Abstract
The role of an expansin gene (IbEXP1) in the formation of the storage root (SR) was investigated by expression pattern analysis and characterization of IbEXP1-antisense sweetpotato (Ipomoea batatas cv. Yulmi) plants in an attempt to elucidate the molecular mechanism underlying SR development in sweetpotato. The transcript level of IbEXP1 was high in the fibrous root (FR) and petiole at the FR stage, but decreased significantly at the young storage root (YSR) stage. IbEXP1-antisense plants cultured in vitro produced FRs which were both thicker and shorter than those of wild-type (WT) plants. Elongation growth of the epidermal cells was significantly reduced, and metaxylem and cambium cell proliferation was markedly enhanced in the FRs of IbEXP1-antisense plants, resulting in an earlier thickening growth in these plants relative to WT plants. There was a marked reduction in the lignification of the central stele of the FRs of the IbEXP1-antisense plants, suggesting that the FRs of the mutant plants possessed a higher potential than those of WT plants to develop into SRs. IbEXP1-antisense plants cultured in soil produced a larger number of SRs and, consequently, total SR weight per IbEXP1-antisense plant was greater than that per WT plant. These results demonstrate that SR development was accelerated in IbEXP1-antisense plants and suggest that IbEXP1 plays a negative role in the formation of SR by suppressing the proliferation of metaxylem and cambium cells to inhibit the initial thickening growth of SRs. IbEXP1 is the first sweetpotato gene whose role in SR development has been directly identified in soil-grown transgenic sweetpotato plants.
Key words: Development, expansin, storage root, sweetpotato, thickening growth
Introduction
The storage roots (SRs) of sweetpotato (Ipomoea batatas) provide high levels of digestible nutrients and fibres. The sweetpotato plant initially produces colourless fibrous roots (FRs), with some of these subsequently acquiring pigmentation and undergoing ‘thickening’ growth to form thick roots (TRs) that ultimately develop into SRs. Three distinct types of sweetpotato roots have been identified on the basis of anatomical studies: the FR, TR, and SR (Kokubu, 1973; Wilson and Lowe, 1973; Nakatani and Komeichi, 1991; Noh et al., 2010). The characteristic features of the TR are active cambium differentiation to form a circular primary cambium around the central metaxylem and anomalous proliferation of the metaxylem cells at the centre part of the stele, while the SR is characterized by anomalous secondary cambial activity inside of a primary cambium.
Many authors have proposed that growth hormones, such as cytokinins, auxins [indole-3-acetic acid (IAA)], and abscisic acid (ABA), play various roles in the formation and thickening growth of SRs (Akita et al., 1962; Matsuo, 1983; Nakatani and Komeichi, 1992; Wang et al., 2006; Eguchi and Yoshida, 2008; Noh et al., 2010). Cytokinin and auxin levels are high during the early stage of SR formation (Akita et al., 1962; Matsuo, 1983; Nakatani and Komeichi, 1992; Noh et al., 2010). Alternatively, secondary thickening growth of SRs is positively correlated with concentrations of ABA and cytokinin—and not with IAA levels (Wang et al., 2006), which actually show a gradual decrease with secondary thickening growth (Akita et al., 1962; Matsuo, 1983; Nakatani and Komeichi, 1992). The results of a recent transcriptome analysis of the SR of sweetpotato suggest a possible involvement of jasmonic acid (JA) in SR development (McGregor, 2006). Furthermore, stele lignification in FRs and TRs may prevent SR development (Togari, 1950). Wilson and Lowe (1973) and Belehu et al. (2004) observed an association between stele lignification and the inability of FRs and TRs to develop into SRs (Firon et al., 2009). In addition, the growth and/or yield of SRs have been shown to be affected by several environmental factors, including soil temperature, humidity, light, photoperiod, carbon dioxide, and drought (Loretan et al., 1994; Hill et al., 1996; Mortley et al., 1996; Eguchi et al., 1998; Pardales et al., 1999; Kano and Ming, 2000; van Heerden and Laurie, 2008).
The molecular mechanism underlying SR development is unknown. Recent advances in various molecular approaches have enabled the mining of genes possibly involved in SR development in sweetpotato, resulting in the identification of a number of genes that are differentially expressed in the developing SR (You et al., 2003; Tanaka et al., 2005). Tanaka et al. (2008) isolated three class I knotted1-like homeobox (KNOX1) genes from sweetpotato and, based on the results of their subsequent comparison of the distribution of the expression of these KNOX1 genes and that of endogenous trans-zeatin riboside (t-ZR) in sweetpotato roots, suggested that these genes could be possible regulators of cytokinin levels in the SR. Ku et al. (2008) isolated IbMADS1 from sweetpotato and analysed its functional role in SR development using IbMADS1-overexpressing (ox) potato (Solanum tuberosum) plants grown in vitro. Very recently, another MADS-box gene, SRD1, from sweetpotato was identified and it was demonstrated that it had 99% nucleotide sequence identity with IbMADS1. Characterization of its role in SRD1-ox sweetpotato plants grown in vitro revealed that SRD1 is involved in the auxin-mediated initial thickening growth of the SR by enhancing proliferation activity in metaxylem and cambium cells (Noh et al., 2010). Further elucidation of the molecular mechanism underlying SR development will require identification of the full set of genes involved in SR development and direct characterization of their functional roles in soil-grown sweetpotato plants. Within this framework, Wang et al. (2010) and Xie et al. (2012) performed large-scale transcriptome sequencing of the SRs of sweetpotato using the Illumina paired-end sequencing technology and characterized the sweetpotato root transcriptome. However, functional characterization of the transcriptome in SR development has not yet been reported.
Expansins have been characterized as cell wall-loosening proteins (McQueen-Mason et al., 1992; McQueen-Mason and Cosgrove, 1995; Li et al., 2003). They are believed to be important regulators of wall extension during plant cell growth (Lee et al., 2001; Cosgrove et al., 2002; Li et al., 2003), and the expression levels of expansin genes have been found to be strongly correlated with elongation growth in the root, internode, flower, and leaf (Wu et al., 1996; Cho and Cosgrove, 2000; Lee and Kende, 2002; Choi et al., 2003; Gookin et al., 2003; Lee et al., 2003; Zenoni et al., 2004; Sloan et al., 2009; Harada et al., 2011), with expansion growth of grain (Lizana et al., 2010), and with fruit ripening (Rose et al., 1997; Brummell et al., 1999; Hayama et al., 2006; Mbéguié-A-Mbéguié et al., 2009). Expansin activities have also been found to be influenced by various abiotic stresses, such as drought (Wu et al., 2001; Jones and McQueen-Mason, 2004; Li et al., 2011; Zhao et al., 2011), flooding (Cho and Kende, 1997; Huang et al., 2000; Kim et al., 2000; Vriezen et al., 2000; Colmer et al., 2004), and salt and osmotic stress (Buchanan et al., 2005; Geilfus et al., 2010). Very recently, using gain- and/or loss-of-function approaches, a number of researchers have demonstrated that expansins are involved in the elongation growth of root hairs (Lin et al., 2011; ZhiMing et al., 2011) and stomatal opening (Wei et al., 2011; Zhang et al., 2011). Expansins are encoded by multifamily genes consisting of four gene subfamilies: α-expansin, β-expansin, expansin-like A, and expansin-like B (Sampedro and Cosgrove, 2005). These genes are similar in size, share a number of conserved motifs, and express similar wall-loosening activities (Cosgrove et al., 1997; Sampedro and Cosgrove, 2005). Expansin-like genes have been identified in tomato (Solanum lycopersicum) (Keller and Cosgrove, 1995), rice (Oryza sativa) (Lee and Kende, 2002), and Dictyostelium discoideum (Darley et al., 2003; Ogasawara et al., 2009), but the roles of expansin genes in SR development have not yet been investigated.
In an earlier study, cDNAs of three expansin genes (IbEXP1, IbEXP2, and IbEXPL1) from the young storage root (YSR) cDNA library of sweetpotato (I. batatas cv. Jinhongmi) were identified (You et al., 2003) and the transcriptional regulation of these three genes in response to chilling temperature was investigated (12–28 °C; Noh et al., 2009). In the study reported here, the functional role of IbEXP1 in SR development was characterized using IbEXP1-antisense sweetpotato plants. The results demonstrate that the initial thickening growth of the SR was enhanced in IbEXP1-antisense plants by an increased multiplication of metaxylem and cambium cells, which consequently promoted SR development, resulting in an increased production of SRs. These finding provide experimental evidence that the functional role of IbEXP1 involves blocking the initial thickening growth of the SR by suppressing metaxylem and cambium cell proliferation in the FR.
Materials and methods
Plant materials and growth conditions
Sweetpotato (I. batatas cv. Yulmi) plants were propagated by cutting and planting apical stems bearing 2–3 leaves in large [27×27×24 (height) cm] pots containing commercial horticultural potting soil (Baroker; Seoul Bio, Chungcheongbuk-do, Korea) in the greenhouse at 25–30 °C under a long-day photoperiod (16/8h, light/dark). No additional fertilizer was added.
RNA gel blot analysis
Total RNA was extracted from various tissues at three different developmental stages [FR (diameter <0.2cm), YSR (diameter 0.5–1.0cm), and mature storage root (MSR; diameter >5.0cm) stages] using the modified method with guanidinium–SDS lysis buffer and the CsCl gradient method as described in You et al. (2003). Total RNA (25 µg) was denatured, electrophoresed, and then transferred onto nylon membranes (Tropilon-Plus; Tropix) using the downward alkaline capillary method. A biotin-labelled probe was prepared by PCR amplification of the full-length IbEXP1 cDNA with T3 and T7 primers. The PCR cycling conditions consisted of pre-denaturation at 95 °C for 5min, followed by 30 cycles of 30 s at 95 °C, 20 s at 58 °C, and 30 s at 72 °C using dNTP mixed with biotin-labelled dCTP (Invitrogen). The labelled probe was purified using a PCR purification kit (Qiagen) according to the manufacturer’s instructions. Hybridization, washing, and detection were performed as described previously (You et al., 2003).
Generation of IbEXP1-antisense sweetpotato plants
Embryogenic calli were induced from I. batatas (L.) Lam. cv. ‘Yulmi’ shoot apical meristems cultured on MS medium (Murashige and Skoog, 1962) supplemented with 1mg l–1 2,4-dichlorophenoxy acetic acid (2,4-D), 3% sucrose, and 0.4% gelite (MS1D), kept at 25 °C in the dark, and proliferated by subculture at 4 week intervals on the same fresh medium. The full-size IbEXP1 cDNA was amplified with IbEXP1-specific primers (Supplementary Fig. S1 available at JXB online) (forward: 5’-gatggtaccCATTCCTCTACCAATTCAACTGAA-3’; reverse: 5’-gatg gatccACTGTCTCCACACTCAGCATT-3’). KpnI and BamHI restriction sites were introduced at the ends of the forward and reverse primers, respectively, in order to facilitate subcloning. The PCR products were then subcloned into the pGEM-T Easy vector (Promega), digested with BamHI and KpnI restriction enzymes, and fused to the Caulifower mosaic virus (CaMV) 35S promoter in an antisense orientation by insertion of the IbEXP1 fragments at the BamHI and KpnI sites of the pMBP1 binary vector. The resulting construct was used to transform sweetpotato plants by particle bombardment. DNA-coated microparticles were prepared using the CaCl2/spermidine method as described by Kikkert (1993). A 1 µg aliquot of the plasmid DNA was mixed with 500 µg of gold particles (1.0 Micron Gold, Bio-Rad) in the presence of 1M spermidine and 16mM CaCl2. A biolistic gun device (PDS-1000/He; Bio-Rad) was used to deliver the plasmid-coated gold particles (500ng per bombardment) at the following parameters: the stopping screen was positioned 3cm below the rupture disk; target calli were positioned 6cm below the stopping screen; helium pressure was 1100 psi. The bombarded embryogenic calli were then kept on MS1D medium containing 100mg l–1 kanamycin (selection medium) at 25 °C in the dark and subcultured under the same conditions every 3 weeks for 4–5 months. Somatic embryos were induced by transferring kanamycin-resistant calli to hormone-free MS medium containing 100mg l–1 kanamycin. Regenerated plants were cultured on the same medium and maintained at 25 °C under a 16/8h (light/dark) photoperiod with light supplied by fluorescent tubes at a light intensity of 40 µmol m–2 s–1. The plantlets were transplanted into pots and grown in the greenhouse.
RT-PCR
Total RNA (5 µg) was used for the first-strand cDNA synthesis using the SuperScript III first-strand synthesis supermix (Invitrogen). The resulting cDNA solution was then diluted with 30 µl of TE (10mM TRIS-HCl, pH 8.0, 1mM EDTA). Primers for reverse transcription-PCR (RT-PCR) were IbEXP1-specific primers (5’-GGCCGGAATTCATCAAACACC-3’; 5’-ACTGTCTCCACACTCAGC-3’), IbEXP2-specific primers (5’-GA GTCTGAATTAAGGAAGAAGGGG-3’; 5’-GGCTACATTCTTGTGC AGTCAC-3’), and IbEXPL1-specific primers (5’-TCCTGATCATCAT CTCTGGCG-3’ and 5’-CAGTTTACTAGGTCGACATGGAATA-3’) (Supplementary Fig. S1 at JXB online). Sweetpotato β-tubulin DNA was amplified with primers (5’-CAACTACCAGCCACCAACTGT-3’ and 5’-CAGATCCTCACGAGCTTCAC-3’) as an internal equal loading control. A 1 µl aliquot of the cDNA reaction mixture and 10 pmol of each oligonucleotide primer were used in a total reaction volume of 20 µl. PCR amplification was performed with an initial denaturation for 5min at 95 °C, followed by 30 s at 94 °C, 30 s at 58 °C, and 30 s at 72 °C, and terminated with a 5min final extension at 72 °C. The numbers of cycles used for each amplification were: β-tubulin, 24 cycles; IbEXP1, 27 cycles; IbEXP2, 35 cycles; and IbEXPL1, 25 cycles. The amplified products were electrophoresed on a 1.2% agarose gel.
Microscopic observation
Apical meristems of sweetpotato plants bearing 2–3 leaves were grown on MS medium (Murashige and Skoog, 1962) at 25 °C for 10 d under long-day conditions (16/8h, light/dark). The thickest primary FR on each plant was collected, and transverse and longitudinal sections of the root were prepared from samples obtained at 5mm from the root tip. The samples were first fixed in 2% (w/v) paraformaldehyde and 2% (v/v) glutaraldehyde in 0.1M sodium phosphate buffer, pH 7.0, at 4 °C for 10 d. After fixation, the tissues were washed twice with 0.1M sodium phosphate buffer (42.3mM NaH2PO4 and 57.7mM Na2HPO4) for 5min, dehydrated stepwise for 30min in a graded series of ethanol (50, 60, 70, 80, 90, 95, and 100%), embedded in acrylic resin (LR White; London Resin Company) for 5 d, and cut into 1.0 µm thick sections on an ultramicrotome (BROMMA 2088; LKB).
For the histochemical analysis of lignin, the maturation zone prepared from the thickest FR of sweetpotato plants cultured in vitro for 3 weeks was used. Hand-prepared sections of fresh samples were treated with 0.01% (w/v) phloroglucinol in 95% ethanol for 10 s, washed with 50% (v/v) HCl for 10 s, and mounted in 5 N HCl. The stained sections were observed by bright-field microscopy (BX51; OLYMPUS).
Hormone treatment
Sweetpotato plantlets bearing a single leaf and petiole (single-leaf plantlets) were collected from sweetpotato plants and incubated in flasks containing distilled water for 3 weeks. After FRs had developed from the distal end of the petiole, the single-leaf plantlets were incubated in various concentrations of IAA, JA, and 6-benzylaminopurine (BA) at 25 °C in the dark for 3h. After the hormone treatment, total RNA was extracted from the FRs using the RNeasy Plant Mini Kit (Qiagen) and used for real-time PCR.
qRT-PCR
The first-strand cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis kit (Roche) according to the manufacturer’s instructions. The primers for real-time PCR were as follows: SRD1-specific primers (5’-AGAGGAGAAATGGGTTGTTTA-3’; 5’-GTGCACGAA ACTCCCCTT-3’) (Noh et al., 2010), IbEXP1-specific primers used in the RT-PCR, and CAD primers (5’-ATTTGTCATCGACGTTGGCAAA-3’; 5’- AGGACATTATTACATTACACACTCATTATTATTATTG-3’). The real-time PCR analysis was performed using the LightCycler® 480 quantification system (Roche Diagnostics) as described in Noh et al. (2010). Expression levels were normalized with β-tubulin expression amplified with the β-tubulin-specific primers used in the RT-PCR. The real-time PCR mixture was prepared with 1× KAPA SYBR® FAST Master mix (KAPABIOSYSTEMS) in a final reaction volume of 16 µl. For the SRD1, IbEXP1, and CAD genes, the PCR was performed by subjecting the samples to an initial denaturation at 95 °C for 10min followed by 45 cycles of 95 °C for 10 s, 58 °C for 20 s, and 72 °C for 30 s. The melting curve and cooling were performed under the same conditions as those described by Noh et al. (2010).
Results
Transcript level of IbEXP1 was down-regulated during SR development
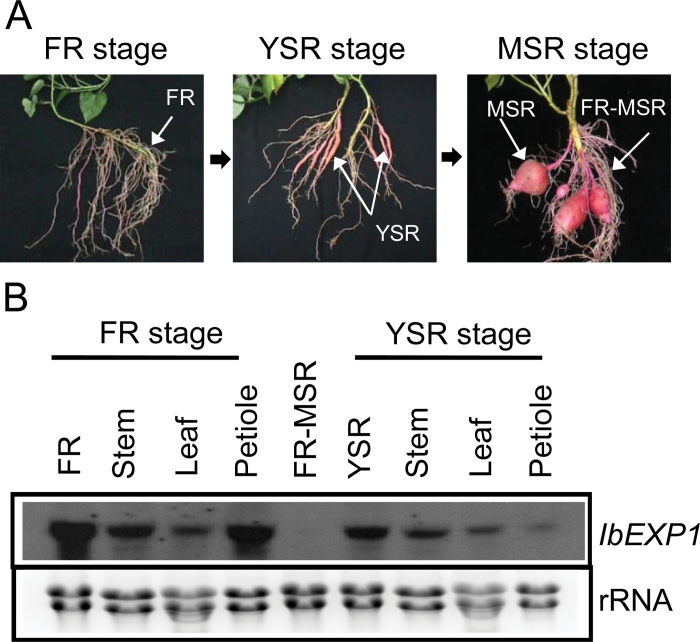
With the aim of elucidating the functional roles of expansin genes in SR development, IbEXP1 was further characterized. Transcriptional regulation of IbEXP1 during SR development was investigated in detail by an RNA gel blot hybridization analysis of RNAs extracted from various tissues (root, stem, leaf, and petiole) at three different root developmental stages, namely, the FR, YSR, and MSR stage, respectively (Fig. 1). Due to relatively low nucleotide sequence identity between IbEXP1 and two other sweetpotato expansin genes (~46% with IbEXP2 and 33% with IbEXPL1) (Noh et al., 2009), the full-length cDNA fragment (1213bp) was used as a probe. At the FR stage, high-level expression of the IbEXP1 transcript was found in the FR and petiole, and relatively low-level expression was found in the stem and leaf. In comparison, at the YSR stage, the expression level of the IbEXP1 transcript had clearly decreased in the YSR, stem, leaf, and petiole, with the extent of the decrease being remarkable in the YSR and petiole. IbEXP1 mRNA was not detected at all in the FR at the MSR stage. These results indicate that the expression of IbEXP1 is transcriptionally down-regulated during SR development.
Fig. 1.
Expression pattern of IbEXP1. (A) Developmental stages of sweetpotato. (B) RNA gel blot analysis of IbEXP1. Full-length IbEXP1 cDNA was used as a probe. The ethidium bromide-stained rRNA is shown as a loading control (lower panel). FR, fibrous root (diameter <0.2cm); YSR, young storage root (diameter 0.5–1.0cm); MSR, mature storage root (diameter >5cm); FR-MSR, fibrous root from mature storage root stage. (This figure is available in colour at JXB online.)
Transcriptional regulation of IbEXP1 in response to exogenous hormones
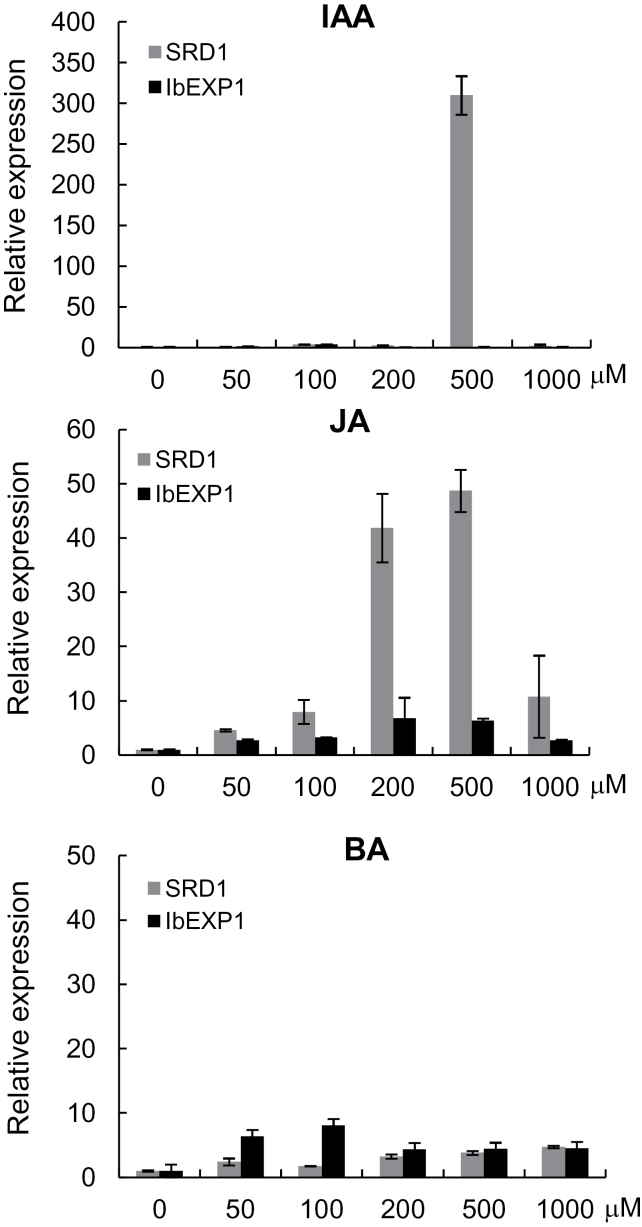
Various authors have suggested that growth hormones, including auxins, JA, and cytokinins, play a role in the formation and thickening growth of SRs (Akita et al., 1962; Matsuo, 1983; Nakatani and Komeichi, 1992; Koda, 1997; McGreger, 2006). To test this notion, the possible transcriptional regulation of IbEXP1 in response to exogenous IAA, JA, and BA was investigated by incubating sweetpotato plantlets bearing a single leaf and petiole (single-leaf plantlets) for 3h in a solution containing various concentrations (0, 50, 100, 200, 500, and 1000 µM) of IAA, JA, or BA (Fig. 2). Total RNAs were then extracted from the FRs of the hormone-treated plants and subjected to analysis by real-time RT-PCR using IbEXP1-specific primers. SRD1 was found to be involved in the initial thickening growth of the SR, and its transcript level increased following treatment with exogenous and endogenous IAA (Noh et al., 2010). The transcriptional regulation of IbEXP1 by IAA, JA, and BA, respectively, was compared with that of SRD1 by evaluating the transcriptional regulation of SRD1 in response to these three hormones using SRD1-specific primers. The transcript level of IbEXP1 remained almost unchanged in response to exogenously applied IAA, although the mRNA level of SRD1 increased remarkably at 500 µM IAA (~300-fold). The transcript levels of IbEXP1 increased gradually in response to increasing concentrations of JA, reaching a maximum transcript level (~10-fold) at 200 µM JA, but the extent of the increase in IbEXP1 transcript level was relatively low compared with the robust increase in SRD1 transcript level (~40- to 50-fold) at 200 µM and 500 µM JA. The transcript levels of both SRD1 and IbEXP1 increased in response to BA treatment, but the sensitivity of these two genes to BA treatment was clearly different in that the maximum increase in their respective transcript levels was detected at different concentrations of BA, with IbEXP1 transcript levels reaching a maximum at 100 µM BA and those of SRD1 at 500 µM BA. These results indicate that IbEXP1 expression is nearly insensitive to IAA treatment and relatively weakly regulated by JA. Thus, transcriptional regulation of IbEXP1 in response to IAA and JA differed from that of SRD1, suggesting that the role of IbEXP1 in SR development is different from that of SRD1.
Fig. 2.
Effect of various hormones on the expression of IbEXP1. Transcript levels of IbEXP1 in response to treatment with various concentrations of exogenous IAA, JA, or BA for 3h. Total RNA was extracted from the fibrous roots treated with the three hormones. Real-time RT-PCR data were normalized to those for the endogenous β-tubulin gene. Error bars indicate the standard deviation (SD) between three technical replicates measured on fibrous roots collected from at least three different sweetpotato plantlets and subsequently pooled for analysis.
IbEXP1-antisense sweetpotato plants showed an earlier thickening growth in FRs
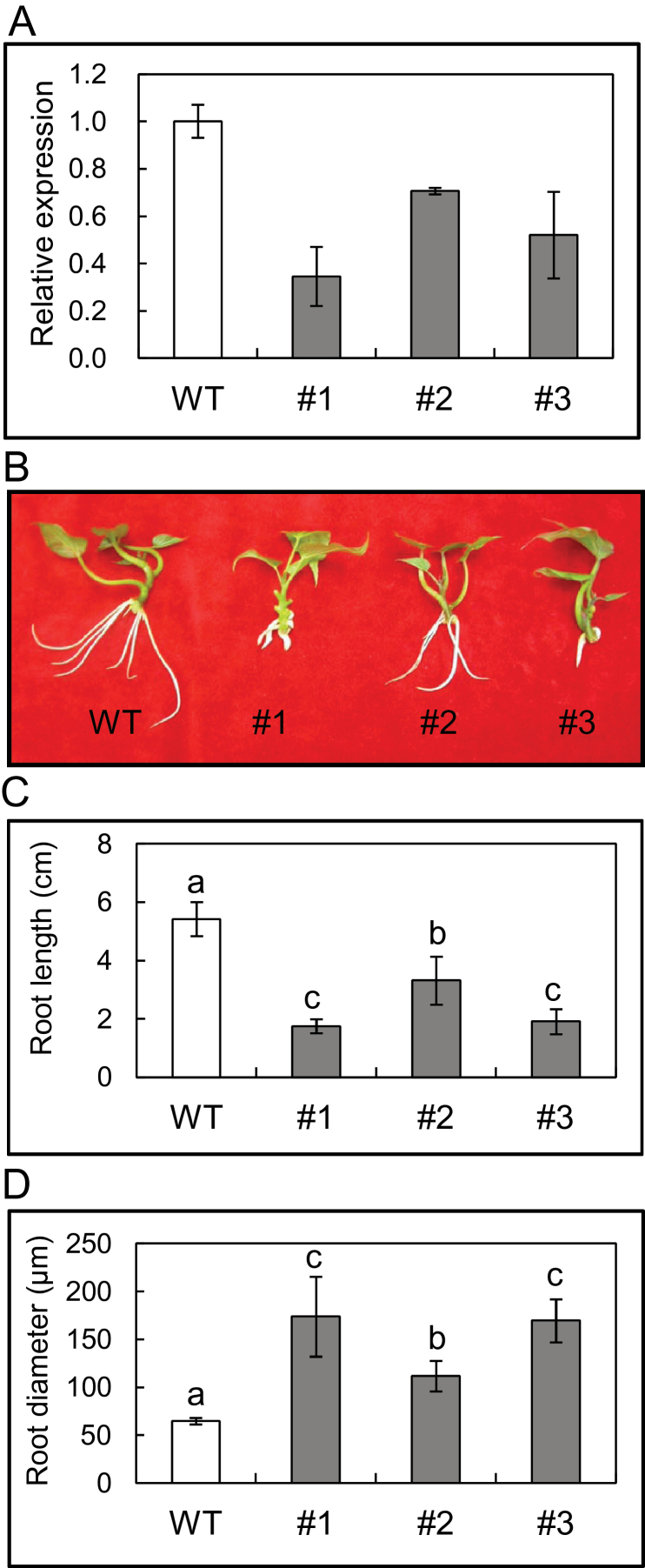
To ascertain the effect of IbEXP1 down-regulation on SR development, IbEXP1-antisense sweetpotato plants with a full-length IbEXP1 fragment under the control of the CaMV35S promoter were generated. Three independent IbEXP1-antisense transgenic lines (#1, #2, and #3) were obtained by in vitro culture. To identify the stable phenotype and expression of IbEXP1 in these transgenic lines, the regenerated IbEXP1-antisense plants of each of these lines were propagated by cutting and planting three times in vitro prior to characterization. Quantitative RT-PCR analysis using IbEXP1-specific primers revealed that IbEXP1 transcript levels in the FRs of IbEXP1-antisense plants of lines #1, #3, and #2 decreased to 35±12, 52±18, and 71±1%, respectively, of that of the wild-type (WT) plants (Fig. 3A). These three IbEXP1-antisense lines were therefore used in subsequent analyses.
Fig. 3.
Characterization of IbEXP1-antisense sweetpotato plants. (A) Transcript levels of IbEXP1 in IbEXP1-antisense plants. Real-time RT-PCR analysis was carried out with total RNAs extracted from fibrous roots grown in vitro. Data were normalized to those for the endogenous β-tubulin gene. Error bars indicate the standard deviation (SD) between three technical replicates measured on fibrous roots collected from at least three different sweetpotato plantlets. (B) Morphology of the IbEXP1-antisense plants. Pictures were taken at 10 d after planting. (C) Root length of IbEXP1-antisense plants. Root length was measured with the three longest roots of each plant. (D) Root diameter of IbEXP1-antisense plants. Root diameter was measured with a dial caliper by measuring the thickest root of each plant. (C and D) Data were collected from sweetpotato plants cultured in vitro for 10 d after planting and are the means ±SD from three separate measurements of three individual plants. Different letters above the bars indicate significantly different means (P < 0.05) as analysed by Duncan’s multiple range test using the SAS statistical program (SAS Institute, Cary, NC, USA). (A–D) Numbers #1-#3 represent IbEXP1-antisense sweetpotato lines #1-#3, respectively. WT, wild type. (This figure is available in colour at JXB online.)
Cuttings of IbEXP1-antisense plants #1, #2, and #3 bearing the apical meristem and 2–3 developing leaves were planted on hormone-free MS medium. After 10 d of growth, the plantlets were examined for any alteration in root morphogenesis. No change in phenotype was observed in the aerial parts of the IbEXP1-antisense plants, but the FRs were shorter and thicker than those of the WT plants (Fig. 3B). Specifically, in the three IbEXP1-antisense lines, the length of the FRs was significantly reduced (Fig. 3C), and the diameter of the FRs had increased markedly in #1 and #3, and moderately in #2 (Fig. 3D). The extent of the reduction in root length and of the increase in root diameter was more severe in IbEXP1-antisense plants #1 and #3 than plants #2, suggesting that the observed root phenotype was attributable to a decrease in IbEXP1 transcript levels.
Three expansin genes (IbEXP1, IbEXP2, and IbEXPL1) are expressed in the YSR of sweetpotato (You et al., 2003; Noh et al., 2009). To verify that the root phenotype of IbEXP1-antisense plants was caused by the observed decrease in IbEXP1 transcript level—and not due to changes in the transcript levels of the other two expansin genes (IbEXP2 and IbEXPL1)— the transcript levels of IbEXP2 and IbEXPL1 in IbEXP1-antisense plants were determined using RNAs extracted from the FRs of IbEXP1-antisense plants #1, #2, and #3 and IbEXP2- and IbEXPL1-specific primers. The results of the RT-PCR analysis on the extracted RNAs demonstrated that the transcript levels of IbEXP2 and IbEXPL1 had remained unchanged even though the IbEXP1 mRNA level was significantly reduced in IbEXP1-antisense plants #1, #2, and #3 (Supplementary Fig. S2 at JXB online). This result suggests that mRNAs of antisense IbEXP1 specifically reduced the transcript level of IbEXP1 but did not affect the transcript levels of IbEXP2 and IbEXPL1.
Cell proliferation was enhanced in FRs of IbEXP1-antisense sweetpotato plants
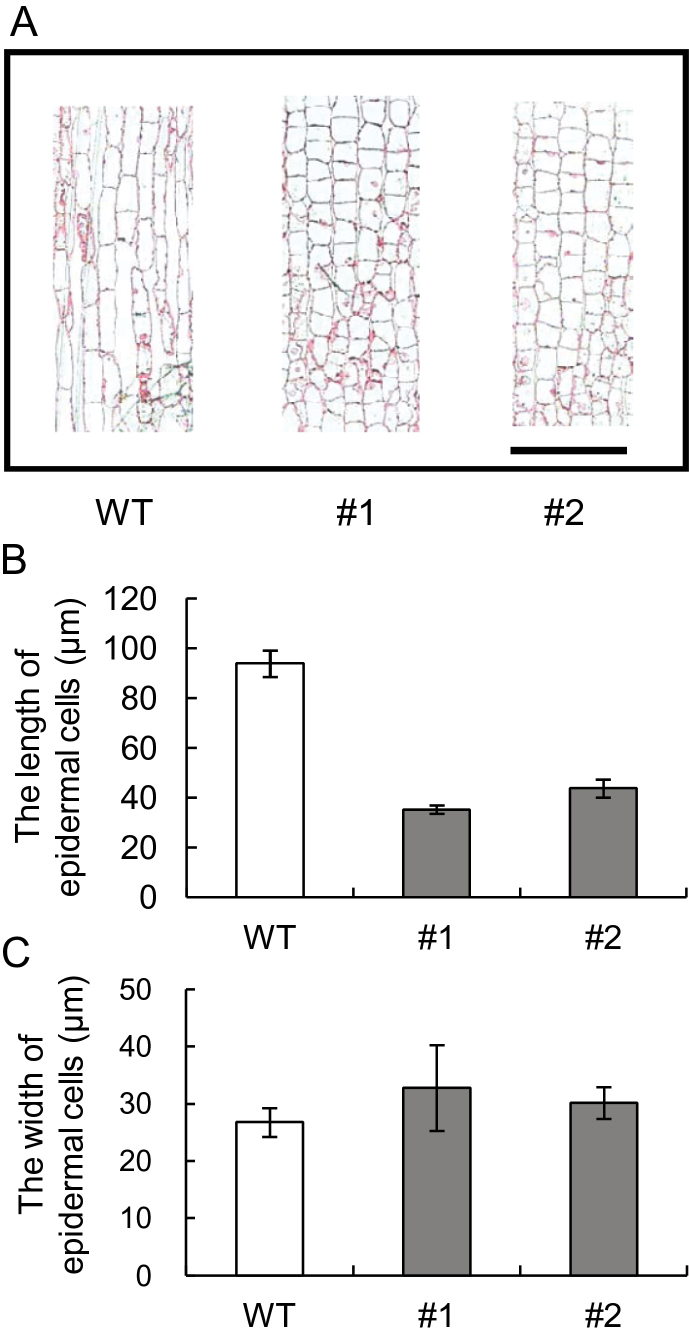
In order to study the root morphology phenotype at the cellular level, longitudinal and transverse sections of the maturation zone (collected at 5mm from the root tip) were prepared from the thickest FR of the WT and IbEXP1-antisense plants cultured in vitro. Cells from IbEXP1-antisense lines #1 and #2, representing the strong and moderate root phenotype, respectively, were then characterized phenotypically. Elongation growth of the epidermal cells was distinctly different between the FRs of WT and IbEXP1-antisense plants (Fig. 4A), with the length of the epidermal cells in the FRs of IbEXP1-antisense plants of lines #1 and #2 being 38% and 47%, respectively, of that of WT plants (Fig. 4B). The extent of this reduction almost accounts for the decreased length of FRs from IbEXP1-antisense plants (32% and 61% of that of WT plants in lines #1 and #2, respectively), suggesting that the decreased length of FRs in IbEXP1-antisense plants was attributable to the reduced elongation growth of the epidermal cells. There was no significant difference in the width of the epidermal cells of WT and IbEXP1-antisense plants (Fig. 4C).
Fig. 4.
Effect of down-regulation of IbEXP1 on the growth of epidermal cells in fibrous roots. (A) Longitudinal sections of epidermis were prepared from fibrous roots of sweetpotato plants cultured in vitro for 10 d after planting. The scale bar represents 50 µm. (B) The length of epidermal cells of the fibrous root. (C) The width of epidermal cells of the fibrous root. (B and C) Between 15 and 30 cells of at least three individual plants were measured. Data are the means ±standard deviation (SD) from three separate experiments. (A–C) Numbers #1 and #2 represent IbEXP1-antisense sweetpotato lines #1 and #2, respectively. WT, wild type. (This figure is available in colour at JXB online.)
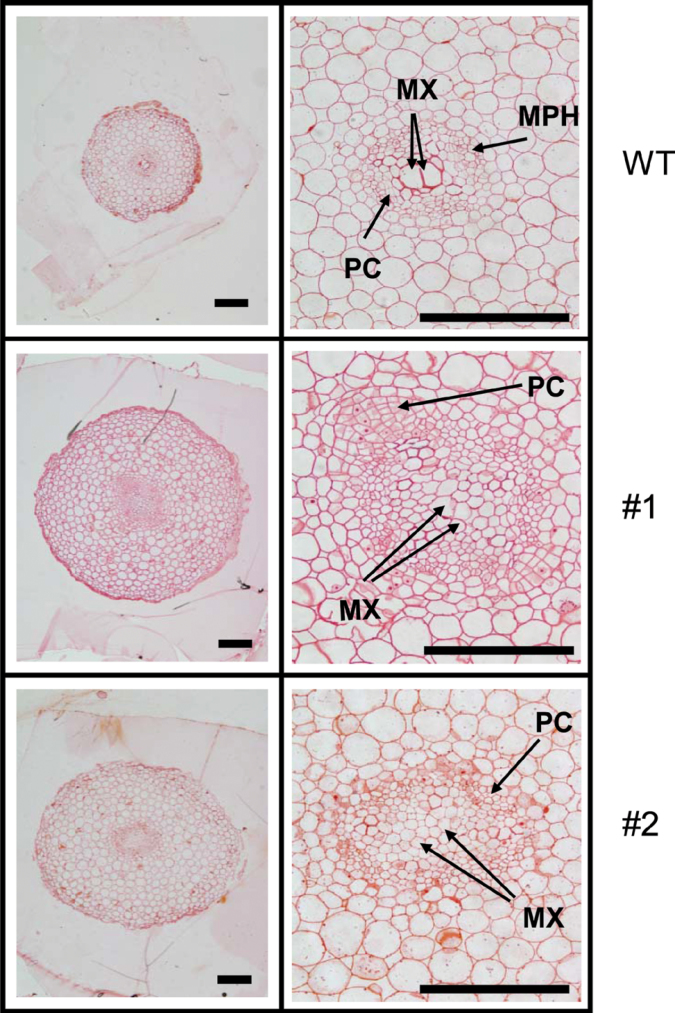
As vigorous proliferation of cambium and metaxylem cells has been shown to be a morphological marker of the initial thickening growth process leading to SR development in sweetpotato (Noh et al., 2010), cell proliferation in the stele was examined in more detail. The cellular structure of the FR of IbEXP1-antisense plants was compared with that of WT plants (Fig. 5). WT plants showed the typical anatomy of FRs, with a relatively small root stele in which metaxylem and metaphloem had recently differentiated. Cell proliferation was, however, significantly enhanced in the IbEXP1-antisense plants, resulting in an increased number of cells in the stele compared with the WT (Table 1). Both metaxylem and cambium cell proliferation were markedly elevated in IbEXP1-antisense plants, with a 2.0- to 2.4-fold increase in the number of metaxylem cells and a 1.4- to 2.0-fold increase in the number of cambium cells relative to WT plants. These results suggest that the enhanced proliferation activity in metaxylem and cambium cells resulted in an earlier thickening growth in the FRs of IbEXP1-antisense plants. In addition, based on differences in the numbers of metaxylem and cambium cells, proliferation activity was higher in IbEXP1-antisense plants #1 than #2, indicating that the proliferation activity was positively correlated with the extent of reduction in IbEXP1 mRNA levels. The increased proliferation activity was therefore attributed to the reduction in IbEXP1 transcripts.
Fig. 5.
Effect of down-regulation of IbEXP1 on cell proliferation in fibrous roots. Transverse sections of fibrous roots were prepared from in vitro cultured sweetpotato plants at 10 d after planting. The right panel is an enlarged image of the left panel. Numbers #1 and #2 represent IbEXP1-antisense sweetpotato lines #1 and #2, respectively. Scale bars: 200 µm. MPH, metaphloem; MX, metaxylem; PC, primary cambium; WT, wild type. (This figure is available in colour at JXB online.)
Table 1.
Effect of IbEXP1 down-regulation on cell proliferation activities in the fibrous root Data are means ±SD from three separate measurements of three sections from three individual plants and are collected from the in vitro cultured sweetpotato plants at 10 d after planting.
| Cell type | Wild type | IbEXP1-antisense #1 | IbEXP1-antisense #2 | ||
|---|---|---|---|---|---|
| No. of cellsa | No. of cells | Rate (fold)b | No. of cells | Rate (fold) | |
| Cambium | 46±6 | 94±5 | (2.0) | 64±7 | (1.4) |
| Metaxylemc | 30±4 | 73±2 | (2.4) | 60±1 | (2.0) |
a Counted on transverse sections of fibrous roots.
b Proliferation rate relative to that of wild-type plants.
c Numbers of metaxylem cells, including mature and immature metaxylem cells.
Lignification was reduced in the FRs of IbEXP1-antisense sweetpotato plants
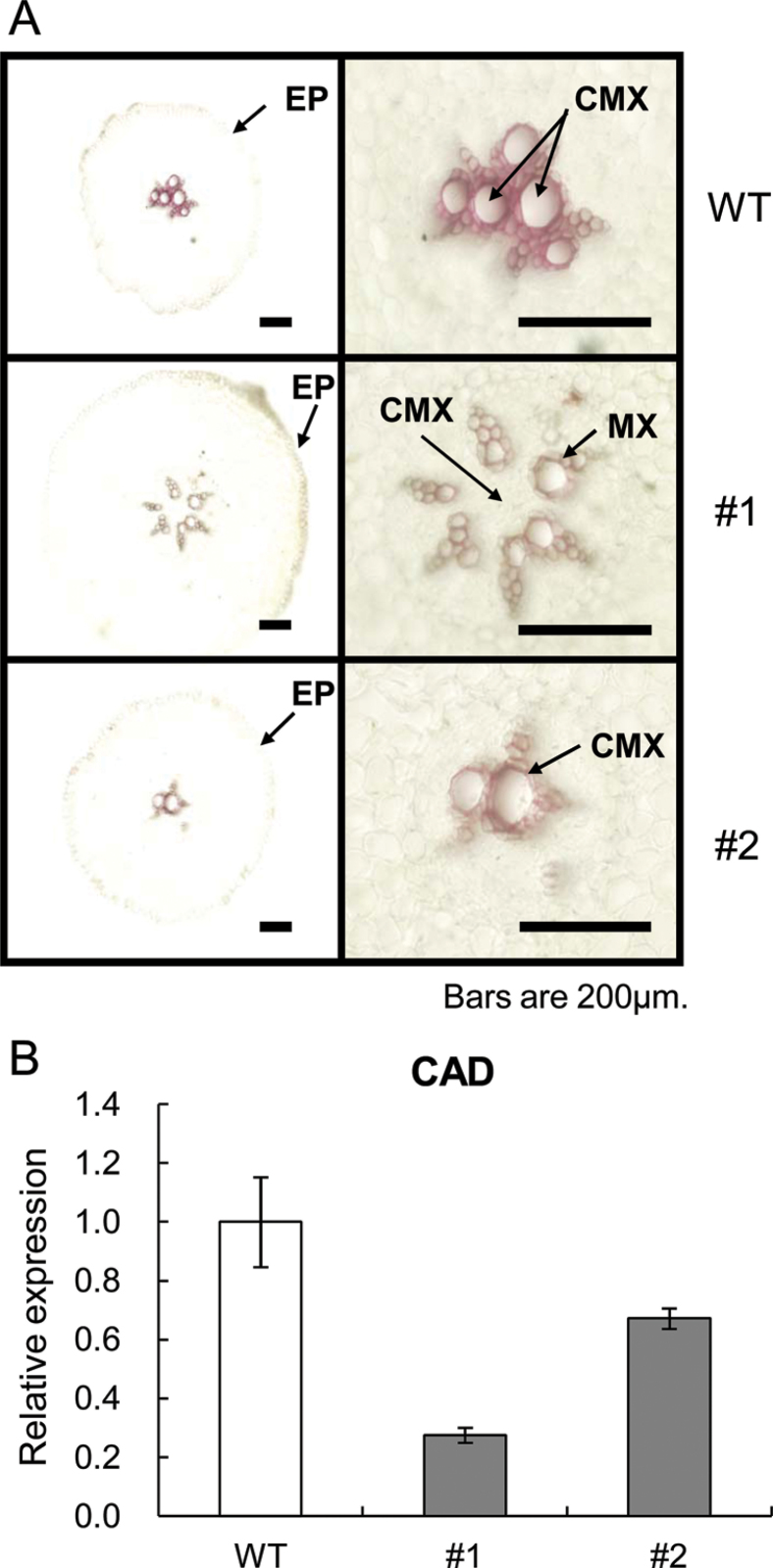
It has been suggested that lignification in the steles of FRs and TRs prevents SR development (Togari, 1950; Wilson and Lowe, 1973; Firon et al., 2009). In addition, Belehu et al. (2004) suggested that the presence of lignified central metaxylem cells is a typical structure of FRs and TRs that lack the potential to develop into SRs. The degree of lignification in the stele of FRs from WT and IbEXP1-antisense sweetpotato plants was therefore examined. Lignification in the FRs was detected by preparing transverse sections of the maturation zone from the thickest FR of WT and IbEXP1-antisense plants cultured in vitro for 3 weeks, staining these with phloroglucinol, and observing the staining intensity microscopically (Fig. 6A). The central metaxylem cells were deeply stained in the FR of WT plants but remained unstained in the hexarch stele of the FR from IbEXP1-antisense plants of line #1. The central metaxylem cells in the stele of FRs from IbEXP1-antisense plants of line #2 were faintly stained, but the lignification level of the metaxylem cells from this line was much lower than that of WT plants. These results indicate that stele lignification was significantly reduced in the FRs of IbEXP1-antisense plants.
Fig. 6.
Altered lignification in the fibrous roots from IbEXP1-antisense sweetpotato plants. (A) Histochemical analysis of lignin deposition in the fibrous roots from IbEXP1-antisense plants. Transverse hand-prepared sections of the fibrous roots from the wild-type (WT) and IbEXP1-antisense plants (#1 and #2) cultured in vitro for 3 weeks were stained with phloroglucinol-HCl. The stele areas of the FRs in the left panel are enlarged in the right panel. Lignified material is stained violet. EP, epidermis; CMX, central metaxylem; MX, metaxylem. (B) Transcript levels of the cinnamyl alcohol dehydrogenase (CAD) gene in IbEXP1-antisense plants. Total RNAs were extracted from fibrous roots of WT and IbEXP1-antisense plants (#1 and #2) cultured in vitro for 3 weeks. Real-time RT-PCR data were normalized to those for the endogenous β-tubulin gene. Error bars indicate the standard deviation (SD) between three technical replicates measured on fibrous roots collected from at least three different sweetpotato plants and subsequently pooled for analysis. (This figure is available in colour at JXB online.)
Cinnamyl alcohol dehydrogenase (CAD) is a key enzyme in the biosynthesis of lignin (Walter et al., 1988; Goujon et al., 2003). Quantitative RT-PCR analysis performed to determine the transcript level of the CAD gene in these transgenic mutants revealed that the transcript levels of CAD were moderately decreased in IbEXP1-antisense plants of line #2 and markedly decreased in those of line #1, as compared with WT plants (Fig. 6B), suggesting that down-regulation of IbEXP1 expression led to a decrease in the transcript abundance of the CAD gene and, consequently, to a reduction in the extent of lignification. This reduced level of stele lignification in the IbEXP1-antisense plants suggests that the FRs of IbEXP1-antisense plants have a higher potential to develop into SRs than those of WT plants. The reduction in stele lignification and CAD transcript abundance was more severe in IbEXP1-antisense plants of line #1 than in those of line #2, indicating that the reduction level was positively correlated with the extent of reduction in IbEXP1 mRNA levels and suggesting that the reduced lignification in these mutants is attributable to the decreased levels of IbEXP1 mRNA accumulation.
IbEXP1-antisense sweetpotato plants produced a higher number of SRs
IbEXP1-antisense plants were transferred to the soil, propagated by cutting and planting three times in the greenhouse, and then examined for growth and phenotypic characteristics. At 6 weeks after planting, compared with WT plants, the aerial parts of IbEXP1-antisense plants grew less vigorously, their stems, petioles, and internodes were shorter, and their leaves were smaller (Supplementary Fig. S3A at JXB online). There were also distinct differences between the WT and the IbEXP1-antisense plants at 6 weeks of growth in terms of root morphology (Supplementary Fig. S3B). The most prominent phenotypic difference was that a higher number of primary roots had already begun thickening growth in the IbEXP1-antisense plants, whereas most of the primary roots in the WT plants had not yet initiated the thickening growth process, resulting in a higher ratio of TRs to total primary roots (TR/PR) in IbEXP1-antisense plants than in the WT (Supplementary Fig. S3C). These TRs in the IbEXP1-antisense plants were observed to develop into SRs at 8 weeks after planting (Supplementary Fig. S3B). These results indicate that the initiation of thickening growth was accelerated in FRs from IbEXP1-antisense plants.
At 10 weeks after planting, the initial growth retardation observed in the leaf and petiole had almost disappeared, but the elongation growth of the stems differed markedly between the WT and IbEXP1-antisense plants (Supplementary Fig. S3D, E at JXB online). In WT plants, the primary stem showed vigorous elongation, but the growth of additional stems was not observed. In contrast, elongation growth of the primary stem was reduced in IbEXP1-antisense plants, resulting in a much shorter primary stem than that formed in WT plants, as well as the production of multiple stems showing similar levels of elongation growth. Consequently, the WT produced a long vine-like stem, while IbEXP1-antisense plants acquired a bush form with multiple stems.
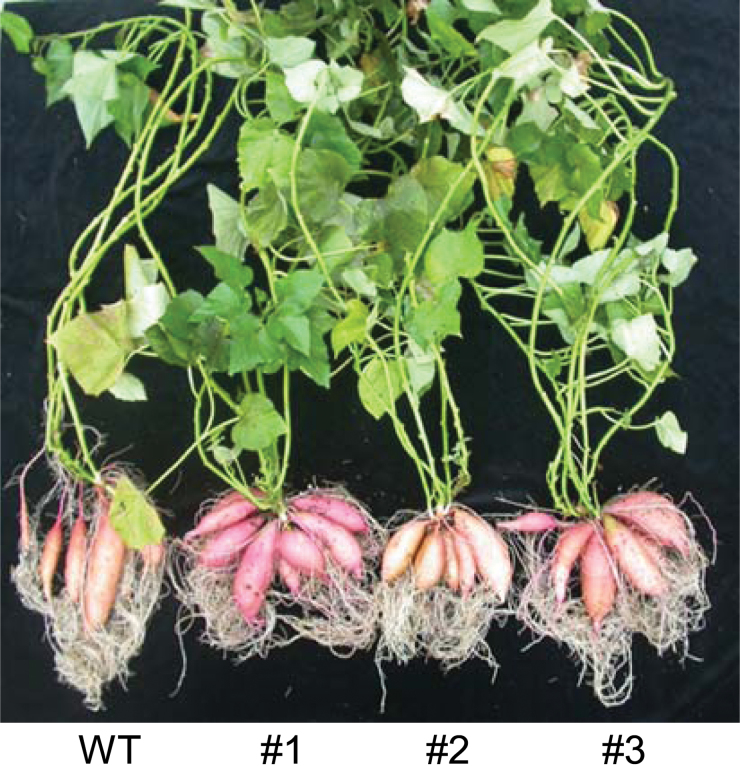
After 5 months of cultivation, WT plants and IbEXP1-antisense plants #1, #2, and #3 were harvested and the growth of both the SR and shoot was examined (Fig. 7, Table 2; Supplementary Fig. S4 at JXB online). Relative to the WT plants, IbEXP1-antisense plants produced a higher number of stems and delayed leaf senescence. Leaf yellowing, which is due to chlorophyll loss, and subsequent leaf loss are typical symptoms of age-dependent senescence. At 5 months post-planting, most of the leaves of the WT plants had fallen off the plants, while those of the IbEXP1-antisense plants were still attached to the stems and had remained green (Supplementary Fig. S4). Consequently, the total shoot weight per IbEXP1-antisense plant was greater than that per WT plant (Table 2). SR development was also altered in IbEXP1-antisense plants, with IbEXP1-antisense plants #1, #3, and #2 producing 7.3±1.5, 7.0±0.6, and 6.0±0.6 SRs, respectively, in comparison with the 4.5±2.4 SRs produced by the WT plants (Fig. 7, Table 2). This elevated number of SRs in each IbEXP1-antisense plant resulted in a significant increase in the total weight of SRs in comparison with that of each WT plant, namely, 393.8±35g (IbEXP1-antisense plant #1), 359.7±44.2g (IbEXP1-antisense plant #3), and 299.8±42.4g (IbEXP1-antisense plant #2) versus 226.1±19.1g (WT plant) (Table 2). The extent of the increase in SR number and total SR weight was greater in IbEXP1-antisense plants #1 and #3 than in IbEXP1-antisense plants #2, suggesting that the observed phenotype in SR development was attributable to a decrease in IbEXP1 transcript level. Another noteworthy phenotype was the difference in level of uniformity in the size of the SRs between WT and IbEXP1-antisense plants. SRs from IbEXP1-antisense plants were relatively uniform in size, while those of WT plants varied in size. These results indicate that although the shoot growth of IbEXP1-antisense plants was retarded during the early developmental stage, the final yield in terms of both SRs and shoots was elevated in IbEXP1-antisense plants.
Fig. 7.
Phenotypic alteration in IbEXP1-antisense sweetpotato plants grown in soil. Sweetpotato plants were grown in soil and harvested at 5 months after planting. Numbers #1–#3 represent IbEXP1-antisense sweetpotato lines #1–#3, respectively. WT, wild type. (This figure is available in colour at JXB online.)
Table 2.
Effect of IbEXP1 down-regulation on storage root and shoot developmentData are means ±SD from three separate measurements of three individual plants grown in the soil.
| Measurementa | Wild type | IbEXP1-antisense #1 | Rateb | IbEXP1-antisense #2 | Rate | IbEXP1-antisense #3 | Rate |
|---|---|---|---|---|---|---|---|
| No. of SRsc | 4.5±2.4 | 7.3±1.5 | (1.6) | 6.0±0.6 | (1.3) | 7.0±0.6 | (1.6) |
| Total weight of SRs (g)d | 226.1±19.1 | 393.8±35.0 | (1.7) | 299.8±42.4 | (1.3) | 359.7±44.2 | (1.6) |
| No. of stems | 3.3±0.5 | 6.3±1.2 | (1.9) | 4.7±1.2 | (1.4) | 5.5±0.7 | (1.7) |
| Total weight of shoots (g)d | 86.4±23.7 | 191.1±16.8 | (2.2) | 108.4±41 | (1.3) | 150.67±23.7 | (1.7) |
a Measured at 5 months after planting.
b Increasing rates (fold) relative to that of wild-type plants.
c Number of storage roots
d Total weight of storage roots or shoots per plant.
SR development was accelerated in IbEXP1-antisense sweetpotato plants
Sweetpotato plants initially produce FRs from underground stems. These FRs undergo elongation growth, followed by thickening growth at certain positions that leads to the development of SRs. Thus, in the FR, there is always a certain distance between the proximal end of the SR and the underground stem.
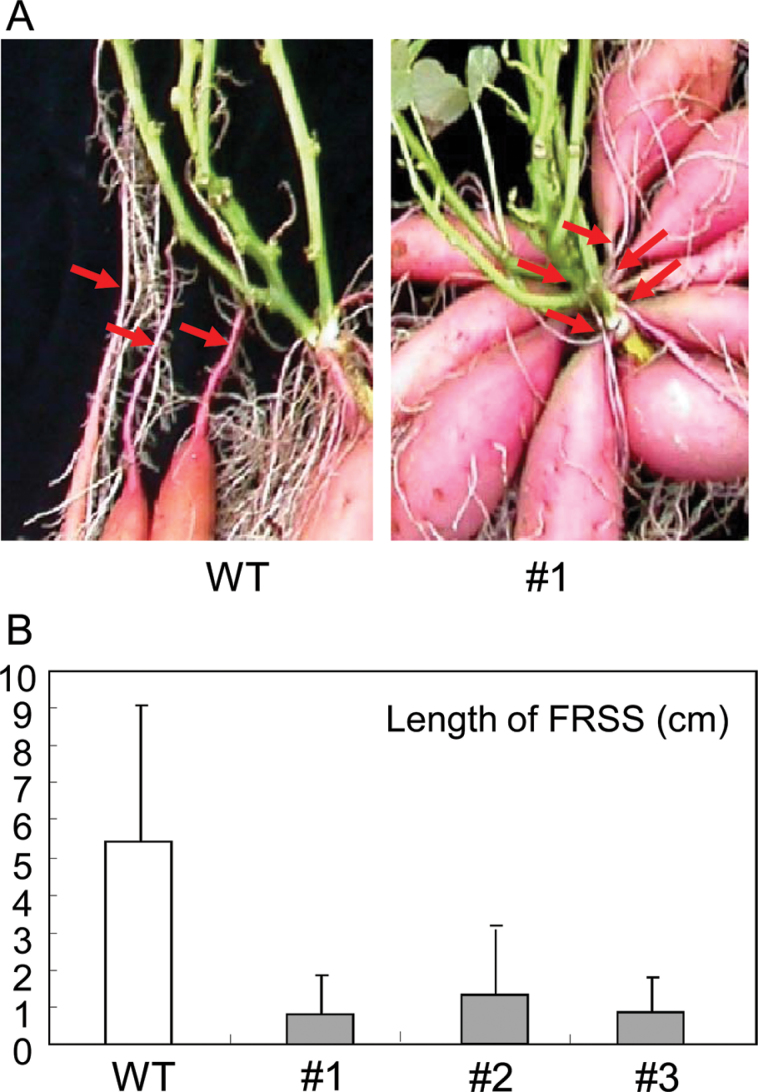
The length of the FRSS [fibrous root between storage root and underground stem (red arrows in Fig. 8A)] decreased markedly in IbEXP1-antisense plants in comparison with WT plants, being <1.5cm in the former and >5.0cm in the latter (Fig. 8B). Consequently, in contrast to the random positions of SRs on FRs in WT plants, all of the SRs in IbEXP1-antisense plants were almost directly attached to the underground stem, indicating that the formation of SRs took placed at identical sites, namely the proximal end of the FR. This result indicates that the thickening growth of the SRs commenced right from the initial stage of FR growth in IbEXP1-antisense sweetpotato plants, further suggesting that SR development was accelerated in IbEXP1-antisense sweetpotato plants.
Fig. 8.
Storage root development in IbEXP1-antisense sweetpotato plants. (A) Site of storage root development. Red arrows represent the FRSS [fibrous root between storage root and underground stem]. (B) Length of the FRSS. Data were collected from the sweetpotato plants grown in soil at 5 months after planting and are the means ±standard deviation (SD) from three separate measurements of three individual plants. (A and B) Numbers #1–#3 represent IbEXP1-antisense sweetpotato lines #1–#3, respectively. WT, wild type. (This figure is available in colour at JXB online.)
Discussion
The expression levels of expansin genes have been found to be strongly correlated with elongation growth in various plant organs (Wu et al., 1996; Lee and Kende, 2002; Choi et al., 2003; Gookin et al., 2003; Lee et al., 2003; Zenoni et al., 2004). Similar to expansin genes in other plants, in the present study, the IbEXP1 transcript level was also found to be high in actively growing tissues, such as the FR and petiole tissues at the FR stage; however, this high transcript level decreased markedly at the YSR stage (Fig. 1). This type of expression pattern suggests the possibility that IbEXP1 plays a negative role in SR development.
It has been suggested that cytokinin and auxin (IAA) have some function during the onset and subsequent primary thickening growth of SRs (Akita et al., 1962; McDavid and Alamu, 1980; Matsuo, 1983; Nakatani and Komeichi, 1992; McGreger, 2006). This hypothesis is supported by recent work in our laboratory demonstrating that SRD1 activates the initial thickening growth of the SR and that its transcript level increases in response to exogenous and endogenous treatment with IAA (Noh et al., 2010). JA has been shown to inhibit root elongation (Staswick et al., 1992; Feys et al., 1994). In sweetpotato, the initiation of SR formation is accompanied by the cessation of root elongation (Kim et al., 2002), suggesting the possible involvement of JA in SR formation (Firon et al., 2009). Based on the transcriptome analysis of the sweetpotato SR, McGregor et al. (2006) also suggested a possible involvement of JA in SR development. In the study reported here, it was found (i) that IAA had almost no effect on regulating the transcript level of IbEXP1 although it did markedly increase that of SRD1; and (ii) that transcriptional induction of IbEXP1 by JA was much weaker than that of SRD1 (Fig. 2). These results suggest a possible negative role for IbEXP1 in SR formation.
IbEXP1-antisense sweetpotato plants cultured in vitro produced shorter FRs than WT plants (Fig. 3), and microscopic observation revealed that the reduced length of FRs could be attributed to the limited elongation growth of epidermal cells (Fig. 4). This observation implies that IbEXP1 plays a role in the elongation growth of epidermal cells in the sweetpotato FR, a role that is supported by the expression patterns of other expansin genes strongly correlating with elongation growth in the root, internode, flower, and leaf (Wu et al., 1996; Cho and Cosgrove, 2000; Lee and Kende, 2002; Choi et al., 2003; Gookin et al., 2003; Lee et al., 2003; Zenoni et al., 2004; Sloan et al., 2009; Harada et al., 2011), and also consistent with the roles of other expansin genes involved in the elongation growth of root hairs (Lin et al., 2011; ZhiMing et al., 2011). In addition, the FRs of IbEXP1-antisense sweetpotato plants cultured in vitro were thicker than those of the WT plants (Fig. 3), and there were significantly more metaxylem and cambium cells in IbEXP1-antisense plants than in WT plants (Fig. 5, Table 1), leading to an earlier thickening growth in the FR. The roots of sweetpotato have been classified into three distinct root types according to their diameters: FR (diameter <2mm), TR (diameter 2–5mm), and SR (diameter >5mm) (Kokubu, 1973; Wilson and Lowe, 1973). The process of SR formation in sweetpotato begins with the formation of colourless FRs, followed by the pigmentation and primary thickening growth of some of these FRs, ultimately leading to the development of SRs by secondary thickening growth. Based on this classification by root type, the formation of the TR from the FR is a prerequisite in the process of SR development, and the proliferation of cambium and metaxylem cells has been determined to be a morphological characteristic of the TR and SR (Noh et al., 2010). Thus, the earlier thickening growth shown by the IbEXP1-antisense plants implies that the potential to develop SRs is enhanced in the FRs of IbEXP1-antisense plants. Moreover, earlier studies have shown that stele lignification in the FRs and TRs in sweetpotato inhibit the thickening growth into SRs (Togari, 1950; Wilson and Lowe, 1973; Belehu et al., 2004). Morphological and anatomical studies of sweetpotato roots have demonstrated that those roots without lignified central metaxylem cells have the potential to develop into SRs, whereas those with lignified central metaxylem cells do not (Wilson and Lowe, 1973; Belehu et al., 2004). A significant reduction was observed in the lignification of the stele, especially that of the central metaxylem cells, of FRs from IbEXP1-antisense plants and a corresponding decrease in the transcript level of the lignin biosynthesis-related gene (CAD gene) (Fig. 6). Therefore, these results also suggest that the potential to develop SRs is enhanced in the FRs of IbEXP1-antisense plants. A correlation between lignification level and expansin gene expression has also been observed in loquat fruit (Yang et al., 2008) and water bamboo shoot (Song et al., 2011). Specifically, Yang et al. (2008) reported that the low expression of EjEXPA1 and EjEXPA4 is associated with the low level of chilling-induced lignification in loquat fruit (cv. Baisha), and Song et al. (2011) demonstrated that the suppression of lignification by 1-methylcyclopropane (an inhibitor of lignification) treatment is accompanied by a decrease in the expression of expansin (ZcExp) in peeled water bamboo shoot (Zizania caduciflora L.).
In this study, the earlier thickening growth in the FRs of IbEXP1-antisense plants cultured in vitro was verified by the observation of the earlier thickening growth in the FRs from soil-grown IbEXP1-antisense plants, leading to an increased ratio of TRs to total primary roots (TR/PR) in the IbEXP1-antisense plants relative to WT plants (Supplementary Fig. S3 at JXB online). Ultimately, IbEXP1-antisense plants produced a higher number of SRs than WT plants (Fig. 7, Table 2), resulting in an elevation in the total weight of SRs per IbEXP1-antisense plant. The most striking phenotype in IbEXP1-antisense plants was a remarkable decrease in the length of the FRSS (Fig. 8). In WT sweetpotato plants, SR development took place at various positions along the FR at different time points, but in IbEXP1-antisense plants most of the SRs developed simultaneously at the proximal ends of the FRs. Therefore, it seems likely that, in contrast to the situation in WT plants in which the elongation growth phase of FRs precedes the thickening growth of SRs, in IbEXP1-antisense plants, thickening growth and elongation growth of FRs proceeded simultaneously at the proximal ends of the FR. This result indicates that the thickening growth of FRs was initiated earlier in IbEXP1-antisense plants than in those of the WT and that, consequently, SR development was accelerated in IbEXP1-antisense sweetpotato plants. Taken together, these results demonstrate that down-regulation of the IbEXP1 gene caused an acceleration of SR development through an enhancement of thickening growth in FRs, suggesting that IbEXP1 plays a negative role in the formation of SRs by inhibiting the initial thickening growth of SRs.
Not only the SR of IbEXP1-antisense plants exhibited significant phenotypic alterations, but also the shoot. Although IbEXP1-antisense plants did show shoot growth retardation and restricted elongation growth in the primary stem during the early stage of development (Supplementary Fig. S3 at JXB online), they eventually produced higher number of stems than WT plants (Fig. 7, Table 2). This increased number of stems may be attributable to reduced apical dominance in IbEXP1-antisense plants. Sweetpotato normally produces a single primary stem in the early stage of development that grows vigorously to form a long vein-like stem. The apical dominance of this primary stem prevents additional stems from growing. In IbEXP1-antisense plants, however, the elongation growth of the primary stem was significantly reduced, possibly weakening the apical dominance of the primary stem to such a degree that it was unable to inhibit the growth of other stems. Consequently, IbEXP1-antisense plants produced more stems than WT plants. Another distinctive phenotype associated with the shoots was the delayed leaf senescence in IbEXP1-antisense plants compared with that of WT plants (Fig. 7; Supplementary Fig. S4). This resulted in an increase in total shoot weight per IbEXP1-antisense plant at harvest (Table 2). With this increased shoot weight, IbEXP1-antisense plants may be able to supply much more photosynthate to SRs undergoing thickening growth than WT plants. Thus, it is likely that the increased shoot weight also contributes—at least partially—to the increased total SR weight in the IbEXP1-antisense plants by providing a larger source of the energy needed for thickening growth of SRs. However, it was observed that during the early developmental stage IbEXP1-antisense plants exhibited an earlier and enhanced formation of SRs relative to WT plants, although they did show retarded shoot growth (Supplementary Fig. S3B, C). This observation strongly suggests that unlike the increased total SR weight, the enhanced SR formation in IbEXP1-antisense plants is not affected by the increased shoot weight.
To date, little progress has been made in elucidating the molecular mechanism underlying SR development in sweetpotato. Although expression analysis studies have identified a number of genes possibly involved in SR development (You et al., 2003; Tanaka et al., 2005; 2008), further functional characterization of these genes has been hindered due to a difficulty in generating transgenic sweetpotato plants. Ku et al. (2008) employed an alternative approach to characterize the role of IbMADS1 in SR development by generating IbMADS1-ox potato plants. These authors observed that the FRs of IbMADS1-ox potato plants cultured in vitro were partially swollen and that the metaxylem cells had undergone enhanced multiplication. However, this phenotype was not confirmed in sweetpotato. Very recently, SRD1-ox transgenic sweetpotato plants were successfully generated and it was demonstrated that SRD1 plays a role in the formation of SRs by activating the proliferation of cambium and metaxylem cells to induce the initial thickening growth of SRs in an auxin-dependent manner (Noh et al., 2010). This gain-of-function study, however, was performed with FRs from SRD1-ox sweetpotato plants cultured in vitro, but the proposed role of SRD1 was not verified in mature SRD1-ox sweetpotato plants grown in soil. To date, researchers have been unable to verify whether a sweetpotato gene is directly involved in regulating SR development in soil-grown sweetpotato plants. As such, IbEXP1 is the first sweetpotato gene whose role in SR development has been directly identified in soil-grown transgenic sweetpotato. Not only can IbEXP1-antisense plants be effectively used for elucidating the molecular mechanism involved in SR development, but, as a promising commercial cultivar, they have an increased productivity in terms of both the SR and shoot.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Nucleotide positions of IbEXP1-, IbEXP2-, and IbEXPL1-specific primers.
Figure S2. Transcript levels of IbEXP2 and IbEXPL1 in IbEXP1
Figure S3. Growth of IbEXP1-antisense sweetpotato plants grown in soil.
Figure S4. Phenotypic alteration in IbEXP1-antisense sweetpotato plants grown in soil.
Acknowledgements
This work was supported by grants from the Next-Generation BioGreen 21 Program (no. PJ00807603 and no. PJ00810302), Rural Development Administration, Republic of Korea, and a grant (no. 2010-0002903) from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology.
References
- Akita S, Yamamoto F, Ono M, Kusuhara M, Kobayashi H, Ikemoto S. 1962. Studies on the small tuber set method in sweetpotato cultivation. Bulletin of the Chugoku National Agricultural Experiment Station. 8, 75–128 (in Japanese with English summary) [Google Scholar]
- Belehu T, Hammes PS, Robbertse PJ. 2004. The origin and structure of adventitious roots in sweetpotato (Ipomoea batatas). Australian Journal of Botany. 52, 551–558 [Google Scholar]
- Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P. 1999. Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. The Plant Cell. 11, 2203–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan CD, Lim S, Salzman RA, et al. 2005. Sorghum bicolor’s transcriptome response to dehydration, high salinity and ABA. Plant Molecular Biology. 58, 699–720 [DOI] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. 2000. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA. 97, 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. 1997. Expansins in deepwater rice internodes. Plant Physiology. 113, 1137–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Lee Y, Cho HT, Kende H. 2003. Regulation of expansin gene expression affects growth and development in transgenic rice plants. The Plant Cell. 15, 1386–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer TD, Peeters AJM, Wagemaker CAM, Vriezen WH, Ammerlaan A, Voesenek L. 2004. Expression of α-expansin genes during root acclimations to O2 deficiency in Rumex palustris . Plant Molecular Biology. 56, 423–437 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. 1997. Group I allergens of grass pollen as cell wall-loosening agents. Proceedings of the National Academy of Sciences, USA. 94, 6559–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D. 2002. The growing world of expansins. Plant and Cell Physiology. 43, 1436–1444 [DOI] [PubMed] [Google Scholar]
- Darley CP, Li Y, Schaap P, McQueen-Mason SJ. 2003. Expression of a family of expansin-like proteins during the development of Dictyostelium discoideum . FEBS Letters. 546, 416–418 [DOI] [PubMed] [Google Scholar]
- Eguchi T, Kitano M, Eguchi H. 1998. Growth of sweetpotato tuber as affected by the ambient humidity. Biotronics. 27, 93–96 [Google Scholar]
- Eguchi T, Yoshida S. 2008. Effects of application of sucrose and cytokinin to roots on the formation of tuberous roots in sweetpotato (Ipomoea batatas (L.) Lam.). Plant Root. 2, 7–13 [Google Scholar]
- Feys BF, Benedetti CE, Penfold CN, Turner JG. 1994. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. The Plant Cell. 6, 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firon N, LaBonte D, Villordon A, McGregor C, Kfir Y, Pressman E. 2009. Botany and physiology: storage root formation and development. In: Loebenstein G, Thottappilly G, eds. The sweetpotato Heidelberg: Springer SBM, 13–26
- Geilfus CM, Zörb C, Mühling KH. 2010. Salt stress differentially affects growth-mediating β-expansins in resistant and sensitive maize (Zea mays L.). Plant Physiology and Biochemistry. 48, 993–998 [DOI] [PubMed] [Google Scholar]
- Gookin TE, Hunter DA, Reid MS. 2003. Temporal analysis of alpha and beta-expansin expression during floral opening and senescence. Plant Science. 164, 769–781 [Google Scholar]
- Goujon T, Sibout R, Eudes A, MacKay J, Jouanin L. 2003. Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana . Plant Physiology and Biochemistry. 41, 677–687 [Google Scholar]
- Harada T, Torii Y, Morita S, Onodera R, Hara Y, Yokoyama R, Nishitani K, Satoh S. 2011. Cloning, characterization, and expression of xyloglucan endotransglucosylase/hydrolase and expansin genes associated with petal growth and development during carnation flower opening. Journal of Experimental Botany. 62, 815–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama H, Shimada T, Fujii H, Ito A, Kashimura Y. 2006. Ethylene-regulation of fruit softening and softening-related genes in peach. Journal of Experimental Botany. 57, 4071–4077 [DOI] [PubMed] [Google Scholar]
- Hill J, Douglas D, David P, Mortley D, Trotman A, Bonsi C. 1996. Biomass accumulation in hydroponically grown sweetpotato in a controlled environment: a preliminary study. Acta Horticulturae. 440, 25–30 [DOI] [PubMed] [Google Scholar]
- Huang J, Takano T, Akita S. 2000. Expression of α-expansin genes in young seedlings of rice (Oryza sativa L.). Planta. 211, 467–473 [DOI] [PubMed] [Google Scholar]
- Jones L, McQueen-Mason S. 2004. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum . FEBS Letters. 559, 61–65 [DOI] [PubMed] [Google Scholar]
- Kano Y, Ming ZJ. 2000. Effects of soil temperature on the thickening growth and the quality of sweetpotato during the latter part of their growth. Environment Control in Biology. 38, 113–120 [Google Scholar]
- Keller E, Cosgrove DJ. 1995. Expansins in growing tomato leaves. The Plant Journal. 8, 795–802 [DOI] [PubMed] [Google Scholar]
- Kikkert JR. 1993. The biolistic PDS-1000/He device. Plant Cell, Tissue and Organ Culture. 33, 221–226 [Google Scholar]
- Kim JH, Cho HT, Kende H. 2000. α-Expansins in the semiaquatic ferns Marsilea quadrifolia and Regnellidium diphyllum: evolutionary aspects and physiological role in rachis elongation. Planta. 212, 85–92 [DOI] [PubMed] [Google Scholar]
- Kim SH, Mizuno K, Fujimura T. 2002. Regulated expression of ADPglucose pyrophosphorylase and chalcone synthase during root development in sweetpotato. Plant Growth Regulation. 38, 173–179 [Google Scholar]
- Koda Y. 1997. Possible involvement of jasmonates in various morphogenic events. Physiologia Plantarum. 100, 639–646 [Google Scholar]
- Kokubu T. 1973. Thermatological studies on the relationship between the structure of tuberous root and its starch accumulating function in sweetpotato varieties. Bulletin of the Faculty of Agriculture, Kagoshima University. 23, 1–126 (in Japanese with English summary) [Google Scholar]
- Ku AT, Huang YS, Wang YS, Ma D, Yeh KW. 2008. IbMADS1 (Ipomoea batatas MADS-box 1 gene) is involved in tuberous root initiation in sweetpotato (Ipomoea batatas). Annals of Botany. 102, 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Choi D, Kende H. 2001. Expansins: ever-expanding numbers and functions. Current Opinion in Plant Biology. 4, 527–532 [DOI] [PubMed] [Google Scholar]
- Lee Y, Kende H. 2002. Expression of α-expansin and expansin-like genes in deepwater rice. Plant Physiology. 13, 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Ahn JH, Song SK, Choi YD, Lee JS. 2003. Expression of an expansin gene is correlated with root elongation in soybean. Plant Physiology. 131, 985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Xing S, Guo Q, Zhao M, Zhang J, Gao Q, Wang G, Wang W. 2011. Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. Journal of Plant Physiology. 168, 960–966 [DOI] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S. 2003. Expansins and cell growth. Current Opinion in Plant Biology. 6, 603–610 [DOI] [PubMed] [Google Scholar]
- Lin C, Choi HS, Cho HT. 2011. Root hair-specific EXPANSIN A7 is required for root hair elongation in Arabidopsis. Molecules and Cells. 31, 393–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizana XC, Riegel R, Gomez LD, Herrera J, Isla A, McQueen-Mason SJ, Calderini DF. 2010. Expansins expression is associated with grain size dynamics in wheat (Triticum aestivum L.). Journal of Experimental Botany. 61, 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loretan PA, Bonsi CK, Mortley DG, Wheeler RM, Mackowiak CL, Hill WA, Morris CE, Trotman AA, David PP. 1994. Effects of several environmental factors on sweetpotato growth. Advances in Space Research. 14, 277–280 [DOI] [PubMed] [Google Scholar]
- Matsuo T, Yoneda T, Itoo S. 1983. Identification of free cytokinins and the changes in endogenous levels during tuber development of sweetpotato (Ipomoea batatas Lam.). Plant and Cell Physiology. 24, 1305–1312 [Google Scholar]
- Mbéguié-A-Mbéguié D, Hubert O, Baurens FC, Matsumoto T, Chillet M, Fils-Lycaon B, Sidibé-Bocs S. 2009. Expression patterns of cell wall-modifying genes from banana during fruit ripening and in relationship with finger drop. Journal of Experimental Botany. 60, 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDavid CR, Alamu S. 1980. The effect of growth regulators on tuber initiation and growth in rooted leaves of two sweetpotato cultivars. Annals of Botany. 45, 363–364 [Google Scholar]
- McGreger CE. 2006. Differential expression and detection of transcripts in sweetpotato (Ipomoea Batatas (L.) Lam.) using cDNA microarrays. PhD thesis, Louisiana State University, USA
- McQueen-Mason S, Cosgrove D. 1995. Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiology. 107, 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. 1992. Two endogenous proteins that induce cell wall extension in plants. The Plant Cell. 4, 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortley D, Hill J, Loretan P, Bonsi C, Hill W, Hileman D, Terse A. 1996. Elevated carbon dioxide influences yield and photosynthetic responses of hydroponically-grown sweetpotato. Acta Horticulturae. 440, 31–36 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiologia Plantarum. 15, 473–497 [Google Scholar]
- Nakatani M, Komeichi M. 1991. Changes in the endogenous level of zeatin riboside, abscisic acid and indole acetic acid during formation and thickening of tuberous roots in sweetpotato. Japanese Journal of Crop Science. 60, 91–100 [Google Scholar]
- Nakatani M, Komeichi M. 1992. Changes in endogenous indole acetic acid level during development of roots in sweetpotato. Japanese Journal of Crop Science. 61, 683–684 (in Japanese) [Google Scholar]
- Noh SA, Lee HS, Huh EJ, Huh GH, Paek KH, Shin JS, Bae JM. 2010. SRD1 is involved in the auxin-mediated initial thickening growth of storage root by enhancing proliferation of metaxylem and cambium cells in sweetpotato (Ipomoea batatas). Journal of Experimental Botany. 61, 1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh SA, Park SH, Huh GH, Paek KH, Shin JS, Bae JM. 2009. Growth retardation and differential regulation of expansin genes in chilling-stressed sweetpotato. Plant Biotechnology Reports. 3, 75–85 [Google Scholar]
- Ogasawara S, Shimada N, Kawata T. 2009. Role of an expansin-like molecule in Dictyostelium morphogenesis and regulation of its gene expression by the signal transducer and activator of transcription protein Dd-STATa. Development, Growth and Differentiation. 51, 109–122 [DOI] [PubMed] [Google Scholar]
- Pardales JR, Bañoc DM, Yamauchi A, Iijima M, Kono Y. 1999. Root system development of cassava and sweetpotato during early growth stage as affected by high root zone temperature. Plant Production Science. 2, 247–251 [Google Scholar]
- Rose JKC, Lee HH, Bennett AB. 1997. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proceedings of the National Academy of Sciences, USA. 94, 5955–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. 2005. The expansin superfamily. Genome Biology. 6, 242–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan J, Backhaus A, Malinowski R, McQueen-Mason S, Fleming AJ. 2009. Phased control of expansin activity during leaf development identifies a sensitivity window for expansin-mediated induction of leaf growth. Plant Physiology. 151, 1844–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Gao H, Chen W, Chen H, Mao J, Zhou Y, Duan X, Joyce DC. 2011. The role of 1-methylcyclopropene in lignification and expansin gene expression in peeled water bamboo shoot (Zizania caduciflora L.). Journal of the Science of Food and Agriculture. 91, 2679–2683 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. 1992. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proceedings of the National Academy of Sciences, USA. 89, 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kato N, Nakayama H, Nakatani M, Takahata Y. 2008. Expression of class 1 Knotted1-like homeobox genes in the storage roots of sweetpotato (Ipomoea batatas). Journal of Plant Physiology. 165, 1726–1735 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takahata Y, Nakatani M. 2005. Analysis of genes developmentally regulated during storage root formation of sweetpotato. Journal of Plant Physiology. 162, 91–102 [DOI] [PubMed] [Google Scholar]
- Togari Y. 1950. A study of tuberous root formation in sweetpotato. Bulletin of Tokyo National Agricultural Experimental Station. 68, 1–96 [Google Scholar]
- van Heerden PDR, Laurie R. 2008. Effects of prolonged restriction in water supply on photosynthesis, shoot development and storage root yield in sweetpotato. Physiologia Plantarum. 134, 99–109 [DOI] [PubMed] [Google Scholar]
- Vriezen WH, De Graaf B, Mariani C, Voesenek L. 2000. Submergence induces expansin gene expression in flooding-tolerant Rumex palustris and not in flooding-intolerant R. acetosa . Planta. 210, 956–963 [DOI] [PubMed] [Google Scholar]
- Walter MH, Grima-Pettenati J, Grand C, Boudet AM, Lamb C. 1988. Cinnamyl-alcohol dehydrogenase, a molecular marker specific for lignin synthesis: cDNA cloning and mRNA induction by fungal elicitor. Proceedings of the National Academy of Sciences, USA. 85, 5546–5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang L, Guan Y, Wang Z. 2006. Endogenous hormone concentration in developing tuberous roots of different sweetpotato genotypes. Agricultural Science in China. 5, 919–927 [Google Scholar]
- Wang Z, Fang B, Chen J, Zhang X, Luo Z, Huang L, Chen X, Li Y. 2010. De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweetpotato (Ipomoea batatas). BMC Genomics. 11, 726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei PC, Zhang XQ, Zhao P, Wang XC. 2011. Regulation of stomatal opening by the guard cell expansin AtEXPA1. Plant Signaling and Behavior. 6, 740–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson LA, Lowe SB. 1973. The anatomy of the root system in West Indian sweetpotato (Ipomoea batatas (L.) Lam.) cultivars. Annals of Botany. 37, 633–643 [Google Scholar]
- Wu Y, Sharp RE, Durachko DM, Cosgrove DJ. 1996. Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiology. 111, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Meeley RB, Cosgrove DJ. 2001. Analysis and expression of the α-expansin and β-expansin gene families in maize. Plant Physiology. 126, 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Burklew CE, Yang Y, Liu M, Xiao P, Zhang B, Qiu D. 2012. De novo sequencing and a comprehensive analysis of purple sweetpotato (Impomoea batatas L.) transcriptome. Planta. 236, 101–113 [DOI] [PubMed] [Google Scholar]
- Yang S, Sun C, Wang P, Shan L, Cai C, Zhang B, Zhang W, Li X, Ferguson I, Chen K. 2008. Expression of expansin genes during postharvest lignification and softening of ‘Luoyangqing’ and ‘Baisha’ loquat fruit under different storage conditions. Postharvest Biology and Technology. 49, 46–53 [Google Scholar]
- You MK, Hur CG, Ahn YS, Suh MC, Shin JS, Bae JM. 2003. Identification of genes possibly related to storage root induction in sweetpotato. FEBS Letters. 536, 101–105 [DOI] [PubMed] [Google Scholar]
- Zenoni S, Reale L, Tornielli GB, et al. 2004. Downregulation of the Petunia hybrida α-expansin gene PhEXP1 reduces the amount of crystalline cellulose in cell walls and leads to phenotypic changes in petal limbs. The Plant Cell. 16, 295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Wei PC, Xiong YM, Yang Y, Chen J, Wang XC. 2011. Overexpression of the Arabidopsis α-expansin gene AtEXPA1 accelerates stomatal opening by decreasing the volumetric elastic modulus. Plant Cell Reports. 30, 27–36 [DOI] [PubMed] [Google Scholar]
- Zhao MR, Li F, Fang Y, Gao Q, Wang W. 2011. Expansin-regulated cell elongation is involved in the drought tolerance in wheat. Protoplasma. 248, 313–323 [DOI] [PubMed] [Google Scholar]
- ZhiMing Y, Bo K, XiaoWei H, ShaoLei L, YouHuang B, WoNa D, Ming C, Hyung-Taeg C, Ping W. 2011. Root hair-specific expansins modulate root hair elongation in rice. The Plant Journal. 66, 725–734 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.