Abstract
Six wheat cultivars were grown at Rothamsted (UK) with three levels of nitrogen fertilizer (100, 200 and 350kg N/ha) in 2009 and 2010. Gene expression in developing caryopses at 21 days post-anthesis (DPA) was profiled using the Affymetrix Wheat GeneChip®. Four of 105 transcripts which were significantly upregulated by nitrogen level were annotated as γ-3 hordein and the identification of corresponding expressed sequence tags showed that they differed in sequence from previously described (typical) γ-gliadins and represented a novel form of γ-gliadin. Real-time reverse transcriptase PCR at 14, 21, 28 and 35 DPA revealed that this transcript was most abundant and most responsive to nitrogen at 21 DPA. Four novel γ-gliadin genes were isolated by PCR amplification from wheat cv. Hereward and the related species Aegilops tauschii and Triticum monococcum while three were assembled from the genomic sequence database of wheat cv. Chinese Spring (www.cerealsdb.uk.net). Comparison of the deduced amino acid sequences of the seven genes showed that they shared only 44.4–46.0% identity with the sequence of a typical γ-gliadin (accession number EF15018), but 61.8–68.3% identity with the sequence of γ-3 hordein from the wild barley species Hordeum chilense (AY338065). The novel γ-gliadin genes were localized to the group 1 chromosomes (1A, 1B, 1D).
Key words: gliadin genes, grain quality, nitrogen, nutrition, protein, wheat.
Introduction
The gluten proteins of wheat account for up to 80% of the total protein in the mature grain (Wrigley and Bietz, 1988) and are of particular interest because of their role in determining the processing properties of the grain. In particular, they form a continuous viscoelastic network in dough that traps and retains the carbon dioxide is produced by fermentation during dough proofing, allowing the production of leavened bread (Shewry et al., 2009a ). Attention has therefore focused on the structure of the gluten network and on the biochemical and molecular bases for its unique biomechanical properties.
Wheat gluten comprises a complex mixture of proteins that are classified into two groups which are usually present in approximately equal amounts. This classification is based on whether the protein subunits are present as monomers (gliadins) or assembled into polymers stabilized by inter-chain disulphide bonds (glutenins). Both groups can be further subdivided, the gliadins into α-gliadins, γ-gliadins and ω-gliadins (based originally on their order of mobility on electrophoresis at low pH), and the reduced glutenin subunits into low- and high-molecular-weight groups according to separation by sodium dodecylsulphate/polyacrylamide gel electrophoresis. This classification has proved to be remarkably durable because these groups of proteins differ in their functional properties, with the glutenins being the major determinants of dough strength (elasticity) and the gliadins of dough viscosity and extensibility. A balance between these properties is required for good breadmaking performance, with insufficient elasticity being the most widespread cause of poor breadmaking quality. Hence, the glutenins have been a major focus of research over the last three decades (reviewed by Shewry et al., 2004, 2006).
The protein content of wheat is determined by genetic and environmental factors. Breadmaking wheats are selected for high protein, with a minimum of 13% (dry matter basis) being specified for the Chorleywood Breadmaking Process, which is used for over 80% of the ‘factory-produced’ bread in the UK. Breadmaking wheats have therefore been bred to have higher protein contents (by about 2% dry weight) when grown under the same conditions of nitrogen fertilization than low-protein wheats bred for livestock feed and industrial uses (Snape et al., 1993; Monaghan et al., 2001). However, environmental factors, particularly the availability of nitrogen, have a much greater impact on grain protein content than genotype, and the protein content may vary by up to 2-fold (from about 7 to 15% dry weight) when the same cultivar is grown with different levels of nitrogen fertilization (Dupont and Altenbach, 2003; Kindred et al., 2008; Godfrey et al., 2010). Farmers, therefore, select the appropriate level of fertilizer to maximize the grain yield and to optimize the protein content for the requirements of the market.
The increase in grain protein under conditions of high nitrogen fertilization results largely from greater synthesis and accumulation of storage proteins with a recent study showing an increase in the yield of wet gluten from about 10 to 30% flour weight when the grain N increased from 1.2 to 2.2% dry weight (Godfrey et al., 2010). However, the gliadins and glutenins differ in their response to nitrogen with the gliadins increasing disproportionally, resulting in increased dough extensibility (Kindred et al., 2008; Jia et al., 1996; Panozzo and Eagles, 2000; Zhu and Khan, 2001). The response may also vary between cultivars (Pechanek et al., 1997), with effects on glutenin composition as well as on the ratio of gliadin to glutenin (Pechanek et al., 1997; Wieser and Seilmeier, 1998; Zhu et al., 1999). More detailed studies of the effects of post-anthesis nitrogen fertilizer on the proportions of individual gluten and non-gluten proteins were reported by Altenbach et al. (2011), who used two-dimensional electrophoresis and mass spectrometry to identify individual components. They also showed an increase in the ratio of gliadin to glutenin with post-anthesis fertilizer, from 1.02 to 1.30.
There is considerable interest in reducing the levels of nitrogen fertilizer required for producing bread wheat, to reduce the environmental footprint, the energy requirement for fertilizer production and the cost to the farmer (Foulkes et al., 2009). Increasing nitrogen availability has both positive and negative effects on breadmaking quality, by increasing the total gluten protein content but also increasing the ratio of gliadin to glutenin to increase extensibility but reduce strength. This represents a challenge for breeders as eliminating the effect on gliadin accumulation could result in improved quality at lower grain protein contents. This study therefore examined the effects of nitrogen fertilization on the expression of gluten proteins, and has led to the characterization of novel family of γ-gliadin genes which are highly responsive to nitrogen fertilization.
Materials and methods
Six wheat cultivars – Cordiale, Hereward, Istabraq, Malacca, Marksman and Xi 19 – were grown in replicated field trials at Rothamsted Research (Harpenden, UK) in 2009 and 2010. Nitrogen was applied at three levels (100, 200 and 350kg N/ha) (Barraclough et al., 2010). Ears were tagged at anthesis and whole caryopses were harvested from the middle part of the ear at 14, 21, 28 and 35 days post-anthesis (DPA) and frozen at −80 °C for RNA extraction.
RNA was extracted using a method based on Chang et al. (1993). About 1.5g of whole caryopses were ground in a cooled mill and RNA extracted in hexadecyltrimethylammonium bromide (CTAB) buffer (2% CTAB, 2% polyvinylpyrrolidinone K30, 100mM Tris/HCl, pH 8.0, 25mM ethylene diaminetetra-acetic acid (EDTA), 2.0M NaCl, 0.5g/l spermidine, 2% (w/v) 2-mercaptoethanol) with chloroform/isoamyl alcohol (IAA) (24:1) to remove proteins. RNA was precipitated with 10M LiCl and incubation on ice overnight, dissolved in buffer (1.0M NaCl, 0.5% (w/v) sodium dodecyl sulphate, 10mM Tris/HCl pH 8.0, 1mM EDTA) to remove polysaccharides and extracted once with 24:1 chloroform/IAA. After ethanol precipitation, total RNA was dissolved in diethylpyrocarbonate-treated water and stored at −80 °C. Primers for PCR amplification are given in Supplementary Table S1. PCR reactions were performed at 95 °C for 2min, followed by 40 cycles at 95 °C for 30 s, 48 °C for 30 s and 72 °C for 2min, with a final extension step at 72 °C for 7min using PfuTaq polymerase (Promega, Madison, WI, USA). PCR products were separated on 1% (w/v) agarose gel and the bands excised, purified and sequenced by Eurofins MWG Operon (London, UK).
For reverse transcriptase PCR (RT-PCR), total RNA was cleaned with a mini RNeasy RNA isolation kit (Qiagen, Hilden, Germany) and treated with RNase-free TURBO DNase (Ambion, Austin, TX, USA). Then 5 µg of total RNA was used for reverse transcription with SuperScript™ III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) using anchored oligo-(dT)23 primers (Sigma, St Louis, MO, USA). Diluted cDNA (1:10) was used for RT-PCR in a 25 µl reaction with 1×SYBR Green PCR master mix (Invitrogen). All time points had three technical and three biological replicates. RT-PCR was performed on an ABI 7500 Real Time PCR system (Applied Biosystems, Foster City, CA, USA).
The transcript Ta.2526.1.S1_at was used as an internal control as it showed the most stable expression in developing caryopses of cv. Hereward between 6 and 42 DPA (Wan et al., 2008). Primers for RT-PCR were designed using Primer Express software and are shown in Supplementary Table S1. The PCR product sizes of the γ-gliadin and internal control were 101 and 50bp, respectively. The specificity of the primers was verified by the amplification of a single band resolved on 3% (w/v) agarose gels and dissociation melting curves. The efficiencies of PCR were estimated using LinRegPCR software (Ramakers et al., 2003) with 101.3% for the γ-gliadin and 101.4% for the internal control. The relative expression was calculated using 7500 sequence detection software version 1.4 (Applied Biosystems) in the formula ratio=2−ΔΔCt.
The He-1 γ-gliadin sequence was blasted from the genome sequence database of cv. Chinese Spring (www.cerealsdb.uk.net) and contigs were aligned and assembled using software Geneious.
Transcriptional profiling was performed by the School of Biological Sciences at the Univeristy of Bristol using the Affymetrix GeneChip® Wheat Genome Array with the standard one-cycle cDNA synthesis protocol and hybridization (as described in the GeneChip® Expression Analysis Technical Manual). Transcriptome data analysis was carried out using GeneSpring® version 11 (Agilent Technologies, Stockport, Cheshire, UK), following the standard workflow for two-factor experiments (nitrogen level, variety).
Results
Grain protein content of six cultivars
Six UK wheat cultivars were grown in randomized field experiments at Rothamsted in 2009 and 2010 with three replicate plots at each of three levels of nitrogen fertilization: 100kg/ha, which is below the usual rate of application in the UK, 200kg/ha, which is similar to the optimal level for yield response and 350kg/ha, which is above the optimum for yield response but results in increased grain protein. The cultivars were five breadmaking wheats (Cordiale, Hereward, Malacca, Marksman and Xi 19) and one feed wheat (Istabraq) and the grain protein contents ranged from 1.49 to 2.44% (2009) and 1.52 to 2.44% (2010) dry weight in the former and from 1.40 to 2.06% (2009) and 1.44 to 1.99% dry weight (2010) in the latter. These differences are consistent with the fact that breadmaking and feed wheats are selected for high and low protein contents, respectively. The grain nitrogen contents of all cultivars increased with higher applications of nitrogen, from a mean of 1.54% at 100kg N/ha to a mean of 2.25% at 350kg N/ha (see Supplementary Table S2 for details).
Identification of a novel type of γ-gliadin gene and its response to nitrogen application
Developing caryopses were harvested from each plot at 21 DPA which corresponds to the most active part of the grain filling period. RNA fractions were isolated and gene expression profiles determined using Affymetrix Wheat GeneChip microarrays for each of the three biological replicates for all treatments (N level × variety, for 2 years).
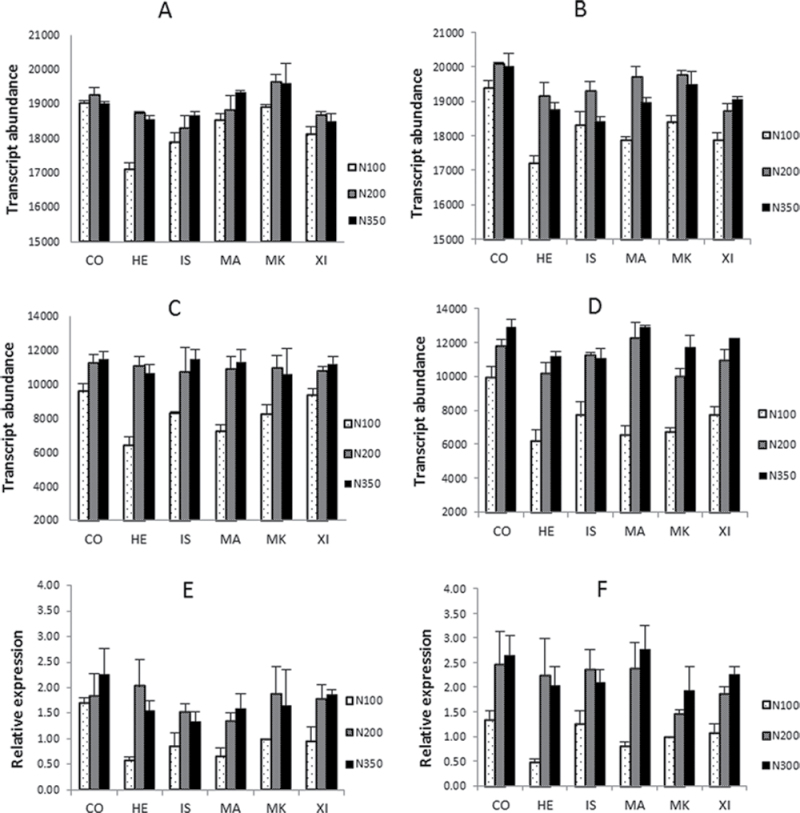
The Affymetrix microarrays contain 18 probe sets annotated as related to γ-gliadins (see Supplementary Table S3). Fourteen correspond to typical γ-gliadin sequences as described by Shewry et al. (2009b) and these also showed broadly similar expression profiles. However, because gluten proteins are highly polymorphic between genotypes not all of the probe sets corresponded to sequences present in all six cultivars. Similarly, the high degree of sequence identity within families of gluten protein genes means that many probe sets would cross-hybridize with related transcripts. The expression profiles of transcripts hybridizing to these 14 probe sets were therefore averaged, and they showed that the expression of the γ-gliadin gene family responded to nitrogen application for all cultivars in 2009 (Fig. 1A) and to an even greater extent in 2010 (Fig. 1B), particularly between the 100 and 200kg N/ha treatments. Furthermore, the cv. Hereward showed the greatest response. The remaining four γ-gliadin probe sets derived from expressed sequence tags (ESTs) BJ235128 (Ta.9940.1.A1_at), CD906599 (Ta.6175.1.S1_at, and Ta.6175.1.S1_x_at) and BJ240530 (Ta.6175.2.A1_x_at) differed in their sequences from typical γ-gliadins and shared high identity with γ-3 hordein from the wild barley species Hordeum chilense (accession number AY338065). They are therefore considered to represent a novel form of wheat γ-gliadin. These sequences also showed a consistently greater response to nitrogen application, as seen in the averaged expression profiles (Fig. 1C and 1D). This strong response of the novel γ-gliadin gene(s) to nitrogen application was confirmed by RT-PCR using the same RNA fractions as those used for the Affymetrix analysis (Fig. 1E, F).
Fig. 1.
Expression of transcripts for typical and novel forms of γ-gliadin. The expression of γ-gliadin transcripts on Affymetrix (A–D) and real-time RT-PCR (E and F) at 21 DPA in 2009 (A, C and E) and 2010 (B, D and F). (A, B) Average expression of sequences hybridizing to 14 probe sets for typical γ-gliadins in 2009 (A) and 2010 (B). (C, D) Average expression of sequences hybridizing to four probe sets for novel γ-gliadins in 2009 (C) and 2010 (D). (E, F) Validation of novel γ-gliadin expression at 21 DPA by real-time RT-PCR in 2009 (E) and 2010 (F). CO, Cordiale; HE, Hereward; IS, Istabraq; MA, Malacca; MK, Marksman; XI, Xi-19. N100 etc., 100kg N/ha, etc. Bars show standard errors.
In order to determine the abundances of the novel γ-gliadin transcripts and their response to nitrogen at a wider range of developmental stages, RT-PCR was carried out on cDNA fractions from cv. Hereward at 14, 21, 28 and 35 DPA in 2010 (Fig. 2). These time stages correspond to the start of grain filling, the most active phase of grain filling, the final phase of grain filling and the start of the desiccation/maturation phase, respectively. There was little difference between the expression profiles of the novel γ-gliadin transcripts in caryopses grown at the three nitrogen levels at 14 DPA. Thereafter, the abundances of the novel γ-gliadin transcripts increased more in the caryopses grown at 200 and 350kg N/ha than in those grown at 100kg N/ha, by 3- to 4-fold at 21 DPA and 4- to 6-fold at 28 DPA. Little effect of N treatment was observed at 35 DPA.
Fig. 2.
Abundance of transcripts related to the novel γ-gliadin in developing caryopses of wheat cv. Hereward grown in 2010, determined by real-time RT-PCR. N100 etc., 100kg N/ha, etc. This figure is available in colour at JXB online. Bars show standard errors.
Characterization of novel γ-gliadin genes from wheat and related species
The EST sequences from which the four probe sets were derived were used to design primers to amplify transcripts from cDNA of developing grain of wheat cv. Hereward at 21 DPA. This identified two sequences, one of which (called He-1, accession number HE819390) consisted of a 981bp open reading frame which covered the four Affymetrix probe sets at different positions (Supplementary Fig. S1). The second gene (He-2, JX081267) consisted of 952bp, but contained an in-frame stop codon and an extra nucleotide insert which resulted in a frame shift. It was therefore concluded that although both genes are transcribed, He-1 and He-2 correspond to functional and non-functional novel γ-gliadin genes, respectively.
The sequences of related novel γ-gliadin genes were then identified from the genomic database for wheat cv. Chinese Spring (www.cerealsdb.uk.net), allowing the assembly of three full-length sequences called CS-1, CS-2 and CS-3 (Supplementary Table S4). CS-1 corresponds to He-1 and is putatively functional (with no stop codons or insertions/deletions). Similarly, CS-2 corresponds to He-2 but lacks the stop codon and insertion present in He-2 and hence is also considered to be putatively functional. CS-3 does not correspond to either He-1 or He-2 and contains two in-frame stop codons; hence it is considered to be non-functional. The same primer sets were then used to amplify related sequences from genomic DNA of diploid species with genomes related to those of hexaploid bread wheat, Aegilops tauschii, which is the progenitor of the D genome, and Triticum monococcum, which is related to the progenitor of the A genome of bread wheat. Single transcripts from each species were identified, called Tm (accession number JX081266) and Ae (JX081265), respectively.
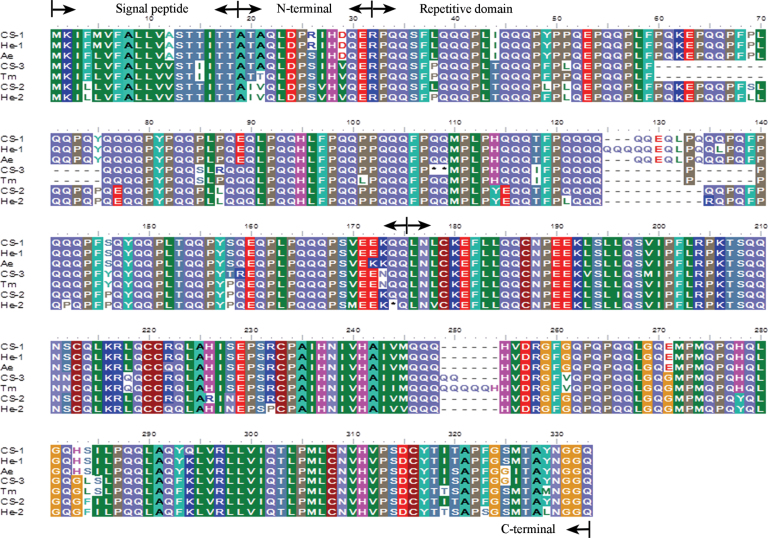
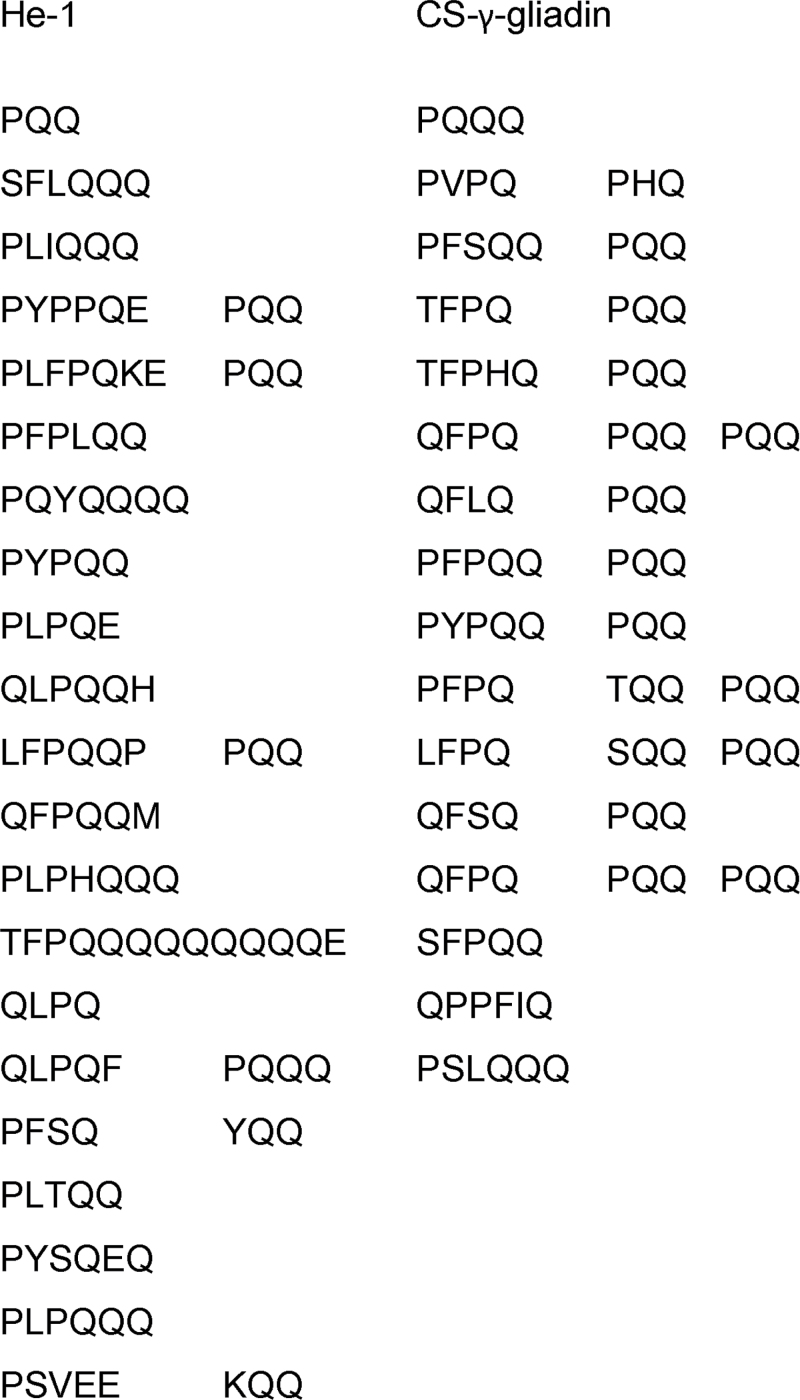
The seven novel γ-gliadin proteins encoded by these sequences are aligned in Fig. 3. The proteins all have a typical γ-gliadin structure with a signal peptide, a short N-terminal domain, a repetitive domain based on short motifs rich in proline and glutamine and a C-terminal domain with eight cysteine residues which are known to form four intra-chain disulphide bonds (see Shewry et al., 2009a ). However, the sequence of the novel γ-gliadin greatly differs from that of the typical γ-gliadin. The repeat sequence motif of the repetitive domains of typical γ-gliadins is PFPQ1–2(PQQ)1–2, while the consensus repeat sequence motif in the novel γ-gliadin is PLPQ3–4 with very few PQQ sequences (Fig. 4).
Fig. 3.
Alignment of novel γ-gliadin proteins. Alignment was performed with ClustalW using the BLOSUM matrix of Geneious Pro5.5.6 version software. CS-1, CS-2 and CS-3 (pseudogene, predicted protein), novel γ-gliadins from Chinese Spring; He-1 and He-2 (pseudogene), novel γ-gliadins from cv. Hereward; Ae and Tm, novel γ-gliadins from Ae. tauschii and T. monococcum, respectively. This figure is available in colour at JXB online.
Fig. 4.
Comparison of repetitive motifs between novel and typical γ-gliadin proteins (EF15018).
Sequence comparison of γ-gliadin proteins
The mature forms of the novel γ-gliadin proteins comprise between 280 and 308 amino acid residues, with calculated molecular masses ranging from 32.2 to 36.0kDa (Supplementary Table S5), and the variation in size resulting mainly from the insertion/deletion of repeat motifs. The novel γ-gliadins showed between 79.7 and 98.7% sequence identity with each other, between 61.8 and 68.3% sequence identity with the γ-3 hordein sequence, but only between 44.4 and 46.0% sequence identity with the typical γ-gliadin encoded by EF15108 (Supplementary Table S6).
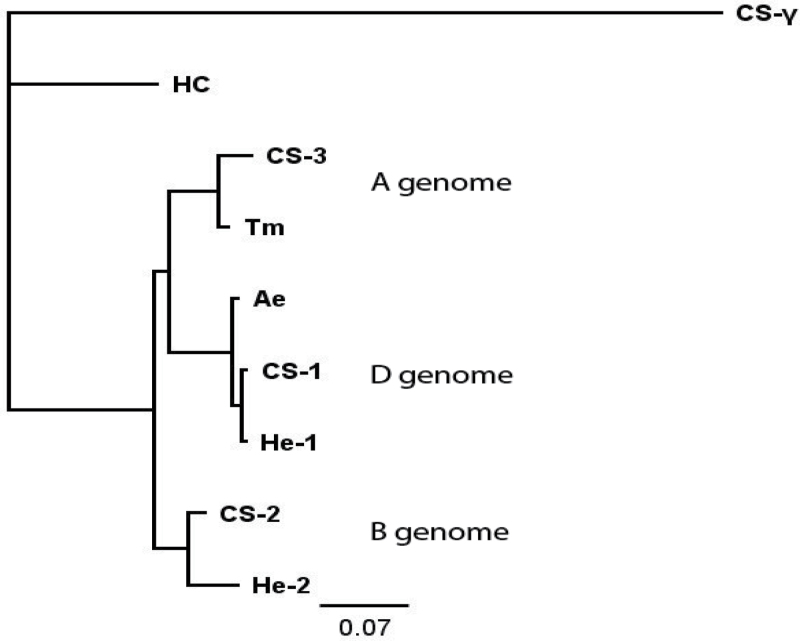
The seven novel γ-gliadin proteins may be separated into three groups based on their sequence similarity (Fig. 5). CS-1 and He-1 are grouped together with Ae (from Ae. tauschii, the progenitor of the D genome of bread wheat) and hence may be putatively assigned to the D genome, while CS-3 is grouped together with Tm (from T. monococcum, which is related to the progenitor of the A genome) and may be similarly assigned to the A genome. Genome sequence blasting in URGI (http://urgi.versailles.inra.fr) confirmed that CS-1 and He-1 are located on chromosome 1DS and also showed that CS-2 and He-2 are located on 1BS. CS-3 could not be assigned by this approach as the sequence of the short arm of wheat chromosome 1A is not yet available. Only single copies of the CS-1, CS-2 and CS-3 genes are present and they are therefore considered to be homoeologues.
Fig. 5.
Neighbour-joining tree of the novel γ-gliadin proteins (based on full-length protein sequence). This figure is available in colour at JXB online.
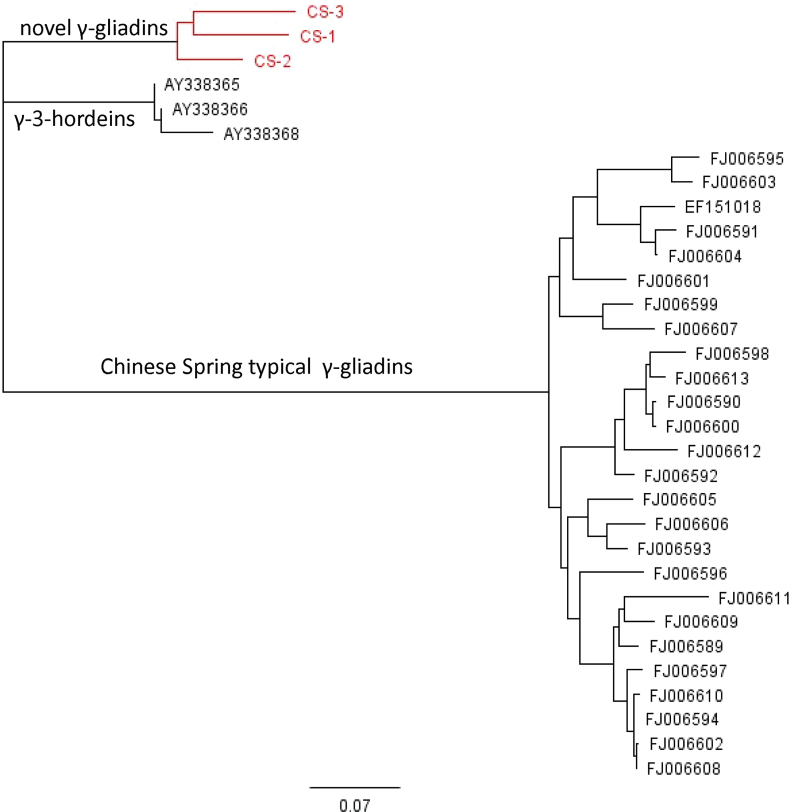
Comparisons between the sequences in Fig. 5 and those encoded by the 26 typical γ-gliadin genes identified in the genomic sequence of cv. Chinese Spring by Bartels et al. (1986) and Qi et al. (2009) showed a clear distinction between the novel and typical types (Fig. 6), with the novel type being closer to the γ-3 hordeins in sequence. A similar degree of separation was also shown when the sequences encoded by 129 γ-gliadin genes (also identified by Qi et al., 2009) were included in the analysis (Supplementary Fig. S2). Whether the whole sequences of the proteins were used for the analysis or only the sequences of the non-repetitive N-terminal sequence and C-terminal domain not significantly affect the relationship (data not shown).
Fig. 6.
Neighbour-joining tree of three novel and 26 typical γ-gliadin proteins from cv. Chinese Spring (based on full-length protein sequences).
Discussion
Seven novel γ-gliadin genes were identified by PCR amplification and sequence assembly from wheat cultivars Hereward and Chinese Spring and the related species Ae. tauschii and T. monococcum. These novel γ-gliadin genes encode proteins comprising signal peptides, short N-terminal sequences, repetitive domains and long non-repetitive C-terminal domains with eight cysteine residues. This structural organization is similar to that of typical γ-gliadins, where the cysteine residues have been shown to form four intra-chain disulphide bonds (Rafalski, 1986; Shewry et al., 2009a ). This is consistent with the novel proteins being monomeric, rather than incorporated into glutenin polymers. However, the sequences of the novel γ-gliadin proteins showed only about 45% identity with those of a typical γ-gliadin. The novel and typical γ-gliadins also differ in their repeat motifs, with consensus motifs of PFPQ1–2(PQQ)1–2 in typical γ-gliadins (Qi et al., 2009; Altenbach et al., 2010) and PLPQ3–4 (with very few PQQ tripeptides) in the novel γ-gliadins.
The genes encoding γ-gliadins and ω-gliadins are mainly located at the Gli-1 loci on the short arms of the group 1 chromosomes while α-type gliadins are encoded by multigene families at the Gli-2 loci on the short arms of the group 6 chromosomes (García-Olmedo et al., 1982; Payne et al., 1984). In this study, He-1 and CS-1 were assigned to the short arm of chromosome 1D, He-2 and CS-2 to the short arm of chromosome 1B and CS-3 provisionally assigned to the A genome.
Southern blotting analyses indicated the presence of 15–40 copies of γ-gliadin genes in Chinese Spring (Sabelli and Shewry, 1991) while a total of 26 putatively functional γ-gliadin genes from Chinese Spring have been sequenced (Bartels et al., 1986; Qi et al., 2009). Our results showed that CS-1, CS-2 and CS-3 clearly differ from all other known γ-gliadins in Chinese Spring, and from more than 340 γ-gliadins from Triticum and Aegilops species encoded by sequences in the NCBI database. Moreover, the sequences of the seven novel γ-gliadin genes are clustered with γ-3 hordein from the barley wild species H. chilense, indicating that the families of genes encoding the novel and typical forms of γ-gliadin may have separated before the separation of Triticum and Hordeum.
The novel γ-gliadin gene(s) in cv. Hereward are expressed at low levels at 14 DPA. Expression increases rapidly to reach to a maximum at 21 DPA, and falls at 35 DPA. This expression pattern is consistent with that of other gliadins (Pistόn et al., 2006; Shewry et al., 2009b ). Their expression is also strongly affected by nitrogen availability, particularly between 21 and 28 DPA. However, comparison of the six cultivars at 21 DPA using the Affymetrix GeneChip and RT-PCR also showed clear differences between cultivars in the response of the novel γ-gliadin genes to nitrogen, with Cordiale and Xi-19 showing higher relative expression at low nitrogen, but less response to increased nitrogen. By contrast, Hereward and Malacca showed lower expression at low nitrogen, but greater responses to increased nitrogen. All six cultivars also showed greater responses to nitrogen in 2010 than in 2009, both in terms of γ-gliadin expression and total grain percentage of nitrogen. The differences in grain percentage of nitrogen between 2009 and 2010 resulted from greater dilution of grain protein in 2009 due to the higher grain yields (wet cool summer), in comparison to low grain yields in 2010 (low spring rainfall) (data not shown). The interaction with γ-gliadin expression may reflect differences in timing of protein synthesis during grain development due to the different weather patterns.
The regulation of prolamin gene expression by nitrogen has been studied in barley by measuring the expression of the UidA (Gus) reporter gene driven by the C hordein (ω-gliadin homologue) promoter under different nitrogen regimes (Müller and Knudsen, 1993). Most prolamin genes of wheat and barley have two conserved motifs 5’ upstream of the transcriptional start site, called the endosperm box and GCN4-like box (Hammond-Kosack et al., 1993), with the GCN4-like motif regulating the response to nitrogen by conferring high expression at high nitrogen, but reducing gene expression at low nitrogen. However, the protein which binds to GCN4-like motif has not been identified in plants and the sensing and signal transduction mechanisms are not understood. Nevertheless, it is likely that these mechanisms will be affected by both the genotype and the wider environment, which would explain the differences observed between the six cultivars and the 2 years of growth.
In practical terms, the identification of differences in the nitrogen responsiveness of individual γ-gliadin genes both within and between cultivars indicates that it may be possible to use targeted plant breeding to modify the effect of nitrogen on gluten protein composition, to reduce the disproportional increase in gliadins which occurs at high nitrogen availability. This would allow the protein content required for breadmaking to be reduced and hence lower amounts of nitrogen fertilizer to be applied.
Supplementary material
Supplementary material is available at JXB online.
Supplementary Table S1. Primers for PCR sequencing and real-time RT- PCR.
Supplementary Table S2. Nitrogen contents of grain grown at Rothamsted in 2009 and 2010. All means of three replicate plots, with standard deviation of the mean in parenthesis.
Supplementary Table S3. Probe sets for γ-gliadin transcripts on the Affymetrix GeneChip Wheat Genome Array.
Supplementary Table S4. Assembled DNA sequences of three novel γ-gliadin genes from Chinese Spring.
Supplementary Table S5. Numbers of amino acid residues and calculated molecular weights of the mature γ-gliadin proteins.
Supplementary Table S6. Sequence similarity of the mature γ-gliadin proteins.
Supplementary Fig. S1. Sequence of novel γ-gliadin He-1 and probe set positions.
Supplementary Fig. S2. Neighbour-joining tree of 129 γ-gliadin genes from Triticum, Aegilops species and H. chilense (based on full length protein sequences).
Acknowledgements
This research was funded by the Biotechnology and Biological Sciences Research Council (BBSRC). The work reported here was supported by the BBSRC Industrial Partnership Award BB/G022437 with support from the Home-Grown Cereals Authority grant RD-2007–3409 “Sustainability of UK-grown wheat for breadmaking”. The authors thank Dr Dhan Bhandari (HGCA) and Dr Gemma Chope (Campden BRI) for critical comments on the manuscript and the IWGSC (International Wheat genome sequencing Consortium) for permission to use the URGI website to assign γ-gliadin genes.
Glossary
Abbreviations:
- CTAB
hexadecyltrimethylammonium bromide
- DPA
days post-anthesis
- EDTA
ethylene diaminetetra-acetic acid disodium
- EST
expressed sequence tags
- IAA
isoamyl alcohol
- RT-PCR
reverse transcriptase PCR.
References
- Altenbach SB, Vensel WH, Dupont FM. 2010. Analysis of expressed sequence tags from a single wheat cultivar facilitates interpretation of tandem mass spectrometry data and discrimination of gamma gliadin proteins that may play different functional roles in flour. BMC Plant Biology. 10, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach SB, Tanaka CK, Hurkman WJ, Whitehand LC, Vensel WH, Dupont FM. 2011. Differential effects of post-anthesis fertilizer regime on the wheat flour proteome determined by quantitive 2-DE. Proteome Science. 9, 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough PB, Howarth JR, Jones J, Lopez-Bellido R, Parmar S, Shepherd CE, Hawkesford MJ. 2010. Nitrogen efficiency of wheat: genotypic and environmental variation and prospects for improvement. European Journal of Agronomy. 33, 1–11 [Google Scholar]
- Bartels D, Altosaar I, Harberd NP, Barker RF, Thompson RD. 1986. Molecular analysis of gamma-glaidin gene families at the complex Gli-1 locus of bread wheat (T. aestivum L). Theoretical and Applied Genetics. 72, 845–853 [DOI] [PubMed] [Google Scholar]
- Chang SJ, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 11, 113–116 [Google Scholar]
- Dupont FM, Altenbach SB. 2003. Molecular and biochemical impacts of environmental factors on wheat grain development and protein synthesis. Journal of Cereal Science. 38, 133–146 [Google Scholar]
- Foulkes MJ, Hawkesford MJ, Barraclough PB, Holdsworth M, Kerr S, Kightly S, Shewry PR. 2009. Identifying traits to improve the nitrogen economy of wheat: recent advances and future prospects. Field Crops Research. 114, 329–342 [Google Scholar]
- Godfrey D, Hawkesford M, Powers S, Millar S, Shewry PR. 2010. Nutritional effects on wheat grain composition and end use quality. Journal of Agricultural and Food Chemistry. 58, 3012–3021 [DOI] [PubMed] [Google Scholar]
- García-Olmedo F, Carbonero P, Jones BL. 1982. Chromosomal locations of genes that control wheat endosperm proteins. Advances in Cereal Science and Technology. 5, 1–47 [Google Scholar]
- Hammond-Kosack MCU, Holdsworth MJ, Bevan MW. 1993. In vivo footprinting of a low molecular weight glutenin gene (LMWG-1D1) in wheat endosperm. EMBO Journal. 12, 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y-Q, Masbou V, Aussenac T, Fabre J-L, Deaeke P. 1996. Effects of nitrogen fertilization and maturation conditions on protein aggregates and on the breadmaking quality of Soissons, a common wheat cultivar. Cereal Chemistry. 73, 123–130 [Google Scholar]
- Kindred DR, Verhoeven TMO, Weightman RM, Swanston JS, Agu RC, Brosnan JM, Sylvester-Bradley R. 2008. Effects of variety and fertiliser nitrogen on alcohol yield, grain yield, starch and protein content, and protein composition of winter wheat. Journal of Cereal Science. 48, 46–57 [Google Scholar]
- Monaghan JM, Snape JW, Chojecki JS, Kettlewell PS. 2001. The use of grain protein deviation for identifying wheat cultivars with high grain protein concentration and yield. Euphytica. 122, 309–317 [Google Scholar]
- Müller M, Knudsen S. 1993. The nitrogen response of a barley C-hordein promoter is controlled by positive and negative regulation of the GCN4 and endosperm box. The Plant Journal. 4, 343–355 [DOI] [PubMed] [Google Scholar]
- Panozzo JF, Eagles HA. 2000. Cultivar and environmental effects on quality characters in wheat. II. Protein. Australian Journal of Agricultural Research. 51, 629–636 [Google Scholar]
- Payne PI, Jackson EA, Holt MA, Law CN. 1984. Genetic linkage between endosperm storage protein genes on each of the short arms of chromosomes 1a and 1B in wheat. Theoretical and Applied Genetics. 67, 235–243 [DOI] [PubMed] [Google Scholar]
- Pechanek U, Karger A, Gröger S, Charvat B, Schöggl G, Lelley T. 1997. Effect of nitrogen fertilization on quality of flour protein components, dough properties, and breadmaking quality of wheat. Cereal Chemistry. 74, 800–805 [Google Scholar]
- Pistόn F, Dorado G, Martin A, Barro F. 2006. Cloning of nine γ-gliadin mRNAs (cDNA) from wheat and the molecular characterization of comparative transcript levels of γ-gliadin subclasses. Journal of Cereal Science. 43, 120–128 [Google Scholar]
- Qi P-F, Wei Y-M, Ouellet T, Chen Q, Tan X, Zheng Y-L. 2009. The γ-gliadin multigene family in common wheat (Triticum aestivum) and its closely related species. BMC Genomics. 10, 168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski A. J. 1986. Structure of wheat gamma-gliadin genes. Gene. 43, 221–229 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JMM, Deprez RHl, Moorman AFM. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters. 339, 62–66 [DOI] [PubMed] [Google Scholar]
- Sabelli P, Shewry PR. 1991. Characterization and organization of gene families at the Gli-1 loci of bread and durum wheats by restriction fragment analysis. Theoretical and Applied Genetics. 83, 209–216 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Halford NG, Tatham AS, Popineau Y, Lafiandra L, Belton PS. 2004. The high molecular weight subunits of wheat glutenin and their role in determining wheat processing properties. In Taylor SL, ed. Advances in Food and Nutrition Research Vol 45 Academic Press; San Diego: pp 220–302 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Halford NG, Lafiandra D. 2006. The high-molecular-weight subunits of glutenin. In Wrigley C, eds.Gliadins and Glutenins. AACC; St Paul: pp 143–169 [Google Scholar]
- Shewry PR, D’Ovidio R, Lafiandra D, Jenkins J, Mills ENC, Békés F. 2009. a Wheat grain proteins. In Khan K, Shewry PR, eds. Wheat Chemistry and Technology Ed 4 AACC; St Paul: pp 223–298 [Google Scholar]
- Shewry PR, Underwood C, Wan Y, Lovegrove A, Bhandari D, Toole G, Mills ENC, Denyer K, Mitchell RAC. 2009. b Storage product synthesis and accumulation in developing grains of wheat. Journal of Cereal Science. 50, 106–112 [Google Scholar]
- Snape JW, Hyne V, Aitken K. 1993. Targeting genes in wheat using marker mediated approaches. In Proceedings of the 8th International Wheat Genetics Symposium. Beijing, China: China Agricultural Scientech Press; pp 749–759 [Google Scholar]
- Wan YF, Poole RL, Huttly AK, et al. 2008. Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics. 9, 121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser H, Seilmeier W. 1998. The influence of nitrogen fertilisation on quantities and proportions of different protein types in wheat flour. Journal of Science of Food and Agriculture. 76, 49–55 [Google Scholar]
- Wrigley CW, Bietz JA. 1988. Proteins and amino acids. In Pomeranz Y, ed. Wheat Chemistry and Technology Vol 1 AACC; St Paul: pp 159–273 [Google Scholar]
- Zhu J, Khan K. 2001. Effects of genotype and environment on glutenin polymers and breadmaking quality. Cereal Chemistry. 78, 125–130 [Google Scholar]
- Zhu J, Khan K, Huang S, O’Brien L. 1999. Allelic variation at Glu-D1 locus for high molecular weight (HMW) glutenin subunits: quantification by multistacking SDS-PAGE of wheat grown under nitrogen fertilization. Cereal Chemistry. 76, 915–919 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.