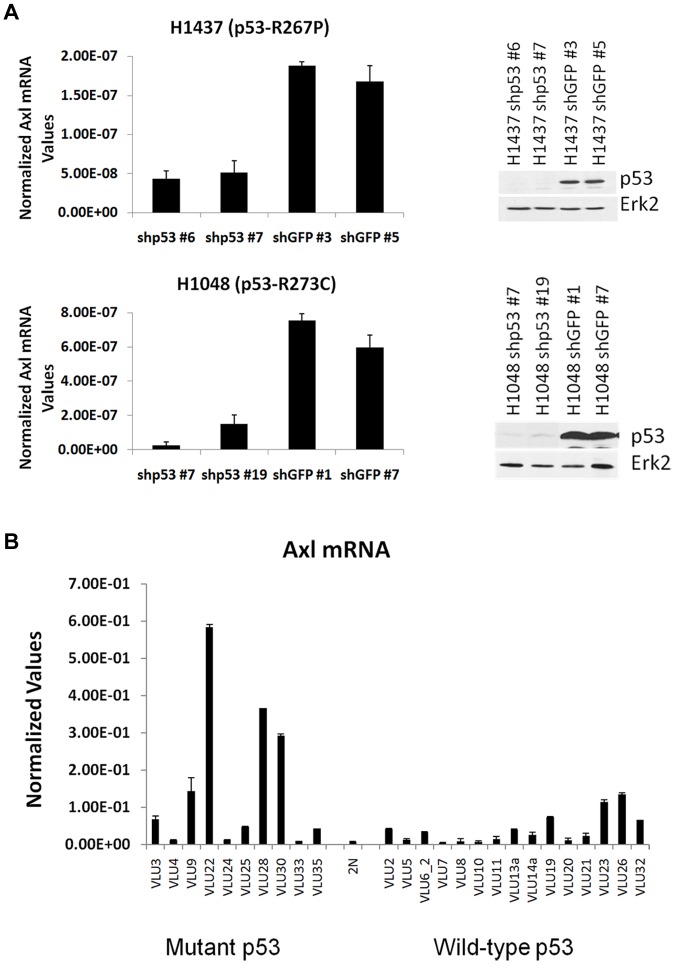
We investigated that if the converse were true, reduced expression of endogenous mutant p53 would result in reduced expression of Axl. We studied the Axl RNA level in 2 lung cancer cell lines with endogenous mutant p53, H1437 (p53-R267P), and H1048 (p53-R273C) after knockdown of the mutant p53 levels in these lines by generating stable cell clones using lentivirus vectors expressing p53 shRNA in comparison to control cells expressing green fluorescence protein (GFP) shRNA. Data depicted in Figure 2A show that a reduction of mutant p53 levels is accompanied by a reduction in Axl levels, suggesting that along with the mutant p53 induction studies, mutant p53 regulates Axl expression.
Figure 2.
Lung tumor cells expressing p53 mutations show higher Axl levels. (A) RT-qPCR of Axl mRNA levels in lung cancer cell lines H1437 and H1048. Lung cancer cell lines H1048 and H1437 were infected with control shRNA or p53 shRNA lentivirus to generate mutant p53 knockdown cell lines. Western blot analysis was performed on isolated clones to identify p53 knockdown clones. Erk2 was used as a loading control. These clones were further analyzed for Axl mRNA expression by qPCR using primers specific for the Axl gene and normalized by GAPDH. The data indicate that a decrease in mutant p53 levels lowers Axl levels. (B) RT-qPCR of Axl levels in lung tumors. cDNA was prepared from human lung tumor RNA (labeled “VLU” to protect patient identity) and a normal tissue sample (labeled “2N”) using the SuperScript III cDNA synthesis kit (Invitrogen) and qPCR performed using primers specific for Axl. The degree of expression was quantitated using a relative standard curve and normalized to GAPDH corresponding to the cDNA batch. Experiments were performed in technical triplicates as described in the text. Error bars showing standard deviations are indicated.
Lung tumor cells expressing mutant p53 show higher Axl levels
Since we observed that mutant p53 up-regulates Axl expression in lung cancer cell lines studied, we investigated whether this is also valid in human lung tumors expressing mutant p53. Figure 2B depicts the Axl level of different human lung tumors collected in the Virginia Commonwealth University cancer tissue repository. On average, there was significantly more Axl expression in samples with mutant p53 versus samples with wild-type p53 (average 5.63-fold; P = 0.013), corroborating our cell culture data that mutant p53 up-regulates Axl expression.
Axl up-regulation mediates GOF activities of mutant p53
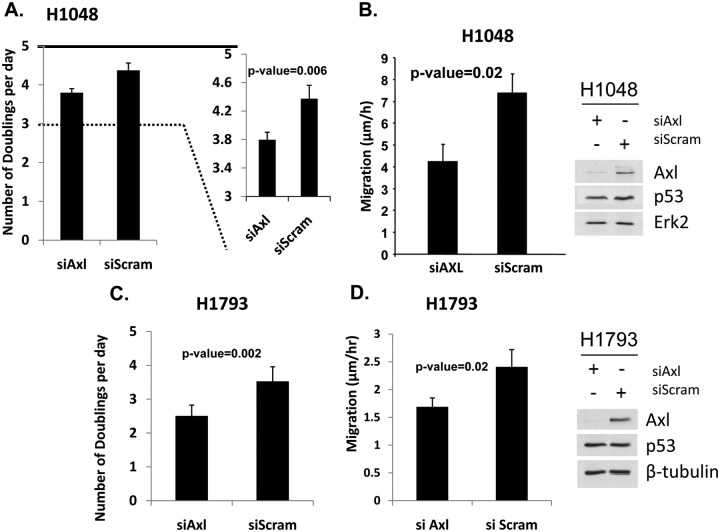
Axl is known to be involved in promoting the growth and movement of cells.37,39,43,44,47-49 Therefore, we tested whether mutant p53–induced enhancement of growth and motility has any relationship with the fact that mutant p53 up-regulates the expression of Axl. We used the lung cancer cell line H1048 expressing a mutant p53 (-R273C) for testing its growth rate and motility as described in Materials and Methods. To test whether the level of Axl affects the properties affected by GOF of mutant p53 expression, we transfected this cell line with siRNA against Axl (or control-scrambled siRNA) and performed the growth and motility assays. Figure 3A shows a representative example of the growth effect, showing a reduction of the growth rate (shown as cell doubling per day) when the Axl level is reduced (P = 0.0065). This suggests a correlation between mutant p53–mediated up-regulation of the Axl level and the growth rate enhancement induced by mutant p53. We also tested the relationship between cell motility rate and Axl level in these cells. As shown in Figure 3B, there is a significant reduction in cell mobility rate as measured by scratch assays when the Axl level is reduced by Axl siRNA, even though the mutant p53 level remains unchanged. Similar data have been obtained from H1793 cells as well (Fig. 3C and 3D). Thus, mutant p53 may induce some of its GOF activities via the induction of Axl.
Figure 3.
Axl up-regulation by mutant p53 has physiological significance. (A, C) Growth rate of H1048 and H1793 lung cancer cells depends on the Axl level. H1048 and H1793 cells were transfected with control or Axl-specific siRNA, plated in equal numbers, and harvested each day for 5 days to determine the rate of doubling. In parallel, p53, Axl, and Erk2 levels were determined by immunoblotting (right). (B, D) Motility of H1048 and H1793 cells depends on the Axl level. H1048 and H1793 cells were transfected as above. Forty-eight hours later, standard scratch assays were carried out as described in Materials and Methods and migrating cells stained and counted. Bar = ±1 standard deviation. Experiments were performed in triplicate. Error bars showing standard deviations are indicated.
Mutant p53 up-regulates the Axl promoter
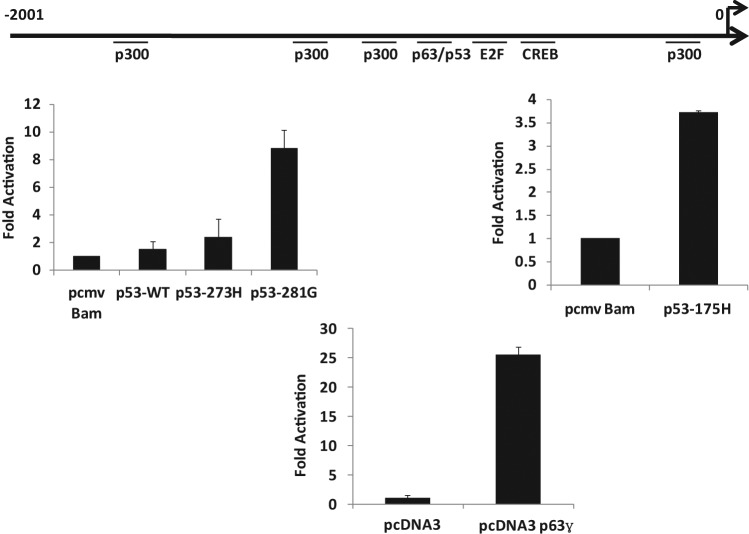
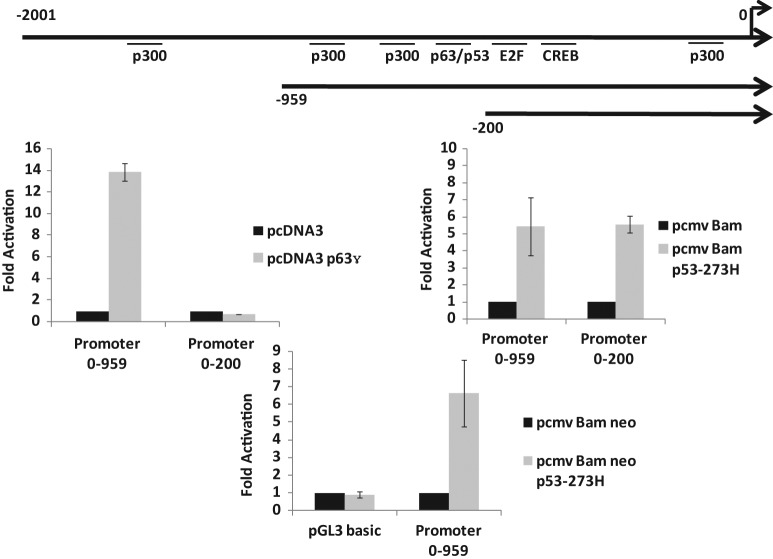
We have evidence that mutant p53–expressing cells have higher Axl levels both at RNA and protein levels (see above); therefore, we determined whether the upstream sequences of the human Axl gene can act as a faithful regulatory promoter in a transient promoter assay. We have cloned a 2,000-bp-long fragment encompassing the upstream sequences of Axl into the pGL3 basic luciferase reporter vector (Promega, Madison, WI) and tested its promoter activity by transient transfection analysis. We transfected p53-null human lung cancer H1299 cells with Axl–pGL3 basic in the presence and absence of different p53 expression plasmids and, after 48 hours, performed luciferase assays as described.9,50 The data shown in Figure 4 indicate that mutant p53 up-regulates the Axl upstream sequences as expected. The Axl promoter sequence contains a putative p53/p63 binding site on the Axl upstream sequences, raising the possibility of its involvement in mutant p53–mediated transactivation since some studies of GOF functions have implicated p63–mutant p53 interactions.27-29,51 Data shown in Figures 4 and 5 also show that p63 (full-length p63γ) and wild-type p53 could transactivate the Axl promoter depending on the presence of the p53/p63 binding site.
Figure 4.
Mutant p53 up-regulates the Axl promoter in H1299 cells. H1299 cells were transfected with 200 ng of a pGL3 basic vector containing the Axl promoter upstream of the luciferase reporter gene and 1 ug of the indicated p53 expression plasmid. Cell lysates were prepared 48 hours after transfection, and luciferase activity was determined. Data are shown as fold activation over the control. Experiments were performed in triplicate. Error bars showing standard deviations are indicated.
Figure 5.
Mutant p53 up-regulates the Axl promoter in H1299 cells independent of the p63 site. Two deletion mutants of the Axl promoter were constructed and tested for promoter activity in H1299 cells in the presence and absence of mutant p53 and p63 to determine if the putative p63 binding site is required for mutant p53–mediated transactivation. The conditions of the assay were similar to those described in the caption of Figure 4. The data indicate that mutant p53 does not require the p63 binding site to transactivate the Axl promoter. H1299 cells were also co-transfected with pGL3 basic or the larger Axl deletion mutant and an empty vector or mutant p53 to ensure the transactivation capability was not lost. Data are shown as fold activation over the control. Experiments were performed in triplicate. Error bars showing standard deviations are indicated.
Mutant p53 up-regulates the Axl promoter in H1299 cells independent of the p63 binding site
We generated 2 Axl promoter deletion mutants with and without the p63 binding site and tested whether transactivation by mutant p53 requires the p63 binding site. The data presented in Figure 5 show that p63-mediated transactivation is lost by a deletion of the p63 binding site; however, mutant p53–induced transactivation remained undisturbed even in the absence of the p63 binding site, while the negative control (pGL3 basic alone) did not get activated at all. This suggests that GOF mutant p53 transactivates the Axl promoter independent of the p63 binding sites in transient transfection assays.
Mutant p53 induces acetylation of histones on the Axl promoter
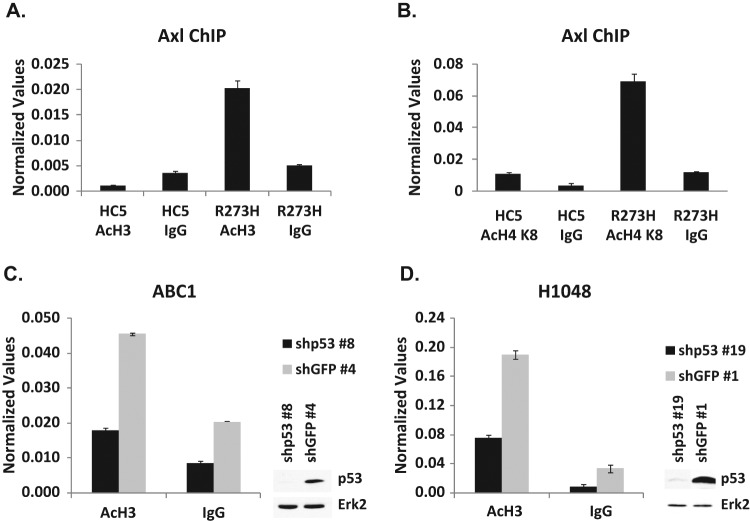
We used H1299 cells expressing mutant p53-R273H as a model to study the mechanism of up-regulation of Axl expression by mutant p53. We performed chromatin immunoprecipitation (ChIP) assays using H1299 cells stably transfected with an empty vector alone or H1299 cells expressing mutant p53-R273H using an antibody directed against acetylated histone to assay for the extent of histone modification as a measure of transcriptional activity at this gene. The data shown in Figure 6A and 6B demonstrate that mutant p53 induces the enhanced formation of acetylated histones H3 and H4 on the Axl promoter in H1299 cells expressing mutant p53-R273H, suggesting that the up-regulation of Axl expression by mutant p53 seen in these cells may occur via a transactivation role of mutant p53 through chromatin modification at the Axl promoter location.
Figure 6.
ChIP assay demonstrates the increased acetylation of histones on the Axl promoter. ChIP analysis was performed on H1299 cells stably transfected with a vector (HC5) or mutant p53-R273H (R273H) to test whether histones are preferentially acetylated on the Axl promoter. As described in Materials and Methods, cells were cross-linked with formaldehyde, harvested, and DNA sonicated to generate 500-bp to 2-kb fragments. Designated proteins were immunoprecipitated using respective antibodies. Cross-linking was reversed and the DNA isolated and purified. qPCR was performed using gene-specific primers. Normalized values represent qPCR values normalized to a fragment 5 kb upstream of the Axl gene not affected by mutant p53. (A) ChIP was performed with an antibody directed against acetylated histone H3 (acetylated at K9 and K14). (B) ChIP was performed with an antibody directed against acetylated histone H4 (acetylated at K8). (C, D) ChIP performed on ABC-1 and H1048 p53 knockdown cell lines using an antibody directed against acetylated histone H3 (acetylated at K9 and K14) shows a reduction in histone acetylation on the Axl promoter. Experiments were performed in triplicate. Error bars showing standard deviations are indicated.
Knockdown of mutant p53 reduces histone acetylation on the Axl promoter
Since the overexpression of mutant p53 induces the acetylation of histones on the Axl promoter, we wanted to see if the reduction of mutant p53 would produce the opposite effect. We used 2 lung cancer cell lines, ABC-1 (p53-P278S) and H1048 (p53-R273C), after knockdown of their endogenous mutant p53 using shRNA lentivirus directed against p53 or control GFP. The data shown in Figure 6C and 6D show a decrease of histone acetylation upon mutant p53 reduction compared to GFP control in both ABC-1 (Fig. 6C) and H1048 (Fig. 6D) cell lines. This further supports the idea that mutant p53 up-regulates Axl expression via chromatin modification.
Nucleation of mutant p53 on the Axl promoter
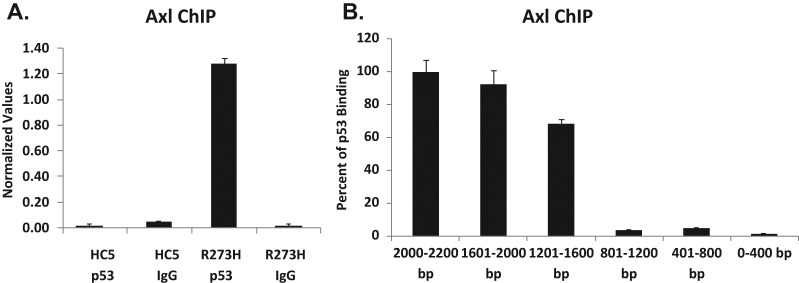
Next, we examined if we could locate mutant p53 on the Axl promoter sequences using the ChIP and H1299 cell systems described above (Fig. 6). Here, chromatins were immunoprecipitated using p53 antibodies D01 and FL-393 (Santa Cruz Biotechnology, Santa Cruz, CA), or corresponding antibody controls, as described in Materials and Methods. Immunoprecipitated DNA was quantified by qPCR, and data are shown in Figure 7A. It is clear that mutant p53 was localized on the sequences upstream to the Axl gene, either directly or indirectly binding to DNA. To locate the site or sites of mutant p53 binding to the Axl promoter, the full-length promoter was divided into 5 fragments, and primers were designed. Fold activation by mutant p53 on the Axl promoter was determined, and the percentage of p53 binding was calculated (Fig. 7B). The data shown indicate major mutant p53 interactions distal to ATG.
Figure 7.
ChIP data showing p53-R273H binding in H1299 cells expressing this mutant to the Axl promoter region. (A) ChIP was performed on H1299 cells stably transfected with a vector (HC5) or mutant p53-R273H (R273H) to test whether mutant p53 is nucleated on the promoter. As described in Materials and Methods, cells were cross-linked with formaldehyde, harvested, and DNA sonicated to generate 500-bp to 2-kb fragments. Mutant p53 was immunoprecipitated using D01 and FL-393 from Santa Cruz Biotechnology. Cross-linking was reversed, and DNA was isolated and purified. qPCR was performed using gene-specific primers. Normalized values represent qPCR values normalized to a fragment 5 kb upstream of the Axl gene not affected by mutant p53. (B) Primers were designed to amplify the full-length Axl promoter in 5 regions: 2,000-1,601 bp, 1,201-1,600 bp, 801-1,200 bp, 401-800 bp, and 1-400 bp. qPCR was performed for each of these fragments on H1299 cells expressing either an empty vector (HC5) and p53-R273H, which were used for ChIP with antibodies against p53 (D01 and FL393), or IgG as a control.
Transcription factor binding is enhanced by mutant p53 on the Axl promoter
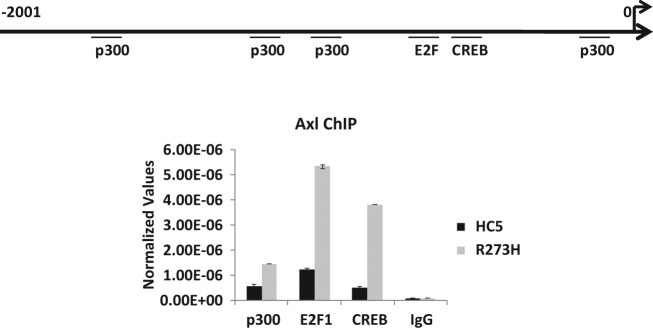
Next, we wanted to check whether we could determine the transcription factor(s) that bind more efficiently on the Axl promoter in the presence of mutant p53. For that purpose, we performed ChIP assays using antibodies against transcription factors p300, E2F1, and CREB, which putatively may interact with the Axl promoter. ChIP assay data carried out using vector-transfected cells and mutant p53 (-R273H) expressing H1299 cells and corresponding antibodies against these transcription factors show that binding all transcription factors is increased: p300, 2.5-fold; E2F1, 4.3-fold; and CREB, 8-fold. Increased binding of transcription factors is likely to contribute to the induced Axl expression mediated by mutant p53.
Discussion
We and others have shown that the expression of tumor-derived mutant p53 in cells leads to the up-regulation of expression of a set of genes, some of which are involved in oncogenesis; this is accompanied with GOF phenotypes. However, the mechanism of up-regulation of gene expression and the relationship to the GOF phenotype are only starting to be defined. Earlier, we had suggested that the GOF activities observed in H1299 cells after the expression of mutant p53 can be explained in part by mutant p53’s ability to enhance the expression of NF-κB2.9 Axl has been identified as another player, which has previously been implicated in oncogenesis35 and is significantly up-regulated by tumor-derived p53 mutants (Figs. 1 and 2) in lung cancer cell lines and human lung tumors. One interesting and important aspect of Axl being up-regulated by p53 mutants is the fact that p53 mutants do not apparently require an intact transactivation domain for this up-regulation (Fig. 1). Thus, this mutant p53 target seems to be a good candidate for a gene that has been up-regulated by mutant p53 perhaps through interaction with p63/p73.52 We note that there is a second transactivation subdomain that may contribute to transactivaton, or alternatively, mutant p53 may be utilizing another transcriptional activator subdomain as a surrogate for transactivation. Using human lung cancer cell lines and lung tumor samples expressing mutant p53, we demonstrate that the expression of GOF mutant p53 induces Axl expression (Fig. 2). We found that knocking down Axl levels by RNAi in H1048 cells led to a decrease in cell motility and growth rate (Fig. 3) while mutant p53 levels remained constant. This suggests that GOF mutant p53 may induce part of its GOF activity via the induction of Axl.
We have examined the promoter sequences of Axl and identified putative transcription factor binding sites (Fig. 4, top) and have undertaken an in vivo transient transcriptional analysis to determine the mechanism of activation of the promoter by GOF mutant p53. Analysis showed that mutant p53 indeed transactivated the Axl promoter in H1299 cells (Fig. 4). Interestingly, our promoter deletion analysis indicated that the p53/p63 binding site present is not needed for in vivo transactivation by mutant p53 in H1299 cells (Fig. 5). This shows that GOF p53 mutants transactivate the Axl promoter without the necessity of p63 binding to the promoter, suggesting that mutant p53 is not activating the Axl promoter by p63 interaction.
ChIP assay analysis demonstrated that GOF mutant p53 induces histone acetylation at the Axl promoter (Fig. 6), suggesting that mutant p53 causes chromatin modifications on the promoter, indicative of increased transcriptional activity. Transcription factor ChIP analyses indicated that mutant p53 induced interactions between p300, CREB, and E2F1 and the Axl promoter (Fig. 8), suggesting their involvement, in general, in the transactivation of Axl by mutant p53. The observed increased histone acetylation may lead to increased interactions of the transcription factors p300, CREB, and E2F1 with their consensus binding sites on the Axl promoter, which may lead to increased Axl promoter activity. Since CBP/p300 has histone acetylase activity associated with it, these data suggest that mutant p53 may be enhancing histone acetylation through the use of CBP/p300. Interestingly, interactions of CBP/p300 and E2F1 and CREB53 may further nucleate p300 on the Axl promoter and lead to further acetylation of histones, increasing its promoter activity.
Figure 8.
ChIP assay shows the mutant p53–mediated increase in transcription factor binding on the Axl promoter. ChIP analysis was performed on H1299 cells stably transfected with a vector (HC5) or mutant p53-R273H (R273H) as described for Figure 6 to test whether transcription factors are preferentially binding to the Axl promoter. Cells were cross-linked with formaldehyde, harvested, and DNA sonicated to generate 500-bp to 2-kb fragments. Designated proteins were immunoprecipitated using respective antibodies. Cross-linking was reversed and the DNA isolated. qPCR was performed using gene-specific primers. Normalized values represent qPCR values normalized to a fragment 5 kb upstream of the Axl gene not affected by mutant p53. ChIP was performed with antibodies directed against CREB, E2F1, and p300. Experiments were performed in triplicate. Error bars showing standard deviations are indicated.
ChIP assays using p53 antibodies demonstrate unequivocally that mutant p53 in H1299 cells expressing the p53-R273H protein is nucleated on the Axl regulatory sequences (Fig. 7). We have demonstrated that there is more mutant p53–mediated activation of the Axl promoter further away from the transcription start site, indicating that mutant p53 may bind to an enhancer-like region to regulate transcription. Our transient transcriptional analysis (Figs. 4 and 5) demonstrates that Axl promoter activation by mutant p53 does not require the p53/p63 binding site. When we examined the in vivo interaction of p63 with the Axl promoter by ChIP assays, we could detect a marginal increase in binding over the background in the chromatin setting in the presence of mutant p53 (data not shown). This may suggest that mutant p53 helps to open up the Axl promoter region, enhancing the interaction of transcription factors with the promoter. Thus, one has to speculate the involvement of other transcription factors in this transactivation unless it is assumed (an unlikely event) that mutant p53 itself interacts with the DNA. Further in vivo and in vitro work is needed to clarify the mechanism of transactivation by mutant p53.
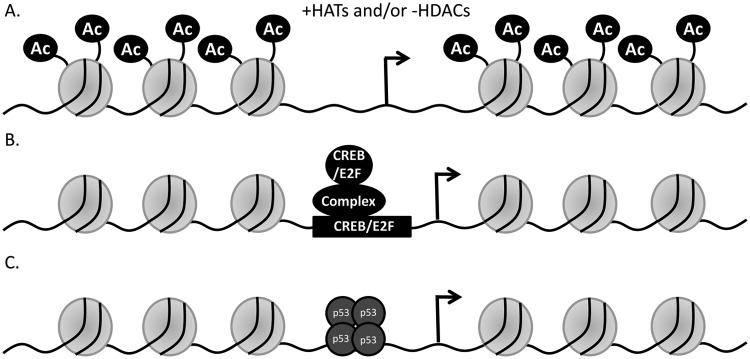
At present, we envision 3 ways that mutant p53 may induce the up-regulation of target gene expression (Fig. 9). 1) Mutant p53 may induce histone acetylase(s) and/or inhibit histone deacetylase(s) and thereby cause the acetylation of histones (Fig. 9A) and chromatin reorganization on the promoters of mutant p53 target genes; this would lead to activation of the promoter and transactivation. 2) Mutant p53 may induce interactions of one or more transcription factor(s) with the promoter, leading to transactivation of the target gene (Fig. 9B). 3) Mutant p53 itself is nucleated on the target promoter directly or indirectly (Fig. 9C) through another transcription factor and then modulates the recruitment of other factors on the promoter with eventual promoter activity enhancement. A combination of these possible mechanisms may also be at work.
Figure 9.
Model of up-regulation of gene expression by mutant p53. Three different molecular mechanisms of how mutant p53 may activate transcription from its target promoter are depicted. (A) Mutant p53 may induce histone acetylase(s) and/or inhibit histone deacetylase(s) and thereby cause the acetylation of histones and chromatin reorganization on the promoters of mutant p53 target genes; this would lead to activation of the promoter and transactivation. (B) Mutant p53 may induce the interaction of one or more transcription factor(s) with the promoter, leading to transactivation of the target gene. (C) The third possibility is that mutant p53 itself is nucleated on the target promoter and then modulates the recruitment of other factors on the promoter with eventual promoter activity enhancement. These 3 possibilities are not mutually exclusive.
Materials and Methods
Cell lines
Five human lung cancer cell lines, H1299 (p53-null, non–small cell lung cancer [NSCLC]), H1048 (p53-R273C, small cell lung cancer), H1437 (p53-R267P, NSCLC), ABC-1 (p53-P278S, adenocarcinoma), and H1793 (p53-R273H, NSCLC), were used in these studies. H1299, H1048, and H1437 were grown in RPMI media supplemented with 10% fetal bovine serum, while H1793 was grown in DMEM supplemented with 5% fetal bovine serum.
Generation of H1299 cells expressing GOF p53 mutants
To determine whether the expression of mutant p53 leads to GOF phenotypes in human cells, we have used the H1299 (p53-null) lung cancer cell line and generated cells expressing mutant p53-R175H, -R273H, and -D281G.54 These GOF p53 mutants are commonly found in human cancer (http://www.iarc.fr/p53/Index.html).
Generation of mutant p53 knockdown cell lines
The independent cell lines H1048, ABC-1, and H1437 were used to make stable p53 knockdown cell lines. They were generated by using lentivirus expressing short hairpin RNA (shRNA) against p53 utilizing lentivirus systems (Open Biosystems, Huntsville, AL) following the manufacturer’s protocol. Clones were isolated using puromycin selection at 1 µg/mL.
Expression analysis
Microarray analysis was carried out as described previously9,55 after isolating RNA from H1299 cells expressing p53 mutants or stably transfected with a vector alone. We analyzed the expression of different genes using Affymetrix U95Av2 arrays as described earlier.9,55 qPCR analyses of RNA levels were also performed as described previously.9,55
Tumor RNA analysis and p53 sequencing
Tumor RNA was provided by the Tissue and Data Acquisition and Analysis Core repository (Virginia Commonwealth University) under an Institutional Review Board–approved protocol; cDNA was prepared using the SuperScript III cDNA synthesis kit (Invitrogen, Carlsbad, CA) and qPCR performed using primers specific for Axl (F: 5′-TGT TTG GTG TTT CTG GGA CA-3′ and R: 5′-TCG CAG GAG AAA GAG GAT GT-3′). The degree of expression was quantitated using a relative standard curve and normalized to GAPDH (F: 5′-GTC AAC GGA TTT GGT CGT ATT-3′ and R: 5′-GAT CTC GCT CCT GGA AGA TGG-3′) corresponding to the cDNA batch. The p53 gene was sequenced following the method described by Sjogren et al.56 Whenever a mutation was found, a new PCR reaction was performed and the fragment resequenced to verify the sequence information obtained previously. The AXL levels in the 2 populations of tumors, one with wild-type p53 and one with mutant p53, were compared and found to be different with a statistically significant difference based on the Student t test.
siRNA transfection
Lung cancer cells were transfected with Lipofectamine 2000 (Invitrogen) 2 times (once every 24 hours) with RNAi directed against a specific or nonspecific gene (luciferase) following the manufacturer’s protocol. Sequences used were the following: control (C): 5′-CAU GUC AUG UGU CAC AUC ACT T-3′ and 5′-GAG AUG UGA CAC AUG ACA UGT T-3′ and Axl siRNA, which was purchased from Santa Cruz Biotechnology. Forty-eight hours after transfection, cells were trypsinized, counted, and plated, and growth or migration assays were carried out as described below.
Growth assay
Growth assays were carried out as described by us earlier with slight modifications.9,10 Cells were plated at 50,000 cells/6-cm dish in triplicate for 5 time points and harvested after incubation with trypsin and counted using a Coulter Counter (Beckman Coulter, Brea, CA). Each set of triplicate plates was counted 3 times, giving a total of 9 counts per siRNA treatment per day. To determine the number of cell doublings, the ratio of Axl and control (Scram) siRNA-treated cells was calculated to be the number of cells counted on day 2 divided by day 1 and so on for the 5 time points. The ratios throughout the assay were averaged and plotted, and the standard deviation was calculated to be the difference in ratios from the triplicate counts per time point. All experiments were performed in triplicate and repeated multiple times. The cell lines were analyzed for the growth rate using replicates based on exponential growth. The statistical difference between growth rates was determined by the Student t test.
Axl promoter cloning and transient promoter assays
The Axl promoter and its deletion mutants were cloned in a pGL3 basic vector using sequence information available at the National Center for Biotechnology Information (Bethesda, MD). The primers used to generate the Axl promoter deletions were the following: 0-959: 5′-CCG GGG TAC CCG CAG GCA GCA GAT CTG CAA TAA C-3′ and 0-200: 5′-CCG GTT ACC GGG AGT GAG GGA AGG AGG CAG GGG TGC TGA-3′. Transient transfection was performed with 200 ng of the promoter and 1 ug of the expression plasmid using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Luciferase analysis was carried out using the luciferase assay system (E1500) and instructions from Promega. Both transfection and luciferase assays were as described previously in triplicate.57
Chromatin immunoprecipitation
ChIP was performed as described.55 To cross-link protein and DNA, cell cultures were incubated in 2% formaldehyde for 10 minutes at 37°C, and then 200 mM of glycine was added for a further 10 minutes. Cells were washed in cold phosphate buffered saline (PBS), scraped, and centrifuged. Pellets were resuspended in lysis buffer containing 1% protease inhibitors and then sheared by 6 passages through a 27.5-gauge needle followed by 25 minutes of sonication on ice such that the chromatin was fragmented to 500 to 2,000 bp in length. Following centrifugation, the protein content of the supernatants was determined and equal amounts used for immunoprecipitation overnight at 4°C with gentle tilting with protein-specific antibodies or IgG as a control. Immune complexes were captured using protein A agarose and then washed sequentially in RIPA buffer (150 mM NaCl, 50 mM Tris, pH 8, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40), high salt buffer (500 mM NaCl, 50 mM Tris, pH 8, 0.1% SDS, 1% NP-40), twice in LiCl buffer (250 mM LiCl, 50 mM Tris, pH 8, 0.5% sodium deoxycholate, 1% NP-40), and then twice in TE buffer. Protein was eluted from beads in fresh elution buffer (20% SDS, 10 mM DTT, 100 mM NaHCO3), cross-linking was reversed overnight at 65°C in the presence of NaCl, and then samples were ethanol precipitated. Following centrifugation, pellets were resuspended in TE buffer and incubated sequentially with 10 mg/mL RNase A (30 minutes) and 20 mg/mL proteinase K (1 hour). Samples were phenol extracted and ethanol precipitated, and the pellets were washed in 70% ethanol, dried, and resuspended in sterile water. Acetyl histone H3 (17-615), acetyl histone H4 K8 (07-328), and normal rabbit IgG were from Millipore (Billerica, MA). p300 (sc-585), CREB (sc-186), E2F1 (sc-22820), p53 DO1 (sc-126), and p53 FL393 (sc-6243) were obtained from Santa Cruz Biotechnology. PCR primers used to analyze ChIP samples were the following: Axl (5 kb) F: 5′-CCT TGA CTG AGG CTT TAC CA-3′ and R: 5′-TTT TCA AAG TGC ACC GAC AT-3′, and Axl F: 5′-GAT GCA GCA GTT CCC AAA AT-3′ and R: 5′-TAT CAT CCC TTC TCC ATC GC-3′. The Axl promoter scanning primers were the following: Axl 2,200-2,000 bp are the same primers indicated above; 1-400 bp F: 5′-CCC CGT CTC TAC CAA AAA TA-3′ and R: 5′-GGC CCT TCA CCG TTG T-3′; 401-800 bp F: 5′-GAA GGG GCA GGT AGA AGA GA-3′ and R: 5′-AGC CCT GAT CAT TCC ACT G-3′; 801-1,200 bp F: 5′-AGC GAT CCT CCC ACC TT-3′ and R: 5′-ATC TTC AGA CAC GCC AAA AC-3′; 1,201-1,600 bp F: 5′-TCT GCG TGT CTC TGC TTG TC-3′ and R: 5′-TCT GGG CTC TGT GTC TGG TA-3′; and 1,601-2,000 bp F: 5′-GGT CCC CTT CCC CCT CCT CA-3′ and R: 5′-CCC AGC AGC CGC CTT CTC A-3′. Axl (5 kb) corresponds to a sequence 5 kb upstream of the Axl gene without any regulatory sequences in it. This was used to normalize the qPCR values. The percentage of p53 binding was calculated by determining the fold activation of the Axl promoter by mutant p53 (v. control) for each region of the promoter. Five sets of ChIP experiments were averaged, and the percentage of binding was calculated by setting the Axl ChIP primers (2,200-2,000 bp) as equal to 100%. The standard deviation was calculated between the 5 sets of experiments and plotted.
Western blotting
Immunoblotting was carried out as described.9 Axl levels were detected using an antibody from Abnova (H00000558-M01, Taipei, Taiwan). Erk2 (sc-154) and β-tubulin (sc-5274) levels were detected by antibodies from Santa Cruz Biotechnology. p53 was detected using the p53 antibody PAb 1801.9 Western blots were developed by the ECL method (GE Healthcare, Piscataway, NJ).
Migration assays
Cell migration was carried out using wound-healing (scratch) assays, as previously described.58 Briefly, cells were trypsinized, plated in quadruplicate in 12-well cell culture plates, and incubated at 37°C until cells were completely confluent. At this time, a sterile pipette tip was used to scratch across the surface of the plate, removing the complete layer of cells within the scratch area. Following cell removal, each well was washed once with PBS and then replaced with growth medium. Immediately following, the width of the scratch was measured at 6 specific points under a 5× objective using a light microscope and AxioVision software (Carl Zeiss Microimaging, Thornwood, NY). Cells were incubated at 37°C from 20 to 60 hours depending on the cell line under study, at which time the scratch width was measured at the same position as at time 0.
Acknowledgments
The authors thank Arnold Levine and Bert Vogelstein for providing them with reagents.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the National Institutes of Health to S.D. (CA70712 and CA121144) and S.P.D. (CA74172); Pilot Project Awards from the Massey Cancer Center to S.D., S.P.D., A.Y., and B.W.; and a grant from the Jeffress Trust Foundation to B.W.
References
- 1. Ahrendt SA, Hu Y, Buta M, et al. p53 mutations and survival in stage I non-small-cell lung cancer: results of a prospective study. J Natl Cancer Inst. 2003;95(13):961-70 [DOI] [PubMed] [Google Scholar]
- 2. Miyatake K, Gemba K, Ueoka H, et al. Prognostic significance of mutant p53 protein, P-glycoprotein and glutathione S-transferase-pi in patients with unresectable non-small cell lung cancer. Anticancer Res. 2003;23(3C):2829-36 [PubMed] [Google Scholar]
- 3. Thompson AM, Crichton DN, Elton RA, Clay MF, Chetty U, Steel CM. Allelic imbalance at chromosome 17p13.3 (YNZ22) in breast cancer is independent of p53 mutation or p53 overexpression and is associated with poor prognosis at medium-term follow-up. Br J Cancer. 1998;77(5):797-800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Skaug V, Ryberg D, Kure EH, et al. p53 mutations in defined structural and functional domains are related to poor clinical outcome in non-small cell lung cancer patients. Clin Cancer Res. 2000;6(3): 1031-7 [PubMed] [Google Scholar]
- 5. Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schimmelpenning H, Eriksson ET, Zetterberg A, Auer GU. Association of immunohistochemical p53 tumor suppressor gene protein overexpression with prognosis in highly proliferative human mammary adenocarcinomas. World J Surg. 1994;18(6):827-32, discussion 832-3. [DOI] [PubMed] [Google Scholar]
- 7. Campling BG, El-Deiry WS. Clinical implication of p53 mutation in lung cancer. Mol Biotechnol. 2003;24(2):141-56 [DOI] [PubMed] [Google Scholar]
- 8. Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2):a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scian MJ, Stagliano KE, Anderson MA, et al. Tumor-derived p53 mutants induce NF-kappaB2 gene expression. Mol Cell Biol. 2005;25(22):10097-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scian MJ, Stagliano KE, Ellis MA, et al. Modulation of gene expression by tumor-derived p53 mutants. Cancer Res. 2004;64(20):7447-54 [DOI] [PubMed] [Google Scholar]
- 11. Cadwell C, Zambetti GP. The effects of wild-type p53 tumor suppressor activity and mutant p53 gain-of-function on cell growth. Gene. 2001;277(1-2):15-30 [DOI] [PubMed] [Google Scholar]
- 12. Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60(24):6788-93 [PubMed] [Google Scholar]
- 13. Singer S, Ehemann V, Brauckhoff A, et al. Protumorigenic overexpression of stathmin/Op18 by gain-of-function mutation in p53 in human hepatocarcinogenesis. Hepatology. 2007;46(3):759-68 [DOI] [PubMed] [Google Scholar]
- 14. Strano S, Dell’Orso S, Di Agostino S, Fontemaggi G, Sacchi A, Blandino G. Mutant p53: an oncogenic transcription factor. Oncogene. 2007;26(15):2212-9 [DOI] [PubMed] [Google Scholar]
- 15. Weisz L, Damalas A, Liontos M, et al. Mutant p53 enhances nuclear factor kappaB activation by tumor necrosis factor alpha in cancer cells. Cancer Res. 2007;67(6):2396-401 [DOI] [PubMed] [Google Scholar]
- 16. Lin J, Teresky AK, Levine AJ. Two critical hydrophobic amino acids in the N-terminal domain of the p53 protein are required for the gain of function phenotypes of human p53 mutants. Oncogene. 1995;10(12):2387-90 [PubMed] [Google Scholar]
- 17. Lanyi A, Deb D, Seymour RC, Ludes-Meyers JH, Subler MA, Deb S. ‘Gain of function’ phenotype of tumor-derived mutant p53 requires the oligomerization/nonsequence-specific nucleic acid-binding domain. Oncogene. 1998;16(24):3169-76 [DOI] [PubMed] [Google Scholar]
- 18. Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192(2):209-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taylor WR, Egan SE, Mowat M, Greenberg AH, Wright JA. Evidence for synergistic interactions between ras, myc and a mutant form of p53 in cellular transformation and tumor dissemination. Oncogene. 1992;7(7):1383-90 [PubMed] [Google Scholar]
- 20. Shi XB, Nesslinger NJ, Deitch AD, Gumerlock PH, deVere White RW. Complex functions of mutant p53 alleles from human prostate cancer. Prostate. 2002;51(1):59-72 [DOI] [PubMed] [Google Scholar]
- 21. Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2):a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blandino G, Levine AJ, Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18(2):477-85 [DOI] [PubMed] [Google Scholar]
- 23. Lowe SW, Bodis S, McClatchey A, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266(5186):807-10 [DOI] [PubMed] [Google Scholar]
- 24. Lai SL, Perng RP, Hwang J. p53 gene status modulates the chemosensitivity of non-small cell lung cancer cells. J Biomed Sci. 2000;7(1):64-70 [DOI] [PubMed] [Google Scholar]
- 25. Bristow RG, Peacock J, Jang A, Kim J, Hill RP, Benchimol S. Resistance to DNA-damaging agents is discordant from experimental metastatic capacity in MEF ras-transformants-expressing gain of function MTp53. Oncogene. 2003;22(19):2960-6 [DOI] [PubMed] [Google Scholar]
- 26. Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74(6):957-67 [DOI] [PubMed] [Google Scholar]
- 27. Strano S, Blandino G. p73-mediated chemosensitivity: a preferential target of oncogenic mutant p53. Cell Cycle. 2003;2(4):348-9 [PubMed] [Google Scholar]
- 28. Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG. Chemosensitivity linked to p73 function. Cancer Cell. 2003;3(4): 403-10 [DOI] [PubMed] [Google Scholar]
- 29. Muller M, Schleithoff ES, Stremmel W, Melino G, Krammer PH, Schilling T. One, two, three: p53, p63, p73 and chemosensitivity. Drug Resist Updat. 2006;9(6):288-306 [DOI] [PubMed] [Google Scholar]
- 30. Neilsen PM, Noll JE, Suetani RJ, et al. Mutant p53 uses p63 as a molecular chaperone to alter gene expression and induce a pro-invasive secretome. Oncotarget. 2011;2(12):1203-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18(9):1487-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61(2):203-12 [DOI] [PubMed] [Google Scholar]
- 33. Hafizi S, Dahlback B. Gas6 and protein S: vitamin K-dependent ligands for the Axl receptor tyrosine kinase subfamily. FEBS J. 2006;273(23):5231-44 [DOI] [PubMed] [Google Scholar]
- 34. O’Bryan JP, Frye RA, Cogswell PC, et al. axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol. 1991;11(10):5016-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Janssen JW, Schulz AS, Steenvoorden AC, et al. A novel putative tyrosine kinase receptor with oncogenic potential. Oncogene. 1991;6(11):2113-20 [PubMed] [Google Scholar]
- 36. Li Y, Ye X, Tan C, et al. Axl as a potential therapeutic target in cancer: role of Axl in tumor growth, metastasis and angiogenesis. Oncogene. 2009;28(39):3442-55 [DOI] [PubMed] [Google Scholar]
- 37. Sawabu T, Seno H, Kawashima T, et al. Growth arrest-specific gene 6 and Axl signaling enhances gastric cancer cell survival via Akt pathway. Mol Carcinog. 2007;46(2):155-64 [DOI] [PubMed] [Google Scholar]
- 38. Rochlitz C, Lohri A, Bacchi M, et al. Axl expression is associated with adverse prognosis and with expression of Bcl-2 and CD34 in de novo acute myeloid leukemia (AML): results from a multicenter trial of the Swiss Group for Clinical Cancer Research (SAKK). Leukemia. 1999;13(9):1352-8 [DOI] [PubMed] [Google Scholar]
- 39. Wimmel A, Glitz D, Kraus A, Roeder J, Schuermann M. Axl receptor tyrosine kinase expression in human lung cancer cell lines correlates with cellular adhesion. Eur J Cancer. 2001;37(17):2264-74 [DOI] [PubMed] [Google Scholar]
- 40. Shieh YS, Lai CY, Kao YR, et al. Expression of axl in lung adenocarcinoma and correlation with tumor progression. Neoplasia. 2005;7(12):1058-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meric F, Lee WP, Sahin A, Zhang H, Kung HJ, Hung MC. Expression profile of tyrosine kinases in breast cancer. Clin Cancer Res. 2002;8(2):361-7 [PubMed] [Google Scholar]
- 42. Wu CW, Li AF, Chi CW, et al. Clinical significance of AXL kinase family in gastric cancer. Anticancer Res. 2002;22(2B):1071-8 [PubMed] [Google Scholar]
- 43. Holland SJ, Powell MJ, Franci C, et al. Multiple roles for the receptor tyrosine kinase axl in tumor formation. Cancer Res. 2005;65(20):9294-303 [DOI] [PubMed] [Google Scholar]
- 44. Vajkoczy P, Knyazev P, Kunkel A, et al. Dominant-negative inhibition of the Axl receptor tyrosine kinase suppresses brain tumor cell growth and invasion and prolongs survival. Proc Natl Acad Sci U S A. 2006;103(15):5799-804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2(4):466-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chung BI, Malkowicz SB, Nguyen TB, Libertino JA, McGarvey TW. Expression of the proto-oncogene Axl in renal cell carcinoma. DNA Cell Biol. 2003;22(8):533-40 [DOI] [PubMed] [Google Scholar]
- 47. Ou WB, Corson JM, Flynn DL, et al. AXL regulates mesothelioma proliferation and invasiveness. Oncogene. 2011;30(14):1643-52 [DOI] [PubMed] [Google Scholar]
- 48. Nielsen-Preiss SM, Allen MP, Xu M, et al. Adhesion-related kinase induction of migration requires phosphatidylinositol-3-kinase and ras stimulation of rac activity in immortalized gonadotropin-releasing hormone neuronal cells. Endocrinology. 2007;148(6):2806-14 [DOI] [PubMed] [Google Scholar]
- 49. Sainaghi PP, Castello L, Bergamasco L, Galletti M, Bellosta P, Avanzi GC. Gas6 induces proliferation in prostate carcinoma cell lines expressing the Axl receptor. J Cell Physiol. 2005;204(1):36-44 [DOI] [PubMed] [Google Scholar]
- 50. Scian MJ, Frum R, Deb SP, Deb S. Transactivation and transrepression studies with p53. Methods Mol Biol. 2003;234:93-110 [DOI] [PubMed] [Google Scholar]
- 51. Martynova E, Pozzi S, Basile V, et al. Gain-of-function p53 mutants have widespread genomic locations partially overlapping with p63. Oncotarget. 2012;3(2):132-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21(5):1874-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang H, Larris B, Peiris TH, et al. C/EBPbeta activates E2F-regulated genes in vivo via recruitment of the coactivator CREB-binding protein/P300. J Biol Chem. 2007;282(34):24679-88 [DOI] [PubMed] [Google Scholar]
- 54. Mitsudomi T, Steinberg SM, Nau MM, et al. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene. 1992;7(1): 171-80 [PubMed] [Google Scholar]
- 55. Scian MJ, Stagliano KE, Deb D, et al. Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene. 2004;23(25):4430-43 [DOI] [PubMed] [Google Scholar]
- 56. Sjogren S, Inganas M, Norberg T, et al. The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. J Natl Cancer Inst. 1996;88(3-4):173-82 [DOI] [PubMed] [Google Scholar]
- 57. Vaughan CA, Singh S, Windle B, et al. p53 mutants induce transcription of NF-kappaB2 in H1299 cells through CBP and STAT binding on the NF-kappaB2 promoter and gain of function activity. Arch Biochem Biophys. 2012;518(1):79-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang D, Baldwin AS. Activation of nuclear factor-kappaB-dependent transcription by tumor necrosis factor-alpha is mediated through phosphorylation of RelA/p65 on serine 529. J Biol Chem. 1998;273(45):29411-6 [DOI] [PubMed] [Google Scholar]