Abstract
Viperidae snakes containing various venomous proteins also have several anti-toxic proteins in their sera. However, the physiological function of serum protein has been elucidated incompletely. Small serum protein (SSP)-1 is a major component of the SSPs isolated from the serum of a Japanese viper, the habu snake (Trimeresurus flavoviridis). It exists in the blood as a binary complex with habu serum factor (HSF), a snake venom metalloproteinase inhibitor. Affinity chromatography of the venom on an SSP-1-immobilized column identified HV1, an apoptosis-inducing metalloproteinase, as the target protein of SSP-1. Biacore measurements revealed that SSP-1 was bound to HV1 with a dissociation constant of 8.2 × 10−8 M. However, SSP-1 did not inhibit the peptidase activity of HV1. Although HSF alone showed no inhibitory activity or binding affinity to HV1, the SSP-1–HSF binary complex bound to HV1 formed a ternary complex that non-competitively inhibited the peptidase activity of HV1 with a inhibition constant of 5.1 ± 1.3 × 10−9 M. The SSP-1–HSF complex also effectively suppressed the apoptosis of vascular endothelial cells and caspase 3 activation induced by HV1. Thus, SSP-1 is a unique protein that non-covalently attaches to HV1 and changes its susceptibility to HSF.
Keywords: apoptosis, proteinase inhibitor, small serum protein, snake serum, snake venom metalloproteinase
The venom of snakes belonging to the Viperidae family contains metalloproteinases, many of which cause hemorrhage (1). Although snakebites are serious problems for humans and other animals, snakes themselves exhibit a remarkable resistance to their own venom (2, 3). As naturally occurring factors that neutralize hemorrhagins, three anti-hemorrhagic proteins have been purified from the sera of venomous snakes: habu serum factor (HSF) from the serum of Trimeresurus flavoviridis (4), BJ46a from Bothrops jararaca (5) and mamushi serum factor from Gloydius blomhoffi brevicaudus (6). These compounds are acidic glycoproteins with no proteolytic activity. Based on their primary structures, they have been classified as members of the fetuin family that display a double-headed cystatin-like domain and an extra domain. HSF inhibits the protease activity of several snake venom metalloproteinases (SVMPs) (7). It is also resistant to heating and stable in solutions with extreme pH.
Small serum proteins (SSPs) are low-molecular-mass proteins isolated from T. flavoviridis serum (8). At present, five homologues—namely SSP-1 through SSP-5—have been isolated (9). Structural analysis has indicated that they belong to the prostatic secretory protein of 94 amino acids (PSP94) family, which is characterized by a low molecular mass of ∼10 kDa and 10 conserved cysteine residues (10, 11). Although SSP-1, SSP-2 and SSP-5 are composed of ∼90 amino acids, SSP-3 and SSP-4 have only 60, as they lack the 30 C-terminal residues.
All the SSPs exist in high-molecular-mass forms in serum (12), and because they do not self-associate in physiological buffers, they may be present in protein complexes. Similar to the SSPs in vipers, human PSP94 exists in complex with a specific protein (PSP94-binding protein) in the blood and with cysteine-rich secretory protein-3 (CRISP-3) in prostate fluid (13). In a search for SSP-binding proteins in T. flavoviridis serum, we isolated a novel protein called serotriflin that shows significant sequence similarity to triflin, a CRISP family protein in T. flavoviridis venom (14). Although serotriflin was isolated as a binding protein candidate for SSPs, it showed affinity only to SSP-2 and SSP-5 (12). Recently, we have reported that HSF is the carrier protein for all SSPs (15).
We know little about the physiological functions of SSPs. SSP-2 and SSP-5 bind triflin and serotriflin (12). Although SSP-1 and SSP-3 suppress the proteolytic activity of brevilysin H6 (16), an SVMP isolated from the venom of Chinese viper (G. blomhoffi brevicaudus), the inhibition is weak compared with that by HSF (8). As SSPs and brevilysin H6 are present in different animals, H6 cannot be a physiological target of SSP. Furthermore, we have found no other SVMPs that are sensitive to SSP-1 in the venom of T. flavoviridis.
In this study, we determined the target molecules of SSP-1 using affinity chromatography on an SSP-1-immobilized column. We found that HV1 in T. flavoviridis venom is the binding protein of SSP-1. HV1 is a homodimeric protein with a molecular mass of 110 kDa that induces apoptosis in vascular endothelial cells (VECs) (17). Although HV1 is a typical P-III class dimeric SVMP composed of metalloproteinase/disintegrin/cysteine-rich (MDC) domains (18), its biochemical properties have yet to be reported. We also examined the interaction of SSP-1 and HV1 and the effects of SSP-1 on the proteolytic and apoptosis-inducing activity of HV1.
Materials and Methods
Materials
Blood of habu snake T. flavoviridis from the Amami island was collected by decapitation. The serum was separated by centrifugation and stored at −20°C. The venom of T. flavoviridis was also collected, lyophilized and stored at −20°C. SSPs were purified as described earlier (8, 19). Bovine trypsin and protein substrates (bovine fibrinogen, vitronectin, collagen type IV and human fibronectin) were obtained from Sigma Chem. Co. (St. Louis). The peptide substrates were from Peptide Institute, Inc. (Osaka). SP-Sepharose, Sephacryl S-200 HR and S-300 HR, N-hydroxysuccimide-activated HiTrap, and Superdex 75 columns were purchased from GE Healthcare. EBM-2 medium was purchased from Sanko Junyaku Co. Ltd (Tokyo). All other chemicals were purchased from Wako Pure Chemical Industries Ltd (Osaka). Human umbilical endothelial cells were obtained from Lonza (Walkersville), and maintained on gelatin-coated plastic dishes in EBM-2 medium supplemented with EBM-2 SingleQuots (Lonza) containing 10% fetal bovine serum, several growth factors, hydrocortisone, ascorbic acid and heparin.
Quantification of proteins
The concentration of pure samples was determined using a V-530 spectrophotometer (Jasco), and the molar extinction coefficients at 280 nm were calculated for SSP-1 (9,105 M–1 cm–1), HSF (23,670 M–1 cm–1) and HV1 (36,965 M–1 cm–1) (20).
SDS–PAGE
SDS–PAGE was carried out on 10% gels using Laemmli’s method (21). Prestained XL-ladder broad kit (APRO Life Science Institute, Inc., Naruto) was used for a molecular weight marker. After running the gels under a constant current, they were stained with 0.1% Coomassie brilliant blue R-250 and destained with 10% acetic acid.
Isolation of SSP-1 binding protein
Affinity adsorbent was prepared by reacting an N-hydroxysuccimide-activated HiTrap column (1 ml) with purified SSP-1 (5.5 mg) following the manufacturer’s instructions. Crude venom (100 mg) dissolved in 20 mM phosphate buffer, 0.15 M NaCl at pH 7.4 was loaded onto the resulting SSP-1-HiTrap column and washed with the same buffer. Absorbed materials on the column were then eluted using 0.1 M glycine-HCl buffer, 0.5 M NaCl at pH 3.0, desalted by dialysis, and purified by a reverse-phase HPLC on a SepaxBio-C8 column (0.46 × 25 cm, Sepax Technologies Inc.) with a linear gradient of acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 1.0 ml/min. Elution was monitored at 220 nm.
Identification of SSP-1 binding protein
The N-terminal and internal amino acid sequences of SSP-1 binding protein were determined by Edman degradation with protein sequencer PPSQ 21 (Shimadzu). To analyse the internal sequences, the purified protein (400 μg) was dissolved in 25 μl of 4 M urea, diluted with 100 μl of 100 mM NH4HCO3 and digested at 37°C for 24 h with trypsin (E/S = 1 : 50). The resulting peptides were purified by reverse-phase HPLC using a YMC-Pack ODS AM-303 column (0.46 × 25 cm, YMC) with a linear gradient of acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 1.0 ml/min.
Purification of HV1
HV1 was purified from T. flavoviridis venom as described (17) with some modifications. Briefly, 1.26 g of crude venom dissolved in 5 ml of 50 mM Tris-HCl, 50 mM NaCl, 5 mM CaCl2 at pH 7.4 was loaded onto a Sephacryl S-300 HR column (5.0 × 90 cm) and then eluted with the same buffer, and 15 ml fractions were collected. The presence of HV1 was assayed by fluorescein thiocarbamoyl (FTC)-caseinolytic activity and by detecting a 110-kDa band on SDS–PAGE. Proteins in the second peak were desalted by dialysis and subjected to ion-exchange chromatography using a SP-Sepharose column (2.6 × 15 cm) equilibrated with 10 mM Tris-HCl, 1 mM CaCl2, 10 mM NaCl at pH 7.0. Elution was performed at 4°C with a linear gradient of NaCl from 10 to 400 mM in the same buffer, and 10 ml fractions were collected. Active fractions containing HV1 eluted at 120 mM NaCl were collected, desalted by dialysis and subjected to gel filtration on a Sephacryl S-200 HR column (2.6 × 92 cm) in 50 mM phosphate buffer, 150 mM NaCl at pH 7.0. Proteins recovered from the first peak were collected, concentrated by ultrafiltration using a ultrafiltration tube with a molecular weight cut-off of 30,000 (Amicon Ultra-15, Millipore) and applied to a column of Superdex 75 HiLoad (1.6 × 60 cm) equilibrated with 20 mM Tris-HCl, 200 mM NaCl at pH 8.0. Active fractions were pooled, concentrated and stored at –20°C.
Surface plasmon resonance analysis of binding kinetics
The binding studies were performed using Biacore 3000 instrument (Biacore BA) equipped with a CM5 sensor chip. SSP-1 in 10 mM sodium acetate buffer (pH 4.0) was covalently immobilized onto the chip at a density of ∼500 resonance units (RU) using standard primary amine coupling procedure. HSF was immobilized similarly. Albumin was used as a negative control protein. The analyte dissolved in running buffer (10 mM HEPES buffer, 0.15 M NaCl, 3 mM EDTA, 0.005% surfactant P20 at pH 7.4) was injected for 120 s and the dissociation from the surface was monitored for 150 s.
Kinetic measurements of the interaction between SSP-1 and HV1 were performed using SSP-1 chip. The analyte (HV1) diluted to various concentrations in running buffer was injected for 90 s during the association phase at a constant flow rate of 20 µl/min at 25°C. The dissociation was subsequently followed for 180 s at the same flow rate. The surface of the sensor chip was regenerated using 10 mM Gly–HCl buffer (pH 2.0) after the binding of analyte. The sensorgrams were corrected by subtraction of the signal from the negative control surface and used to calculate the rate and affinity constants by using BiaEvaluation 4.1 (Biacore AB) and Origin 5.0 (Microcal).
Measurement of proteolytic activity
Proteolytic activity was measured in 5 mM CaCl2-50 mM Tris-HCl (pH 8.5) using FTC-casein as the substrate (22). The increase in fluorescence was analysed on a fluorescence spectrophotometer FP-750 (Jasco) at 520 nm with excitation at 490 nm. Fibrinogen, vitronectin, collagen type IV and fibronectin were also used as the substrates and the reaction was followed by SDS–PAGE using a 5–20% gradient gel (ATTO). Human fibrinogen (100 μg), vitronectin (50 μg), collagen type IV (100 μg) or fibronectin (50 μg) was incubated with purified HV1 (0.2 μg) in 50 μl of 20 mM Tris-HCl (pH 8.0) containing 0.15 M NaCl and 1 mM CaCl2 at 37°C for 10 min to 22 h. To terminate the reaction, 1 mM EDTA (5 μl) was added to a 5 μl aliquot, and the mixture was lyophilized.
Measurement of peptidase activity
Substrates were prepared as 2 mM stock solutions in dimethyl sulfoxide. A typical assay was carried out by incubating 90 μl of various concentrations of a substrate with 10 μl of HV1 (final 10 nM) in 50 mM Tris-HCl, 0.1 M NaCl, 0.05% Brij at pH 8.0 at 37°C for 10 min. The reaction was stopped by the addition of 1,900 μl of 0.1 M acetate (pH 4), and the fluorescence of the mixture was measured at 393 nm with excitation at 328 nm. The amount of substrate hydrolysis was calculated based on the fluorescence values of the Mca-Pro-Leu-Gly standard solution after subtraction of the reaction blank value (stopping solution added before the enzyme). Kinetic parameters (kcat and Km) were determined over a substrate concentration range of 3–20 μM and calculated by double-reciprocal plots. To determine the site of hydrolysis in substrate III (Table II), two peptides produced by the action of HV1 were purified by a reversed-phase HPLC on YMC-Pack ODS-A column (0.46 × 25 cm, YMC). Their N-terminal sequences were determined as described earlier.
Table II.
Hydrolysis of several fluorogenic peptide substrates by HV1.
| Substrate | Fluorogenic peptidea | Hydrolysis (μM/min)b |
|---|---|---|
| I | Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2 | 0.0071 |
| II | Mca-Arg-Pro-Lys-Pro-Tyr-Ala-Nva-Trp-Met-Lys(Dnp)-NH2 | 0.052 |
| III | Mca-Arg-Pro-Lys-Pro-Val-Glu-Nva-Trp-Arg-Lys(Dnp)-NH2 | 0.733 |
| IV | MocAc-Asp-Glu-Val-Asp-Ala-Pro-Lys(Dnp)-NH2 | 0.0016 |
| V | Mca-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(Dnp)-D-Arg-NH2 | 0.040 |
| VI | MocAc-Ser-Glu-Val-Asn-Leu-Asp-Ala-Glu-Phe-Arg-Lys(Dnp)-Arg-Arg-NH2 | 0.000 |
| VII | Mca-Ala-Pro-Ala-Lys-Phe-Phe-Arg-Leu-Lys(Dnp)-NH2 | 0.0021 |
| VIII | Mca-Gly-Ser-Pro-Ala-Phe-Leu-Ala-Lys(Dnp)-D-Arg-NH2 | 0.026 |
| IX | Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2 | 0.024 |
aDnp, 2,4-dinitrophenyl; Dpa, N-3-(2,4-dinitrophenyl)-l-2,3-diaminopropionyl; Nva, norvaline.
bEnzyme concentration, 50 nM. Measured at 37°C in 0.1 M NaCl-0.05% Brij 50 mM Tris-HCl (pH 8.5).
Apparent inhibition constant (Ki) was determined by measuring the initial rate of hydrolysis of substrate at pH 8.0 in the presence or absence of inhibitor. Nonlinear regression method was applied to analyse inhibition data according to the Morrison’s equation (23). For the non-competitive mechanism of inhibition, the true inhibition constants are equal to the apparent Ki (24).
Analysis of VEC Apoptosis
VEC (1 × 105) was cultured in 24-well culture plates at 37°C in 5% CO2 and 95% air for 24 h. The medium was replaced with EBM-2 after cells had been washed once with phosphate-buffered saline (PBS). Cells were further treated with or without HV1, SSP-1 and HSF for 24 h. After microscopic examination, the culture cells were trypsinized and washed twice with PBS. Cell viability was determined by the MTT method (25). Caspase-3 activity was also determined after the cells were incubated for 24 h. Cells recovered after trypsinization were washed twice with PBS, and solubilized in 1% Triton X-100 and 1% NP-40 at 0°C for 30 min. After centrifugation at 4°C and 15,000 rpm for 5 min, the supernatant was mixed with Ac-DEVD-MCA (final 1 mM) in 100 mM HEPES, 5 mM DTT, 10% sucrose (pH 7.4) and incubated at 37°C for 1 h. The reaction was stopped by adding 1 M acetate buffer (pH 4.2) and the fluorescence was measured at 440 nm with excitation at 360 nm. To observe nuclear fragmentation, cells were fixed in 70% ethanol overnight at 4°C, centrifuged, washed twice with PBS and stained with 0.05 mg/ml propidium iodide (Sigma) for 5 min. One drop of stained cells was placed on a glass slide and nuclei were examined by fluorescence microscopy.
Results
Identification of SSP-1-binding protein
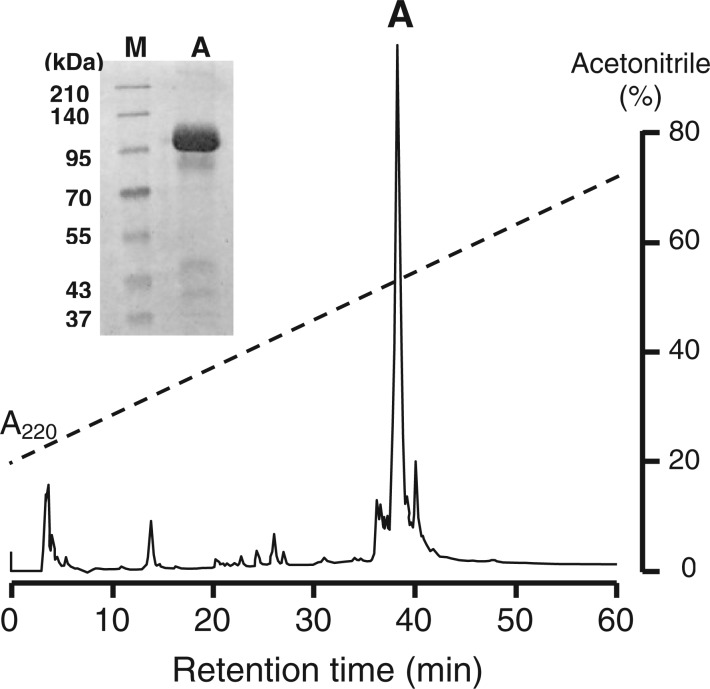
To determine the target molecules of SSP-1 in the venom of T. flavoviridis, we used affinity chromatography with an SSP-1-immobilized column. A 110-kDa protein remained specifically bound on the column when crude T. flavoviridis venom was applied (data not shown). The absorbed material was further purified using reverse-phase HPLC (peak A). The inset in Fig. 1 shows the SDS–PAGE analysis of peak A in Fig. 1. The exact mass of the 110-kDa protein determined by MALDI-TOF (Matrix Assisted Laser Desorption Ionization-Time of Flight) mass spectrometry was 102,259.3 Da (data not shown). As the N-terminus of this protein seemed to be blocked, the internal sequences were determined after tryptic digestion. We could assign 12 peptides to the amino acid sequence of HV1 (17) as shown in Table I. HV1 is a dimeric metalloproteinase with apoptosis-inducing activity in VECs.
Fig. 1.
Reverse phase HPLC of SSP-1 binding protein purified by affinity chromatography of T. flavoviridis venom. The absorbed protein fraction on a SSP-1-HiTrap column was applied to a SepaxBio-C8 column (0.46 × 25 cm) and eluted with a linear gradient of acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 1.0 ml/min. Elution was monitored at 220 nm. Peak A was recovered. Inset, the analysis of peak A by SDS–PAGE (10% acrylamide gel). M, molecular weight markers.
Table I.
Assignment of peptide fragments to the primary structure of a vascular apoptosis-inducing protein, HV1, from T. flavoviridis.
| Tryptic fragmenta | Sequence | Residue in Hv1b |
|---|---|---|
| T2 | YVK | 204–206 |
| T3 | YGR | 219–221 |
| T4 | LR | 440–441 |
| T5 | YLNAK | 198–202 |
| T8a | IAXEPQNVKc | 550–558 |
| T8b | QGNYYGDYXRKc | 534–544 |
| T9 | ESVLLK | 279–284 |
| T10 | XADGMAXNSNGQXVDVNRc | 595–612 |
| T11 | IIVQSVPDVTLK | 261–272 |
| T13 | LFATWR | 273–278 |
| T16a | KNPXNIIYSPNDEDKGMVLPGTKc | 572–594 |
| T16b | TDVVSPPVXGNYFVEVGEDXDXGSPATXRc | 401–429 |
Purification of HV1
To confirm that HV1 binds to SSP-1, we isolated HV1 from the venom of T. flavoviridis via gel filtration on a Sephacryl S-300 HR column, ion exchange chromatography on a SP-Sepharose column and two successive gel filtrations on Sephacryl S-200 HR and Superdex 75 columns. A pure protein (0.7 mg) was obtained from 1.26 g crude venom. It gave a single band at 110 kDa on SDS–PAGE, but no N-terminal sequence was obtained with direct sequencing. Tryptic digestion of the purified sample yielded several peptide fragments that corresponded to those of HV1 (see Table I).
Biding of SSP-1 to HV1
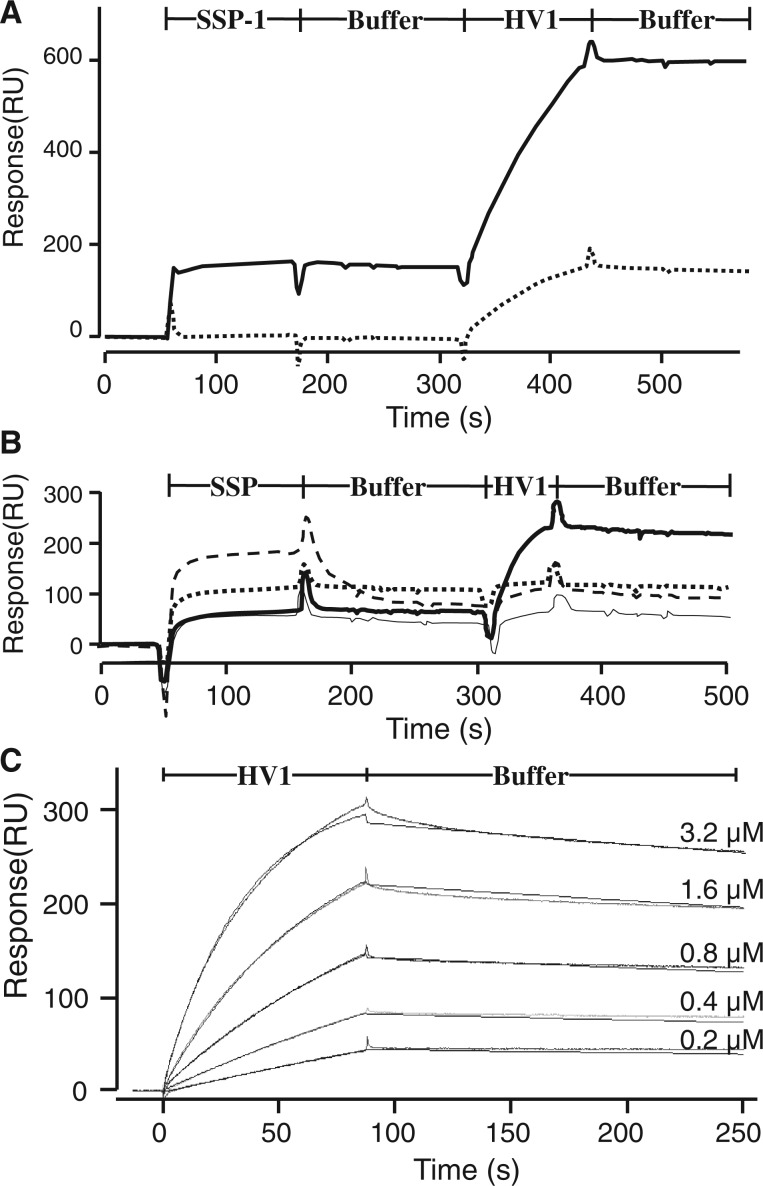
The interaction of SSP-1 and HV1 was examined with surface plasmon resonance. SSP-1 was injected for 120 s into an HSF-immobilized or SSP-1-immobilized chip and the dissociation from the surface was monitored for 150 s (Fig. 2A). The sensorgrams showed the predicted rapid binding of SSP-1 to HSF and apparent slow dissociation rate (15). Injection of the second analyte (HV1) resulted in a resonance signal of 200 response units, which indicated the direct binding of HV1 to SSP-1. When HV1 was injected into an SSP-1-bound chip with HSF as a capturing molecule, a much higher response was observed (full line in Fig. 2A). This result clearly indicated the formation of a ternary complex composed of HSF, SSP-1 and HV1. Moreover, HV1 did not bind to HSF in the absence of SSP-1 (data not shown). The binding of SSPs other than SSP-1 was examined similarly because all the SSPs have a high affinity to HSF. SSP-2, SSP-3 and SSP-5 did not interact with HV1, but SSP-4 showed weak binding (Fig. 2B).
Fig. 2.
Surface plasmon resonance analysis of SSP binding to HV1. (A) Binding property of SSP-1. The first analyte (SSP-1, 30.8 µM) was injected for 120 s to HSF-immoilized chip (full line) or SSP-1-immoilized chip (dotted line) and the dissociation from the surface was monitored for 150 s. Then, the second analyte (HV1, 3.8 µM) was injected similarly. (B) Binding analysis of other SSPs to HV1. SSP-2 (11.9 µM, dotted line), SSP-3 (17.7 µM, narrow full line), SSP-4 (14.0 µM, bold full line) or SSP-5 (34.5 µM, broken line) was injected similarly to HSF-immoilized chip and the dissociation from the surface was monitored. Then, the second analyte (HV1, 3.8 µM) was injected. (C) Direct binding of SSP-1 to HV1. Serially diluted HV1 (3.2–0.2 μM) was injected at a flow rate of 20 μl/min through flow cells with the immobilized SSP-1.
Serially diluted HV1 (3.2–0.2 μM) was injected into SSP-1-coated sensor chips and the binding was measured (Fig. 2C). Global evaluation using the 1 : 1 Langmuir binding model yielded kinetic binding parameters. The rate constants for association, dissociation and Kd value were 8.7 × 103 M−1 s−1, 7.2 × 10−4 s−1 and 8.2 × 10−8 M, respectively.
Characterization of HV1
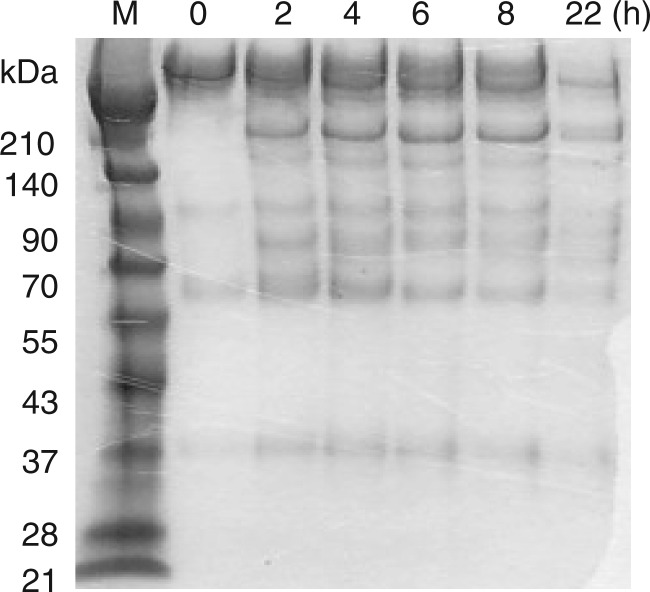
The purified HV1 degraded casein slowly. As many SVMPs hydrolyse fibrinogen, the fibrinogenolytic activity of HV1 was investigated by reacting the enzyme with human fibrinogen at 37°C. When the enzyme reaction products were analysed using SDS–PAGE, HV1 did not degrade the protein (data not shown). The proteolytic activity of HV1 was also examined with some extracellular matrix proteins. Only fibronectin was slowly degraded by HV1 (Fig. 3). The enzyme could not degrade type IV collagen or vitronectin.
Fig. 3.
Degradation of fibronectin by HV1. Fibronectin (50 μg) was incubated with HV1 (0.2 μg) in 50 μl of 20 mM Tris-HCl (pH 8.0) containing 0.15 M NaCl and 1 mM CaCl2 at 37°C for 10 min and 22 h. The reaction was stopped by the addition of 1 mM EDTA (5 μl) to a 5 μl aliquot. The mixture was lyophilised and analysed by SDS–PAGE using a 5–20% gradient gel.
Some fluorogenic substrates for matrix metalloproteinases (MMPs) were next examined to measure the peptidase activity of HV1. Table II shows peptide substrates with (7-methoxycoumarin-4-yl)acetyl (Mca) as a fluorophore group and dinitrophenyl (Dnp) as a quenching moiety (26, 27). Among nine peptides, Mca-Arg-Pro-Lys-Pro-Val-Glu-Nva-Trp-Arg-Lys(Dnp)-NH2 (substrate III), known as a substrate for MMP-3 (stromelysin 1) (27), was one of the best substrates for HV1. A reverse-phase HPLC of a 24-h digest followed by sequence analysis of the resulting fragments showed that the enzyme cleaved the Pro-Val bond of substrate III. Kinetic parameters for the hydrolysis of substrate III were determined as follows: Km = 8.2 ± 2.1 μM; kcat = 1.7 ± 0.9 s−1; and kcat/Km = 203,000 M−1 s−1. The kcat/Km value was almost equal to that of MMP-3 (27). Other peptides listed in Table II were poor substrates for HV1.
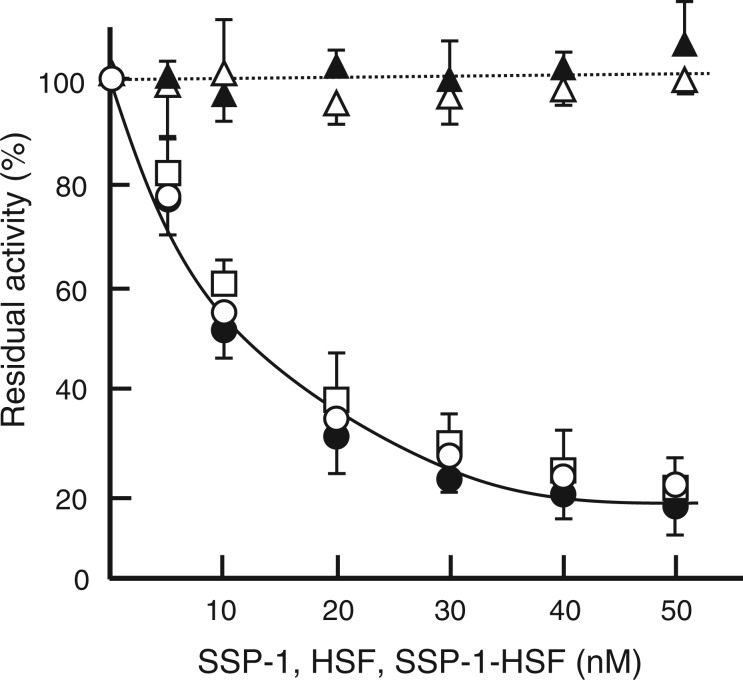
Inhibition of the peptidase activity of HV1 by the SSP-1–HSF complex
Using substrate III, we investigated the effect of SSP-1 on the peptidase activity of HV1 (10 nM). Despite its high affinity to HV1, SSP-1 showed no inhibition even at 50 nM (Fig. 4, a dotted line). HSF, an effective inhibitor of several SVMPs, was not inhibitory to the enzyme either. Conversely, the SSP-1–HSF complex showed a dose-dependent suppression of HV1 activity. The influence of substrate concentration on apparent Ki was measured to determine the mode of inhibition at substrate concentrations ranging from 5 to 20 µM. The inhibition was independent of substrate concentration, and the apparent Ki values were 6.2, 5.3 and 7.0 nM at 5, 10 and 20 µM, respectively (Fig. 4). This indicates that HV1 inhibition by the complex is non-competitive (24). For the non-competitive fit to the data shown by a solid line in Fig. 4, the average Ki values at three substrate concentrations with their standard error were 5.1 ± 1.3 × 10−9 M.
Fig. 4.
Effect of SSP-1 and HSF on the peptidase activity of HV1. Peptidase activity of HV1 was measured in the presence of SSP-1 (open triangles) or HSF (closed triangles) with 20 μM substrate at pH 8.0 and 37°C for 10 min. The final concentration of HV1 was 10 nM. HV1 activity was measured similarly in the presence of SSP-1–HSF complex at various substrate concentrations; 5 μM (open circle), 10 μM (closed circle) and 20 μM (open square).
Inhibition of the apoptosis-inducing activity of HV1 by the SSP-1–HSF complex
In accordance with the results of Masuda et al. (17), HV1 induced apoptosis in VECs (Fig. 5A and B). After a 36-h exposure of VECs to HV1, cell viability decreased in a dose-dependent manner with a half-lethal dose of ∼0.3 μg/ml which is almost identical to the value (0.2 μg/ml) in the reference (17). When VECs were treated with HV1 (18 nM) for 36 h, typical apoptotic morphological changes—namely cell shrinkage, bleb formation on cell surfaces and the generation of apoptotic bodies—were observed under a light microscope (Fig. 5E). In contrast, when HV1 was mixed with various amounts of SSP-1–HSF complex before addition to cell cultures, dose-dependent suppression of apoptosis was detected (Fig. 6A). The addition of 20 nM of the complex greatly enhanced cell viability (Fig. 5C), and no morphological changes were observed (Fig. 5F), similar to the response in control cells (Fig. 5D). Notably, SSP-1, HSF and the combination of the two had no effect on the growth of VECs in the absence of HV1 (data not shown). Moreover, SSP-1 and HSF alone did not suppress the apoptosis-inducing activity of HV1.
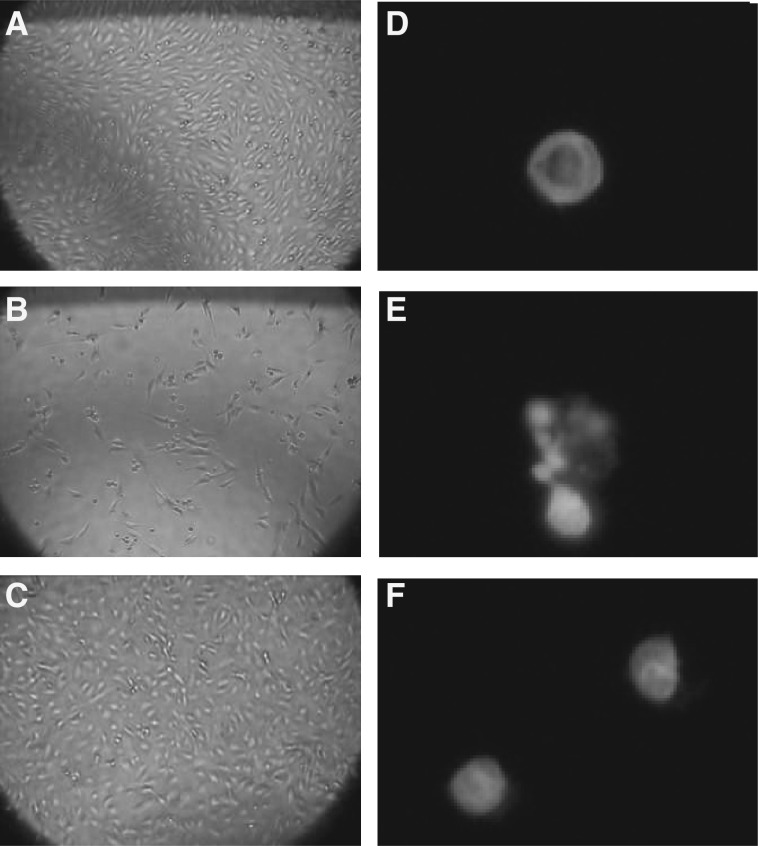
Fig. 5.
Effects of SSP-1–HSF complex on morphological change and nuclear-fragmentation induced by HV1. The complete medium was replaced by medium that had been supplemented with 1.5 μg/ml HV1. (A, D) Control VEC; (B, E) cells incubated with HV1 (20 nM); (C, F) cells incubated with HV1 and SSP-1–HSF (20 nM each). Left panels (A–C): after treatment for 24 h, cells were photographed under a light microscope. Right panels (D–F): after treatment for 34 h, cells were fixed with glutaraldehyde and stained with propidium iodide. Nuclear fragmentation was examined using fluorescence microscopy.
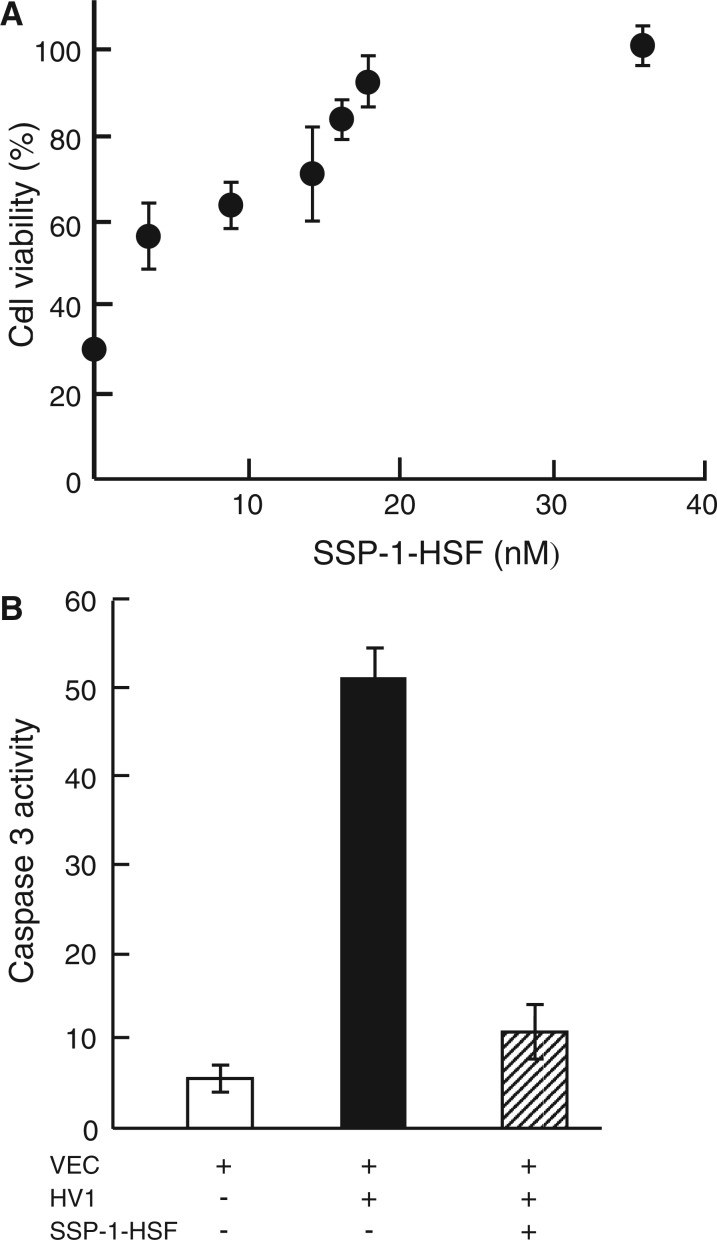
Fig. 6.
Effects of SSP-1–HSF complex on viability and caspase-3 activation induced by HV1. (A) After VEC was treated with SSP-1–HSF complex at the indicated concentrations in the presence of HV1 (18 nM) for 24 h, viabilities were determined by MTT assay. Results are shown by relative values (%). (B) Caspase 3 activity. VEC was treated with or without 50 nM HV1 or HV1 plus SSP-1–HSF complex (50 nM each) for 24 h. Caspase 3 activities were assessed by cleavage of Ac-DEVDl-MCA as described in ‘Materials and Methods’ section.
To assess the apoptotic events evoked by HV1 (50 nM) treatment, we examined the activities of caspase-3 in VECs. After 24 h of incubation, the caspase-3 activity in HV1-treated cells was increased ∼10-fold compared with that in control cells (see open and closed bars in Fig. 6B). This caspase-3 activation was greatly reduced by the addition of SSP-1–HSF complex (50 nM; see a hatched bar in Fig. 6B).
Discussion
Hemorrhagic metalloproteinases and toxic phospholipase A2 are present in the venom of snakes belonging to the Crotalidae and Viperidae families (28, 29). To prepare for accidental self-poisoning, some snakes have endogenous serum proteins that neutralize the hemorrhagic factors or toxins. These proteins can be classified as inhibitors of myotoxic/neurotoxic toxins bearing a phospholipase A2 structure (30) and inhibitors of SVMPs (6, 7). The examples of the latter are T. flavoviridis HSF and B. jararaca BJ46a. We have added SSPs as a third class of self-defense proteins (8). As SSPs are small proteins with a molecular mass of 10 kDa or less, they exist as a protein complex of ∼50–70 kDa in blood. The binding protein for SSP-2 and SSP-5 was first identified as serotriflin, a CRISP-family glycoprotein with 65% identity to triflin (12). Recently, we have showed that HSF is another SSP-binding protein (15). All the SSPs bind tightly to HSF, indicating that they are present in the complex with HSF. Interestingly, HSF has a dual role in habu serum—as a defense protein (namely, anti-hemorrhagin) and as an SSP carrier.
A putative role for SSP-2 was suggested because it has high affinity for triflin, a CRISP with L-type calcium channel-blocking activity excreted by the venom gland of T. flavoviridis (31). SSP-2 may thus work defensively against self-poisoning, suggesting a possible role for SSP-2 as an antidote to triflin. SSP-5 also binds to triflin. In this study, we have proved that SSP-1 has high affinity to HV1, an apoptosis-inducing metalloproteinase in the venom of T. flavoviridis (17).
Although HV1 is a P-III class dimeric SVMP, its proteolytic properties remain to be reported because it has been isolated only through monitoring the viability of cultured VECs. To examine the interaction between HV1 and SSP-1 as well as the biochemical nature of the enzyme, we purified native HV1 from the crude venom of T. flavoviridis using conventional methods. To ascertain the identity of our preparation to the desired protein, we carried out N-terminal sequencing. However, the purified 110-kDa protein could not be directly sequenced via Edman degradation, presumably because it has a blocked N-terminal residue. The blocked residue may be a pyrrolidone carboxylate group because most mature SVMPs have a N-terminal glutamate residue that can attack itself to form a cyclic pyroglutamate group. Thus, the N-terminal sequence of HV1 might start at the fourth residue (Gln192) of the reported sequence T189PAQQKYL (17).
Although we used FTC caseinolytic activity to detect proteinases during the purification of HV1, the purified enzyme degraded casein slowly. Therefore, we examined other proteins as substrates. Among the extracellular matrix proteins tested, only fibronectin was degraded. Thus, HV1 seemed to have a low proteolytic activity. The true target molecule of VEC apoptosis remains to be elucidated.
Fluorescence-quenched peptide substrates are widely used for the quantification of enzymatic activity of various endoproteinases, particularly for the measurement of MMPs (32). The fluorescence-quenched MMP substrate Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2 (33) has been used extensively for monitoring MMP activity (34). Because of its high kcat/Km value, this peptide is extraordinarily useful for such measurements, but it was a poor substrate for HV1 (Table II). After screening for a better substrate, we found that Mca-Arg-Pro-Lys-Pro-Val-Glu-Nva-Trp-Arg-Lys(Dnp)-NH2 (substrate III), an excellent substrate for MMP-3 (kcat/Km = 65,700 M−1 s−1) (27), was also the best substrate for HV1. The kcat/Km value was 203,000 M−1 s−1. HV1 cleaved substrate III at the Pro-Val bond. Replacements of Pro-Val to Pro-Tyr (substrate II), Pro-Ile (substrate V) and Pro-Ala (substrate VIII) greatly reduced susceptibility to the enzyme, indicating a high dependence on P1' site (Table II).
Using fluorescently labelled and quenched substrate III, we investigated the effect of SSP-1 and HSF on the peptidase activity of HV1. HSF, a SVMP inhibitor, had no effect on the activity, which is in accord with its inability to interact with HV1. Interestingly, SSP-1 did not suppress the activity either despite the tight binding potency to HV1. Conversely, HSF and SSP-1 inhibited the peptidase activity of HV1 (Fig. 4). SSP-1 can bind HSF to form a tight binary complex (15), which in turn interacts to form a ternary complex as shown in Fig. 2A. These results imply that SSP-1 has a dual binding region—one for HSF and another for HV1—that do not overlap. Moreover, the binding affinity of the SSP-1–HSF complex to HV1 was 5.1 ± 1.3 × 10−9 M, which was ∼10 times higher than that of SSP-1 alone (8.2 × 10−8 M). This result indicates that the additional interaction between HV1 and HSF may be induced by the biding of SSP-1 to HV1 and contribute to the inhibition by the metalloproteinase inhibitor HSF. Thus, SSP-1 is a unique modulating protein that renders HV1 susceptible to HSF.
Several P-III class SVMPs with apoptosis-inducing activities have been reported (35–37). Their activity is abolished by pretreatment with EDTA because they are zinc-dependent proteinases. The apoptosis-inducing activity of HV1 also depends on proteolytic activity, and EDTA can suppress this activity as well (17). In fact, the SSP-1–HSF complex suppressed the apoptosis induced by HV1 and its proteolytic activity. Addition of the complex to an HV1-containing culture of VECs increased the viability of cells dose dependently and suppressed morphological changes, nuclear fragmentation, bleb formation and elevated caspase-3 activity—all of which are the characteristics of the apoptosis elicited by HV1.
A disintegrin and metalloproteinase (ADAM) family proteins belong to a class of mammalian-membrane-bound glycoproteins with functions implicated in cell–cell and cell–matrix adhesion and signalling (38, 39). ADAMs have two additional domains at the C-terminus of the complete MDC structure. They are also related to the ADAM with thrombospondin type-1 motif (ADAMTS) family of proteinases. Phylogenetically, SVMPs are closely related to these families of proteins. Recent crystallographic studies of P-III SVMPs and ADAM/ADAMTS proteins have shed new light on their structure–function properties (40). These proteins have a common C-shaped MDC domain. The cysteine-rich domain at the distal end points towards the catalytic site of the metalloproteinase domain. The structures of the peripheral loops in the cysteine-rich differ markedly among these proteins, providing extended surface areas, and this region has been designated as the hypervariable region (HVR) (41). Each P-III SVMP has a distinct HVR sequence, which results in distinct surface features. They might, therefore, function in specific protein–protein interactions, explaining the diversity of biological activities characteristic of P-III SVMPs. The HVR may constitute an exosite that captures target molecules directly or through an ‘associated’ protein (40, 41).
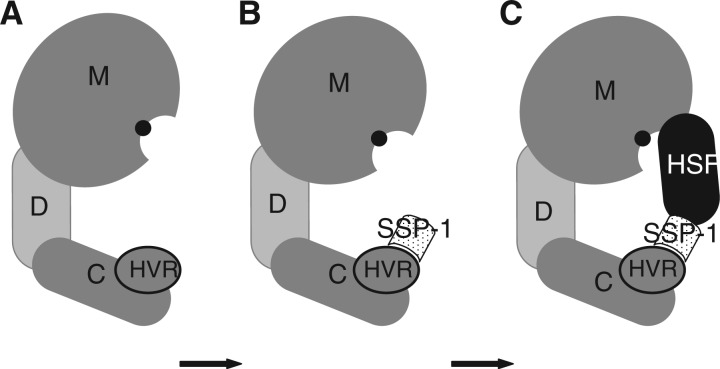
As shown in Fig. 4, the independence of inhibition from substrate concentration suggests that the mode of inhibition of the SSP-1–HSF complex is non-competitive and that SSP-1 binds to a non-catalytic site of the enzyme. As mentioned earlier, HVR (residues 563–583 in HV1) seems to be an exosite for interaction with SSP-1. Figure 7 shows a schematic model for HV1 inhibition by the complex. The binding of SSP-1 to the HVR of the enzyme as an ‘associated’ protein (42) elicits a conformational change in a cryptic site for HSF binding. HSF, which is brought by SSP-1, may attack the enzyme and convert its conformation to a non-productive form. The HSF-binding site must reside in the metalloproteinase domain because HSF inhibits P-I SVMPs and P-III class enzymes. Regardless, HSF gains access to the vicinity of the active site via SSP-1 and behaves like a substrate, resulting in the inhibition.
Fig. 7.
Schematic model of HV1 inhibition by SSP-1–HSF complex. (A) Native HV1; (B) fully active intermediate; (C) non-productive form of HV1. M, metalloproteinase domain; D, disintegrin-like domain; C, cysteine-rich domain. A Zn2+ ion at the active site is shown by a black circle. Note that SSP-1 plays a role of an ‘associated’ protein (42), which binds to HVR in C domain and assists the binding of substrate/inhibitor.
Brevilysin H6 is an SVMP from Gloydius halys brevicaudus venom (16). Initially, SSP-1 was discovered as a weak inhibitor of this enzyme (8). Recently, we have found that the inhibition of H6 by SSP-1 or HSF is non-competitive (unpublished data). HSF also inhibits many SVMPs irrespective of snake source (10), although HV1 and some SVMPs are insensitive to HSF. We speculate that these inhibitors bind to their target enzymes through the HVR. SSP-1 is a low-molecular weight protein that may target the exosites of SVMPs. It may become a promising scaffold for the design of specific inhibitors of SVMPs and ADAM/ADAMTS proteins.
Acknowledgements
The authors wish to express their thanks to the stuffs in the Radioisotope Center of Fukuoka University for the assistance in protein sequencing analysis. This work was supported in part by MEXT (Ministry of Education, Culture, Sports, Science and Technology)-Supported, and the Grant-in-Aid for Young Scientists (B) 22770134 of Japan.
Conflict of interest
None declared.
Glossary
Abbreviations
- ADAM
a disintegrin and metalloproteinase
- ADAMTS
ADAM with thrombospondin type-1 motif
- CRISP-3
cysteine-rich secretory protein-3
- Dnp
dinitrophenyl
- HSF
habu serum factor
- HVR
hypervariable region
- Ki
inhibition constant
- Mca
(7-methoxycoumarin-4-yl)acetyl
- MDC
metalloproteinase/disintegrin/cysteine-rich
- MMP
matrix metalloproteinase
- PSP94
prostatic secretory protein of 94 amino acids
- SSP
small serum protein
- SVMP
snake venom metalloproteinase
- VEC
vascular endothelial cell
References
- 1.Bjarnason JB, Fox JW. Hemorrhagic metalloproteinases from snake venoms. Pharmacol. Ther. 1994;62:325–372. doi: 10.1016/0163-7258(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 2.Pérez JC, Sánchez EE. Natural protease inhibitors to hemorrhagins in snake venoms and their potential use in medicine. Toxicon. 1999;37:703–728. doi: 10.1016/s0041-0101(98)00205-0. [DOI] [PubMed] [Google Scholar]
- 3.Perales J, Neves-Ferreira AG, Valente RH, Domont GB. Natural inhibitors of snake venom hemorrhagic metalloproteinases. Toxicon. 2005;45:1013–1020. doi: 10.1016/j.toxicon.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 4.Yamakawa Y, Omori-Satoh T. Primary structure of the antihemorrhagic factor in serum of the Japanese Habu: a snake venom metalloproteinase inhibitor with a double-headed cystatin domain. J. Biochem. 1992;112:583–589. doi: 10.1093/oxfordjournals.jbchem.a123944. [DOI] [PubMed] [Google Scholar]
- 5.Valente RH, Dragulev B, Perales J, Fox JW, Domont GB. BJ46a, a snake venom metalloproteinase inhibitor. Isolation, characterization, cloning and insights into its mechanism of action. Eur. J. Biochem. 2001;268:3042–3052. doi: 10.1046/j.1432-1327.2001.02199.x. [DOI] [PubMed] [Google Scholar]
- 6.Aoki N, Tsutsumi K, Deshimaru M, Terada S. Properties and cDNA cloning of antihemorrhagic factors in sera of Chinese and Japanese mamushi (Gloydius blomhoffi) Toxicon. 2008;51:251–261. doi: 10.1016/j.toxicon.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Deshimaru M, Tanaka C, Fujino K, Aoki N, Terada S, Hattori S, Ohno M. Properties and cDNA cloning of an antihemorrhagic factor (HSF) purified from the serum of Trimeresurus flavoviridis. Toxicon. 2005;46:937–945. doi: 10.1016/j.toxicon.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Aoki N, Sakiyama A, Deshimaru M, Terada S. Identification of novel serum proteins in a Japanese viper: homologs of mammalian PSP94. Biochem. Biophys. Res. Commun. 2007;359:330–334. doi: 10.1016/j.bbrc.2007.05.091. [DOI] [PubMed] [Google Scholar]
- 9.Aoki N, Matsuo H, Deshimaru M, Terada S. Accelerated evolution of small serum proteins (SSPs)—the PSP94 family proteins in a Japanese viper. Gene. 2008;426:7–14. doi: 10.1016/j.gene.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 10.Seidah NG, Arbatti NJ, Rochemont J, Sheth AR, Chretien M. Complete amino acid sequence of human seminal plasma beta-inhibin. Prediction of post Gln-Arg cleavage as a maturation site. FEBS Lett. 1984;175:349–355. doi: 10.1016/0014-5793(84)80766-8. [DOI] [PubMed] [Google Scholar]
- 11.Mäkinen M, Valtonen-Andre C, Lundwall A. New world, but not Old World, monkeys carry several genes encoding β-microseminoprotein. Eur. J. Biochem. 1999;264:407–414. doi: 10.1046/j.1432-1327.1999.00614.x. [DOI] [PubMed] [Google Scholar]
- 12.Aoki N, Sakiyama A, Kuroki K, Maenaka K, Kohda D, Deshimaru M, Terada S. Serotriflin, a CRISP family protein with binding affinity for small serum protein-2 in snake serum. Biochim. Biophys. Acta. 2008;1784:621–628. doi: 10.1016/j.bbapap.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Udby L, Lundwall A, Johnsen AH, Fernlund P, Valtonen-Andre C, Blom AM, Lilja H, Borregaard N, Kjeldsen L, Bjartell A. β-Microseminoprotein binds CRISP-3 in human seminal plasma. Biochem. Biophys. Res. Commun. 2005;333:555–561. doi: 10.1016/j.bbrc.2005.05.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki Y, Koike H, Sugiyama Y, Motoyoshi K, Wada T, Hishinuma S, Mita M, Morita T. Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur. J. Biochem. 2002;269:2708–2715. doi: 10.1046/j.1432-1033.2002.02940.x. [DOI] [PubMed] [Google Scholar]
- 15.Shioi N, Narazaki M, Terada S. Novel function of antihemorrhagic factor HSF as an SSP-binding protein in Habu (Trimeresurus flavoviridis) serum. Fukuoka Univ. Sci. Rep. 2011;41:177–184. [Google Scholar]
- 16.Fujimura S, Oshikawa K, Terada S, Kimoto E. Primary structure and autoproteolysis of brevilysin H6 from the venom of Gloydius halys brevicaudus. J. Biochem. 2000;128:167–173. doi: 10.1093/oxfordjournals.jbchem.a022737. [DOI] [PubMed] [Google Scholar]
- 17.Masuda S, Hayashi H, Atoda H, Morita T, Araki S. Purification, cDNA cloning and characterization of the vascular apoptosis-inducing protein, HV1, from Trimeresurus flavoviridis. Eur. J. Biochem. 2001;268:3339–3345. doi: 10.1046/j.1432-1327.2001.02246.x. [DOI] [PubMed] [Google Scholar]
- 18.Fox JW, Serrano SM. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008;275:3016–3030. doi: 10.1111/j.1742-4658.2008.06466.x. [DOI] [PubMed] [Google Scholar]
- 19.Aoki N, Matsuo H, Deshimaru M, Terada S. Accelerated evolution of small serum proteins (SSPs)―the PSP94 family proteins in a Japanese viper. Gene. 2008;426:7–14. doi: 10.1016/j.gene.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 20.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Twining SS. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal. Biochem. 1984;143:30–34. doi: 10.1016/0003-2697(84)90553-0. [DOI] [PubMed] [Google Scholar]
- 23.Morrison JF. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim. Biophys. Acta. 1969;185:269–286. doi: 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]
- 24.Cheng YC, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 25.Mosmann T. Rapid colorimetric assay for cellular growth and cell survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 26.Knight CG, Willenbrock F, Murphy G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- 27.Nagase H, Fields CG, Fields GB. Design and characterization of a fluorogenic substrate selectively hydrolyzed by stromelysin 1 (matrix metalloproteinase-3) J. Biol. Chem. 1994;269:20952–20957. [PubMed] [Google Scholar]
- 28.Stocker K. Snake venom proteins affecting hemostasis and fibrinolysis in Medical Use of Snake Venom Proteins. In: Stocker K, editor. Boca Raton, Ann Arbor, Boston: CRC Press; 1990. pp. 97–160. [Google Scholar]
- 29.Matsui T, Fujimura Y, Titani K. Snake venom proteases affecting hemostasis and thrombosis. Biochim Biophys Acta. 2000;1477:146–156. doi: 10.1016/s0167-4838(99)00268-x. [DOI] [PubMed] [Google Scholar]
- 30.Thwin MM, Samy RP, Satyanarayanajois SD, Gopalakrishnakone P. Venom neutralization by purified bioactive molecules: synthetic peptide derivatives of the endogenous PLA2 inhibitory protein PIP (a mini-review) Toxicon. 2010;56:1275–1283. doi: 10.1016/j.toxicon.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 31.Yamasaki Y, Koike H, Sugiyama Y, Motoyoshi K, Wada T, Hishinuma S, Mita M, Morita T. Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur. J. Biochem. 2002;269:2708–2715. doi: 10.1046/j.1432-1033.2002.02940.x. [DOI] [PubMed] [Google Scholar]
- 32.Stack MS, Gray RD. Comparison of vertebrate collagenase and gelatinase using a new Xuorogenic substrate peptide. J. Biol. Chem. 1989;264:4277–4281. [PubMed] [Google Scholar]
- 33.Knight CG, Willenbrock F, Murphy G. A novel coumarinlabelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- 34.Wells GM, Catlin G, Cossins JA, Mangan M, Ward GA, Miller KM, Clements JM. Quantitation of matrix metalloproteinases in cultured rat astrocytes using the polymerase chain reaction with a multi-competitor cDNA standard. Glia. 1996;18:332–340. [PubMed] [Google Scholar]
- 35.Masuda S, Hayashi H, Araki S. Two vascular apoptosis-inducing proteins from snake venom are members of the metalloprotease/disintegrin family. Eur. J. Biochem. 1998;253:36–41. doi: 10.1046/j.1432-1327.1998.2530036.x. [DOI] [PubMed] [Google Scholar]
- 36.Tanjoni I, Weinlich R, Della-Casa MS, Clissa PB, Saldanha-Gama RF, de Freitas MS, Barja-Fidalgo C, Amarante-Mendes GP, Moura da Silva AM. Jararhagin, a snake venommetalloproteinase, induces a specialized form of apoptosis (anoikis) selective to endothelial cells. Apoptosis. 2005;10:851–861. doi: 10.1007/s10495-005-2945-1. [DOI] [PubMed] [Google Scholar]
- 37.You W, Seo H, Chung K, Kim D. A novel metalloprotease from Gloydius halys venom induces endothelial cell apoptosis through its protease and disintegrin-like domains. J. Biochem. 2003;134:739–749. doi: 10.1093/jb/mvg202. [DOI] [PubMed] [Google Scholar]
- 38.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol. Aspects Med. 2009;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 40.Takeda S, Takeya H, Iwanaga S. Snake venom metalloproteinases: structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim. Biophys. Acta. 2011;1824:164–176. doi: 10.1016/j.bbapap.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 41.Takeda S, Igarashi T, Mori H, Araki S. Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold. EMBO J. 2006;25:2388–2396. doi: 10.1038/sj.emboj.7601131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeda S. Three-dimensional domain architecture of the ADAM family proteinases. Semin. Cell. Dev. Biol. 2009;20:146–152. doi: 10.1016/j.semcdb.2008.07.009. [DOI] [PubMed] [Google Scholar]