Abstract
The adult form of Sandhoff disease with the motor neuron disease phenotype is a rare neurodegenerative disorder caused by mutations in HEXB encoding the β-subunit of β-hexosaminidase, yet the properties of mutant β-subunits of the disease have not been fully determined. We identified a novel mutation (H235Y) in the β-sheet of the (β/α)8-barrel domain, in addition to the previously reported P417L mutation that causes aberrant splicing, in a Japanese patient with the motor neuron disease phenotype. Enzyme assays, gel filtration studies and immunoprecipitation studies with HEK293 cells transiently expressing mutant β-subunits demonstrated that the H235Y mutation abolished both α–β and β–β dimer formation without increasing β-hexosaminidase activity, whereas other reported mutant β-subunits (Y456S, P504S or R533H) associated with the motor neuron disease phenotype formed dimers. Structural analysis suggested that the H235Y mutation in the β-sheet of the (β/α)8-barrel domain changed the conformation of the β-subunit by causing a clash with the E288 side chain. In summary, H235Y is the first mutation in the β-sheet of the (β/α)8-barrel domain of the β-subunit that abolishes α–β and β–β dimer formation; the presented patient is the second patient to exhibit the motor neuron disease phenotype with P417L and a non-functional allele of HEXB.
Keywords: adult form of Sandhoff disease, dimer formation, β-hexosaminidase, missense mutation, motor neuron disease phenotype
Human β-hexosaminidase (EC 3.2.1.52) is a lysosomal hydrolase that cleaves the terminal β-N-acetylgalactosamine or β-N-acetylglucosamine moieties of gangliosides, glycolipids, glycoproteins and glycosaminoglycans (1). This hydrolase has three isoenzymes: β-hexosaminidase A (Hex A), β-hexosaminidase B (Hex B) and β-hexosaminidase S (Hex S), composed of an α-subunit and a β-subunit, a homodimer of β-subunits and a homodimer of α-subunits, respectively. Hex A is the only enzyme responsible for GM2 ganglioside degradation. Defects in the α- or β-subunit cause the accumulation of GM2 ganglioside in lysosomes, in particular in neurons, and lead to the neurodegenerative disorders of GM2 gangliosidoses. Mutations in the genes encoding the α-subunit (HEXA) and β-subunit (HEXB) of Hex result in the autosomal recessive disorders Tay–Sachs disease and Sandhoff disease, respectively. GM2 gangliosidoses also result from mutations in genes encoding the GM2 activator required for GM2 ganglioside hydrolysis. Classic Sandhoff disease and Tay–Sachs disease are caused by non-functional Hex A; prognosis is usually fatal within 4 years and is associated with extensive GM2 accumulation in the central nervous system (1). In contrast, a variety of less severe and chronic phenotypes of Sandhoff disease with a small percentage of residual Hex A activity have been reported, including motor neuron disease, spinocerebellar ataxia, intellectual disability and dysfunction of the autonomic nervous system (2–10). Only four missense mutations (Y456S, P504S, R533H and P417L) associated with the adult form of Sandhoff disease presenting the motor neuron disease phenotype have been reported (3, 7, 9, 11–13). In this study, we report the molecular and biochemical study of a Japanese male patient with the adult form of Sandhoff disease with the motor neuron disease phenotype. We identified compound heterozygous mutations in this patient, including a novel H235Y mutation and further analysed the Hex activities as well as the α–β and β–β dimer formation in HEK293 cells expressing mutant β-subunits associated with the adult form of Sandhoff disease with the motor neuron disease phenotype.
Materials and Methods
Patient
The clinical features of the patient have been described in a previous report (14). Briefly, a 46-year-old Japanese male presented with a slowly progressing motor neuron disease phenotype. The patient’s parents are non-consanguineous, and there is no history of motor neuron disease in any family member. At the age of 42 years, the patient noticed weakness in both legs. He was admitted to the Akita Red Cross Hospital at the age of 46 years, and neurological examination revealed muscle weakness, atrophy of the upper and lower extremities and hyperreflexia of the upper extremities. His Wechsler Adult Intelligence Scale Revised Score and funduscopic findings were normal. Electromyography in the upper and lower limbs demonstrated a decreased number of motor units, many of which showed high amplitude and long duration. Conduction velocities of motor and sensory nerves were normal. The differential heat inactivation method revealed that the Hex A and Hex B activities of the patient’s leukocytes decreased to 20.9 and 0.6% of those of the controls, respectively. A membranous cytoplasmic body in the submucosal plexus in the rectum was observed. Thus, he was diagnosed with the adult form of Sandhoff disease with the motor neuron disease phenotype.
Bioethics approval
The experiments were conducted after approval by the institutional review board at the Institute for Developmental Research, Aichi Human Service Center. Written informed consent was obtained from the patient and normal controls participating in this study.
Amplification of HEXB from genomic DNA and DNA sequencing
Genomic DNA was isolated from the peripheral blood leukocytes of the patient and normal controls by phenol/chloroform extraction. Part of the promoter region, 14 exons, splice junctions and parts of the introns were amplified from the genomic DNA by polymerase chain reaction (PCR) with HEXB-specific primers according to Wakamatsu et al. (4). Amplified DNA fragments were sequenced directly with the same PCR primers (15).
Construction of wild-type and mutant HEXB cDNA expression vectors
Lymphoblastoid cell lines were established by the Institute for Developmental Research, Aichi Human Service Center, through Epstein–Barr virus transformation of peripheral blood samples obtained from the patient and normal controls. A fibroblast cell line from the patient was not available. Wild-type and H235Y HEXB cDNA were amplified from first-strand cDNA prepared from the patient and normal control lymphoblastoid cells using a specific primer pair: S1 (sense primer in exon 1), 5′-aagcactcgagcggccatgga-3′ and A1 (antisense primer in exon 14), 5′-ttcccctggatcctttacat-3′. An XhoI recognition site (ctcgag) or a BamHI recognition site (ggatcc) was introduced into S1 or A1, respectively. After confirming the nucleotide sequence, the XhoI/BamHI fragments of the wild-type and H235Y HEXB cDNA were subcloned into the EcoRV/BamHI site of a mammalian expression vector, p3 × FLAG-CMV (Sigma-Aldrich, St Louis, MO, USA) after the XhoI site had been blunt-ended by a Klenow reaction (pHEXB and pHEXB-H235Y). pHEXB and pHEXB-H235Y contained a termination codon (TAA) before a 3 × FLAG-tag sequence. To construct HEXB-FLAG expression vectors, the 3′-end of the HEXB cDNA was amplified with a primer pair S2–A2 (S2, exon 10: 5′-ccataaacaagggatccattg-3′ and A2, exon 14: 5′-tccatggatcccatgttctcatggt-3′). A BamHI recognition site (ggatcc, underlined) was introduced at the site of the termination codon (TAA). The PCR product was digested with BamHI, and a 468-bp BamHI fragment was exchanged into pHEXB and pHEXB-H235Y (pHEXB-FLAG and pHEXB-H235Y-FLAG). To generate the Y456S HEXB expression vector, two parts of HEXB cDNA containing the Y456S-encoding mutation (TAT > TCT) were amplified with primer pairs S3–A4 (S3, exon 9: 5′-ggcacagattttaagaaactag-3′ and A4, exon 11: 5′-caatcttgtccagagctaatcaaat-3′) and S4–A2 (S4, exon 11: 5′-atttgattagctctggacaagattg-3′) (codons subjected to mutagenesis, underlined). The PCR was performed for 30 cycles consisting of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 60 s using KOD-plus Polymerase (Toyobo, Osaka, Japan). Two PCR products were mixed, diluted 1/500 and subjected to a second PCR with the S3–A2 primer pair using KOD-plus Polymerase. The PCR product was digested with BamHI and exchanged into the BamHI site of pHEXB-FLAG (pHEXB-Y456S-FLAG). FLAG-tagged pHEXB-P504S and pHEXB-R533H were generated using the same in vitro mutagenesis method.
Construction of HEXA, HEXA-MYC and HEXB-MYC cDNA expression vectors
First, we constructed the 2 × MYC expression vector by adding 2 × MYC-encoding sequences (EQKLISEEDLEQKLISEEDL) at the EcoRV/BamHI site of p3 × FLAG-CMV (p2 × MYC-CMV). This MYC expression vector contained a termination codon (TAG) after the MYC sequence but before the 3 × FLAG sequence. Thus, p2 × MYC-CMV is useful for the expression of MYC-tagged proteins. Using the method described above, we produced a HEXA expression construct by subcloning HEXA cDNA containing a termination codon (TGA) at the 3′-end into the HindIII/XbaI site of p3 × FLAG-CMV (pHEXA). The HEXA-MYC expression construct was generated by subcloning HEXA cDNA into the HindIII/EcoRV site of p2 × MYC-CMV (pHEXA-MYC). The HEXB-MYC expression construct was also generated by subcloning HEXB cDNA into the HindIII/EcoRV site of p2 × MYC-CMV (pHEXB-MYC).
Determination of the Hex activity of transiently expressed wild-type and H235Y β-subunits in HEK293 cells
HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal calf serum. Seeded HEK293 cells were grown to 80–90% confluence in 24-well dishes at 37°C with 5% carbon dioxide. HEK293 cells were transfected with 0.8 µg of human HEXB expression vectors (pHEXB, pHEXB-FLAG, pHEXB-H235Y, pHEXB-H235Y-FLAG) or the control vector (p3 × FLAG-CMV) with 10 ng of pCMV-β-gal (an Escherichia coli β-galactosidase expression vector) using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). The medium was replaced with fresh medium after 12 h, and the culture growth was continued for an additional 48 h. The cells were harvested, washed twice with phosphate buffered saline (PBS), suspended in ice-cold 20 mM sodium phosphate buffer (pH 6.0) containing 0.5% Triton X-100 and 1:1,000 diluted Protease Inhibitor Cocktail (Sigma-Aldrich) and homogenized with pestles (AS ONE, Osaka, Japan). The Hex activities of the cell extracts were assayed using the common substrate of 2 mM 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide (MUG) (Sigma-Aldrich) (4), α-subunit-specific substrate of 3 mM 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide 6-sulphate (MUGS) (EMD Chemicals, San Diego, CA, USA) (16) and the α-mannosidase was determined using 2 mM 4-methylumbelliferyl-α-d-mannopyranoside (MUM) (Sigma-Aldrich) (17) as substrates. The enzyme assay was performed in a total volume of 150 µl containing 0.1 M sodium citrate buffer (pH 4.5) and was terminated by the addition of 1 ml of 0.2 M glycine–NAOH buffer (pH 10.6). Enzyme activities were determined by the increase in fluorescence measured by a FP-6200 Fluorescence Spectrophotometer (JASCO, Tokyo, Japan) with an excitation wavelength at 365 nm and emission wavelength at 448 nm using 4-methylumbelliferone sodium salt (Sigma-Aldrich) as a standard. The enzyme activities using the MUG, MUGS and MUM substrates are referred to as MUG activity, MUGS activity and MUM activity, respectively. The efficiency of the DNA transfection was verified by measuring the E. coli β-galactosidase activity using o-nitrophenyl-β-d-galactopyranoside as the substrate (17). The protein concentration was determined using the Advanced Protein Assay Reagent (Cytoskeleton Inc., Denver, CO, USA).
Gel filtration profiles of the wild-type and H235Y β-subunits
HEK293 cells were grown to 80–90% confluence in 75-cm2 tissue culture flasks. Three 75-cm2 flasks were transfected with 15.0 µg of HEXB expression vector (p3 × FLAG-CMV, pHEXB-FLAG or pHEXB-H235Y-FLAG, respectively). Three days after transfection, the cells were harvested and washed twice with PBS. The cells were suspended in a buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl and 1:1,000 diluted Protease Inhibitor Cocktail and were then sonicated with a Branson 250 Sonifier (Danbury, CT, USA). Protein extracts were centrifuged at 12,000 × g for 10 min and then re-centrifuged at 100,000 × g for 20 min. Supernatants (2.5 ml) were loaded onto a Cellufine GCL-2000 gel filtration column (2.8 × 90 cm; Chisso Corporation, Tokyo, Japan) equilibrated with 20 mM Tris–HCl (pH 7.5) and 0.15 M NaCl. Each fraction (3 ml) was collected, and 50-µl aliquots were subjected to MUG activity assays and western blotting with anti-FLAG M2 antibody.
Analysis of the association between the wild-type or mutant β-subunits and the wild-type α- or β-subunit
To analyse the association between the H235Y β-subunit and the wild-type α- or β-subunit, various combinations of 0.4 µg of FLAG expression vectors (p3 × FLAG-CMV, pHEXB-FLAG and pHEXB-H235Y-FLAG) and 0.4 µg of MYC expression vectors (p2 × MYC-CMV and pHEXA-MYC) or 0.8 µg of wild-type pHEXB-MYC were transfected into HEK293 cells in 24-well dishes using the Lipofectamine 2000 reagent. Similarly, to analyse the association between wild-type or various mutant β-subunits and the wild-type α- or β-subunit, pHEXB-FLAG, pHEXB-H235Y-FLAG, pHEXB-Y456S-FLAG, pHEXB-P504S-FLAG, or pHEXB-R533H-FLAG and pHEXA-MYC or pHEXB-MYC were co-transfected into HEK293 cells. The cells were washed twice with PBS 60 h after transfection and were then solubilized with lysis buffer containing 20 mM Tris–HCl (pH 7.0), 150 mM NaCl, 1% Nonidet P-40 (Sigma-Aldrich) and 1:1,000 diluted Protease Inhibitor Cocktail. Insoluble material was removed by centrifugation at 4°C for 10 min at 10,000 × g. The resulting supernatants were subjected to immunoprecipitation using anti-FLAG M2 antibody conjugated to agarose (Sigma-Aldrich) for 2 h at 4°C with gentle mixing. After washing the gels three times with lysis buffer, the precipitates were analysed by western blotting with the anti-FLAG M2 antibody for the β-subunit (1:6,000 dilution, Sigma-Aldrich) and with the anti-MYC antibody for the α- and β-subunits (kindly provided by Dr K. Nagata, Aichi Human Service Center). Immunoreactive bands were visualized with the enhanced chemiluminescence western blotting detection system (GE Healthcare, Tokyo, Japan).
Structural analysis of Hex A
H235 of the β-subunit of the Hex A coordinate (PDB code: 2GJX) was replaced with a tyrosine using Swiss PDB Viewer (18) in order to observe whether this replacement caused clashes with surrounding residues.
Results
Identification of HEXB mutations
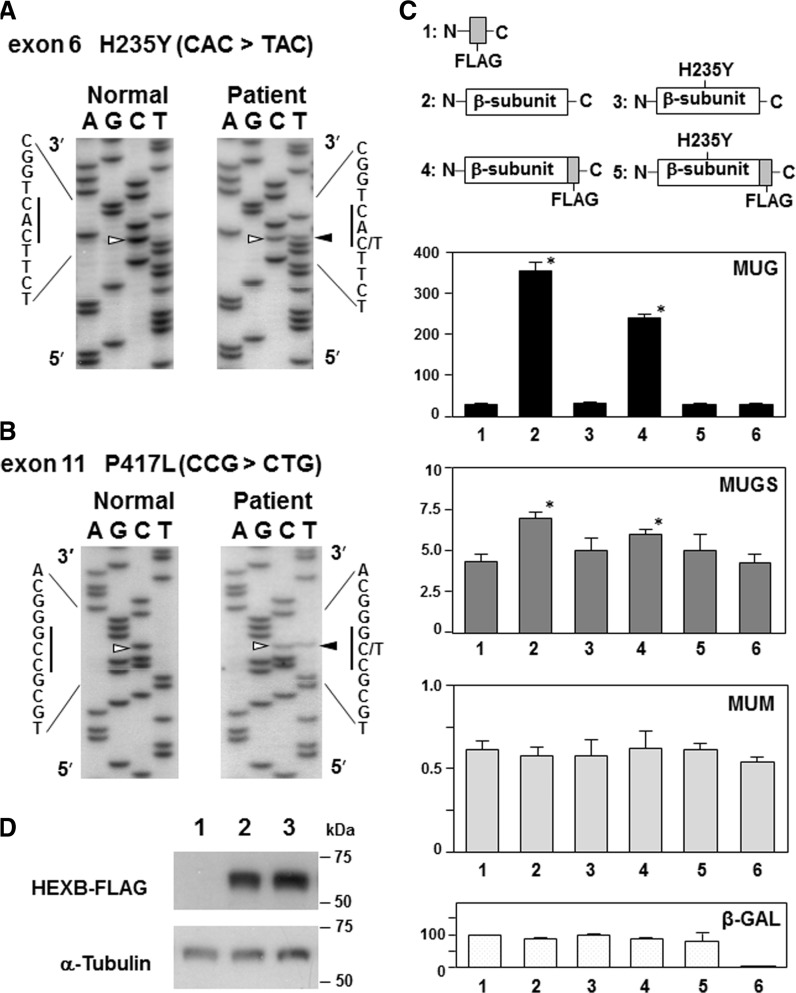
We determined the nucleotide sequence of part of the promoter, 14 exons, splice junctions and parts of the introns of the patient’s HEXB by DNA sequencing. Two single-base substitutions were identified. The first mutation was a 703C > T (CAC to TAC) substitution in exon 6, resulting in the H235Y amino acid substitution (Fig. 1A). This nucleotide change is not a reported single-nucleotide polymorphism in the NCBI Human Genome Reference Assembly and was not found in 200 alleles from normal Japanese individuals (data not shown). The second mutation was the previously reported 1250C > T (CCG to CTG) substitution in exon 11, resulting in a P417L substitution (Fig. 1B). This mutation leads to aberrant HEXB mRNA by introducing a cryptic splice acceptor site in exon 11, which results in decreased levels of HEXB mRNA (4). To confirm aberrant splicing, HEXB mRNAs from the patient and control lymphoblastoid cells were analysed by reverse transcriptase (RT)–PCR. An abnormal DNA fragment generated by a 112-bp deletion in exon 11 was identified in the patient, in addition to the normal PCR product (data not shown). Because the DNA of the patient’s parents was not available, we performed RT–PCR to amplify both mutation sites. The DNA fragment was subcloned into pGEM-T Easy vector (Promega, Madison, WI, USA), and the nucleotide sequences of 12 clones were determined. The results demonstrated that all clones contained the H235Y mutation and that no other mutations were observed (Supplementary Fig. S1). Thus, the patient is a compound heterozygote for the H235Y and P417L mutations.
Fig. 1.
Identification of missense mutations in HEXB and Hex activity of the transiently expressed wild-type or H235Y β-subunit in HEK293 cells. (A and B) Partial nucleotide sequences of amplified HEXB genomic DNA from the patient and controls, confirming single-base substitutions in exon 6 (c.703C > T, [p.His235Tyr]) (A) and exon 11 (c.1250C > T, [p.Pro417Leu]) (B). Arrowheads indicate the position of the nucleotide substitutions. (C) Hex, α-mannosidase and relative E. coli β-galactosidase (β-gal) activities in HEK293 cells after transfection with each HEXB expressing vector. The mean enzyme activities (nmol min−1 mg−1 protein) with MUG, MUGS and MUM as well as the relative β-gal activity (%) are shown. The vertical bars indicate the standard error of the mean (n = 3). Lane 1, p3 × FLAG-CMV; lane 2, pHEXB; lane 3, pHEXB-H235Y; lane 4, pHEXB-FLAG; lane 5, pHEXB-H235Y-FLAG and lane 6, untransfected cells. Asterisks (*) mean the significance of differences (P-value < 0.05), analysed with one-way analysis of variance followed by Bonferroni correction for multiple comparisons. (D) Western blot analysis of HEK293 cells transfected with each of the HEXB-expressing vectors using antibodies specific for FLAG and α-tubulin. Lane 1, p3 × FLAG-CMV; lane 2, pHEXB-FLAG and lane 3, pHEXB-H235Y-FLAG.
The H235Y β-subunit mutation abolishes Hex activity
We first analysed the effect of the FLAG-tag at the end of the β-subunit on Hex activity. A transfection study showed that pHEXB increased the MUG activity 13-fold compared with the Mock transfection using p3 × FLAG-CMV. Transfection of pHEXB-FLAG retained ∼70% of the MUG activity of pHEXB (Fig. 1C), confirming that the C-terminal 3 × FLAG tag of the β-subunit did not significantly affect Hex activity. Next, we analysed the effect of the H235Y β-subunit on Hex activity. Transient expression of pHEXB-H235Y and pHEXB-H235Y-FLAG did not increase MUG activity (Fig. 1C). Similarly, transient expression of pHEXB and pHEXB-FLAG resulted in a small increase in MUGS activity (∼1.6- and ∼1.4-fold, respectively); however, transient expression of pHEXB-H235Y and pHEXB-H235Y-FLAG did not increase MUGS activity (Fig. 1C). Thus, the H235Y β-subunit does not form catalytically active dimers with the wild-type α- and β-subunits, and the transiently expressed wild-type β-subunit preferentially forms a dimer with the β-subunit responsible for the remarkably increased MUG activity in HEK293 cells. Western blot analysis using anti-FLAG M2 antibody revealed that the H235Y β-subunit protein was stably expressed (Fig. 1D), indicating that the defective Hex activity of the H235Y-mutant enzyme was not caused by an unstable β-subunit.
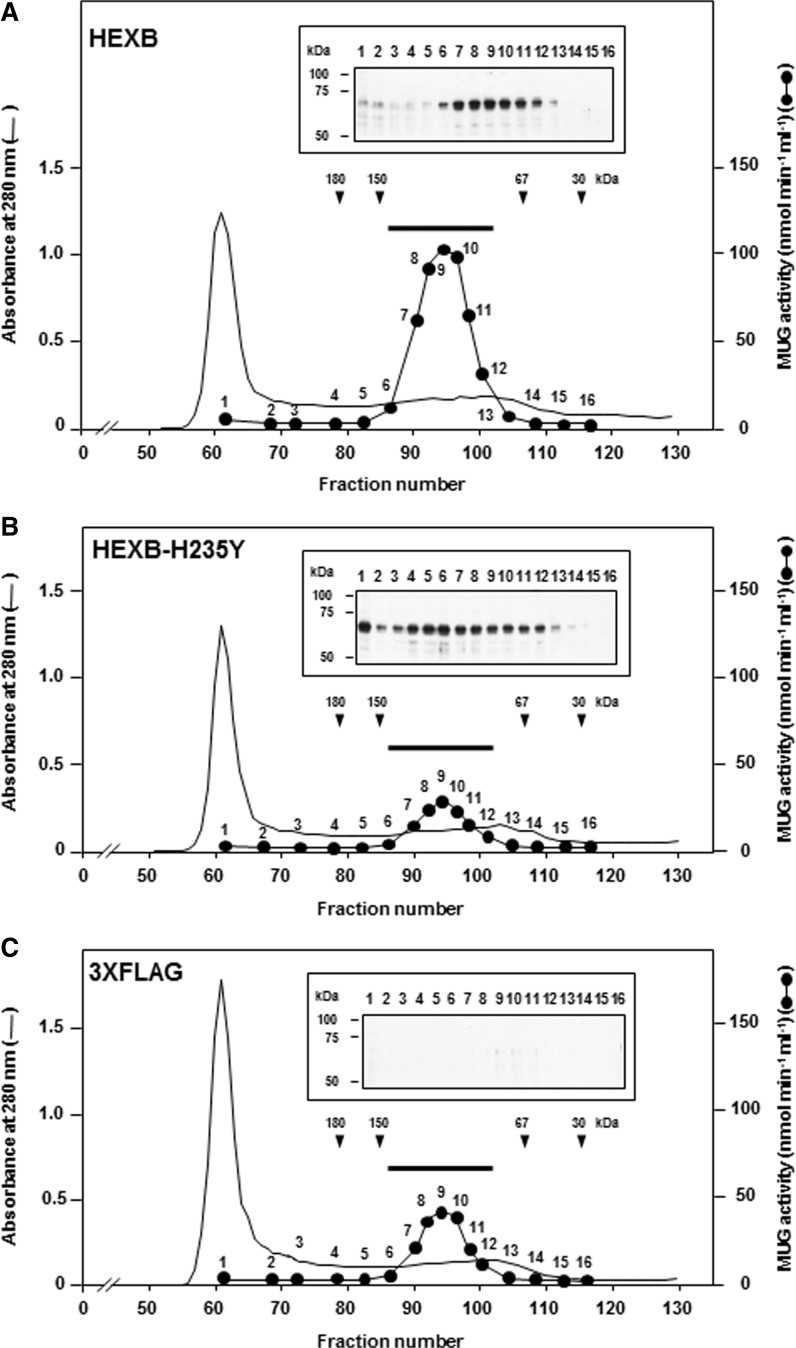
The H235Y β-subunit is not eluted in the same gel filtration fraction as the wild-type β-subunit
To analyse the effect of the H235Y β-subunit on dimer formation (Hex A and Hex B), pHEXB-FLAG or pHEXB-H235Y-FLAG was transfected into HEK293 cells, and the corresponding proteins were separated by gel filtration chromatography (Fig. 2). The transiently expressed, FLAG-tagged, wild-type β-subunit was eluted in fractions corresponding to 100–150-kDa proteins with increased activity towards MUG (Fig. 2A). The molecular weight of the pre-processed Hex A (heterodimer of α- and β-subunits) and Hex B (homodimer of β-subunits) is 124 and 126 kDa, respectively. Both enzymes hydrolyzed the MUG substrate. Thus, increased Hex A and Hex B proteins from HEK293 cells were eluted as a single peak by this gel filtration method. In contrast, the transiently expressed, FLAG-tagged, H235Y β-subunit did not show a single protein peak corresponding to an increased amount of Hex A and Hex B (Fig. 2B), and only endogenous MUG activity was detected (Fig. 2C). The recovery of the MUG activities in the 17 pooled gel filtration fractions of the HEK293 cells transiently expressed with wild-type β-subunit, H235Y β-subunit and 3 × FLAG was 42.1, 60.5 and 79.5% (Fig. 2A–C), respectively.
Fig. 2.
Elution profiles of the transiently expressed wild-type β-subunit, H235Y β-subunit and 3 × FLAG in HEK293 cells. Chromatography of soluble proteins from transiently expressed wild-type (WT) β-subunit (A), H235Y β-subunit (B) or 3 × FLAG (C) in HEK293 cells. The protein extracts were loaded onto the Cellufine GCL-2000 gel filtration column, and the elution of the proteins was monitored by measuring the absorbance at 280 nm. The Hex activity (filled circles, 1–16) was determined using MUG as the substrate (nmol min−1 ml−1). Arrowheads indicate the fractions in which the molecular weight markers eluted. The results of the western blotting using anti-FLAG M2 antibody are shown in the corresponding numbers used to measure MUG activity. Aliquots (5 µl) of the fractions, for which the Hex activity was measured (1–16), were separated on a 10% SDS–PAGE gel, transferred to a PVDF membrane and subjected to western blotting with anti-FLAG M2 antibody. The MUG activities used for chromatography of (A)–(C) were 4363, 632.8 and 808.8 nmol min−1, respectively. To measure the recovery of the Hex activity of the chromatography, 17 fractions (no. 86-102, indicated by bars) with Hex activity were collected and assessed with MUG as the substrate.
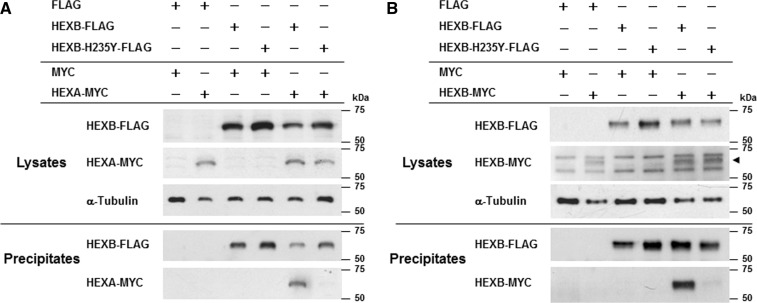
The H235Y β-subunit does not associate with the wild-type α- and β-subunits of Hex
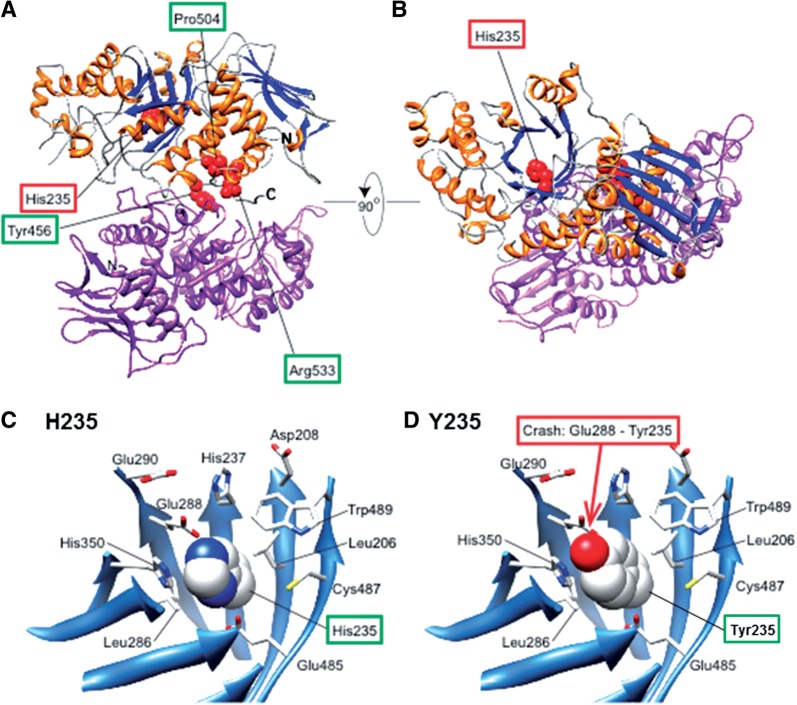
We studied the effect of the H235Y β-subunit on wild-type α- or β-subunit binding by immunoprecipitation. Transfection of pHEXA-MYC increased the Hex activity in HEK293 cells compared with the mock (p2 × MYC-CMV) transfection (data not shown). Thus, the MYC-tagged α-subunit binds to the endogenous α- or β-subunit and produces catalytically active Hex S or Hex A in HEK293 cells. Western blot analysis of the immunoprecipitation products using anti-MYC antibody demonstrated that the MYC-tagged α-subunit co-precipitated with the FLAG-tagged wild-type β-subunit, whereas a barely detectable level of the α-subunit co-precipitated with the H235Y β-subunit (Fig. 3A). Similarly, the MYC-tagged wild-type β-subunit co-precipitated with the FLAG-tagged wild-type β-subunit, whereas the majority of the MYC-tagged β-subunit did not co-precipitate with the H235Y β-subunit (Fig. 3B). Thus, the H235Y mutation of the β-subunit abolished the formation of Hex A (α–β-subunit) and Hex B (β–β-subunit) dimers. Swiss PDB Viewer visualization showed that H235 is tightly packed in the barrel, and the replacement of histidine with tyrosine presumably causes a clash with the E288 side chain (Fig. 4C and D).
Fig. 3.
Association of the wild-type or the H235Y β-subunit with the wild-type α- or β-subunit of Hex. (A) Immunoprecipitation of the β-subunit with the α-subunit. HEK293 cells were transfected with a combination of FLAG-tagged HEXB expression vectors (wild-type, H235Y or FLAG control) and MYC-tagged protein expression vectors (pHEXA-MYC or MYC control). Plus symbol indicates the FLAG- or MYC-tagged vectors used in each study. The anti-FLAG M2 antibody was used for immunoprecipitation. Lysates and precipitates were immunoblotted with antibodies as indicated (FLAG, MYC and α-tubulin). (B) Immunoprecipitation of the β-subunit with the β-subunit. Same as (A), but wild-type pHEXB-MYC was used instead of pHEXA-MYC. The results of western blotting are shown. The arrowhead indicates MYC-tagged β-subunits.
Fig. 4.
Dimer configuration of Hex A. The α-subunit is shown in purple, and the α-helices and β-strands of the β-subunit are shown in orange and blue, respectively. The red spheres represent the residues mentioned in this article, including mutated residues such as H235. This figure was generated using UCSF Chimera (19). (A) View to distinguish the residues at the dimer interface (Y456, P504 and R533) from H235. (B) View to show that H235 is located in the (β/α)8-barrel. (C and D) Enlarged views around H235 of the β-subunit of Hex A. β-Strands are shown as ribbons, and the residues around H235 are depicted as sticks. H235 is depicted as a sphere (C). (D) The same view as (C). H235 replaced with Tyr, depicted as a sphere, where Tyr clashes with Glu288 (E288).
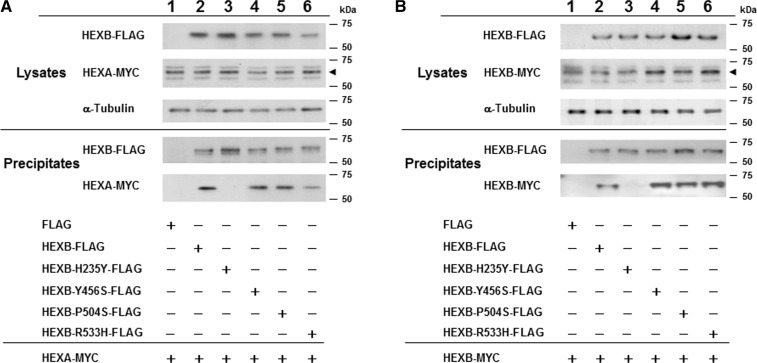
Y456S-, P504S- and R533H-mutant β-subunits identified from the adult form of Sandhoff disease with the motor neuron disease phenotype form dimers with the wild-type α- and β-subunits of Hex
Three missense mutations (Y456S, P504S and R533H) in HEXB have been identified in patients with the adult form of Sandhoff disease with the motor neuron disease phenotype (3, 9, 11–13). To analyse dimer formation between these wild-type or mutant β-subunits and the wild-type α- or β-subunit, we performed immunoprecipitation using anti-FLAG M2 antibody-conjugated gels as depicted in Fig. 5. The results demonstrated that H235Y abolished α–β- and β–β-subunit dimerization, whereas the Y456S-, P504S- and R533H-mutant β-subunits formed dimers with the wild-type α- and β-subunits similar to dimer formation with the wild-type β-subunit (Fig. 5).
Fig. 5.
Association of the wild-type α- or β-subunit of Hex with mutant β-subunits identified in patients with the adult form of Sandhoff disease with the motor neuron disease phenotype. (A) Immunoprecipitation of mutant β-subunits with the wild-type α-subunit. HEK293 cells were transfected with various combinations of FLAG-tagged HEXB expression vectors (wild-type, H235Y, Y456S, P504S, R533H or FLAG control) and MYC-tagged protein expression vectors (pHEXA-MYC or MYC control). Plus symbol indicates the FLAG- or MYC-tagged vectors used in each study. The anti-FLAG M2 antibody was used for immunoprecipitation. Lysates and precipitates were immunoblotted with the indicated antibodies (FLAG, MYC and α-tubulin). The arrowhead indicates MYC-tagged α-subunits. (B) Immunoprecipitation of mutant β-subunits with the wild-type β-subunit. Same as (A), but wild-type pHEXB-MYC was used instead of pHEXA-MYC. The results of western blotting are shown. The arrowhead indicates MYC-tagged β-subunits.
Discussion
We report that compound heterozygous mutations, H235Y and P417L, in HEXB are associated with the adult form of Sandhoff disease presenting with the motor neuron disease phenotype. To elucidate the specific molecular defect associated with HEXB, we analysed the H235Y β-subunit in detail. Because the dimerization of the α- and β-subunits of Hex occurs before proteolytic processing events (20), we detected signals from unprocessed α- and β-subunits in the gel filtration studies and immunoprecipitation assay to characterize the dimerization of mutant β-subunits with wild-type α- and β-subunits. This study demonstrates that the newly identified H235Y mutation in the β-sheet of the (β/α)8-barrel domain abolished Hex activity by disrupting the dimerization with the wild-type α- and β-subunits.
The (β/α)8-barrel proteins account for ∼10% of all proteins with known three-dimensional structures (21, 22). The active-site residues are located on the catalytic face of the barrel, and the remainder of the fold, including the opposite face of the barrel (stability face), is important for conformational stability (23). X-ray crystallographic analysis demonstrated that H212, D494 and N497 of the β-subunit interact by hydrogen bonding with N518, R504 and Q515 of the α-subunit, respectively, to form a α–β dimer (24). Similarly, R211, H212, D494, N497 and R501 of one β-subunit interact by hydrogen bonding with G549, Y547, R533, Q544 and Q544 of another β-subunit, respectively, to form a β–β dimer (25, 26). These nine amino residues of the β-subunit, responsible for α–β and β–β dimer formation, are located on the catalytic face. The newly identified H235Y replacement in the β-sheet of the (β/α)8-barrel is proximal to the stability face (Fig. 4). Therefore, a clash between the E288 and Y235 side chains could affect conformational stability, possibly dramatically changing the structure of the mutant β-subunit, which results in the impairment of three pairs and five pairs of hydrogen bonds with the wild-type α- and β-subunits, respectively (24–26). Three missense mutations (Y456S, P504S and R533H) have been reported to associate with the adult form of Sandhoff disease with the motor neuron disease phenotype. Each missense mutation is a compound heterozygous mutation with the null allele or dramatically decreased HEXB mRNA levels by aberrant splicing of HEXB (3, 9, 11–13). X-ray crystallographic analysis of Hex A demonstrated that these three missense mutations are located at the dimer interface (Fig. 4A and B), where the mutations likely impair catalytic activity and/or dimer formation (24, 25). This study clearly showed that the Y456S, P504S and R533H missense mutations did not abolish α–β-subunit dimer formation (based on immunoprecipitation analysis). Because the three mutations are located at the dimer interface (catalytic face) of the barrel, the mutations likely have only mild effects on conformational stability. Thus, the decreased Hex A activity caused by these mutations is likely due to an impairment of the catalytic active site.
The P417L mutation in exon 11 is a previously reported mutation that affects 3′-splice-site selection and reduces HEXB mRNA to a much lower level than that observed in the controls (4). In GM2 gangliosidoses, the critical threshold of Hex A activity, inferred from the maximal activity of the respective enzyme in late-onset patients and the minimal activity of healthy probands, is ∼5–10% of the average activity (27). The P417L allele has been reported to be associated with a variety of late-onset phenotypes; a P417L mutation and a 16-kb deletion at the 5′-end were noted in a patient with the motor neuron disease phenotype (7). Both autonomic nervous system dysfunction and normal phenotype were observed in one family with the same combination of HEXB mutations (5). These findings confirm that the patient with P417L and null alleles of HEXB has a range of critical thresholds of Hex A activity, and tissue-specific differential splicing of the HEXB mRNA may be associated with a variety of neurological features. In summary, the presented case is the second case of the adult form of Sandhoff disease with the motor neuron disease phenotype, with a P417L mutation and a non-functional allele of HEXB.
The presented study highlights the importance of biochemical characterization of mutant β-subunits, in particular investigating the impairment of the catalytic active site and the large conformational changes responsible for impaired dimer formation for understanding the molecular basis of mutant β-subunits underling adult form of Sandhoff disease with the motor neuron disease phenotype.
Supplementary Data
Supplementary Data are available at JB Online.
Funding
This study was supported by the Takeda Science Foundation, a Health Labor Sciences Research Grant and Scientific Research (B) (21390319) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N.W.).
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We are grateful to the patient who participated in this study and to his family.
Glossary
Abbreviations
- Hex
β-hexosaminidase
- MUM
4-methylumbelliferyl-α-d-mannopyranoside
- MUG
4-methylumbelliferyl-N-acetyl-β-d-glucosaminide
- MUGS
4-methylumbelliferyl-N-acetyl-β-d-glucosaminide 6-sulphate
- PBS
phosphate buffered saline
- RT–PCR
reverse transcriptase–polymerase chain reaction
References
- 1.Gravel RA, Kaback MM, Proia RL, Sandhoff K, Suzuki K. The Metabolic and Molecular Bases of Inherited Disease. 2001. The GM2-gangliosidosis in. (Scriver, C.R., Beaudet, A.L., Sly, W.S., Valle, D., Childs, B., Kinzler, K.W., and Vogelstein B, eds.) Vol. 3, pp. 3827–3876, McGraw-Hill, New York. [Google Scholar]
- 2.Bolhuis PA, Oonk JG, Kamp PE, Ris AJ, Michalski JC, Overdijk B, Reuser AJ. Ganglioside storage, hexosaminidase lability, and urinary oligosaccharides in adult Sandhoff’s disease. Neurology. 1987;37:75–81. doi: 10.1212/wnl.37.1.75. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee P, Siciliano L, Oliveri D, McCabe NR, Boyers MJ, Horwitz AL, Li SC, Dawson G. Molecular basis of an adult form of beta-hexosaminidase B deficiency with motor neuron disease. Biochem. Biophys. Res. Commun. 1991;181:108–115. doi: 10.1016/s0006-291x(05)81388-9. [DOI] [PubMed] [Google Scholar]
- 4.Wakamatsu N, Kobayashi H, Miyatake T, Tsuji S. A novel exon mutation in the human beta-hexosaminidase beta subunit gene affects 3′ splice site selection. J. Biol. Chem. 1992;267:2406–2413. [PubMed] [Google Scholar]
- 5.McInnes B, Potier M, Wakamatsu N, Melancon SB, Klavins MH, Tsuji S, Mahuran DJ. An unusual splicing mutation in the HEXB gene is associated with dramatically different phenotypes in patients from different racial backgrounds. J. Clin. Invest. 1992;90:306–314. doi: 10.1172/JCI115863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolhuis PA, Ponne NJ, Bikker H, Baas F, Vianney de Jong JM. Molecular basis of an adult form of Sandhoff disease: substitution of glutamine for arginine at position 505 of the beta-chain of beta-hexosaminidase results in a labile enzyme. Biochim. Biophys. Acta. 1993;1182:142–146. doi: 10.1016/0925-4439(93)90134-m. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Lira M, Sangalli A, Mottes M, Perusi C, Pignatti PF, Rizzuto N, Salviati A. A common beta hexosaminidase gene mutation in adult Sandhoff disease patients. Hum. Genet. 1995;96:417–422. doi: 10.1007/BF00191799. [DOI] [PubMed] [Google Scholar]
- 8.Hara A, Uyama E, Uchino M, Shimmoto M, Utsumi K, Itoh K, Kase R, Naito M, Sugiyama E, Taketomi T, Sukegawa K, Sakuraba H. Adult Sandhoff’s disease: R505Q and I207V substitutions in the HEXB gene of the first Japanese case. J. Neurol. Sci. 1998;155:86–91. doi: 10.1016/s0022-510x(97)00299-2. [DOI] [PubMed] [Google Scholar]
- 9.Yoshizawa T, Kohno Y, Nissato S, Shoji S. Compound heterozygosity with two novel mutations in the HEXB gene produces adult Sandhoff disease presenting as a motor neuron disease phenotype. J. Neuro. Sci. 2002;195:129–138. doi: 10.1016/s0022-510x(02)00007-2. [DOI] [PubMed] [Google Scholar]
- 10.Delnooz CC, Lefeber DJ, Langemeijer SM, Hoffjan S, Dekomien G, Zwarts MJ, Van Engelen BG, Wevers RA, Schelhaas HJ, van de Warrenburg BP. New cases of adult-onset Sandhoff disease with a cerebellar or lower motor neuron phenotype. J. Neurol. Neurosurg. Psychiatry. 2010;81:968–972. doi: 10.1136/jnnp.2009.177089. [DOI] [PubMed] [Google Scholar]
- 11.Rubin M, Karpati G, Wolfe LS, Carpenter S, Klavins MH, Mahuran DJ. Adult onset motor neuronopathy in the juvenile type of hexosaminidase A and B deficiency. J. Neurol. Sci. 1988;87:103–119. doi: 10.1016/0022-510x(88)90058-5. [DOI] [PubMed] [Google Scholar]
- 12.Redonnet-Vernhet I, Mahuran DJ, Salvayre R, Dubas F, Levade T. Significance of two point mutations present in each HEXB allele of patients with adult GM2 gangliosidosis (Sandhoff disease) homozygosity for the Ile207–>Val substitution is not associated with a clinical or biochemical phenotype. Biochim. Biophys. Acta. 1996;1317:127–133. doi: 10.1016/s0925-4439(96)00044-0. [DOI] [PubMed] [Google Scholar]
- 13.Hou Y, Mclnnes B, Hinek A, Karpati G, Mahuran D. A Pro504 –> Ser substitution in the beta-subunit of beta-hexosaminidase A inhibits alpha-subunit hydrolysis of GM2 ganglioside, resulting in chronic Sandhoff disease. J. Biol. Chem. 1998;273:21386–21392. doi: 10.1074/jbc.273.33.21386. [DOI] [PubMed] [Google Scholar]
- 14.Takado Y, Koide T, Yoshikawa K, Amaya N, Yoshida Y, Ishiguro H. A patient with GM2 gangliosidosis presenting with motor neuron disease symptom in his forties. Clin. Neurol. (Rinsho Shinkeigaku) 2007;47:37–41. [PubMed] [Google Scholar]
- 15.Yamada Y, Goto H, Suzumori K, Adachi R, Ogasawara N. Molecular analysis of five independent Japanese mutant genes responsible for hypoxanthine guanine phosphoribosyltransferase (HPRT) deficiency. Hum. Genet. 1992;90:379–384. doi: 10.1007/BF00220463. [DOI] [PubMed] [Google Scholar]
- 16.Brown CA, Mahuran DJ. beta-Hexosaminidase isozymes from cells cotransfected with alpha and beta cDNA constructs: analysis of the alpha-subunit missense mutation associated with the adult form of Tay–Sachs disease. Am. J. Hum. Genet. 1993;53:497–508. [PMC free article] [PubMed] [Google Scholar]
- 17.Gotoda Y, Wakamatsu N, Kawai H, Nishida Y, Matsumoto T. Missense and nonsense mutations in the lysosomal α-mannosidase gene (MANB) in severe and mild forms of α-mannosidosis. Am. J. Hum. Genet. 1998;63:1015–1024. doi: 10.1086/302048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 19.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 20.Proia RL, d’Azzo A, Neufeld EF. Association of alpha- and beta-subunits during the biosynthesis of beta-hexosaminidase in cultured human fibroblasts. J. Biol. Chem. 1984;259:3350–3354. [PubMed] [Google Scholar]
- 21.Gerlt JA, Babbitt PC. Barrels in pieces? Nat. Struct. Biol. 2001;8:5–7. doi: 10.1038/83048. [DOI] [PubMed] [Google Scholar]
- 22.Wierenga RK. The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett. 2001;492:193–198. doi: 10.1016/s0014-5793(01)02236-0. [DOI] [PubMed] [Google Scholar]
- 23.Höcker B, Jürgens C, Wilmanns M, Sterner R. Stability, catalytic versatility and evolution of the (βα)8-barrel fold. Curr. Opin. Biotechnol. 2001;12:376–381. doi: 10.1016/s0958-1669(00)00230-5. [DOI] [PubMed] [Google Scholar]
- 24.Lemieux MJ, Mark BL, Cherney MM, Withers SG, Mahuran DJ, James MN. Crystallographic structure of human beta-hexosaminidase A: interpretation of Tay–Sachs mutations and loss of GM2 ganglioside hydrolysis. J. Mol. Biol. 2006;359:913–929. doi: 10.1016/j.jmb.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier T, Strater N, Schuette CG, Klingenstein R, Sandhoff K, Saenger W. The X-ray crystal structure of human beta-hexosaminidase B provides new insights into Sandhoff disease. J. Mol. Biol. 2003;328:669–681. doi: 10.1016/s0022-2836(03)00311-5. [DOI] [PubMed] [Google Scholar]
- 26.Mark BL, Mahuran DJ, Cherney MM, Zhao D, Knapp S, James MN. Crystal structure of human beta-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay–Sachs disease. J. Mol. Biol. 2003;327:1093–1109. doi: 10.1016/s0022-2836(03)00216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leinekugel P, Michel S, Conzelmann E, Sandhoff K. Quantitative correlation between the residual activity of beta-hexosaminidase A and arylsulfatase A and the severity of the resulting lysosomal storage disease. Hum. Genet. 1992;88:513–523. doi: 10.1007/BF00219337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.