Abstract
Tertiary dentin is deposited inside teeth after various stimuli and serves as a major defensive wall to preserve pulp cells. However, the molecular mechanisms of the activation of quiescent odontoblasts, immature pulp cells and tertiary dentin formation are still unclear. Therefore, we performed a comprehensive gene expression analysis of pulp cells after cavity preparation of 9-week-old rat molars to clarify the critical molecules in tertiary dentinogenesis. As a result, mRNA expression of various molecules was up- or down-regulated. Notably, several members of the matrix metalloprotease family and their endogenous inhibitors were up-regulated after cavity preparation. In situ hybridization showed that tissue inhibitor of metalloprotease 1 (Timp1) was widely and continuously distributed in the pulp beneath the cavity in vivo. We also observed accumulation of β-catenin in the pulp cells beneath the cavity by fluorescence immunohistochemistry. Furthermore, Timp1 transcription was repressed by a dominant-negative TCF4 in immature undifferentiated mesenchymal cells, but not altered in mature odontoblast-like cells. These results indicate that cavity preparation may activate the Wnt/β-catenin pathway and the Wnt/β-catenin pathway and Timp1 may be correlatively involved in pulp repair. Timp1 might play crucial roles in reactivation of immature pulp cells for tertiary dentinogenesis.
Keywords: matrix metalloprotease, pulp cell, tertiary dentin, tissue inhibitor of metalloprotease 1, Wnt, β-catenin pathway
The basic concept for caries treatment is removal of infected enamel and dentin, followed by replacement with restorative material. Conventional restorative materials are inert and not functional for healing of the tissue. Therefore, current clinical dentistry requires newly developed materials that promote healing such as promotion of tertiary dentin secretion for pulp capping. Primary dentin is formed by the interaction between epithelial and mesenchymal tissues (1–4), whereas tertiary dentin, which is generated in response to non-physiological stimuli such as caries or cavity preparation, is formed only by mesenchymal tissue (5, 6). Thus, these two processes should be initiated by different stimuli and may undergo different molecular mechanisms to form primary and tertiary dentin. Previous studies have mainly focused on primary dentin formation, and little information is available on tertiary dentin formation.
Odontoblasts are similar to osteoblasts in many aspects. Both cell types are of mesenchymal origin, secrete abundant amounts of extracellular matrix (ECM) and eventually form similar mineralized tissue, namely dentin and bone, respectively. For bone, immature mesenchymal cells differentiate into osteochondroprogenitor cells by expression of specific transcription factors. Then, β-catenin and other signalling molecules induce osteoblastogenesis. In addition, it has recently been reported that mechanical stimuli, such as vibration, of cartilage tissue results in Wnt/β-catenin pathway activation and increased new cartilage tissue formation (7). Osteoblast differentiation during the healing process of bone fractures is controlled by the Wnt/β-catenin pathway (8, 9). However, involvement of the Wnt/β-catenin pathway in the healing process of the dentin-pulp complex is still unclear. Based on this information, we hypothesized that the Wnt/β-catenin pathway played important roles in tertiary dentin formation.
To clarify the mechanisms underlying the formation of tertiary dentin, we performed microarray analysis of in vivo dental pulp cells after cavity preparation of rat molars. In addition, the localization of significantly up-regulated genes during tertiary dentinogenesis was analyzed. We also investigated the relationships between the stimulus of cavity preparation and the canonical Wnt/β-catenin pathway.
Materials and Methods
Animals and tissue preparation
The experimental animal protocol was approved by the Ethical Guidelines Committee for Animal Care of Osaka University Graduate School of Dentistry (No. 19-047-0). Nine-week-old male Wistar rats (CLEA Japan Inc., Tokyo, Japan) were intraperitoneally anaesthetized with 5% chloral hydrate (Nacalai Tesque, Kyoto, Japan). Then, a saucer-shaped cavity (the remaining dentin thickness was 200–300 μm) with 1 mm diameter was prepared using a round steel bur on the mesial surface of the upper first molar. The cavity was sealed with glass ionomer cement (Fuji IX; GC, Tokyo, Japan) after cleaning with 2.5% sodium hypochlorite solution.
RNA extraction
Teeth were extracted immediately (Day 0) and on Days 1 and 3 after cavity preparation. Connective tissues around the extracted teeth were removed and homogenized using a TissueLyser (Qiagen, Hilden, Germany). Then, total RNA was extracted from pulp cells using an RNeasy Lipid Tissue Mini Kit (Qiagen).
Microarray analysis
Gene expression levels were comprehensively compared between cavity-induced pulp and normal pulp using GeneChip Rat Genome 230 2.0 arrays (Affymetrix, Santa Clara, CA, USA) according to the manufacturer’s instructions. Up- or down-regulated genes with at least a 2-fold change were determined using DNA MicroArray Viewer Software (Kurabo Industries, Osaka, Japan) and genes continuously up-regulated at two or three consecutive time points were selected. In addition, genes associated with inflammation, wound healing and cell differentiation were selected based on the Gene Ontology classification.
qRT–PCR
qRT–PCR was performed using TaqMan Fast Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Total RNA was reverse transcribed to cDNA using a High Capacity RNA-to-cDNA Master Mix (Applied Biosystems). TaqMan probes to detect matrix metalloproteases 3 and 13 (Mmp3, Mmp13) and tissue inhibitor of metalloprotease 1 (Timp1) expressions were also purchased from Applied Biosystems as follows: Mmp3, Rn00591740_m1; Mmp13, Rn01448199_m1 and Timp1, Rn00587558_m1. PCR was performed using a 7500 Fast Real-Time PCR system (Applied Biosystems) and all samples were run as quintuplets. PCR conditions were as follows: 40 cycles of 95°C for 10 s and 60°C for 60 s. Quantifications were normalized to the housekeeping gene β-actin, Rn00667869_m1. Relative quantification was calculated by the ΔΔCt method (10).
Preparation of histological sections
Samples were collected on Days 0, 1 and 3. At each time point, animals were perfused with 4% paraformaldehyde in phosphate-buffered saline. The maxillae, including the prepared teeth, were dissected and immersed in the same fixative for an additional 12 h at 4°C. Samples were decalcified with formic acid/sodium citrate for 1 week and then embedded in OCT compound (Sakura Finetechnical Co., Tokyo, Japan). Specimens were sectioned at 12 μm using a cryostat (CM3050S; Leica, Wetzlar, Germany) for in situ hybridization and immunohistochemistry.
In situ hybridization
In situ hybridization was performed as described elsewhere (11). Mmp3, Mmp13, Timp1 and Axin2 probes were used in the present study. Mmp3, Mmp13 and Timp1 rat anti-sense and sense RNA probes were prepared by amplifying DNA fragments using specific primers as follows: Mmp3, 5′-ttccttgggctgaagatgaca-3′ and 5′tcatgagcagcaaccaggaat-3′; Mmp13, 5′-caggagatgaagaccccaacc-3′ and 5′- cacacgtggttccctgagaag-3′; Timp1, 5′- ctcaaaggattcgacgctgtg-3′ and 5′-tgttcaggcttcagcttttgc-3′. The Axin2 probe (Plasmid 21277) was purchased from Addgene (Cambridge, MA, USA). In vitro cRNA transcription was performed using a DIG RNA Labeling Kit (Roche, Basel, Switzerland). Details of restriction enzymes and RNA polymerases are listed in Table V.
Table V.
Restriction enzymes and RNA polymerases used to prepare RNA probes for in situ hybridization.
| Restriction enzyme | RNA Polymerase | |
|---|---|---|
| Mmp3 | ||
| Antisense | SpeI | T7 |
| Sense | SacII | SP6 |
| Mmp13 | ||
| Antisense | PstI | T7 |
| Sense | SacII | SP6 |
| Timp1 | ||
| Antisense | SalI | T7 |
| Sense | SacII | SP6 |
| Axin2 | ||
| Antisense | SalI | T7 |
| Sense | NotI | T3 |
Immunohistochemistry
Immunohistochemistry of β-catenin was performed as described elsewhere (11) using an anti-β-catenin rabbit IgG (Abcam, Cambridge, UK) and a secondary Cy3-labelled goat polyclonal antibody against rabbit IgG-H&L (Abcam). Nuclei were counterstained with 4,6-diamidino-2-phenylindole, dihydrochloride (DAPI; Molecular Probes, Carlsbad, CA, USA).
Cell culture
Odontoblast-like MDPC-23 cells (12) and undifferentiated OD-21 mesenchymal cells (13) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% foetal bovine serum (FBS; Sigma, St Louis, MO, USA) and 10 μg/ml penicillin-streptomycin (Sigma) at 37°C in a humidified incubator with 5% CO2.
DNA transfection and luciferase assay
The 2.8-kb mouse Timp1 proximal promoter was amplified by high fidelity Taq polymerase (TOYOBO, Osaka, Japan) using primers with introduced restriction sites (5′-ttgagctcaaccttccactctcagaagcaa-3′ and 5′-gccctctcccgttatattcctct-3′). The PCR product was subcloned into pGL3-basic (Promega, Madison, WI, USA) to generate the Timp1 luciferase reporter. T41A-β-catenin (0.5 μg/well; a kind gift from Dr Christine Neuveut, Institut Pasteur, Paris, France) (14), DN-TCF4 (0.5 μg/well, Addgene) (15) and LacZ (0.5 μg/well; pcDNA3.1/Hygro/LacZ, Invitrogen) vectors were used for the following transfection.
MDPC-23 and OD-21 cells, seeded into 12-well plates at 7 × 104 cells/well, were transfected with 0.5 μg luciferase reporter vector, 0.5 μg each expression vector and 0.05 μg pRL-TK Vector (Promega) using Lipofectamine 2000 reagent (Invitrogen). After 40 h, cells were harvested and firefly and renilla luciferase activities were simultaneously measured with a Dual-Luciferase Reporter Assay System (Promega) using a Lumat LB 9507 (Berthold, Wildbad, Germany). Reporter gene activity, as indicated by firefly luciferase activity, was normalized to that of renilla luciferase to show relative luciferase activity.
Statistical analysis
Statistical significance was determined using the Mann–Whitney U-test, and a value of P < 0.05 was considered significant.
Results
Comprehensive gene expression analysis of dental pulp
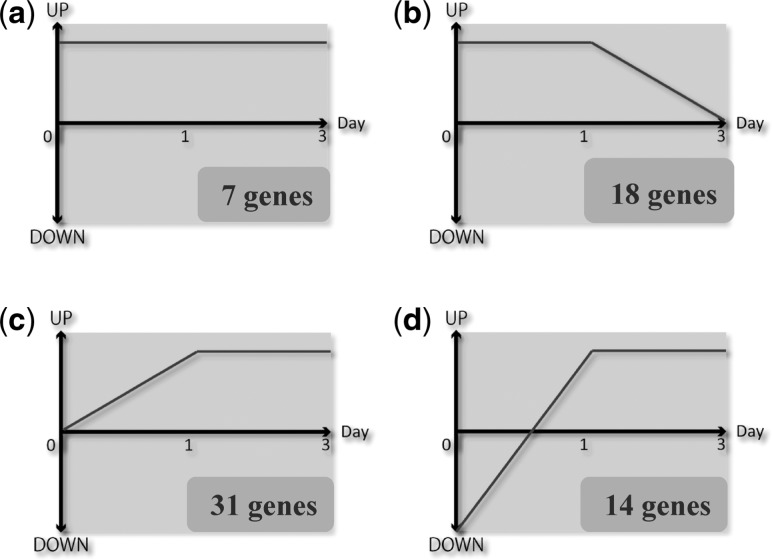
We compared the gene expression profiles of normal pulp without a cavity (control) and wounded pulp by cavity preparation using microarray analysis. The number of genes that showed increased or decreased expression by >2-fold, compared with that of the control, is shown in Table I. Among these genes, we chose candidate genes based on microarray data as follows. Seven genes were up-regulated by >2-fold from Day 0 to 3 (Fig. 1a), 18 genes were up-regulated on Days 0 and 1 with no change on Day 3 (Fig. 1b), 31 genes were unchanged on Day 0 and up-regulated on Days 1 and 3 (Fig. 1c) and 14 genes were down-regulated on Day 0 and up-regulated on Days 1 and 3 (Fig. 1d). In total, 70 genes showed continuously increased expression from immediately after cavity preparation to 3 days post-operation (Fig. 1 and Table II). Next, we focused on genes involved in inflammation, wound healing, cell development and differentiation, based on the Gene Ontology classification (16). As a result, nine genes were selected as shown in Table III. Because two matrix metalloproteases, namely Mmp3 and Mmp13 and Timp1 were among the nine candidate genes, we performed the following experiments.
Table I.
Number of genes that were up- or down-regulated by >2-fold in pulp cells after cavity preparation, compared with those in normal pulp (control).
| Day 0 (immediately after cavity preparation) | Day 1 | Day 3 | |
|---|---|---|---|
| Up-regulated | 308 | 329 | 337 |
| Down-regulated | 203 | 123 | 169 |
Total number of genes analyzed: 31,099.
Fig. 1.
Continuously up-regulated genes after cavity preparation as determined by microarray. Seven genes showed continuously increased expression on Days 0, 1 and 3. (a) In total, 18 genes were up-regulated on Days 0 and 1. (b) About 31 genes were highly expressed on Days 1 and 3. (c) About 14 genes showed continuously increased expression on Days 1 and 3, despite decreased expression on Day 0 (d) In total, 70 genes showed continuously higher expression compared with those of the control.
Table II.
List of genes continuously up-regulated at the two or three consecutive time points.
| Up-regulated on Days 0,1 and 3: 7 transcripts |
| 1. Mt1a (Metallothionein 1a) |
| 2. Ka13 (Type I keratin KA13) |
| 3. Kb4 (Type II keratin Kb4) |
| 4. LOC683295 (Similar to keratin complex 2, basic, gene 6a) |
| 5. LOC686203 (Similar to S100 calcium-binding protein A14 (S114)) |
| 6. Sprr3_predicted (Small proline-rich protein 3 (predicted)) |
| 7. Tfpi2 (Tissue factor pathway inhibitor 2) |
| Up-regulated on Days 0 and 1: 18 transcripts |
| 1. Aldh1a7 (aldehyde dehydrogenase family 1, subfamily A7) |
| 2. Angptl4 (Angiopoietin-like 4) |
| 3. Anxa8 (Annexin A8) |
| 4. Drp2 (Dystrophin-related protein 2) |
| 5. Fgfbp1 (Fibroblast growth factor binding protein 1) |
| 6. Fos (FBJ murine osteosarcoma viral oncogene homologue) |
| 7. Il6 (Interleukin 6) |
| 8. Krt1-12 (Keratin complex 1, acidic, gene 12) |
| 9. Lamc2 (Laminin, gamma 2) |
| 10. LOC682651 /// LOC689415 (Similar to Metallothionein-2 (MT-2) (Metallothionein-II) (MT-II)) |
| 11. Mall (Mal, T-cell differentiation protein-like) |
| 12. Myt1_predicted [Myelin transcription factor 1 (predicted)] |
| 13. RGD1563577_predicted [Similar to cornifelin (predicted)] |
| 14. S100a8 [S100 calcium binding protein A8 (calgranulin A)] |
| 15. S100a9 [S100 calcium binding protein A9 (calgranulin B)] |
| 16. Slc38a4 (Solute carrier family 38, member 4) |
| 17. Tacstd2 (Tumour-associated calcium signal transducer 2) |
| 18. Tmprss11d (Transmembrane protease, serine 11d) |
| Up-regulated on Days 1 and 3: 31 transcripts |
| 1. Adamts5 [A disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2)] |
| 2. Asgr1 (Asialoglycoprotein receptor 1) |
| 3. C6 (Complement component 6) |
| 4. Cd24 (CD24 antigen) |
| 5. Ctrc [Chymotrypsin C (caldecrin)] |
| 6. Cxcl1 [Chemokine (C-X-C motif) ligand 1] |
| 7. Dsp (Desmoplakin) |
| 8. Fabp5 (Fatty acid binding protein 5, epidermal) |
| 9. Fetub (Fetuin beta) |
| 10. Il1a (Interleukin 1 alpha) |
| 11. Kng1 /// LOC25087 /// MGC108747 (Kininogen 1 /// K-kininogen /// similar to alpha-1 major acute phase protein prepeptide) |
| 12. Krtdap (Keratinocyte differentiation-associated protein) |
| 13. Lbp (Lipopolysaccharide binding protein) |
| 14. Lgals3 (Lectin, galactose binding, soluble 3) |
| 15. LOC363060 (Similar to RIKEN cDNA 1600029D21) |
| 16. LOC683295 /// LOC683313 (Similar to keratin complex 2, basic, gene 6a) |
| 17. LOC684841 (Similar to CG31613-PA) |
| 18. LOC685912 [Similar to Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B)] |
| 19. Mal2 (Mal, T-cell differentiation protein 2) |
| 20. Mmp13 (Matrix metallopeptidase 13) |
| 21. Mmp3 (Matrix metallopeptidase 3) |
| 22. Perp_predicted [PERP, TP53 apoptosis effector (predicted)] |
| 23. RGD1560880_predicted [Similar to RIKEN cDNA 2310002J15 (predicted)] |
| 24. RGD1561521_predicted [Similar to 1110014F24Rik protein (predicted)] |
| 25. RGD1562059_predicted [Similar to RIKEN cDNA 1110038F21 (predicted)] |
| 26. RGD1562868_predicted (Similar to squamous cell carcinoma antigen 2 (predicted)) |
| 27. Scel_predicted (Sciellin (predicted)) |
| 28. Serpinb12_predicted (Serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 12 (predicted)) |
| 29. Smr2 (SMR2) |
| 30. Spink5_predicted [Serine protease inhibitor, Kazal type 5 (predicted)] |
| 31. Timp1 (Tissue inhibitor of metallopeptidase 1) |
| Down-regulated on Day 0, up-regulated on Days 1 and 3: 14 transcripts |
| 1. Aqp3 (Aquaporin 3) |
| 2. Calm4_predicted [Calmodulin 4 (predicted)] |
| 3. Cxcl2 [Chemokine (C-X-C motif) ligand 2] |
| 4. Dsg1c_predicted /// LOC679010 (Desmoglein 1 gamma (predicted) /// similar to Desmoglein-1 alpha precursor (Dsg1-alpha,Desmoglein-1) [Desmosomal glycoprotein I,(DG1) (DGI)] |
| 5. Gjb2 (Gap junction membrane channel protein beta 2) |
| 6. Il1b (Interleukin 1 beta) |
| 7. Ka17 (Type I keratin KA17) |
| 8. Kb1 (Type II keratin Kb1) |
| 9. Lgals7 (Lectin, galactose binding, soluble 7) |
| 10. LOC290595 (Hypothetical gene supported by AF152002) |
| 11. RGD1559993_predicted [RGD1559993 (predicted)] |
| 12. RGD1562305_predicted [Similar to suprabasal-specific protein suprabasin (predicted)] |
| 13. RGD1562885_predicted [Similar to RIKEN cDNA 2300002M23 (predicted)] |
| 14. RGD1565500_predicted [RGD1565500 (predicted)] |
Table III.
Nine candidate genes from microarray results based on the Gene Ontology classification.
| Cxcl1 [Chemokine (C-X-C motif) ligand 1] |
| Dsp (Desmoplakin) |
| Fabp5 (Fatty acid binding protein 5) |
| Mmp3 (Matrix metalloprotease 3) |
| Mmp13 (Matrix metalloprotease 13) |
| Spink5 (Serine protease inhibitor, kazal type 5) |
| Sprr3 (Small proline-rich protein 3) |
| Timp1 (Tissue inhibitor of metalloprotease1) |
| Tmprss11d (Transmembrane protease, serine 11d) |
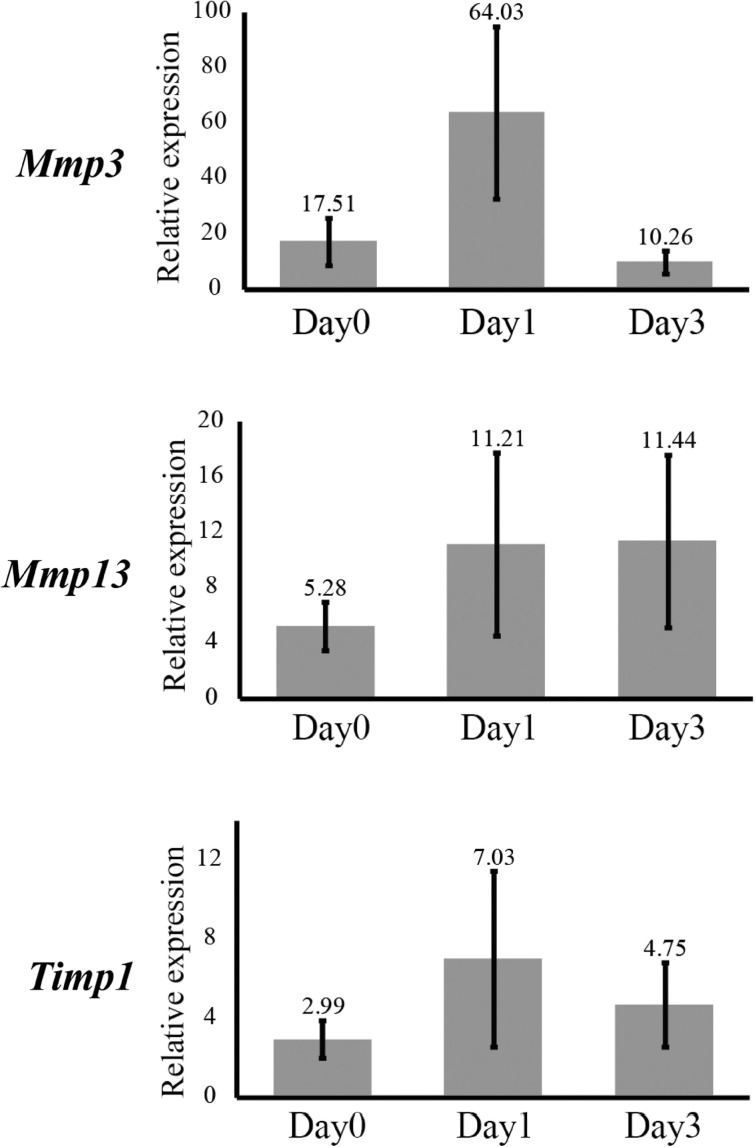
We confirmed microarray results by quantitative reverse-transcriptase polymerase chain reaction (qRT–PCR). The expression levels of Mmp3, Mmp13 and Timp1 were similar to those detected by microarray analysis, which showed the relative expression levels in the pulp after cavity preparation were >2-fold higher than those of the control (Fig. 2 and Table IV).
Fig. 2.
Expression levels of Mmp3, Mmp13 and Timp1 in the pulp after cavity preparation as determined by qRT–PCR. The expression levels of Mmp3, Mmp13 and Timp1 were similar to those obtained by microarray analysis and >2-fold higher than that of the control. Numbers on bars indicate the actual mean result. Expression levels were normalized to that of β-actin and data are the mean ± standard deviation (SD) of five independent experiments.
Table IV.
The relative expression of Mmp3, Mmp13 and Timp1 from microarray analysis.
| Day 0 | Day 1 | Day 3 | |
|---|---|---|---|
| Mmp3 | 1.8 | 7.6 | 3.5 |
| Mmp13 | 1.0 | 9.8 | 13.0 |
| Timp1 | 1.7 | 2.5 | 3.8 |
Localization of Mmp3, Mmp13 and Timp1 in the pulp after cavity preparation
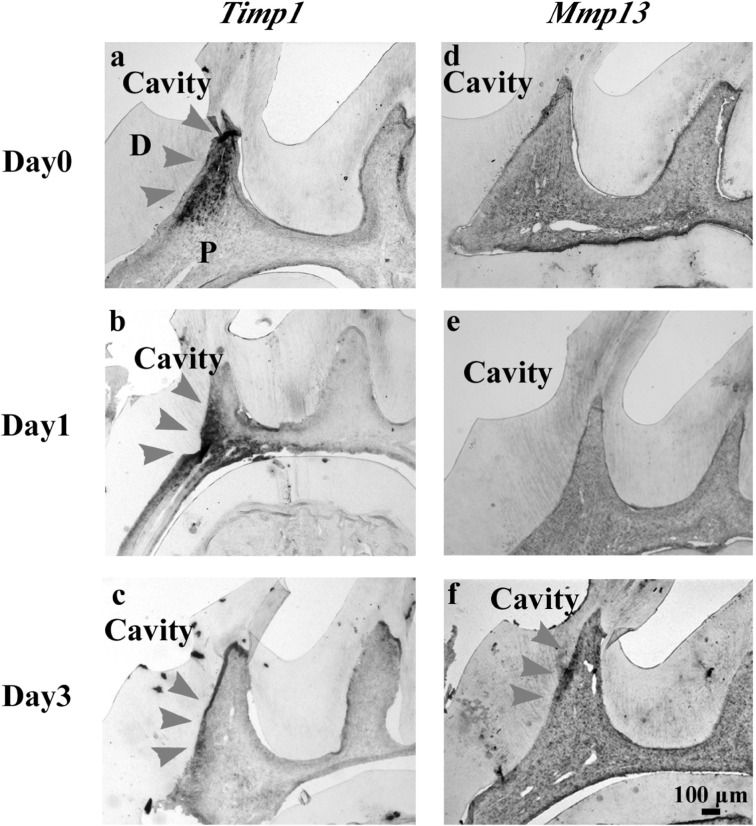
In situ hybridization analysis of the pulp after cavity preparation was performed to clarify the localization of Mmp3, Mmp13 and Timp1 that showed relatively higher expression in microarray and qRT–PCR analyses. Timp1 was observed on Days 0, 1 and 3 (Fig. 3a, b and c). Mmp13 was observed in the pulp beneath the cavity on Day 3 (Fig. 3f). Timp1 showed broad distribution and continuous expression, compared with those of Mmp13. Mmp3 was not detected, even though we cloned three gene fragment sites and attempted detection by in situ hybridization (data not shown).
Fig. 3.
Localization of Timp1- and Mmp13-positive cells in the pulp after cavity preparation as indicated by in situ hybridization. (a–f) Timp1-positive cells are indicated by arrowheads in (a), (b) and (c). Mmp13-positive cells are indicated by arrowheads in (f). Timp1 showed broad distribution and continuous expression compared with those of Mmp13. D, dentin; P, pulp.
Activation of the canonical Wnt/β-catenin pathway after cavity preparation
The canonical Wnt/β-catenin pathway is known to induce osteogenesis. To investigate whether the Wnt/β-catenin pathway was activated in pulp cells during tertiary dentinogenesis induced by cavity preparation, we examined the localization of β-catenin by fluorescence immunohistochemistry. A gross light microscopic image of a specimen is shown in Fig. 4a. Figure 4b shows the localization of β-catenin (red) and cell nuclei (blue). β-Catenin was accumulated in pulp cells beneath the cavity compared with that on the opposite unaffected side of the pulp and this accumulation facing the cavity showed broader and deeper distributions together with translocation of β-catenin into the nuclei (Fig. 4b). To confirm activation of the canonical Wnt/β-catenin pathway in pulp cells, expression of Axin2, which is induced by the canonical Wnt/β-catenin pathway, was examined by in situ hybridization. Axin2 expression was observed in the surface layer of pulp cells beneath the cavity (Fig. 5).
Fig. 4.
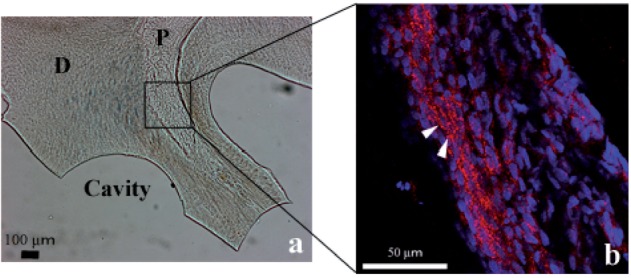
Localization of β-catenin in the pulp after cavity preparation as determined by fluorescence immunohistochemistry. Gross image under a light microscope (a) and a laser scanning microscopy image (b) of the boxed region in (a). β-Catenin is accumulated in the pulp cells beneath the cavity compared with that on the opposite side of the pulp and some cells show translocation of β-catenin into their nuclei (arrowheads). (b) Red: Cy3 (β-catenin), blue: DAPI (nucleus).
Fig. 5.
Localization of Axin2-positive cells in the pulp after cavity preparation as determined by in situ hybridization. Axin2-positive cells are indicated by arrowheads. Axin2 expression was observed in the surface layer of pulp cells beneath the defective dentin.
Regulation of Timp1 transcription by the Wnt/β-catenin pathway
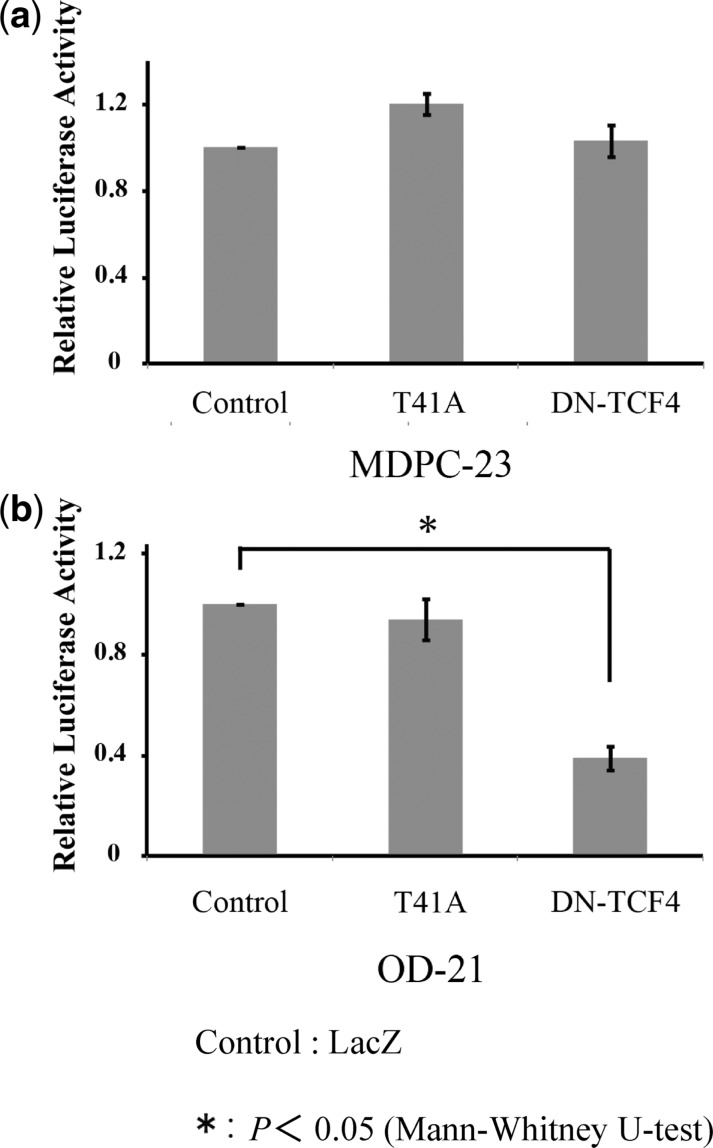
To evaluate whether activation of the canonical Wnt/β-catenin pathway in the pulp contributed to Timp1 transcription, we investigated the effects of β-catenin on the mouse Timp1 proximal promoter. In mature MDPC-23 cells, both constitutively active β-catenin (T41A-β-catenin) and dominant-negative TCF4 (DN-TCF4) showed no effect on Timp1 transcription (Fig. 6a). In contrast, using immature undifferentiated OD-21 mesenchymal cells in the same assay, T41A-β-catenin showed no effect on Timp1 transcription, whereas transcription was strongly inhibited by DN-TCF4 (Fig. 6b) (P < 0.05).
Fig. 6.
Effect of β-catenin on the mouse Timp1 proximal promoter. Co-transfection of constitutively active β-catenin (T41A) and dominant negative TCF-4 (DN-TCF4) stimulated the Timp1 luciferase construct in MDPC-23 and OD-21 cells. Firefly luciferase activity was normalized to the protein amount. In MDPC-23 cells, both T41A and DN-TCF4 showed no effect on Timp1 transcription. (a) In OD-21 cells, T41A showed no effect on Timp1 transcription, whereas DN-TCF4 strongly inhibited Timp1 transcription. (b) This result indicates that Timp1 transcription depends on the Wnt/β-catenin pathway, particularly in immature cells rather than differentiated cells at an advanced stage of differentiation. Results are the mean ± SD. Similar results were obtained in four independent experiments. Asterisks indicate statistically significant differences (P < 0.05).
Discussion
Despite numerous reports that describe the molecular mechanisms of primary dentin formation (1–4), the mechanisms of tertiary dentin formation after various stimuli are unclear. Some researchers have investigated gene expression patterns in the pulp after cavity preparation in vivo (17–19). However, these studies focused on specific molecules that are expressed specifically in odontoblasts or osteoblasts during development. Thus, we considered that there might be unreported molecules involved in tertiary dentin formation. Our preliminary experiments showed that tertiary dentin was formed in the pulp beneath the cavity 7 days after cavity preparation (data not shown). Therefore, we performed microarray analysis to globally analyse gene expression changes in the pulp on Days 0, 1 and 3 after cavity preparation. Based on the Gene Ontology classification, three genes, namely Mmp3, Mmp13 and Timp1, were analyzed in this study.
Mmp3 and Mmp13 have been reported to be induced by inflammatory stimuli (20–22) and play roles in wound healing such as blood vessel formation and remodelling of the ECM (23–25). On the other hand, TIMPs have been reported to inhibit the activity of MMPs by forming complexes with them (26). In addition, recent evidence suggests that TIMPs perform various growth factor-like functions such as the induction of cell proliferation and differentiation (27, 28). Based on these facts, we considered that MMP3, MMP13 and TIMP1 may be involved in the healing process of pulp.
We performed in situ hybridization to investigate mRNA localization of these three molecules in the pulp after cavity preparation. Mmp13 expression was observed on Day 3 (Fig. 3f) and Timp1 expression was observed on Days 0, 1 and 3 (Fig. 3a, b and c). Notably, because Timp1 showed continuous expression in a larger area, compared with that of Mmp13, we considered that Timp1 may have a greater effect on the healing process of pulp than that of Mmp13. The cause of the earlier induction of Timp1 expression, compared with that of the Mmps, is unclear, but this result indicates that TIMP1 may prevent excess MMP activity in advance or promote healing on Days 0 and 1. Although it has been reported that MMPs and TIMPs work together in a balanced manner during tissue repair (29, 30), decreased expression of Timp1 (Fig. 3c) and induction of Mmp13 (Fig. 3f) on Day 3 indicates that these molecules work correlatively to heal pulp. We also attempted to detect Mmp3 expression in the same assay, even though we prepared three probes to detect rat Mmp3, we were unable to obtain a reliable signal (data not shown). In qRT–PCR analysis, we showed higher expression of Mmp3 than those of Mmp13 and Timp1, although Mmp3 required 10 additional PCR cycles for detection, compared with that of the other two molecules (data not shown). Therefore, Mmp3 expression may be less than the detection limit of in situ hybridization. We considered that widespread and continuous expression of Timp1, compared with those of Mmp3 and Mmp13, in the pulp after cavity preparation could be significantly involved in the healing process of pulp.Taken together, TIMP1 may be immediately induced under the non-physiological condition to inhibit baseline MMP activity and prevent ECM degradation, which can be a scaffold for initial tertiary dentin formation. This theory supports that TIMP1 might promote healing process of the pulp both as an MMP inhibitor and a growth factor.
Following Wnt/β-catenin pathway activation, secreted β-catenin accumulates within the cytoplasm and enters the nucleus during bone formation and wound healing. Then, β-catenin forms a complex with T cell/lymphoid enhancer factors and promotes transcription of target genes (31, 32). We observed accumulation of β-catenin in pulp cells beneath the cavity by fluorescence immunohistochemistry (Fig. 4). Moreover, in situ hybridization showed the expression of Axin2, a negative feedback factor of the Wnt/β-catenin pathway (33) in pulp cells (Fig. 5). Microarray results showed that Axin2 expression was increased by ∼2-fold, compared with that of the control and some other genes involved in the Wnt/β-catenin pathway were also up-regulated (data not shown). These results suggest Wnt/β-catenin pathway activation by stimuli for cavity preparation in pulp cells. It is interesting to note that dentinal tubules in dentin have a similar structure to that of Haversian and Volkmann’s canals in bone tissue (34). Therefore, the mechanical stress produced during cavity preparation may be transferred to pulp cells via dentinal tubules and these stimuli may activate the Wnt/β-catenin pathway that regulates the healing of bone fractures.
We also investigated the effect of β-catenin on the proximal promoter of the mouse Timp1 gene. In mature MDPC-23 cells, T41A-β-catenin and DN-TCF4 showed no significant effect on Timp1 transcription (Fig. 6a). In contrast, in undifferentiated OD-21 mesenchymal cells, whereas T41A-β-catenin did not show a significant effect on Timp1 transcription, presumably owing to relatively high basal β-catenin-mediated transcriptional activity, DN-TCF4 significantly inhibited Timp1 transcription (P < 0.05) (Fig. 6b). Because of the effect of a dominant negative form of TCF4, an important molecule in the Wnt/β-catenin pathway, Timp1 transcription was inhibited in undifferentiated mesenchymal cells. These results indicate that Timp1 transcription depends on the Wnt/β-catenin pathway, particularly in immature cells rather than cells at an advanced stage of differentiation (Fig. 7).
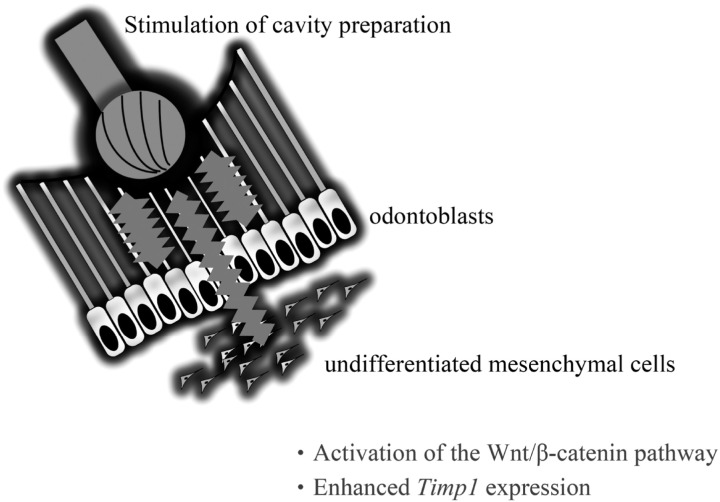
Fig. 7.
Cavity preparation not only affects peripheral odontoblasts beneath the cavity, but also undifferentiated mesenchymal cells inside the pulp. Cavity preparation may activate the Wnt/β-catenin pathway and enhance Timp1 expression.
Notably, Timp1 expression (in situ hybridization, Fig. 3a–c) and accumulation of β-catenin in pulp cells (immunohistochemistry, Fig. 4) were mainly observed inside the peripheral odontoblastic layer, where undifferentiated mesenchymal and pulp stem cells exist. In addition, the effect of β-catenin on Timp1 transcription was significantly detected in undifferentiated mesenchymal cells, compared with that in differentiated odontoblast-like cells (Fig. 6). These results suggest that cavity preparation affects not only peripheral odontoblasts beneath the cavity, but also undifferentiated mesenchymal cells that exist inside the pulp. In conclusion, we have shown that the expression of multiple genes is induced by stimuli for cavity preparation and Timp1 plays a key role in the formation of tertiary dentin. Moreover, cavity preparation may activate the Wnt/βcatenin pathway (Fig. 7). However, the role of MMP13, which has been reported to play roles in the healing of bone fractures (35) and MMP3, which may promote angiogenesis (36), remains unknown in the present study. Further investigations will be necessary to elucidate the involvement of these molecules in the healing process of the dentin–pulp complex.
Funding
This work was supported by Grants-in-Aid for Scientific Research (No. 19209060, 21592419, 23792170, and 24792021) from the Japan Society for the Promotion of Science.
Glossary
Abbreviations
- ECM
extracellular matrix
- MMP
matrix metalloprotease
- TIMP
tissue inhibitor of metalloprotease
References
- 1.Couve E. Ultrastructural changes during the life cycle of human odontoblasts. Arch. Oral Biol. 1986;31:643–651. doi: 10.1016/0003-9969(86)90093-2. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg M, Septier D, Lécolle S, Chardin H, Quintana MA, Acevedo AC, Gafni G, Dillouya D, Vermelin L, Thonemann B. Dental mineralization. Int. J. Dev. Biol. 1995;39:93–110. [PubMed] [Google Scholar]
- 3.Yoshida S, Ohshima H. Distribution and organization of peripheral capillaries in dental pulp and their relationship to odontoblasts. Anat. Rec. 1996;245:313–326. doi: 10.1002/(SICI)1097-0185(199606)245:2<313::AID-AR14>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 4.Jukka J, Irma T. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki T, Kawamata-Kido H. Providing an environment for reparative dentine induction in amputated rat molar pulp by high molecular-weight hyaluronic acid. Arch. Oral Biol. 1995;40:209–219. doi: 10.1016/0003-9969(95)98810-l. [DOI] [PubMed] [Google Scholar]
- 6.Smith AJ, Cassidy N, Perry H, Bègue-Kirn C, Ruch JV, Lesot H. Reactionary dentinogenesis. Int. J. Dev. Biol. 1995;39:273–280. [PubMed] [Google Scholar]
- 7.Takeuchi R, Saito T, Ishikawa H, Takigami H, Dezawa M, Ide C, Itokazu Y, Ikeda M, Shiraishi T, Morishita S. Effects of vibration and hyaluronic acid on activation of three-dimensional cultured chondrocytes. Arthritis Rheum. 2006;54:1897–1905. doi: 10.1002/art.21895. [DOI] [PubMed] [Google Scholar]
- 8.Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J. Clin. Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Alman BA. Wnt pathway, an essential role in bone regeneration. J. Cell Biochem. 2009;106:353–362. doi: 10.1002/jcb.22020. [DOI] [PubMed] [Google Scholar]
- 10.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, Ståhlberg A, Zoric N. The real-time polymerase chain reaction. Mol. Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Abe M, Michikami I, Fukushi T, Abe A, Maeda Y, Ooshima T, Wakisaka S. Hand2 regulates chondrogenesis in vitro and in vivo. Bone. 2010;46:1359–1368. doi: 10.1016/j.bone.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 12.Hanks CT, Fang D, Sun Z, Edwards CA, Butler WT. Dentin-specific proteins in MDPC-23 cell line. Eur. J. Oral Sci. 1998;106:260–266. doi: 10.1111/j.1600-0722.1998.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 13.Hanks CT, Sun ZL, Fang DN, Edwards CA, Wataha JC, Ritchie HH, Butler WT. Cloned 3T6 cell line from CD-I mouse fetal molar dental papillae. Connect. Tissue Res. 1998;37:233–249. doi: 10.3109/03008209809002442. [DOI] [PubMed] [Google Scholar]
- 14.Lévy L, Wei Y, Labalette C, Wu Y, Renard CA, Buendia MA, Neuveut C. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol. Cell. Biol. 2004;24:3404–3414. doi: 10.1128/MCB.24.8.3404-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Souza RN, Bachman T, Baumgardner KR, Butler WT, Litz M. Characterization of cellular responses involved in reparative dentinogenesis in rat molars. J. Dent. Res. 1995;74:702–709. doi: 10.1177/00220345950740021301. [DOI] [PubMed] [Google Scholar]
- 18.Hirata M, Yamaza T, Mei YF, Akamine A. Expression of osteocalcin and Jun D in the early period during reactionary dentin formation after tooth preparation in rat molars. Cell Tissue Res. 2005;319:455–465. doi: 10.1007/s00441-004-1035-y. [DOI] [PubMed] [Google Scholar]
- 19.Oyama M, Myokai F, Ohira T, Shiomi N, Yamashiro K, Arai H, Nishimura F, Takashiba S. Isolation and expression of FIP-2 in wounded pulp of the rat. J. Dent. Res. 2005;84:842–847. doi: 10.1177/154405910508400912. [DOI] [PubMed] [Google Scholar]
- 20.Herouy Y, Mellios P, Bandemir E, Dichmann S, Nockowski P, Schöpf E, Norgauer J. Inflammation in stasis dermatitis upregulates MMP-1, MMP-2 and MMP-13 expression. J. Dermatol. Sci. 2001;25:198–205. doi: 10.1016/s0923-1811(00)00128-6. [DOI] [PubMed] [Google Scholar]
- 21.Chakraborti S, Mandal M, Das S, Mandal A, Chakraborti T. Regulation of matrix metalloproteinases: an overview. Mol. Cell Biochem. 2003;253:269–285. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- 22.Warner RL, Bhagavathula N, Nerusu KC, Lateef H, Younkin E, Johnson KJ, Varani J. Matrix metalloproteinases in acute inflammation: induction of MMP-3 and MMP-9 in fibroblasts and epithelial cells following exposure to pro-inflammatory mediators in vitro. Exp. Mol. Pathol. 2004;76:189–195. doi: 10.1016/j.yexmp.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Vaalamo M, Mattila L, Johansson N, Kariniemi AL, Karjalainen-Lindsberg ML, Kähäri VM, Saarialho-Kere U. Distinct populations of stromal cells express collagenase-3 (MMP-13) and collagenase-1 (MMP-1) in chronic ulcers but not in normally healing wounds. J. Invest. Dermatol. 1997;109:96–101. doi: 10.1111/1523-1747.ep12276722. [DOI] [PubMed] [Google Scholar]
- 24.Bullard KM, Lund L, Mudgett JS, Mellin TN, Hunt TK, Murphy B, Ronan J, Werb Z, Banda MJ. Impaired wound contraction in stromelysin-1-deficient mice. Ann. Surg. 1999;230:260–265. doi: 10.1097/00000658-199908000-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill SE, Parks WC. Metalloproteinases and their inhibitors: regulators of wound healing. Int. J. Biochem. Cell Biol. 2008;40:1334–1347. doi: 10.1016/j.biocel.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy G, Docherty AJ. The matrix metalloproteinases and their inhibitors. Am. J. Respir. Cell Mol. Biol. 1992;7:120–125. doi: 10.1165/ajrcmb/7.2.120. [DOI] [PubMed] [Google Scholar]
- 27.Baker AH, Edwards DR, Murphy G. Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J. Cell Sci. 2002;115:3719–3727. doi: 10.1242/jcs.00063. [DOI] [PubMed] [Google Scholar]
- 28.Chirco R, Liu XW, Jung KK, Kim HR. Novel functions of TIMPs in cell signaling. Cancer Metastasis Rev. 2006;25:99–113. doi: 10.1007/s10555-006-7893-x. [DOI] [PubMed] [Google Scholar]
- 29.Vignola AM, Riccobono L, Mirabella A, Profita M, Chanez P, Bellia V, Mautino G, D'accardi P, Bousquet J, Bonsignore G. Sputum metalloproteinase-9/tissue inhibitor of metalloproteinase-1 ratio correlates with airflow obstruction in asthma and chronic bronchitis. Am. J. Respir. Crit. Care Med. 1998;158:1945–1950. doi: 10.1164/ajrccm.158.6.9803014. [DOI] [PubMed] [Google Scholar]
- 30.Yang C, Zhu P, Yan L, Chen L, Meng R, Lao G. Dynamic changes in matrix metalloproteinase 9 and tissue inhibitor of metalloproteinase 1 levels during wound healing in diabetic rats. J. Am. Podiatr. Med. Assoc. 2009;99:489–496. doi: 10.7547/0990489. [DOI] [PubMed] [Google Scholar]
- 31.Nusse R. WNT targets. Repression and activation. Trends Genet. 1999;15:1–3. doi: 10.1016/s0168-9525(98)01634-5. [DOI] [PubMed] [Google Scholar]
- 32.Akiyama T. Wnt/beta-catenin signaling. Cytokine Growth Factor Rev. 2000;11:273–282. doi: 10.1016/s1359-6101(00)00011-3. [DOI] [PubMed] [Google Scholar]
- 33.Leung JY, Kolligs FT, Wu R, Zhai Y, Kuick R, Hanash S, Cho KR, Fearon ER. Activation of AXIN2 expression by beta-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J. Biol. Chem. 2002;277:21657–21665. doi: 10.1074/jbc.M200139200. [DOI] [PubMed] [Google Scholar]
- 34.Lu Y, Xie Y, Zhang S, Dusevich V, Bonewald LF, Feng JQ. DMP1-targeted Cre expression in odontoblasts and osteocytes. J. Dent. Res. 2007;86:320–325. doi: 10.1177/154405910708600404. [DOI] [PubMed] [Google Scholar]
- 35.Kosaki N, Takaishi H, Kamekura S, Kimura T, Okada Y, Minqi L, Amizuka N, Chung UI, Nakamura K, Kawaguchi H, Toyama Y, D'Armiento J. Impaired bone fracture healing in matrix metalloproteinase-13 deficient mice. Biochem. Biophys. Res. Commun. 2007;354:846–851. doi: 10.1016/j.bbrc.2006.12.234. [DOI] [PubMed] [Google Scholar]
- 36.Zheng L, Amano K, Iohara K, Ito M, Imabayashi K, Into T, Matsushita K, Nakamura H, Nakashima M. Matrix metalloproteinase-3 accelerates wound healing following dental pulp injury. Am. J. Pathol. 2009;175:1905–1914. doi: 10.2353/ajpath.2009.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]