Abstract
High-throughput low-cost DNA sequencing has emerged as one of the challenges of the post-genomic era. Here we present the proof of concept for a new single-molecule platform that allows for DNA identification and sequencing. In contrast with most present methods, our scheme is not based on the detection of the fluorescence of incorporated nucleotides, but rather on the measurement of a DNA hairpin length. By cyclically modulating the force pulling on small magnetic beads tethered by a hairpin to a surface, one can unzip and rezip the molecule. In the presence of complementary oligonucleotides in solution, reziping may be transiently interrupted by the hybrids they form with the hairpin. By measuring the extension of the blocked hairpin, one can determine the position of the hybrid along the molecule with nearly single base precision. Our approach, well adapted to a high-throughput scheme, can be used to identify a DNA fragment of known sequence among a sample of various fragments and to sequence an unknown DNA fragment by hybridization or ligation.
Introduction
Chain terminator Sanger sequencing has dominated the DNA sequencing field for almost twenty years1. It uses a DNA polymerase to replicate the target molecule in the presence of fluorescently or radioactively labeled chain terminating nucleotides; followed by electrophoresis to sequentially read a large population of partial replicates. The need for faster (i.e. higher throughput) and cheaper methods has driven the development of alternative approaches. These 'next'-generation DNA sequencing (NGS) platforms2–10 achieve a high throughput by monitoring in parallel the successive incorporation of fluorescently labeled nucleotides by DNA polymerase or ligase in a very large number of microscopic vessels (e.g. small beads or droplets) each containing thousands of PCR-copies of a short DNA fragment. However, due to the limited read length as well as the complexity and bias of the required pre-amplification step, so called 'third'-generation sequencing platforms have been developed11–13. By directly monitoring the incorporation of fluorescently labeled nucleotides in an array of single DNA molecules, they do away with pre-amplification and allow for longer read length. However, these single-molecule sequencing methods are still plagued by the high cost of the labeled nucleotides and struggle with high error-rates (from 4 to 15%) resulting from low signal to noise ratios and non- or misdetection of the fluorescent signal11. Although non-fluorescent single-molecule sequencing alternatives have been proposed (nanopore14, Raman-based15, AFM16, and pyrosequencing17), they are not yet competitive with the fluorescence based methods.
In parallel with the invention of novel sequencing technologies, high-throughput methods have been developed for large-scale genome analyses, such as gene identification, SNPs detection and gene expression profiling, in particular cDNA library characterization18. In these instances one seeks to identify and quantify the existence of specific DNA fragments of known sequence in a given sample. The current microarray technology addresses that issue by measuring the fluorescent signals generated by the hybrids between DNA fragments spotted on a surface or in solution and complementary oligonucleotides in solution or arrayed on a surface19, 20. This high-throughput approach suffers from the need to pre-amplify the target DNA. Its quality is limited by non-specific hybridization, adsorption and fluorescent quantification19, 20.
In this work, we present the proof of concept of novel single-molecule identification and sequencing methods which do not rely on fluorescence but on the measurement of the DNA's extension. They are based on the detection of the transient blockage in the rezipping pathway of a hairpin to which a small complementary strand has been hybridized (Fig.1b). A high degree of parallelism is possible achieved by using a magnetic trap21, 22 to apply a constant force on all the hairpin molecules tethering small magnetic beads to a surface.
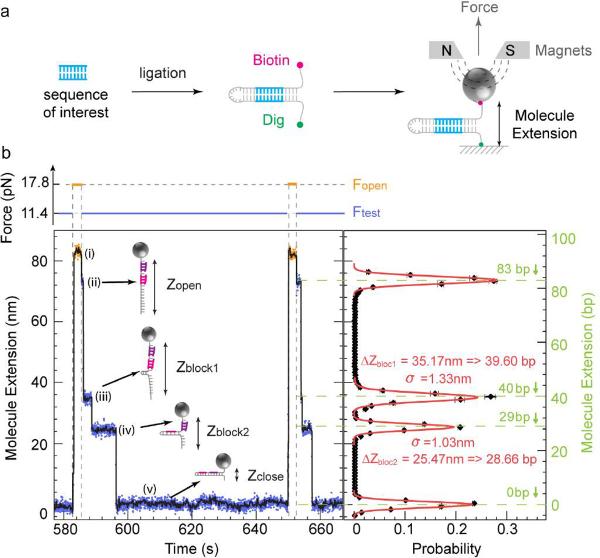
Figure 1.
Detection of oligonucleotide-induced blockages during rehybridization. (a) Hairpin construction design with pre-planted target in the stem. (b) Example of roadblocks due to the hybridization of two oligonucleotides (5'-ACAGCCAGC-3', 5'-ATGACAATCAG-3') on the 83 bps hairpin (see Methods). Left panel: Experimental traces recorded at Fopen = 17.8 pN (orange) and Ftest = 11.4 pN (blue; see force trace at the top). Five different extension levels are observed corresponding from top to bottom to: (i) the open hairpin at Fopen, (ii) the open hairpin at Ftest, (iii) the partially annealed hairpin blocked by the first oligo, (iv) the partially annealed hairpin blocked by the second oligo and (v) the folded hairpin. The black curve corresponds to a 1 s average of the raw data. Right panel: Histogram of blockages (ii)–(iv). The black curve represents the histogram of the number of blockages per cycle at a given extension of the hairpin upon rehybridization at Ftest: ΔZ = Zblock − Zclose in base pairs obtained from ~23 force cycles on a single hairpin. Gaussian fits to the data are shown in red. The variance of these fits (σ ~1 nm) defines the resolution of the apparatus. The roadblocks Zblock1 and Zblock2 are observed at 39.60 and 28.66 bps along the hairpin, in good agreement with the expected position of 40 and 29 bps (shown in green).
Results
Detection of roadblocks in the rezipping pathway of a hairpin
In the present approach a DNA hairpin is attached at one end to a coverslip via a digoxigenin (Dig) - anti-Dig bond and at the other to a magnetic bead via a biotin-streptavidin bond. This DNA hairpin can be generated in various ways. For example it can be formed by ligation of a genomic DNA fragment to a DNA loop at one end and to a DNA fork structure labeled with biotin and Dig at its other end (Fig. 1a). Alternatively this hairpin can be generated from cDNA obtained after trapping mRNAs on poly-T coated beads and reverse transcription (Supplementary Fig. 5).
Small magnets in the vicinity of the sample are used to apply a vertical force on the tethered beads. The distance of a bead to the surface (i.e. the end-to-end distance of the hairpin) can be deduced in real time from the bead's image21. Alternatively, it can be deduced using evanescent field illumination from the intensity of the light scattered by the bead (see Discussion). While the set-up is similar to the DNA unzipping experiment performed with optical tweezers23, the use of a magnetic trap allows for a high degree of parallelism through the simultaneous application of the same force on many molecules21, 22.
We modulate the force to periodically open and close the DNA hairpins in a solution24 containing oligonucleotides complementary to a section of the hairpin. In Figure 1b an 83 bps hairpin is periodically unfolded by applying a force Fopen (> 15 pN) and rezipped by reducing the force to Ftest (~10 pN). In the unfolded or opened state two different oligonucleotides in solution can hybridize to their individual complementary sequences on the hairpin. They transiently block the refolding of the hairpin at low force, which are readily observable as two pauses in the hairpin's extension in the time course of its refolding.
This measurement scheme provides two pieces of valuable information: the position and lifetime of the blockage along the hairpin. The opening of one base pair results in a change in the hairpin extension of about 0.85 nm. With the present resolution of our apparatus (~1 nm), we can thus record the position of a blockage with an accuracy of about one nucleotide. Notice that switching between the opened and closed states of the hairpin provides a differential extension measurement which is insensitive to slow experimental drifts. Moreover the precise value of the applied force is not critical as long as it remains constant (Supplementary Online Materials). Concerning the blockage lifetime, which is related to the stability of the hybrid, we found that it depends on the applied tension, the size of the complementary oligonucleotide and the presence and location of mismatches between the oligonucleotide and the hairpin (data not shown).
Sequence identification by hybridization fingerprinting or single cycle ligation
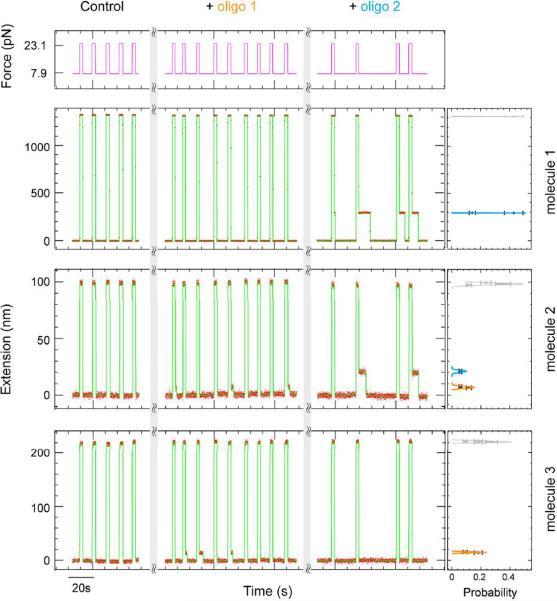
The identification of a desired DNA in a given sample is a relevant issue in many situations, such as the detection of genetic mutations, gene duplications, mRNA expression patterns, etc. With the present scheme this identification issue is quite easy. It requires the detection of a given hybridization fingerprint obtained with a set of chosen short (~10 nt) oligonucleotides on a DNA hairpin sample. This can be achieved by testing for blockages during rehybridization of the hairpins in the presence of one or a few probe oligonucleotide(s). As shown in Figure 2 the identification of three different DNA sequences can be achieved by hybridization of two different oligonucleotides. Notice that since we can detect the exact position of the hybrid it is not necessary to test each probe sequentially as is usually done. Rather one may identify the blockage positions along the hairpin of several selected probes and use this hybridization pattern, instead of the mere presence of hybrids, as a fingerprint of the DNA sequence.
Figure 2.
Sequence identification by hybridization. Three different hairpins (marked as molecule 1, 2, 3) were identified by two different oligos (marked as oligo1 and 2) (see Methods). Left panel: Experimental traces recorded during hairpin open-close cycles. To show the corresponding blockage, oligos 1 and 2 were added sequentially into the solution: oligo 1 hybridized on molecules 2 and 3, while oligo 2 hybridized on molecules 1 and 2. Right panel: Histogram of blockages from oligos 1 (orange) and 2 (blue) on each molecule, while the full extension of the molecule is shaded in grey.
An alternative to fingerprinting by hybridization is to use the ligation between two adjacent short oligonucleotides on the target DNA as an identification tag. The blockage time of the ligated fragment is orders of magnitude longer than the blockage time of each fragment separately. As exemplified in Figure 3, two oligonucleotide sequences (7 and 10 nt long) were chosen to match contiguous parts on the target DNA. As the hairpin is mechanically unzipped both sequences can hybridize to their target and be ligated if they are adjacent. In the absence of ligation, the blockage produced by the small oligonucleotides during rezipping lasts for a few seconds at ~10 pN. However, if the two fragments are ligated the blockage time can last for hours. Reducing the pulling force to below 1 pN reduces the blockage time to ~1 min thus allowing the ligation test to be repeated on the same refolded hairpin.
Figure 3.
Sequence identification by single ligation cycle. By applying a force Fopen (>15 pN) a 1214 bps hairpin DNA is opened periodically for 2 s to allow for the ligation between adjacent 7-nt and 10-nt oligomers hybridized to the hairpin stem (see Methods). In the absence of ligation (i.e. before the 9th cycle, left side of the green dashed line), a short pause (~1 s) during rehybridization (at Ftest) is observed probably due to blockage by the 10-nt oligomer. However, upon ligation (at the 9th cycle) the blockage due to the generated 17-nt oligomer is much longer. A quasi-permanent blockage is observed from the 10th cycle to the 28th cycle, right side of the green dashed line.
Single molecule sequencing by hybridization
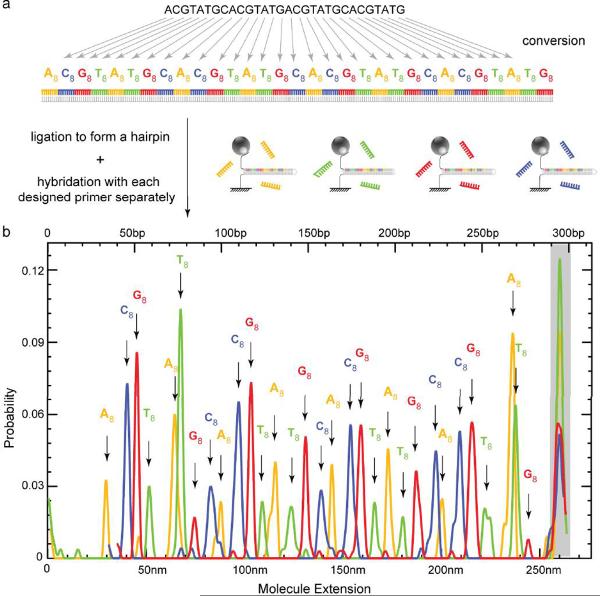
Identification by hybridization can be used for sequencing as demonstrated recently using a complete set of universal, fluorescently-labeled, overlapping oligonucleotide “tiles”6. Since this approach only requires the detection of hybridization, it is compatible with the present platform, which does not require fluorescent labeling and provides additional information on the hybridization position. However the number of different oligonucleotides to test is rather large (>500) making this type of assay difficult to perform without an automated system. To demonstrate sequencing by hybridization we have applied the method to a recoded DNA sample as proposed in the context of nanopore sequencing25.
A DNA fragment can be biochemically converted into a new sequence25 where each original nucleotide is coded by a specific 8 nt sequence (A8, T8, G8 and C8) with balanced GC content (Fig. 4a). The original DNA sequence can then be determined using the present platform by sequential hybridization with a set of four complementary oligomers. We implemented this scheme by recoding a 31 nt DNA fragment into a 248 bps dsDNA ligated with appropriate fragments to form a hairpin. The sequence was then determined by a series of opening/closing cycles done with oligonucleotides A8, T8, G8 and C8 successively added in the solution. As expected, the histogram of the blocking positions reveals the location of each expanded base (Fig. 4b). These positions along the recoded hairpin need to be determined with an accuracy of only 7 nm, well within the characteristics of our current setup.
Figure 4.
Single-molecule sequencing by hybridization. (a) A 31-nt target DNA is recoded by an 8-nt sequence A8, T8, G8 and C8 to yield a 248 bps fragment which is then ligated to form a 295 bps hairpin (see Methods). (b) To sequence the recoded hairpin we successively hybridize its open state with the coding oligonucleotides. The histogram of the hairpin blockage during rehybridization at Ftest is presented for each oligonucleotide: A8 (orange), T8 (green), G8 (red) and C8 (blue), while the full extension of the molecule at Ftest is shaded in grey.
Single-molecule sequencing by periodic ligation cycles
Sequencing DNA via successive ligation reactions is currently used in some NGS schemes26. We have implemented a similar protocol on our set-up using the detection of the ligation of a complementary oligonucleotide to a growing primer to determine the underlying sequence. As shown in Figure 5a, we tested the ligation of a complete set of oligonucleotides to a growing DNA primer. We used a 7 nt oligonucleotide library: 5'-NNNNNNrX-3', where X is the tested base, N represents any of the four deoxyribonucleotides and Nr represents any of the four ribonucleotides. We tested the successive ligation to a primer strand of each of the four bases in a series of hairpin opening and closing cycles. Ligation is attempted in the opened state of the hairpin. If it has occurred (i.e. if X is an expected complementary base) the oligonucleotide prevents rehybridization upon rezipping of the last 7 nucleotides of the hairpin. Such an event results in a detectable increase of ~6 nm in the molecule's extension (Znlig), whereas no change in extension is observed in the absence of ligation. This step is followed by RNase cleavage of the last 6 nucleotides extending the primer strand by a single base. Such cleavage allows rezipping of the hairpin by 6 nucleotides with a concomitant decrease in the molecule's extension (Znrnase) of ~5 nm. Therefore, upon successive injections of one of the four NNNNNNrX oligonucleotides, an increase in extension of 6 nm after ligation followed by a decrease of 5 nm after cleavage is a clear indication of the incorporation of a complementary nucleotide (Fig. 5b).
Figure 5.
Single-molecule sequencing by ligation. (a) Principle of sequencing by cyclic ligation and cleavage reactions. A target hairpin DNA is probed by successive injections of oligonucleotides NNNNNNrA, NNNNNNrG, NNNNNNrC and NNNNNNrT; here the green Nr represents a ribonucleotide. During each cycle, the hairpin is opened with force Fopen (>15 pN) to allow for hybridization and ligation of the proper oligonucleotide, which is tested by measuring the extension of the molecule at Ftest in the ligation buffer (Znlig). Following a successful ligation in cycle n, the extension of the molecule is increased. The buffer is then exchanged by an RNase buffer and the new extension (ZnRNase) is measured. It decreases by ~6 nm as the six nonspecific nucleotides (NNNNNNr) are cleaved by the RNase, leaving the molecule with a single added specific nucleotide (X), which is then phosphorylated for the next cycle. (b) Sequencing of a target DNA (AGCTTCTG…) using the ligation/cleavage protocol. Upon ligation (colored bands, NNNNNNrA (orange), NNNNNNrG (red), NNNNNNrC (blue) and NNNNNNrT (green)) the molecule's extension at Ftest increases by: ΔZ = Znlig − Zn'RNase ~ 6 nm. Upon RNase cleavage of the last six nucleotides (gray bands), the molecule's extension decreases by: ΔZ = Znlig - ZnRNase ~ 5 nm. Lower inset: the injection of a mismatched oligonucleotide (ending with A, G or C) results in no significant change in extension while the injection of a matched oligonucleotide (ending with T) results in an extension increase of 6 nm.
To assess the accuracy of the ligation process we have compared the time t necessary for ligation between a 10-nt matched oligomer and an adjacent 7-nt oligomer possibly displaying a single mismatch. This was done by opening the hairpin for a time t and measuring the probability P(t) of ligation indicated by the presence of a long blockage (>1 min) upon rezipping. With match pairs P(t) saturates exponentially with a typical time τ ~10 sec: P(t) = 1 − exp(−t/τ). On the other hand in the presence of a mismatch, τ increases drastically to hours. This allows for an estimation of the ligation error when the hairpin is opened for a time ttrial : Pmismatch(ttrial)/Pmatch (ttrial). Typical error rates below 1% are routinely achieved (see Table 1).
Table 1.
Ligation error rate on two specific template regions. We tested for the ligation of a matched 10-nt oligomer to a possibly matched, adjacent 7-nt oligomer (of the form NNNXNNrN) at the sequences: AATCAGG (43% GC content) or GAGCGGA (71% GC content). The hairpin was periodically opened for a time t and the ligation was confirmed during the rezipping phase by the presence of a long blockage time. We tested more than 500 ligation runs and counted the number of successful ligations for each possible specific nucleotide (X). We fitted the measured probability of ligation P(t) to an exponential form: 1 − exp(−t/τ). We thus determined the ligation time τmatch and τmismatch for matched and mismatched oligonucleotides. The error rate for a ligation time τtrial is then computed as ε = Pmismatch(τtrial)/Pmatch (τtrial).
| oligonucleotide | Expected: GAGCGGA | Lifetime (τlig) | Error rate (ttrial = 40s) |
|---|---|---|---|
| NNNGNNrN | Mismatch | 2235 s | 1.77% |
| NNNCNNrN | Matching | 6 s | - |
| NNNTNNrN | Mismatch | 8459 s | 0.47% |
| NNNANNrN | Mismatch | 27302 s | 0.15% |
| oligonucleotide | Expected: AATCAGG | Lifetime (τlig) | Error rate (ttrial = 50s) |
|---|---|---|---|
| NNNGNNrN | Mismatch | 19825 s | 0.25% |
| NNNCNNrN | Matching | 45 s | - |
| NNNTNNrN | Mismatch | 3091 s | 1.60% |
| NNNANNrN | Mismatch | 22275 s | 0.22% |
Discussion
We have presented here a palette of tools for DNA identification and sequencing based on the detection of blockages during the rezipping phase of a mechanically opened hairpin.
In the hybridization fingerprinting method rather than detecting the mere presence of a hybrid, maximal information is obtained by determining the hybridization position along the hairpin. This allows for DNA identification with much fewer different hybridization assays. Identification by a single ligation cycle is simpler as it does not require the measurement of the extension but only the detection of a long blockage during rezipping. It also presents various advantages.
-
-
It allows for easy detection of SNPs via the ligation of matched adjacent oligomers. A 7-nt oligomer possessing the desired base mutation at its center will not bind long enough to be ligated with an adjacent 10-nt oligomer if it doesn't match the substrate (see below). On the other hand ligation will be achieved in a few seconds for a matched sequence and will be detected as a very long pause in the rehybridization pathway of the underlying hairpin.
-
-
Sequencing by unchained base reads has recently been shown to provide for a very efficient means to sequence genomic DNA2, 3. This approach amounting to a series of single ligation cycles can be implemented directly here without the need for a pre-amplification step (e.g. rolling circle amplification) and fluorescent detection.
-
-
The measured error rate (~1%) is smaller than all other single molecule methods.
With our present instrumentation buffer exchanges have been done manually which has limited our sequencing by the ligation method to twelve cycles of successive ligation tests leading to the identification of only eight consecutive bases. However, in a series of experiments we have shown that much longer read-lengths are possible with an automated apparatus. We have performed a series of 9 consecutive ligations of an 8-nt oligomer spanning 72 nucleotides and probing 9 bases (Supplementary Fig. 6). Full sequencing of these 72 nucleotides would require 7 similar cycles interrogating a different base shifted along the oligomer, which we have not attempted. Removing the strand built by the successive ligations in each cycle can be done by various means, including a denaturing agent, exonuclease or helicase. Here we have used a RecQ helicase27 to efficiently strip off the strand (Supplementary Fig. 7). Further, notice that we can detect the specific ligation of two target oligonucleotides on a 1 kbps DNA (Supplementary Fig. 2), which suggests that even longer reads could be obtained.
Using the molecule's extension to monitor the successive ligation of oligonucleotides on a single hairpin has some major advantages over present NGS schemes. These rely on the detection usually by fluorescence of nucleotide addition through ligation or polymerization in a bulk sample comprised of many identical molecules (a “polony” usually obtained by PCR). Since the yield of ligation (or polymerization) is never 100%, a fraction of the DNA molecules will not incorporate the matching (oligo-) nucleotide and will become unsynchronized from the rest of the “polony”; at the next cycle these molecules will present a sequence shift and incorporate a wrong (oligo-) nucleotide. This desynchronization issue, which worsens as more bases are sequenced, essentially limits the read-length of all bulk methods. In our single-molecule approach, each molecule is sequenced independently and the position of a nucleotide along a given hairpin is determined independently from previously incorporated ones, hence there are no synchronization issues. Moreover in sequencing by ligation, the incorporation of a nucleotide is signaled by both an increase in the molecule's extension upon ligation followed by a decrease upon RNAse treatment. The later step thus provides proof reading of the former.
In contrast with present single-molecule sequencing methods our approach does not rely on fluorescence. The drawback is that the incorporation of each nucleotide has to be tested sequentially, while fluorescence methods can use four different colors to read the nature of the incorporated base. However fluorescence methods suffer from other flaws: since the incorporation of a nucleotide by a DNA polymerase is a stochastic process, the fluorophore associated with the incorporated nucleotide may not be detected either because one of the nearly simultaneous incorporation of two of the same type of nucleotides or because the fluorophore bleached. This is the main cause of the large error rate of these methods. The mechanical measurement of the hairpin extension does not suffer from this problem as it can be repeated many times. Even though, continuously stretching a single molecule may break it, this appears not to be a serious issue: we routinely submit hairpins to more than 20 000 opening and closing cycles without any problems for a significant fraction of them. When the hairpin breaks we suspect that the weak link is between the ligand at the molecule's end (biotin or Dig) and the receptor which could be replaced by a stronger covalent bond if necessary.
The magnetic trap set-up consists of a pair of rare earth magnets that provide a constant force acting simultaneously on all the beads which is an ideal situation for a high-throughput system. Our current set-up, which is implemented on a commercially available magnetic tweezers apparatus, can monitor the distance to the surface of up to 50 beads with a resolution of approximately 1 nm using real-time parallel image processing (Supplementary Online Materials). The measurement of the molecule's extension is presently time consuming and not ideal for a high-throughput platform since it relies on image analysis to deduce the distance of the tethered bead to the surface. However, alternative measurement schemes exist that can readily solve this issue. We are able to measure the distance of the bead to the surface with sub-nanometer precision using evanescent field illumination (data not shown). The intensity of the light diffused by each bead provides a very sensitive proxy for the extension of the tethering molecule. This diffused light could be easily measured and multiplexed on a standard CCD or CMOS camera with each pixel (or small group of pixels) being used as a sensor, thus providing a cheap high-throughput readout. Finally, since all the molecules used on our platform are standard oligonucleotides with no expensive modifications such as fluorescent labeling, the overall cost of this platform should be minimal.
CONCLUSION
We have presented here a proof of principle for a new single-molecule platform for DNA identification and sequencing. We have investigated various schemes that can in principle achieve sequence identification or DNA sequencing with higher accuracy than other current single-molecule approaches.
When the proposed process is automated, one will be able to sequence a genome or quantify a cDNA sample. Because magnetic traps act simultaneously on many beads, the present scheme is inherently parallelizable. The resolution and throughput can be readily improved by detecting the light diffracted by the beads using evanescent wave illumination28, 29. Automating buffer exchanges and implementing a mechanized scanning stage are obvious improvements towards that high-throughput goal. Finally, notice that with a ligation scheme, the change in hairpin extension is permanent allowing for scanning and interrogating the state of a very large number of molecules.
The major advantage of the present approach is in the nature of the detected signal, namely the extension of a DNA hairpin which can be used to measure the distance between two sequences along it. This measure provides for a continuous signal, rather than a binary on/off signal as with most fluorescent methods. It is achieved with regular non-fluorescent imaging and standard cameras, which should provide for a cheaper alternative to current methods. Moreover, the continuous extension signal is more informative than a fluorescence signal. For instance, in the hybridization fingerprinting method, the position of the hybridization events can be detected with almost single base accuracy. As a result, the hybridization pattern along the sequence, rather than the mere presence of hybrids, can be used as a fingerprint for a DNA sequence. This type of information is new and may open the way for novel applications beyond the mere adaptation of the present NGS sequencing schemes demonstrated here.
METHODS
DNA hairpin construction
Short DNA hairpins (83 bps in Fig.1, 2 and 5, or 295 bps in Fig. 4, or 179 bps in Fig. 2 and Supplementary Fig. 7) were constructed by ligating three separate synthetic oligonucleotides (Eurogentec and Integrated DNA technology) as shown in Supplementary Fig. 4. In the first step, oligo A-1 (5′-biotin-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTG GAT TCG CGG GTC TCT-3′) and oligo A-2 (5′-AAC CGT CCT TTA CTT GTC ATG CGC TCT AAT CTC TGG GCA TCT GGC TAT GAT GTT GAT GGA ACT GAC CAA ACG TCG GTG GG-3′) were annealed to the complementary oligo A-3 (5'-phosphorylated-AGG AAG AGA CCC GCG AAT CCC CCA CCG ACG TTT GGT CAG TT-3') in dH2O by heating to 95 °C for 5 min, then rapidly cooling to 80 °C, followed by a slow decrease of 0.7 °C every 10 s until reaching 4 °C. These annealed products, marked part A, were cleaned up with NucleoSpin Extract II kits (Clontech). We repeated the annealing and clean up procedure for oligo B-1 (5'-phosphorylated-TCC TGA TTG TCA TCG GCT GGC TGT TCG GTT AGT TTC GAA GAC TT-3′) and oligo B-2 (5'-phosphorylated-GCG AAA GTC TTC GAA ACT AAC CGA ACA GCC AGC CGA TGA CAA TC-3′), which correspond to the middle section of the 83 bps hairpin, or oligo B'-1 (5'-phosphorylated-TCC TCG TGC GTG AGC GAG CGC GGT CGG TCG GTC GGT AGC GAG CGC GTG CGT GCG TGC GTG GGC TGG CTG GCT GGC TCG GTC GGT CGT GCG TGC GGT CGG TGG CTG GCT AGC GAG CGA GCG AGC GGG CTG GCT GAA GAC TT-3') and oligo B'-2 (5'-phosphorylated-GCG AAA GTC TTC AGC CAG CCC GCT CGC TCG CTC GCT AGC CAG CCA CCG ACC GCA CGC ACG ACC GAC CGA GCC AGC CAG CCA GCC CAC GCA CGC ACG CAC GCG CTC GCT ACC GAC CGA CCG ACC GCG CTC GCT CAC GCA CG-3'), which correspond to the middle section of the 179 bps hairpin, or oligo B”-1 (5'-phosphorylated-TCC TTA GCC AGC CAC CGC GCT AGC CCA CGC GCT ACC GCA CGA GCC ACC GAC CGC ACG CAC GCG CTC GCT AGC CAG CCA CCG CGC TAG CCC ACG CGC TAC CGC ACG AGC CAC CGA CCG CAC GCA CGC GCT CGC TAG CCA GCC ACC GCG CTA GCC CAC GCG CTA CCG CAC GAG CCA CCG ACC GCA CGC ACG CGC TCG CTA GCC AGC CAC CGC GCT AGC CCA CGC GCT ACC GCA CGA GCC ACC GAC CGC ACG CAC GCG C-3′) and oligo B”-2 (5'-phosphorylated-GCG AGC GCG TGC GTG CGG TCG GTG GCT CGT GCG GTA GCG CGT GGG CTA GCG CGG TGG CTG GCT AGC GAG CGC GTG CGT GCG GTC GGT GGC TCG TGC GGT AGC GCG TGG GCT AGC GCG GTG GCT GGC TAG CGA GCG CGT GCG TGC GGT CGG TGG CTC GTG CGG TAG CGC GTG GGC TAG CGC GGT GGC TGG CTA GCG AGC GCG TGC GTG CGG TCG GTG GCT CGT GCG GTA GCG CGT GGG CTA GCG CGG TGG CTG GCT A-3′), which correspond to the middle section of the 295 bps hairpin (due to the length limit of commercial oligonucleotides, this B” part is actually generated by the ligation of four 64nt oligonucleotides). These annealed products are marked part B or B' or B”. We ligated part A, part B (or part B', B”) to oligo C (5'-phosphorylated-TCG CGC CTG ATC GTC CAC TTT TTT TTT AGT GGA CGA TCA GGC-3′), which is the loop of hairpin, using T4 ligase (5 U/μl, Fermentas) in the 1 × T4 ligase reaction buffer at 25 °C for 1.5 h, then stopped the reaction by heating to 65 °C for 20 min. The ligation mixture was cleaned up with NucleoSpin Extract II kits. Finally, the digoxigenin labels were added by a fill-in reaction using Klenow(3′→5′ exo-) (New England Biolabs) in the 1× NEB2 buffer with 1 mM digoxigenin-dUTP (Roche) at 37 °C for 15 min, and stopped by heating to 75 °C for 20 min. The hairpin products were cleaned up with NucleoSpin Extract II kits again.
The preparation method for the 1214 bps hairpin (Fig. 2, 3, Supplementary Fig. 2 and Supplementary Fig. 6) is described in Reference 27.
Single-molecule assay
The constructed hairpin, labeled at one end with digoxigenin and biotin at the other, was attached to the glass surface of a microscope flow chamber previously coated with anti-digoxigenin and passivated with bovine serum albumin (BSA), and to a 1 μm super-paramagnetic bead coated with streptavidin (DYNAL MyOne). Small magnets were used to pull on single DNA molecules attached to the beads. The acquisition of the molecule's extension Z(t) is done with a CCD camera at 31 Hz. The raw data are averaged over 1 s, achieving a resolution of ~1 nm, while the stretching force, F, is measured with 10% accuracy. During data collection we subtract the vertical position of reference beads stuck to the surface (Zref) from the vertical position traces Z(t), which helps to reduce set-up drift. For every opening/closing cycle, we take the center (<Zclose>) of a Gaussian fit to Zclose(Ftest) as the reference position of the closed hairpin (extension 0 nm). Similar Gaussian fits are used to determine the average position in one cycle of Zopen(Ftest) and Zblock(Ftest). The error bars represent the s.e.m.
For the hybridization identification in Figure 2, molecule 1 is the 1214 bps hairpin, molecule 2 is the 83bps hairpin, and molecule 3 is the 179 hairpin, while oligo 1 is 5'-GAAGAGACCC-3' and oligo 2 is 5'-CAGCCGATGAC-3'.
The ligation reaction in Figure 3, Table 1 and Supplementary Figure 2, 6, and 7 was performed in 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 1 mM DTT, 1 mM ATP, 0.2% (w/v) BSA, and 0.02 U/μl T4 DNA ligase (Fermentas). For Figure 3, the 10-nt primer is 5'-phos-ACA GCC AGC A-3' and the 7-nt oligo is 5'-TAACCGA-3'. For Table 1, the expected AATCAGG sequence was tested with an initial primer 5'-phos-ACATACATCAG-3' and ~2 μM oligonucleotide NNNXNNrN, while the expected GAGCGGA sequence was tested with an initial primer 5'-phos-AACTAACCGA-3 and ~2 μM oligonucleotide NNNXNNrN. For Supplementary Figure 7 and 9 the oligonucleotides ligated are 5'-phos-ACCGACCG-3', 5'-phos-CGCTACCG-3', 5'-phos-CACGCGCT-3', 5'-phos-CACGCACG-3', 5'-phos-AGCCCACG-3', 5'-phos-AGCCAGCC-3', 5'-phos-ACCGAGCC-3', 5'-phos-CACGACCG-3' and 5'-phos-ACCGCACG-3'.
The hybridization sequencing in Figure 4 was performed in 3 M TMACL [Tetramethylammonium Chloride, Sigma] (30) and 0.5% (w/v) BSA with ~100 nM oligonucleotides: for 295 bps hairpin in the Fig. 3b, using A8 (GCCACCGA), T8 (GCACGCCA), G8 (TCGCGCAC) and C8 (CCGATCGC) separately. We recorded the blockage position on the complimentary hairpin stem; T = 25 °C.
The ligation sequencing in Figure 5 was performed in 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 1 mM DTT, 1 mM ATP and 0.2% (w/v) BSA for the ligation reaction. We repeatedly opened and closed the hairpin for ~10 min in the presence of ~5 μM oligonucleotide NNNNNrNX and 0.02 U/μl T4 DNA ligase (Fermentas). Then, we changed the buffer to 10 mM Tris-HCl pH 7.5, 1 mM EDTA and 0.02 U/μl RiboShredder (EPICENTRE Biotechnologies) for ~10 min while reducing the stretching force to ~4 pN in order to increase the efficiency of the RNase. Finally, we kept the stretching force at Ftest and flushed the previous buffer with 0.2 U/μl T4 Polynucleotide Kinase (Fermentas) to phosphorylate the 5' end of the ligated and cleaved oligonucleotide. Note, the RNase Blend can also work in the ligase/kinase buffer, only less efficiently. We always kept the stretching force at Ftest when changing the buffers. All experiments were performed at T = 25 °C.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge useful suggestions by M. Volovitch, T. Lionnet and K. Neuman. This work was supported by an ERA-MolMachines grant (to D.B.), a Human Frontier Science Program grant (RGP003/2007 to V.C. and S.J.B) and the ERC grant `MagRepS' 267862 (to V.C., M. M. and S.J.B).
Footnotes
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
REFERENCES
- 1.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 3.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 4.Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc. Natl. Acad. Sci.USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nat. Biotechnol. 2009;27:847–852. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pihlak A, et al. Rapid genome sequencing with short universal tiling probes. Nat. Biotechnol. 2008;26:676–684. doi: 10.1038/nbt1405. [DOI] [PubMed] [Google Scholar]
- 7.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 8.Blow N. DNA sequencing: generation next-next. Nat. Methods. 2008;5:267–274. [Google Scholar]
- 9.Metzker ML. Sequencing technologies - the next generation. Nat. Rev. Genet. 2009;11:31. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 10.Fuller CW, et al. The challenges of sequencing by synthesis. Nat. Biotechnol. 2009;27:1013–1023. doi: 10.1038/nbt.1585. [DOI] [PubMed] [Google Scholar]
- 11.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 12.Greenleaf WJ, Block SM. Single-molecule, motion-based DNA sequencing using RNA polymerase. Science. 2006;313:801. doi: 10.1126/science.1130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munroe DJ, Harris TJR. Third-generation sequencing fireworks at marco island. Nat. Biotechnol. 2010;28:426–428. doi: 10.1038/nbt0510-426. [DOI] [PubMed] [Google Scholar]
- 14.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nano. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 15.Treffer R, Deckert V. Recent advances in single-molecule sequencing. Curr. Opin. Biotechnol. 2010;21 doi: 10.1016/j.copbio.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Husale S, Persson HHJ, Sahin O. DNA nanomechanics allows direct digital detection of complementary DNA and microRNA targets. Nature. 2009;462:1075–1078. doi: 10.1038/nature08626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark MD, et al. An oligonucleotide fingerprint normalized and expressed sequence tag characterized zebrafish cDNA library. Genome. Res. 2001;11:1594–1602. doi: 10.1101/gr.186901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herwig R, et al. Information theoretical probe selection for hybridisation experiments. Bioinformatics. 2000;16:890–898. doi: 10.1093/bioinformatics/16.10.890. [DOI] [PubMed] [Google Scholar]
- 20.Guerasimova A, et al. New tools for oligonucleotide fingerprinting. Biotechniques. 2001;31:490–495. doi: 10.2144/01313st01. [DOI] [PubMed] [Google Scholar]
- 21.Gosse C, Croquette V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys. J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strick TR, Allemand J, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 23.Brower-Toland BD, et al. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Essevaz-Roulet B, Bockelmann U, Heslot F. Mechanical separation of the complementary strands of DNA. Proc. Natl. Acad. Sci. USA. 1997;94:11935–11940. doi: 10.1073/pnas.94.22.11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNally B, et al. Optical recognition of converted DNA nucleotides for Single-Molecule DNA sequencing using nanopore arrays. Nano. Lett. 2010;10:2237–2244. doi: 10.1021/nl1012147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mir KU, Qi H, Salata O, Scozzafava G. Sequencing by cyclic ligation and cleavage (CycLiC) directly on a microarray captured template. Nucl. Acids. Res. 2009;37:e5. doi: 10.1093/nar/gkn906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manosas M, Spiering MM, Zhuang Z, Benkovic SJ, Croquette V. Coupling DNA unwinding activity with primer synthesis in the bacteriophage t4 primosome. Nat. Chem. Biol. 2009;5:904–912. doi: 10.1038/nchembio.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim K, Saleh OA. A high-resolution magnetic tweezer for single-molecule measurements. Nucleic. Acids. Res. 2009;37:e136. doi: 10.1093/nar/gkp725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh-Zocchi M, Dixit S, Ivanov V, Zocchi G. Single-molecule detection of DNA hybridization. Proc. Natl. Acad. Sci. USA. 2003;100:7605–7610. doi: 10.1073/pnas.1337215100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melchior WB, Von Hippel PH., Jr. Alteration of the relative stability of dA - dT and dG* dC base pairs in DNA. Proc. Nat. Acad. Sci. USA. 1973;70:298. doi: 10.1073/pnas.70.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.