Abstract
Matriptase is a type II transmembrane serine protease containing two complement proteases C1r/C1s–urchin embryonic growth factor–bone morphogenetic protein domains (CUB repeat) and four low-density lipoprotein receptor class A domains (LDLRA repeat). The single-chain zymogen of matriptase has been found to exhibit substantial protease activity, possibly causing its own activation (i.e. conversion to a disulfide-linked two-chain fully active form), although the activation seems to be mediated predominantly by two-chain molecules. Our aim was to assess the roles of CUB and LDLRA repeats in zymogen activation. Transient expression studies of soluble truncated constructs of recombinant matriptase in COS-1 cells showed that the CUB repeat had an inhibitory effect on zymogen activation, possibly because it facilitated the interaction of two-chain molecules with a matriptase inhibitor, hepatocyte growth factor activator inhibitor type-1. By contrast, the LDLRA repeat had a promoting effect on zymogen activation. The effect of the LDLRA repeat seems to reflect its ability to increase zymogen activity. The proteolytic activities were higher in pseudozymogen forms of recombinant matriptase containing the LDLRA repeat than in a pseudozymogen without the repeat. Our findings provide new insights into the roles of these non-catalytic domains in the generation of active matriptase.
Keywords: CUB repeat, hepatocyte growth factor activator inhibitor type-1, LDLRA repeat, matriptase, zymogen activation
Matriptase is a member of the type II transmembrane serine protease group, which is characterized by the presence of an N-terminal cytoplasmic domain followed by a signal-anchor transmembrane domain and an extracellular domain, including a C-terminal serine protease catalytic domain (SPCD) (Fig. 1) (1–6). Matriptase is first synthesized as a zymogen comprising 855 amino acid residues in human, mouse and rat enzymes. The matriptase zymogen undergoes cleavage between Arg614 and Val615 (activation cleavage), and the disulfide-linked two-chain fully active enzyme is generated (Fig. 1) (1–6). Two-chain matriptase exhibits activity with trypsin-like specificity and can cleave and activate a number of proteins, including pro-hepatocyte growth factor (HGF) and the precursor of a membrane-bound serine protease, prostasin (7–9). The potential substrates, abundant expression in the epithelial cells and keratinocytes and characteristic phenotypes in matriptase gene-disrupted mice, suggest that the enzyme plays important roles in the establishment and maintenance of epithelial and epidermal barriers (1, 3, 10–12).
Fig. 1.
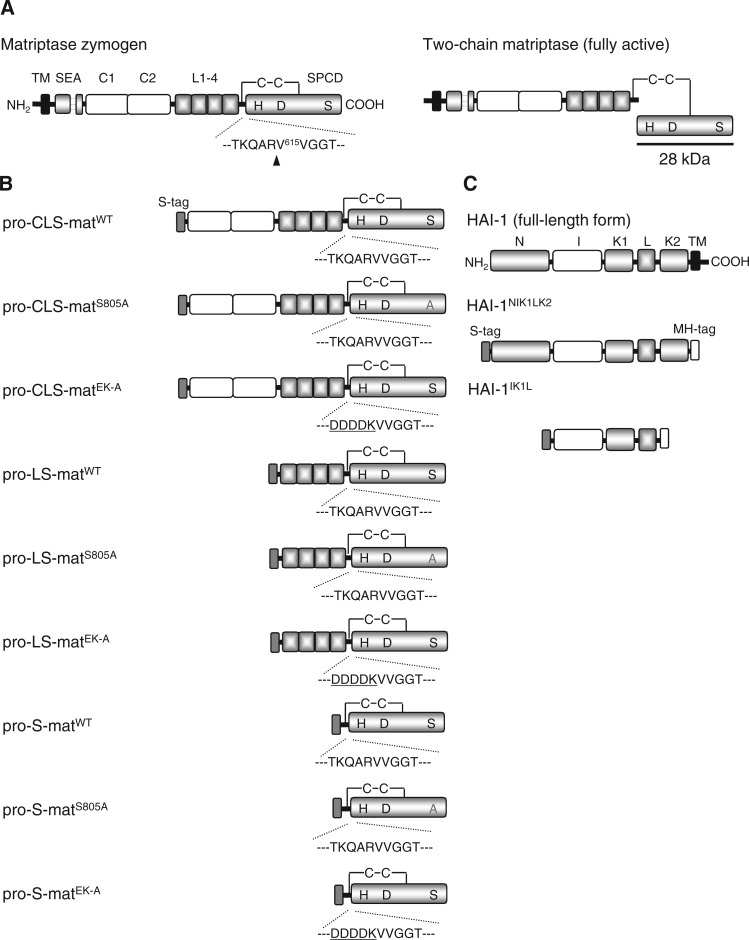
Schematic illustration of the structures of full-length rat matriptase and HAI-1, and their expression constructs. (A) Schematic illustration of the domain structures of full-length rat matriptase (Matriptase zymogen). The amino acid numbering starts from the putative N-terminus of the protein. The amino- and carboxyl-termini are indicated by NH2 and COOH, respectively. The association of the amino-terminal part (Met1–Gly149) with the carboxyl-terminal part (Ser150–Val855) is illustrated with broken lines. The amino acid sequence around the matriptase activation cleavage site is indicated by the single-letter code with amino acid number at the N-terminal valine residue of the SPCD (Val615). The activation cleavage site is indicated by the arrowhead. When cleaved, disulfide-linked two-chain matriptase is generated, shown here as ‘two-chain matriptase (fully active)’. The SPCD of the two-chain matriptase is underlined at the position at which the domain migrates on reducing SDS-PAGE (28 kDa). (B) Schematic illustration of the domain structures of secreted variants of r-matriptase. The expression vector used (pSecTag2/HygroB) contains the sequence of the immunoglobulin κ signal peptide allowing for secretion of translation products from mammalian cells. All variants were fused to the S-Tag at their N-termini. Enterokinase recognition sequences (DDDDK, underlined) and their surrounding sequences are shown in the pro-CLS-, pro-LS- and pro-S-matEK-A variants. (A and B) The predicted disulfide linkages between two cysteine residues corresponding to Cys604 and Cys731 in full-length matriptase and r-matriptase variants are shown as –C–C–, and the catalytic triad, His656, Asp711 and Ser805, are indicated as H, D and S, respectively, in the SPCD except for the pro-CLS-, pro-LS- and pro-S-matS805A variants, in which a serine residue corresponding to matriptase Ser805 is replaced with an alanine residue (denoted by the grey A). TM, transmembrane domain; SEA, SEA domain; C1 and C2, CUB domains I and II; L1-4, LDLRA repeat. (C) Schematic illustration of the domain structures of full-length rat HAI-1 and secreted variants of r-HAI-1. The structure of HAI-1 [HAI-1 (full-length form)] is indicated on the top. The N- and C-termini are indicated by NH2 and COOH, respectively. HAI-1NIK1LK2 and HAI-1IK1L are secreted variants of r-HAI-1. S-tag and Myc-hexahistidine tag (MH-tag) are fused at their N- and C-termini, respectively. N, N-terminal domain; I, internal domain; KD1, Kunitz domain I; L, LDLRA domain; KD2, Kunitz domain II; TM, transmembrane domain.
The activation cleavage of matriptase zymogen is believed to occur through a mechanism requiring its own catalytic triad (His656, Asp711 and Ser805) (1, 2, 13–15). For instance, recombinant (r-) forms of full-length matriptase, in which any residues of the catalytic triad are changed to an alanine residue, are unable to undergo activation when expressed exogenously in BT549 breast cancer cells (13). To date, the zymogen molecules have been postulated to undergo transactivation by which a zymogen interacts with another zymogen, producing the activation cleavage of each zymogen (1, 13). If so, matriptase zymogen should exert protease activity. We have found that (i) a pseudozymogen form of r-matriptase comprising the entire extracellular domain forms a catalytic triad analogous to His–Asp–Ser of trypsin-like serine proteases and (ii) the r-pseudozymogen effectively hydrolyses a peptide substrate, acetyl-l-Lys–l-Thr–l- Lys–l-Gln–l-Leu–l-Arg–4-methyl-coumaryl-7-amide (Ac-KTKQLR-MCA), whose sequence shares some sequence similarities with the activation cleavage site of this protease (Fig. 1) (16). In addition, the r-pseudozymogen exhibited optimal Ac-KTKQLR-MCA-hydrolyzing activity under mildly acidic and low ionic strength conditions (e.g. 5 mM NaCl and pH 6.0), under which matriptase zymogen endogenously expressed in a human mammary epithelial line 184 A1N4 undergoes activation with optimal efficiency (17–19). Our findings reinforce the concepts that the zymogen molecules can activate themselves by self-catalyzed proteolysis and thus that this protease serves as an initiator molecule of a protease cascade that is involved in the maintenance of epithelial and epidermal integrity (1, 3). However, it should be noted that the zymogen activation seems to be mediated predominantly by two-chain molecules. Indeed, the Ac-KTKQLR-MCA-hydrolyzing activity was higher for the corresponding two-chain r-matriptase than for the r-pseudozymogen (16).
Matriptase has an extracellular non-catalytic stem domain comprising a sea urchin sperm protein–enteropeptidase–agrin (SEA) domain, followed by two tandem repeats of complement proteases C1r/C1s–urchin embryonic growth factor–bone morphogenetic protein (CUB) domain and four tandem repeats of a low-density lipoprotein receptor class A (LDLRA) domain (Fig. 1) (1–6). Lin and coworkers have proposed the intimate involvement of the matriptase stem domain in its zymogen activation (13, 19). For instance, a site-directed mutant of full-length matriptase (Gly149Asn), which was designed to escape post-translational cleavage between Gly149 and Ser150 within the SEA domain (Fig. 1), was unable to undergo activation (13). A possible explanation for this is that, on cleavage within the SEA domain, the matriptase zymogen undergoes a conformational change or reorientation on the membrane surface that makes the protease susceptible to activation cleavage. The CUB and LDLRA repeats were also shown to affect the zymogen activation (13, 19). However, the definitive roles of these repeats in the zymogen activation remain unclear. We have shown that (i) the matriptase second CUB domain (CUB domain II) serves as an additional site for interaction with a physiological inhibitor of this protease, HGF activator inhibitor type-1 (HAI-1), which contains two protease-inhibitory Kunitz domains, and (ii) the interaction between CUB domain II and HAI-1 facilitates the primary inhibitory interaction between the enzyme and the inhibitor (i.e. interaction between the SPCD of two-chain matriptase and the first Kunitz domain) (Fig. 1) (20). This finding raises the possibility that CUB domain II may suppress zymogen activation by accelerating the rate of inhibition of the two-chain matriptase by HAI-1.
The aim of this study was to gain insights into the role(s) of CUB and LDLRA repeats in the activation of matriptase zymogen. To date, the structural requirements of matriptase zymogen for its activation cleavage have been studied on transient expression of the membrane-bound forms of r-matriptase in cultured cells (13–15, 21, 22). In the context of membrane-bound forms, internal deletions and point mutations showed differential effects on the activation cleavage (13). For example, full-length r-matriptase bearing a mutation in its LDLRA repeat (tyrosine replacement of aspartic acid residues comprising the calcium cage in each domain) rarely underwent activation, whereas a truncated variant lacking the repeat did it to a similar extent as in full-length, wild-type (WT) r-matriptase (13). Expression studies using soluble, truncated forms of r-matriptase could be expected to solve contradictory conclusions regarding the roles of internal domains (CUB and LDLRA repeats). In addition, the activation behaviours of soluble variants in cellulo would provide a coherent interpretation of the in vitro results obtained using soluble, truncated, pseudozymogen forms of r-matriptase (pro-CLS-, pro-LS- and pro-S-matEK-A variants, Fig. 1) (20). For these reasons, we prepared plasmids for the expression of secreted variants of r-matriptase and investigated whether the r-matriptase variants undergo activation cleavage when expressed in CV-1 in origin and carrying the SV40 genetic material (COS-1) monkey kidney cells. Our in cellulo results provided suggestive evidence that the CUB repeat had an inhibitory effect, whereas the LDLRA repeat had a promoting effect, on zymogen activation. In vitro experiments using the pseudozymogen forms of r-matriptase showed that the LDLRA repeat increased the protease activity of matriptase zymogen. To our knowledge, this is the first report showing how CUB and LDLRA repeats of matriptase participate in its zymogen activation.
Materials and Methods
Expression constructs
Plasmids for the expression of pseudozymogen forms of r-matriptase [pSec-pro-CLS-matEK-A (Gln210–Val855), pSec-pro-LS-matEK-A (Cys453–Val855) and pSec-pro-S-matEK-A (Asp603–Val855)] and of two secreted variants of r-HAI-1 [pSec-HAI-1NIK1LK2 (Pro41–Ser441) and pSec-HAI-1IK1L (Thr154–Ser370)] have already been constructed using pSecTag2/HygroB vector (Invitrogen, Carlsbad, CA, USA) (20, 23).
The plasmid for the expression of a secreted variant of r-matriptase comprising amino acids Gln210–Val855 was created as follows. A DNA fragment was amplified by polymerase chain reaction (PCR) with KOD-plus-DNA polymerase (Toyobo, Osaka, Japan), 5′-TCAGGACAACAGCTGCA-3′ and 5′-CTATACCCCAGTTTGCTCT-3′ as the forward and reverse primers, respectively, and a plasmid for the expression of full-length, WT rat matriptase (pcDNA-WT-matriptase) (15, 22) as the template. The PCR product was phosphorylated with T4 polynucleotide kinase and ligated into the SmaI-linearized pT7blue2 vector (Novagen) (pT7-pro-CLS-mat). pT7-pro-CLS-mat was digested with KpnI and BamHI, and the resulting fragment was ligated into pSec-pro-CLS-matEK-A from which a fragment produced by digestion with KpnI and BamHI had been removed. This plasmid was designated as pSec-pro-CLS-matWT. Plasmids for the expression of secreted variants of r-matriptase comprising amino acids Cys453–Val855 and Asp603–Val855 were made using the same method except that 5′-ATGCCCAGGGATGTTCAT-3′ and 5′-TGACTGTGGGCTGCGATCCTTTAC-3 were used as forward and reverse primers, respectively. The plasmids were designated as pSec-pro-LS-matWT and pSec-pro-S-matWT. The expression plasmids (pSec-pro-CLS-matS805A, pSec-pro-LS-matS805A and pSec-pro-S-matS805A) were created by the same method as those adopted for the creation of pSec-pro-CLS-matWT, pSec-pro-LS-matWT and pSec-pro-S-matWT, except that a plasmid for the expression of a full-length variant of r-matriptase bearing an alanine instead of serine at position 805 (pcDNA-S805A/matriptase) (15) was used as the template in the PCR. The sequences of the expression plasmids constructed in this study were determined in both directions using the dideoxynucleotide chain termination method, as described previously (5).
Transfection of expression plasmids into COS-1 cells and sample preparation
The procedure for the transfection of constructs into COS-1 cells using Lipofectamine2000™ (Invitrogen) has been described previously (15, 21). Note that each of the six plasmids for the expression of r-matriptase variants (2.4 µg) was co-transfected with pSecTag2/hygroB (2.4 µg) or pSec-HAI-1NIK1LK2 (2.4 µg). Cells were incubated for 24 h after transfection. The transfected cells were then washed three times with phosphate-buffered saline (PBS) (8 mM Na2HPO4, 1.5 mM KH2PO4, 136 mM NaCl and 2.7 mM KCl, pH 7.4) and then cultured for an additional 24 h in 1 ml of serum-free medium. After incubation, the conditioned medium was harvested and transferred to a 1.5 ml microcentrifuge tube in which 100 µl of the protease inhibitor cocktail Complete™ (1 × concentrate) (Roche Diagnostics, Mannheim, Germany) was included. After centrifugation at 3,000g for 5 min at 22°C, the resulting supernatant was concentrated to 40 µl by ultrafiltration using Microcon-10 (10,000 NMWL, Millipore, Bedford, MA, USA). After the addition of 10 µl of 5 × Laemmli protein sample buffer (Laemmli buffer) [1 × Laemmli buffer, 0.05 M Tris–HCl (pH 6.8), 10% glycerol, 2% sodium dodecyl sulphate (SDS) and 0.005% bromophenol blue with dithiothreitol at a final concentration of 12 mM] (24), the ultrafiltrates were stored at −20°C until use.
Analysis of expression products of r-matriptase and r-HAI-1 variants by SDS-polyacrylamide gel electrophoresis and western blotting
Samples were thawed, heated to 95°C for 3 min and subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide). A 20-µl portion of the samples was loaded into each lane. After separation, the proteins were transferred by electroblotting onto a polyvinylidene fluoride membrane (Fluorotrans W; Nihon Genetics, Tokyo, Japan), and the blots were rinsed twice with a buffer [50 mM Tris–HCl (pH 7.5) containing 145 mM NaCl and 0.1% Tween 20 (TBST)]. The blots were blocked by incubating in TBST containing 2% Difco™ skim milk (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) for 18 h at 4°C. The signals for r-matriptase variants were visualized as follows. After rinsing with TBST, the blots were incubated with a rabbit anti-matriptase SPCD antibody (Spr992) (22) diluted in an immunoreaction enhancer solution (Can Get Signal® Solution I, Toyobo) (1:20 dilution) for 18 h at 22°C. After washing with TBST, the blots were incubated with a goat anti-rabbit IgG secondary antibody conjugated with horseradish peroxidase (HRP; Dako Japan, Kyoto, Japan) diluted in another immunoreaction enhancer solution (Can Get Signal® Solution II, Toyobo; 1:3,000 dilution) for 2 h at 22°C. The blots were washed with TBST, and the protein bands were visualized using an ECL® detection system (GE Healthcare, Tokyo, Japan). The r-HAI-1 variant and the non-activated forms of r-matriptase variants were probed using HRP-conjugated S-protein (S-protein–HRP) (Novagen, Madison, WI, USA) diluted in Can Get Signal® Solution II (1: 3,000 dilution).
Preparation of pseudozymogen forms of r-matriptase and HAI-1IK1L
We have established Chinese hamster ovary (CHO)-K1 cell lines that stably express pro-CLS-matEK-A, pro-LS-matEK-A or pro-S-matEK-A (20). In this study, we established a CHO-K1 cell line expressing HAI-1IK1L using the same method as described previously (23).
The stably transfected cells were cultured in a 75-cm2 plastic flask (Asahi Techno Glass) as described previously (23). After reaching confluence, cells were washed three times with PBS, and 10 ml of Ham’s F12 without foetal bovine serum was added to the flask. After 48 h of incubation, the conditioned medium was collected, and fresh serum-free medium was added. This was repeated until half of the cells had peeled off. The collected media were centrifuged immediately at 3,000g for 10 min at 22°C, and the resulting supernatants were stored at −20°C until use. For purification, 300 ml of the conditioned media was collected into three flasks. After thawing, the media were pooled and concentrated to 2.5 ml by ultrafiltration using an Amicon® Ultra-15 membrane filter [50,000 molecular weight cut-off (MWCO) for pro-CLS-matEK-A, 30,000 MWCO for pro-LS-matEK-A and HAI-1IK1L, and 10,000 MWCO for pro-S-matEK-A, Millipore]. The concentrated medium was subjected to gel filtration in a buffer [25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.5) containing 145 mM NaCl and 0.1% Triton X-100] (hereinafter called purification buffer) using a PD-10 column (GE Healthcare) with an elution volume of 3.5 ml. The gel filtrate was divided equally, and each part was transferred to a 2.0-ml microcentrifuge tube in which a 62.5-µl slurry of immobilized S-protein was included. The tubes were incubated for 30 min at 22°C with rocking. After brief centrifugation, the supernatants were incubated again with immobilized S-protein as described earlier. The precipitated slurries were pooled (250 µl) and washed three times with 1.5 ml of purification buffer and two times with 1.5 ml of 10 mM HEPES (pH 7.5) containing 5 mM NaCl and 0.01% Triton X-100 (hereinafter called elution buffer). To elute these r-matriptase molecules, the washed slurry was transferred with 500 µl of elution buffer containing 200 µM of synthetic S-Tag peptide (KETAAAKFERQHIDS, BEX, Tokyo, Japan) to a spin column (Attoprep™, 0.22-µm pore size, Atto, Tokyo, Japan) and centrifuged at 5,000g for 1 min at 22°C. The eluate was subjected to gel filtration in elution buffer using an NAP-5 column (GE Healthcare) with an elution volume of 1.0 ml and stored at −20°C until use.
The concentration of each r-matriptase variant was measured semiquantitatively as follows. Various amounts of r-matriptase variants and HAI-1IK1L and Perfect Protein Marker™ protein size marker (Novagen), in which each protein was fused to an S-Tag and the concentration of each marker was known, were subjected to reducing SDS-PAGE (12% polyacrylamide) followed by western transfer. The blots were probed with S-protein–HRP. The X-ray films were scanned digitally, and the signal intensities of protein bands were analysed by densitometry as described previously (23). The concentrations of each r-matriptase variant and HAI-1IK1L were calculated by comparing the signal density of each band of interest with the 50-kDa protein contained as the protein marker.
Preparation of r-matriptase variants corresponding to the two-chain form and determination of their Ac-KTKQLR-MCA-hydrolyzing activities
To generate r-matriptase variants corresponding to the two-chain form of matriptase (hereinafter called CLS-mat, LS-mat and S-mat), a 500-µl portion of the elution buffer containing 50 nM pro-CLS-matEK-A, pro-LS-matEK-A or pro-S-matEK-A was incubated for 24 h at 22°C with a recombinant form of enteropeptidase (hereinafter called r-EK) (Novagen). The final concentration of r-EK was 1 Unit/ml. After incubation, the solution (activation mixture) was stored at −20°C until use. The concentrations of CLS-mat, LS-mat and S-mat in the activation mixture were evaluated by reducing SDS-PAGE (12% polyacrylamide) and western blotting using Spr992 (22). The signal density of a 28-kDa band produced from the SPCD part of CLS-mat, LS-mat or S-mat was compared with that of a 90-kDa band produced from pro-CLS-matEK-A, whose concentration had been determined as described earlier.
Ac-KTKQLR-MCA (Peptide Institute, Osaka, Japan) was dissolved in dimethyl sulfoxide at a concentration of 20 mM and stored at −20°C until use. A portion of CLS-mat, LS-mat or S-mat was incubated with 1 mM of Ac-KTKQLR-MCA in 25 mM HEPES buffer (pH 7.5) containing 145 mM NaCl and 0.1% Triton X-100 for 10 min at 37°C in a volume of 80 µl. The reaction was terminated by adding 350 µl of 0.1 M monochloroacetic acid in 0.1 M sodium acetate buffer, pH 4.3 (stop solution), and the absorbance at 370 nm was measured. The initial rate of hydrolysis was determined using the molar absorption coefficient ε370 of 7.7 mM–1 cm–1 for 4-methyl-coumaryl-7-amine.
Hydrolysis of Ac-KTKQLR-MCA by pro-CLS-matEK-A, pro-LS-matEK-A or pro-S-matEK-A and determination of the kinetic parameters for the hydrolysis
Incubations were performed at 37°C in a 0.5-ml microcentrifuge tube. The reactions were initiated by the addition of 10 µl of elution buffer containing one of the pseudozymogens (3.2 nM for the pro-CLS-matEK-A and pro-LS-matEK-A variants and 16 nM for the pro-S-matEK-A variant) or the buffer alone to 10 µl of buffer [200 mM MES-NaOH (pH 6.0) buffer with 10 mM NaCl and 0.02% Triton X-100] containing 0.1 nM HAI-1IK1L and Ac-KTKQLR-MCA. The final concentrations of the substrate were 0, 1.95, 3.9, 7.8, 15.6, 31.3, 62.5 and 125 µM. The reaction mixtures containing the pseudozymogen forms of r-matriptase variants were incubated for 72 h. To prevent water evaporation within the tube, the incubation was conducted using a temperature controller (PC805-MI, ASTEC, Fukuoka, Japan). The reactions were terminated by adding 400 µl of stop solution. One hundred microlitres of aliquots of the final reaction mixtures were added to the wells of a 96-well assay plate, and the fluorescence of 4-methyl-coumaryl-7-amine liberated from Ac-KTKQLR-MCA was measured using a plate reader (PowerScan, Dainippon Sumitomo Pharma, Osaka, Japan) at an excitation wavelength of 380 nm and an emission wavelength of 460 nm. The concentration of the product was assessed by comparing the fluorescence value with that of stop solution containing 4-methyl-coumaryl-7-amine at various concentrations. To determine the enzyme-catalyzed reaction rate, the non-enzymatic spontaneous reaction rate of the absorbance increase was subtracted from the reaction rate observed in the presence of the enzyme. The Michaelis constant (Km) and the rate of maximum turnover (kcat) for the hydrolysis of Ac-KTKQLR-MCA were calculated by fitting the rate data to the Michaelis–Menten equation using KaleidaGraph (Synergy Software, Reading, PA, USA).
Results
Identification of secreted variants of r-matriptase and the detection methods
To assess the roles of CUB and LDLRA repeats in the activation of matriptase zymogen by in cellulo experiments, we prepared plasmids for the expression of secreted variants of r-matriptase: pro-CLS-matWT comprising the CUB and LDLRA repeats and the SPCD; pro-LS-matWT comprising the LDLRA repeat and SPCD; and pro-S-matWT comprising only the SPCD (Fig. 1). The superscript WT in these r-matriptase variants reflects that each of them has both the activation cleavage site and the catalytic triad, identical to full-length, WT rat matriptase. In this study, the expression plasmids for their respective active site mutants (i.e. mutant in which a serine residue corresponding to the active site residue Ser805 of matriptase was changed to an alanine residue), pro-CLS-matS805A, pro-LS-matS805A and pro-S-matS805A, were also prepared (Fig. 1).
The co-transfection of cDNAs encoding HAI-1 has been shown to be required for successful detection of full-length, WT r-matriptase in transient expression experiments with cultured cells, including COS-1 cells (13, 15, 21, 22). In the absence of HAI-1, the r-matriptase inadvertently activated in the intracellular environment appears to undergo extensive degradation via a self-catalyzed mechanism, thus being rarely detectable (21). On co-expression with HAI-1, both zymogen and two-chain forms of the r-matriptase were detectable (13, 15, 21, 22). It is also noted on co-expression with HAI-1 in COS-1 cells that (i) the activation cleavage of the r-matriptase can occur on the surface of cells and (ii) both the non-activated and activated forms of r-matriptase frequently undergo ectodomain shedding, thereby being detected predominantly in the conditioned medium (15, 21, 22). In this study, we transfected the plasmids to induce the expression of secreted variants of r-matriptase into the cells with or without a plasmid for the expression of a secreted variant of r-HAI-1 comprising all the extracellular domain (pSec-HAI-1NIK1LK2, Fig. 1) (21, 23) and investigated whether the r-matriptase variants undergoing activation cleavage (i.e. cleavage after an arginine residue corresponding to matriptase Arg614) can be detected in media conditioned by transfected cells. The occurrence of activation cleavage was assessed to determine whether a 28-kDa protein is immunostained on a western blot with an anti-matriptase SPCD antibody (Spr992) following reducing SDS-PAGE (Fig. 1) (21, 22). The r-matriptase variants that did not undergo cleavage after an arginine residue corresponding to matriptase Arg614 were analysed by reducing SDS-PAGE and western blotting with Spr992 or with S-protein–HRP (note that an S-Tag is fused to the N-terminus of each variant). HAI-1NIK1LK2 has been detected as a 59-kDa band by reducing SDS-PAGE and western blotting with S-protein–HRP (21).
Analysis of the expression of pro-CLS-matWT and its respective active site mutant
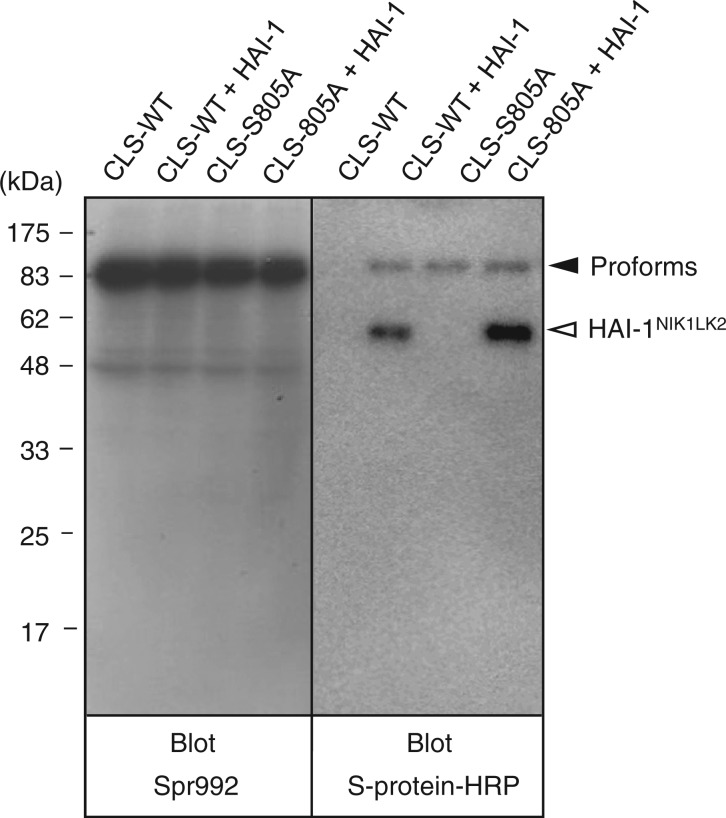
First, we analysed the expression of pro-CLS-matWT and pro-CLS-matS805A (Fig. 2). When a sample of media conditioned by COS-1 cells transfected with pSec-pro-CLS-matWT alone was probed with Spr992, no signal at the position corresponding to 28 kDa was produced (left panel, lane CLS-WT). A similar result was obtained for a medium sample of cells transfected with pSec-pro-CLS-matWT and pSec-HAI-1NIK1LK2 (left panel, lane CLS-WT + HAI-1). Using Spr992 as a probe, several protein bands were produced from medium samples of transfected cells (especially dense at the 80- to 90-kDa positions; also refer to the data for COS-1 cells transfected with pSecTag2/HygroB vector alone shown in Supplementary Fig. S1, lane Mock). These signals are derived from unknown proteins secreted by COS-1 cells (21).
Fig. 2.
Western blot analysis of the pro-CLS-matWT and pro-CLS-matS805A variants. Medium samples of COS-1 cells transfected with pSec-pro-CLS-matWT alone (CLS-WT), pSec-pro-CLS-matWT and pSec-HAI-1NIK1LK2 (CLS-WT + HAI-1), pSec-pro-CLS-matS805A alone (CLS-S805A) and pSec-pro-CLS-matS805A and pSec- HAI-1NIK1LK2 (CLS-S805A + HAI-1) were subjected to reducing SDS-PAGE. After western transfer, the blot was probed with Spr992 or S-protein–HRP. The position to which pro-CLS-matWT and pSec-pro-CLS-matS805A migrated is indicated as ‘Proforms’ to the right of the right panel. The position to which HAI-1NIK1LK2 migrated is indicated to the right of the right panel. The molecular masses of the marker proteins are indicated in kilodaltons to the left of the left panel.
When probed with S-protein–HRP, the medium sample of cells transfected with pSec-pro-CLS-matWT alone produced no visible bands (right panel, lane CLS-WT). In the medium sample of cells co-transfected with pSec-HAI-1NIK1LK2, a protein band at the 90-kDa position, indicative of the pro-CLS-matWT variant, was produced (right panel, lane CLS-WT + HAI-1). From this sample, a 59-kDa signal (indicative of HAI-1NIK1LK2) was also detected. The results obtained using cells transfected with pSec-pro-CLS-matWT and pSec-HAI-1NIK1LK2 suggest that the activation cleavage of pro-CLS-matWT occurs rarely or not at all.
Irrespective of co-transfection of pSec-HAI-1NIK1LK2, the samples of media conditioned by cells transfected with pSec-pro-CLS-matS805A produced a signal at the 90-kDa position in a western blot probed by S-protein–HRP (right panel, lanes CLS-S805A and CLS-S805A + HAI-1). The 90-kDa band may represent pro-CLS-matS805A. The appearance of the 90-kDa protein representing pro-CLS-matWT or pro-CLS-matS805A could not be evaluated in a western blot probed with Spr992 because of the dense signals for unknown proteins around the position (left panel).
Analysis of the expression of pro-LS-matWT and its respective active site mutant
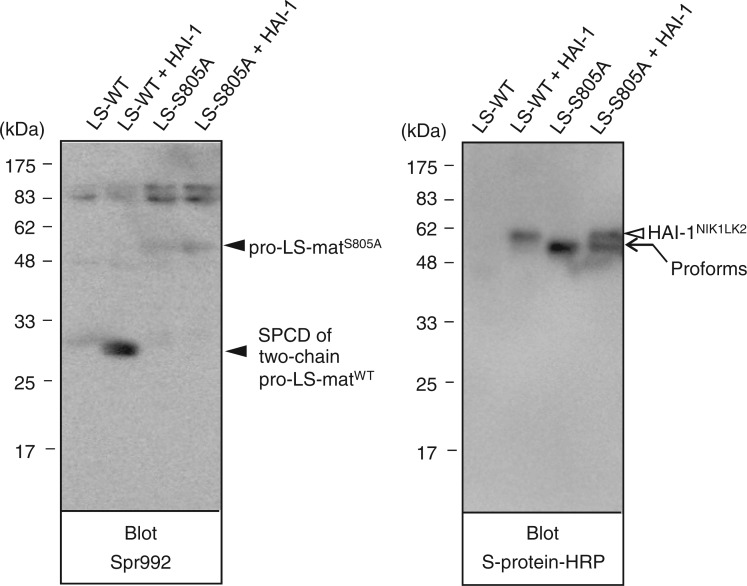
We next analysed the expression of pro-LS-matWT and pro-LS-matS805A (Fig. 3). When a sample of media conditioned by cells transfected with pSec-pro-LS-matWT alone was probed with Spr992, no signal at the 28-kDa position was detected (left panel, lane LS-WT). A protein band at the 28-kDa position was visualized clearly in medium samples of cells transfected with pSec-pro-LS-matWT and pSec-HAI-1NIK1LK2 (left panel, lane LS-WT + HAI-1), demonstrating that the pro-LS-matWT variant undergoes activation cleavage. Probing with S-protein–HRP of the sample of cells transfected with pSec-pro-LS-matWT and pSec-HAI-1NIK1LK2 showed HAI-1NIK1LK2 and a trace of the 57-kDa band representing pro-LS-matWT (right panel, lane LS-WT + HAI-1). In the sample of cells transfected with pSec-pro-LS-matWT alone, no apparent protein bands were visualized (right panel, lane LS-WT).
Fig. 3.
Western blot analysis of the pro-LS-matWT and pro-LS-matS805A variants. Medium samples of COS-1 cells transfected with pSec-pro-LS-matWT alone (LS-WT), pSec-pro-LS-matWT and pSec-HAI-1NIK1LK2 (LS-WT + HAI-1), pSec-pro-LS-matS805A alone (LS-S805A) and pSec-pro-LS-matS805A and pSec-HAI-1NIK1LK2 (LS-S805A + HAI-1) were subjected to reducing SDS-PAGE. After western transfer, the blot was probed with Spr992 or S-protein–HRP. The 28-kDa position to which the SPCD part of two-chain pro-LS-matWT migrated is indicated as ‘SPCD of two-chain pro-LS-matWT’ to the right of the left panel. The 57-kDa position at which pro-LS-matS805A migrated is indicated as ‘pro-LS-matS805A’ to the right of the left panel. The 57-kDa position to which pro-LS-matWT and pro-LS-matS805A migrated is indicated as ‘Proforms’ to the right of the right panel. The position to which HAI-1NIK1LK2 migrated is indicated to the right of the right panel. The molecular masses of the marker proteins are indicated in kilodaltons to the left of both panels.
When probed with Spr992, a protein band at the position corresponding to 57 kDa was visualized in samples of cells transfected with pSec-pro-LS-matS805A alone and samples of cells transfected with pSec-pro-LS-matS805A and pSec-HAI-1NIK1LK2 (left panel, lanes LS-S805A and LS-S805A + HAI-1). Similarly, the 57-kDa band was also visualized when probed with S-protein–HRP (right panel, lanes LS-S805A and LS-S805A + HAI-1). The results obtained using cells transfected with pSec-pro-LS-matS805A indicated that pro-LS-matWT underwent activation cleavage via a mechanism requiring the active site serine residue.
Analysis of the expression of pro-S-matWT and its respective active site mutant
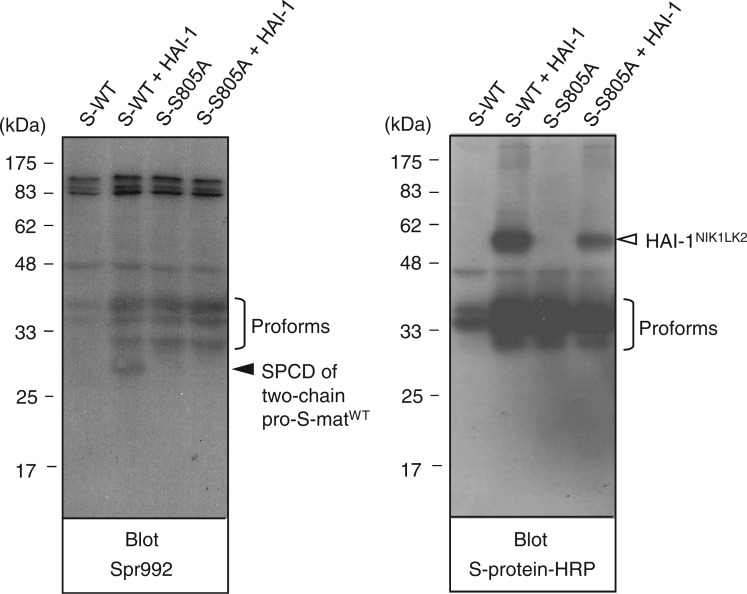
We present the data for the expression of pro-S-matWT and pro-S-matS805A in Fig. 4. When the sample of media conditioned by cells transfected with pSec-pro-S-matWT alone was probed with Spr992, no signal at the 28-kDa position was detected (left panel, lane S-WT), although protein bands at the 40-, 36- and 31-kDa positions were visualized. A signal at the 28-kDa position was produced from the media sample of cells transfected with pSec-pro-S-matWT and pSec-HAI-1NIK1LK2 (left panel, lane S-WT and HAI-1), indicating that the pro-S-matWT variant undergoes activation cleavage. The 40-, 36- and 31-kDa bands were also visualized in the same lane. Irrespective of the co-transfection with pSec-HAI-1NIK1LK2, the samples of media conditioned by cells transfected with pSec-pro-S-matWT produced 40-, 36- and 31-kDa bands in a western blot probed with S-protein–HRP (right panel, lanes S-WT and S-WT and HAI-1). Judging from the molecular masses, the three bands may represent pro-S-matWT species. We also note that the appearance of the three pro-S-matWT species may be explained by cleavage by signal peptidases of COS-1 cells at different sites.
Fig. 4.
Western blot analysis of the pro-S-matWT and pro-S-matS805A variants. Medium samples of COS-1 cells transfected with pSec-pro-S-matWT alone (S-WT), pSec-pro-S-matWT and pSec-HAI-1NIK1LK2 (S-WT + HAI-1), pSec-pro-S-matS805A alone (S-S805A) and pSec-pro-S-matS805A and pSec-HAI-1NIK1LK2 (S-S805A + HAI-1) were subjected to reducing SDS-PAGE. After western transfer, the blot was probed with Spr992 or S-protein–HRP. The 28-kDa position to which the SPCD part of two-chain pro-S-matWT migrated is indicated as ‘SPCD of two-chain pro-S-matWT’ to the right of the left panel. The 40-, 36- and 31-kDa positions to which pro-S-matWT and pro-S-matS805A species migrated are indicated as ‘Proforms’ to the right of the left and right panels. The position to which HAI-1NIK1LK2 migrated is indicated to the right of the right panel. The molecular masses of the marker proteins are indicated in kilodaltons to the left of both panels.
When probed with Spr992 or S-protein–HRP, 40-, 36- and 31-kDa bands were visualized in samples of cells transfected with pSec-pro-LS-matS805A alone and in samples of cells transfected with pSec-pro-LS-matS805A and pSec-HAI-1NIK1LK2 (left and right panels, lanes S-S805A and S-805A + HAI-1). The appearance of the three pro-S-matS805A species indicates that the active site mutant also undergoes cleavage by signal peptidases of COS-1 cells at different sites. The results obtained using cells transfected with pSec-pro-S-matS805A indicate that the pro-S-matWT variant undergoes activation cleavage via a mechanism requiring the active site serine residue.
Comparison of the degrees of activation and levels of expression of pro-CLS-, pro-LS- and pro-S-matWT variants
The degrees of activation cleavage and levels of expression of pro-CLS-, pro-LS- and pro-S-matWT variants were analysed in a western blot (Supplementary Fig. S1). We could not successfully evaluate the levels of expression of r-matriptase variants when each of them was expressed alone. On co-expression with HAI-1NIK1LK2, it is evident that the level of expression of pro-S-matWT was greater than that of pro-LS-matWT. In addition, the level of expression of pro-CLS-matWT was not significantly lower than that of pro-LS-matWT. Therefore, it seems unlikely that the level of expression seriously affects the activation frequency at least in experiments with COS-1 cells. In addition, the levels of HAI-1NIK1LK2 detected did not differ substantially irrespective of the type of r-matriptase variants co-expressed, suggesting that interaction frequencies of the variants with the r-HAI-1 do not differ substantially.
Hydrolysis of Ac-KTKQLR-MCA by pseudozymogen forms of r-matriptase
We have produced pseudozymogen forms of r-matriptase using CHO-K1 cells as the host for expression (20, 23). Of these, a variant comprising all the extracellular domain was found to exhibit substantial Ac-KTKQLR-MCA-hydrolyzing activity (16). In this study, we investigated the Ac-KTKQLR-MCA-hydrolyzing activities of the pro-CLS-, pro-LS- and pro-S-matEK-A variants (Fig. 1). Five amino acid residues needed for activation cleavage of this protease (Thr–Lys–Gln–Ala–Arg614) were changed to those suitable for cleavage by enteropeptidase (Asp–Asp–Asp–Asp–Lys) in any of the variants. This allowed for in vitro generation of an N-terminal valine residue corresponding to matriptase Val615 by treatment with r-EK (20, 23). Each of the r-matriptase variants after treatment with r-EK (CLS-mat, LS-mat and S-mat) hydrolyzed Ac-KTKQLR-MCA with a reaction rate of ∼2 nM·s–1. This is consistent with the observation that the stem domain of matriptase in the two-chain form has no substantial effect on the hydrolysis of peptidyl-MCA substrates (25, 26). The pro-CLS-, pro-LS- and pro-S-matEK-A variants not treated with r-EK can be used as models for matriptase zymogen.
In this study, we prepared a secreted variant of r-HAI-1 comprising the internal, first Kunitz and LDLRA domains using CHO-K1 cells (HAI-1IK1L, Fig. 1). This r-HAI-1 variant was found to be most effective at inhibiting a two-chain r-matriptase variant (23). To avoid the detection of activities derived from CLS-mat, LS-mat and S-mat occurring inadvertently during incubation, the HAI-1IK1L variant was included in the reaction mixtures. We confirmed that the HAI-1IK1L produced in CHO-K1 cells exhibited a matriptase-inhibition activity similar to the variant produced in COS-1 cells (23).
The pro-CLS-, pro-LS- and pro-S-matEK-A variants hydrolyzed Ac-KTKQLR-MCA. The kcat, Km and kcat/Km values are shown in Table I. We note that the kcat/Km values of the pro-CLS- and pro-LS-matEK-A variants were similar but more than 10 times higher than that of the pro-S-matEK-A variant. These results suggest that the CUB repeat has no substantial effect on the expression of zymogen activity and that the LDLRA repeat contributes considerably to the expression of zymogen activity.
Table I.
Kinetic values for hydrolysis of Ac-KTKQLR-MCA catalyzed by pro-CLS-, pro-LS- and pro-S-matEK-A variants.
| r-Matriptase variants | kcat × 103 min–1 | Km (µM) | kcat/Km (mM–1 min–1) |
|---|---|---|---|
| pro-CLS-matEK-A | 44 ± 7 | 7 ± 2 | 6.3 ± 6.1 |
| pro-LS-matEK-A | 146 ± 40 | 26 ± 13 | 5.6 ± 11 |
| pro-S-matEK-A | 8.0 ± 0.4 | 18 ± 5 | 0.43 ± 0.32 |
The kinetic value is expressed as a mean ± SD of triplicate determinations.
Discussion
The CUB domain is a widely occurring module in proteolytic enzymes or proteins that are involved in developmental processes, and the domains in certain proteases seem to be essential for recognition of natural substrates (27, 28). In matriptase, the second CUB domain has been postulated to serve as an additional site for interaction with HAI-1 to downregulate its own catalytic activity (20). r-Matriptase variants with the second CUB domain (e.g. CLS-mat) are inhibited by HAI-1NIK1LK2 with high efficiency, whereas those without the domain (e.g. LS-mat and S-mat) are inhibited with low efficiency (20). In our transient expression experiments, a protein band at the 28-kDa position was detected from a sample of cells transfected with pSec-pro-LS-matWT and pSec-HAI-1NIK1LK2 but not from that of cells transfected with pSec-pro-CLS-matWT and pSec-HAI-1NIK1LK2. Our interpretation is that the activated pro-CLS-matWT (hereinafter called two-chain pro-CLS-matWT) undergoes frequent inactivation by HAI-1NIK1LK2, and thus the activation cleavage of pro-CLS-matWT by two-chain pro-CLS-matWT occurs rarely. By contrast, the activated pro-LS-matWT (hereinafter called two-chain pro-LS-matWT) is inactivated rarely by the r-HAI-1 variant, and thus the activation cleavage of pro-LS-matWT by two-chain pro-LS-matWT occurs efficiently.
It seems likely that the pro-CLS-matWT variant exerts protease activity to generate two-chain pro-CLS-matWT. This is supported by the observations that (i) the sample of cells transfected with pSec-CLS-matWT alone rarely produced a protein band at the 90-kDa position, whereas that of cells transfected with pSec-CLS-matS805A alone did and (ii) pro-CLS-matEK-A hydrolyzed Ac-KTKQLR-MCA to a comparable extent as did pro-LS-matEK-A. In the absence of the r-HAI-1, two-chain pro-CLS-matWT occurring in the intracellular environment might degrade pro-CLS-matWT and two-chain pro-CLS-matWT progressively, and thus, both species would be rarely detected. A similar phenomenon may occur in the cells transfected with pSec-LS-matWT alone and in those transfected with pSec-S-matWT alone. Yet, it is uncertain why an activated form of matriptase was detected from medium samples of COS-1 cells transfected with a plasmid for expression of full-length, WT r-matriptase and pSec-HAI-1NIK1LK2 (21) but not from those transfected with pSec-pro-CLS-matWT and pSec-HAI-1NIK1LK2. One possible explanation is that the CUB domain–accelerated interaction with the r-HAI-1 occurs slowly and inefficiently in a cell membrane–associated state of two-chain matriptase, whereas it does rapidly and efficiently in the soluble state.
The LDLRA domain is ∼40 amino acids, is biologically ubiquitous and is found in several proteins. In general, this domain is thought to be involved in protein–protein interaction (29). In this study, we found that Ac-KTKQLR-MCA-hydrolyzing activity was higher in pro-LS-matEK-A than in pro-S-matEK-A. This strongly suggests that one of the important roles of the LDLRA repeat of matriptase is to increase the zymogen activity, thereby facilitating the zymogen-catalyzed activation cleavage of zymogen. Our transient expression experiments also support this idea. The pro-S-matWT species were detected in the sample from cells transfected with pSec-pro-S-matWT alone, whereas pro-LS-matWT was rarely detected in the sample from cells transfected with pSec-pro-LS-matWT alone. In addition, when co-expressed with HAI-1NIK1LK2, the pro-S-matWT species were detected in a sample from cells transfected with pSec-pro-S-matWT and pSec-HAI-1NIK1LK2, whereas the pro-LS-matWT was hardly detected in the sample from cells transfected with pSec-pro-LS-matWT and pSec-HAI-1NIK1LK2. In other words, the majority of pro-LS-matWT underwent activation cleavage, whereas less than half of pro-S-matWT did.
The LDLRA domain of matriptase has been modelled based on the structure of the fifth low-density lipoprotein (LDL)-binding domain of the LDL receptor (30, 31). Interestingly, the fourth LDLRA domain of matriptase was predicted to interact with the surface of its SPCD opposite to the active site cleft (30). In addition, matriptase zymogen seems to form a catalytic triad analogous to mature trypsin-like serine proteases (16, 32). It is tempting to speculate that the putative intramolecular interaction between the LDLRA repeat and SPCD within a matriptase zymogen facilitates the conformational change required for forming mature form-like structure. The CUB repeat does not appear to exert a substantial effect on the expression of zymogen activity. The Ac-KTKQLR-MCA-hydrolyzing activity did not differ substantially between pro-CLS-matEK-A and pro-LS-matEK-A.
In summary, we have provided insights into the roles of CUB and LDLRA repeats of matriptase in its zymogen activation. The CUB repeat might facilitate interaction of its SPCD of the two-chain enzyme with HAI-1, thereby contributing to the suppression of the activation cleavage of zymogen catalyzed by the active enzyme. The LDLRA repeat seems to have a critical effect on the expression of zymogen activity. These non-catalytic domains of matriptase might be organized in ways that allow this protease to serve as the most upstream regulator of the protease cascade that controls epithelial and epidermal integrity.
Supplementary Data
Supplementary Data are available at JB Online.
Funding
Grants-in-Aid for Scientific Research (to K. Inouye) from the Japan Society for the Promotion of Science.
Conflict of Interest
None declared.
Supplementary Material
Acknowledgements
The authors thank Seiya Mochida for valuable discussions.
Glossary
Abbreviations
- Ac-KTKQLR-MCA
acetyl-l-Lys–l-Thr–l-Lys–l-Gln–l-Leu–l-Arg–4-methyl-coumaryl-7-amide
- COS
CV-1 in origin and carrying the SV40 genetic material
- CUB
complement proteases C1r/C1s–urchin embryonic growth factor–bone morphogenetic protein
- HAI-1
hepatocyte growth factor activator inhibitor type-1
- HGF
hepatocyte growth factor
- HRP
horseradish peroxidase
- LDLRA
low-density lipoprotein receptor class A MWCO, molecular weight cut-off
- r-EK
recombinant enteropeptidase
- SEA
sea urchin sperm protein–enteropeptidase–agrin
- SPCD
serine protease catalytic domain
- WT
wild-type
References
- 1.Lin CY, Tseng IC, Chou FP, Su SF, Chen YW, Johnson MD, Dickson RB. Zymogen activation, inhibition, and ectodomain shedding of matriptase. Front. Biosci. 2008;13:621–635. doi: 10.2741/2707. [DOI] [PubMed] [Google Scholar]
- 2.Darragh MR, Bhatt AS, Craik CS. MT-SP1 proteolysis and regulation of cell-microenvironment interactions. Front. Biosci. 2008;13:528–539. doi: 10.2741/2698. [DOI] [PubMed] [Google Scholar]
- 3.Bugge TH, List K, Szabo R. Matriptase-dependent cell surface proteolysis in epithelial development and pathogenesis. Front. Biosci. 2007;12:5060–5070. doi: 10.2741/2448. [DOI] [PubMed] [Google Scholar]
- 4.Kim MG, Chen C, Lyu MS, Cho EG, Park D, Kozak C, Schwartz RH. Cloning and chromosomal mapping of a gene isolated from thymic stromal cells encoding a new mouse type II membrane serine protease, epithin, containing four LDL receptor modules and two CUB domains. Immunogenetics. 1999;49:420–428. doi: 10.1007/s002510050515. [DOI] [PubMed] [Google Scholar]
- 5.Satomi S, Yamasaki Y, Tsuzuki S, Hitomi Y, Iwanaga T, Fushiki T. A role for membrane-type serine protease (MT-SP1) in intestinal epithelial turnover. Biochem. Biophys. Res. Commun. 2001;287:995–1002. doi: 10.1006/bbrc.2001.5686. [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi T, Shuman MA, Craik CS. Reverse biochemistry: use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue. Proc. Natl Acad. Sci. USA. 1999;96:11054–11061. doi: 10.1073/pnas.96.20.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SL, Dickson RB, Lin CY. Activation of hepatocyte growth factor and urokinase/plasminogen activator by matriptase, an epithelial membrane serine protease. J. Biol. Chem. 2000;275:36720–36725. doi: 10.1074/jbc.M007802200. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi T, Harris JL, Huang W, Yan KW, Coughlin SR, Craik CS. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J. Biol. Chem. 2000;275:26333–26342. doi: 10.1074/jbc.M002941200. [DOI] [PubMed] [Google Scholar]
- 9.Netzel-Arnett S, Currie BM, Szabo R, Lin CY, Chen LM, Chai KX, Antalis TM, Bugge TH, List K. Evidence for a matriptase–prostasin proteolytic cascade regulating terminal epidermal differentiation. J. Biol. Chem. 2006;281:32941–32945. doi: 10.1074/jbc.C600208200. [DOI] [PubMed] [Google Scholar]
- 10.List K, Haudenschild CC, Szabo R, Chen W, Wahl SM, Swaim W, Engelholm LH, Behrendt N, Bugge TH. Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene. 2002;21:3765–3779. doi: 10.1038/sj.onc.1205502. [DOI] [PubMed] [Google Scholar]
- 11.List K, Szabo R, Molinolo A, Nielsen BS, Bugge TH. Delineation of matriptase protein expression by enzymatic gene trapping suggests diverging roles in barrier function, hair formation, and squamous cell carcinogenesis. Am. J. Pathol. 2006;168:1513–1525. doi: 10.2353/ajpath.2006.051071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.List K, Kosa P, Szabo R, Bey AL, Wang CB, Molinolo A, Bugge TH. Epithelial integrity is maintained by a matriptase-dependent proteolytic pathway. Am. J. Pathol. 2009;175:1453–1463. doi: 10.2353/ajpath.2009.090240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oberst MD, Williams CA, Dickson RB, Johnson MD, Lin CY. The activation of matriptase requires its noncatalytic domains, serine protease domain, and its cognate inhibitor. J. Biol. Chem. 2003;278:26773–26779. doi: 10.1074/jbc.M304282200. [DOI] [PubMed] [Google Scholar]
- 14.Désilets A, Béliveau F, Vandal G, McDuff FO, Lavigne P, Leduc R. Mutation of G827R in matriptase causing autosomal recessive ichthyosis with hypotrichosis yields an inactive protease. J. Biol. Chem. 2008;283:10535–10542. doi: 10.1074/jbc.M707012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyake Y, Yasumoto M, Tsuzuki S, Fushiki T, Inouye K. Activation of a membrane-bound serine protease matriptase on the cell surface. J. Biochem. 2009;146:273–282. doi: 10.1093/jb/mvp066. [DOI] [PubMed] [Google Scholar]
- 16.Inouye K, Yasumoto M, Tsuzuki S, Mochida S, Fushiki T. The optimal activity of a pseudozymogen form of recombinant matriptase under the mildly acidic pH and low ionic strength conditions. J. Biochem. 2010;147:485–492. doi: 10.1093/jb/mvp190. [DOI] [PubMed] [Google Scholar]
- 17.Wang JK, Lee MS, Tseng IC, Chou FP, Chen YW, Fulton A, Lee HS, Chen CJ, Johnson MD, Lin CY. Polarized epithelial cells secrete matriptase as a consequence of zymogen activation and HAI-1-mediated inhibition. Am. J. Physiol. Cell Physiol. 2009;297:C459–C470. doi: 10.1152/ajpcell.00201.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tseng IC, Xu H, Chou FP, Li G, Vazzano AP, Kao JP, Johnson MD, Lin CY. Matriptase activation, an early cellular response to acidosis. J. Biol. Chem. 2010;285:3261–3270. doi: 10.1074/jbc.M109.055640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MS, Tseng IC, Wang Y, Kinomiya K, Johnson MD, Dickson RB, Lin CY. Autoactivation of matriptase in vitro: requirement for biomembrane and LDL receptor domain. Am. J. Physiol. Cell Physiol. 2007;293:C95–C105. doi: 10.1152/ajpcell.00611.2006. [DOI] [PubMed] [Google Scholar]
- 20.Inouye K, Tsuzuki S, Yasumoto M, Kojima K, Mochida S, Fushiki T. Identification of the matriptase second CUB domain as the secondary site for interaction with hepatocyte growth factor activator inhibitor type-1. J. Biol. Chem. 2010;285:33394–33403. doi: 10.1074/jbc.M110.115816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyake Y, Tsuzuki S, Yasumoto M, Fushiki T, Inouye K. Requirement of the activity of hepatocyte growth factor activator inhibitor type 1 for the extracellular appearance of a transmembrane serine protease matriptase in monkey kidney COS-1 cells. Cytotechnology. 2009;60:95–103. doi: 10.1007/s10616-009-9219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuzuki S, Murai N, Miyake Y, Inouye K, Hirayasu H, Iwanaga T, Fushiki T. Evidence for the occurrence of membrane-type serine protease 1/matriptase on the basolateral sides of enterocytes. Biochem. J. 2005;388:679–687. doi: 10.1042/BJ20041639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kojima K, Tsuzuki S, Fushiki T, Inouye K. Roles of functional and structural domains of hepatocyte growth factor activator inhibitor type 1 in the inhibition of matriptase. J. Biol. Chem. 2008;283:2478–2487. doi: 10.1074/jbc.M709073200. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Kojima K, Tsuzuki S, Fushiki T, Inouye K. The activity of a type II transmembrane serine protease, matriptase, is dependent solely on the catalytic domain. Biosci. Biotechnol. Biochem. 2009;73:454–456. doi: 10.1271/bbb.80713. [DOI] [PubMed] [Google Scholar]
- 26.Mochida S, Tsuzuki S, Yasumoto M, Inouye K, Fushiki T. Secreted expression of pseudozymogen forms of recombinant matriptase in Pichia pastoris. Enzyme Microb. Technol. 2009;45:288–294. [Google Scholar]
- 27.Bork P, Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J. Mol. Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 28.Thielens NM, Enrie K, Lacroix M, Jaquinod M, Hernandez JF, Esser AF, Arlaud GJ. The N-terminal CUB-epidermal growth factor module pair of human complement protease C1r binds Ca2+ with high affinity and mediates Ca2+-dependent interaction with C1s. J. Biol. Chem. 1999;274:9149–9159. doi: 10.1074/jbc.274.14.9149. [DOI] [PubMed] [Google Scholar]
- 29.Moestrup SK. The α2-macroglobulin receptor and epithelial glycoprotein-330: two giant receptors mediating endocytosis of multiple ligands. Biochem. Biophys. Acta. 1994;1197:197–213. doi: 10.1016/0304-4157(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 30.Friedrich R, Fuentes-Prior P, Ong E, Coombs G, Hunter M, Oehler R, Pierson D, Gonzalez R, Huber R, Bode W, Madison EL. Catalytic domain structures of MT-SP1/matriptase, a matrix-degrading transmembrane serine proteinase. J. Biol. Chem. 2002;277:2160–2168. doi: 10.1074/jbc.M109830200. [DOI] [PubMed] [Google Scholar]
- 31.Fass D, Blacklow S, Kim PS, Berger JM. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature. 1997;388:691–693. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- 32.Williams EB, Krishnaswamy S, Mann KG. Zymogen/enzyme discrimination using peptide chloromethyl ketones. J. Biol. Chem. 1989;264:7536–7545. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.