Abstract
Adipose tissues regulate metabolism, reproduction, and life span. The development and growth of adipose tissue are due to increases of both adipocyte cell size and cell number; the latter is mediated by adipocyte progenitors. Various markers have been used to identify either adipocyte progenitors or mature adipocytes. The fatty acid binding protein 4 (FABP4), commonly known as adipocyte protein 2 (aP2), has been extensively used as a marker for differentiated adipocytes. However, whether aP2 is expressed in adipogenic progenitors is controversial. Using Cre/LoxP-based cell lineage tracing in mice, we have identified a population of aP2-expressing progenitors in the stromal vascular fraction (SVF) of both white and brown adipose tissues. The aP2-lineage progenitors reside in the adipose stem cell niche and express adipocyte progenitor markers, including CD34, Sca1, Dlk1, and PDGFRα. When isolated and grown in culture, the aP2-expressing SVF cells proliferate and differentiate into adipocytes upon induction. Conversely, ablation of the aP2 lineage greatly reduces the adipogenic potential of SVF cells. When grafted into wild-type mice, the aP2-lineage progenitors give rise to adipose depots in recipient mice. Therefore, the expression of aP2 is not limited to mature adipocytes, but also marks a pool of undifferentiated progenitors associated with the vasculature of adipose tissues. Our finding adds to the repertoire of adipose progenitor markers and points to a new regulator of adipose plasticity.—Shan, T., Liu, W., Kuang, S. Fatty acid-binding protein 4 expression marks a population of adipocyte progenitors in white and brown adipose tissues.
Keywords: mouse, stromal vascular fraction
Adipose tissues play important regulatory roles in the metabolism, reproduction, and life span of animals. In mammals, there are two types of adipose tissues: white adipose tissue (WAT) and brown adipose tissue (BAT). WAT and BAT have distinct morphologies and functions. WAT adipocytes store triglycerides as energy source and each has a single large lipid (fat) droplet. By contrast, BAT adipocyte contains multiple small lipid droplets and many mitochondria enriched with uncoupling protein 1 (UCP-1), which uncouples oxidative phosphorylation from ATP synthesis, leading to energy dissipation (1). More recent studies demonstrate that many subcutaneous adipose depots traditionally classified as WAT also express brown adipose-specific markers and are thus referred to as “beige” or “brite” adipose (2–4).
The development and postnatal growth of adipose tissue are due to increases of both adipocyte cell size (hypertrophy) and cell number (hyperplasia); the latter depends on the contribution of adipocyte progenitors (5). Therefore, identification of adipogenic progenitors and elucidation of their differentiation potential are critical for understanding adipose plasticity. Recently, white adipocyte stem or progenitor cells have been identified from WAT using cell-surface marker and lineage-tracing experiments (6, 7). These cells are able to differentiate into mature adipocyte in vitro and form adipose depots in vivo (6, 7). Brown adipocyte progenitors have also been isolated from different adipose depots and skeletal muscles using cell surface markers (8, 9). Presently, several markers have been identified and defined in either adipocyte progenitors or mature adipocytes (10). Among them, CD34, stem cell antigen 1 (Sca1), decorin, and platelet-derived growth factor receptor α (PDGFRα) have been reported as the stem cell markers (7, 10–12), while perilipin, adiponection, and fatty acid synthase (FAS) have been used as the mature adipocyte markers (10).
The fatty acid binding protein 4 (FABP4), commonly known as adipocyte protein 2 (aP2), has been extensively used as a marker for differentiated adipocytes. Whether aP2 is expressed in adipocyte progenitors is controversial. It was reported that there is no aP2 expression in adipose stromal vascular fraction (SVF) cells (7), a population of heterogeneous cells, including the adipose progenitor cells (5, 13). However, other reports suggested that aP2 is expressed by preadipocytes (14, 15). In addition, it has been shown that aP2 is expressed in embryonic day 9.5, long before the formation of adipocytes (16). These results indicated that aP2 may be expressed by adipocyte progenitors, though definitive evidence supporting this notion has been lacking.
Stem cell niche refers to the tissue microenvironment where an adult stem cell resides. Stem cell niche not only regulates the behavior and function of the resident stem cells, but also provides an anatomical and structural basis for stem cell identification. Adipocyte stem and progenitor cells occupy a niche closely associated with blood vessels. Specifically, PPARγ-lineage-tracing experiments demonstrate that PPARγ+ progenitors are located on the surface of adipose vasculatures and coexpress mural cell (pericyte) marker PDGFRβ (7), suggesting the mural cell compartment as a stem cell niche for adipose progenitors. More recent studies reported that a proportion of Zfp423-GFP-labeled adipose progenitors in WAT and BAT are also located in the endothelial layer of blood vessels (17). Ultrastructure analysis and VE-cadherin labeling support the notion that these cells are of endothelial origin and give rise to preadipocytes (18). Therefore, adipose stem cells can be found in the endothelium and pericyte niches of adipose vasculatures.
In this study, we used cell-lineage labeling, lineage ablation, fluorescence-activated cell sorting (FACS), and cell transplantation to demonstrate the adipogenic potential of aP2-lineage progenitors. We first conducted cell-lineage-tracing experiments to dissect the progeny of aP2 progenitors in various tissues, and identified a population of aP2+ adipocyte progenitors in SVF of both WAT and BAT. We also showed that the aP2+ progenitor cells reside in the adipose stem cell niche and express adipocyte progenitor markers, including CD34, Sca1, Dlk1 (Pref-1), and PDGFRα. Finally, using cell-lineage ablation and FACS techniques, we investigated the proliferation and differentiation capacity of the aP2+ SVF cells in vitro and in vivo. Our findings demonstrate that aP2 expression marks a population of adipocyte progenitors in brown and white adipose tissues. These results provide novel insights into adipose tissue development and postnatal plasticity; such knowledge may lead to strategies to control adipose tissue dynamics and treat metabolic diseases.
MATERIALS AND METHODS
Animals
All procedures involving mice were performed in accordance with the Purdue University Animal Care and Use Committee. Mice were housed in the animal facility with free access to standard rodent chow and water. All mice were from Jackson Laboratory (Bar Harbor, ME, USA) under these stock numbers: aP2-Cre, stock no. 005069; Rosa26-iDTR, 007900; Rosa26-tdTomato, 007905; and Rosa26-EYFP, 007903. The PCR genotyping was done using protocols described by the supplier.
SVF cell and stromal vascular particulate (SVP) isolation
SVF cells were isolated using collagenase digestion, followed by density separation. Briefly, subcutaneous WAT depots and interscapular BAT depots were collected and minced into 2- to 5-mm2 pieces. The WAT pieces were then digested in 1.5 mg/ml collagenase at 37°C for 1.5–2 h, and the BAT pieces were digested for 0.5 h. The digestions were stopped with defined minimal essential medium (DMEM) containing 10% fetal bovine serum (FBS); floating cells on the top of the medium were collected as mature adipocytes, while the cell suspensions were filtered through 100-μm filters and centrifuged at 450 g for 5 min. The isolated cells were seeded in tissue culture dishes or subjected to FACS. Adipose SVPs were isolated according to published methods (7). Breifly, WAT depots were digested in 1.5 mg/ml collagenase for 1.5–2 h, and passed through a 100-μm mesh and then a 30-μm mesh. SVPs retained on the 30-μm mesh were rinsed with DMEM and collected. Vasculature debris (tubular debris) was picked for examination of perivascular aP2-lineage cells.
Cell culture
Freshly isolated SVF cells from the WAT and BAT were cultured in growth medium containing DMEM, 20% FBS, and 1% penicillin/streptomycin at 37°C with 5% CO2 for 3 d, followed by feeding with fresh medium every 2 d. Upon confluence, the cells were induced with induction medium containing DMEM, 10% FBS, 2.85 μM insulin, 0.3 μM dexamethasone (DEXA), and 0.63 mM 3-isobutyl-methylxanthine (IBMX) for 3 d and then differentiated in differentiation medium containing DMEM, 200 nM insulin and 10 nM triiodothyronine for 4 d until adipocytes matured. For cell ablation in culture, the SVF cells from WAT and BAT of the aP2-Cre/Rosa26-iDTR mice were treated with a final diphtheria toxin (DT) concentration of 200 ng/ml in culture medium for 48 h. To avoid the effect of cell density on adipogenic differentiation, the control and the DT-treated cells were induced for differentiation when they reached at least 90% confluence, regardless of the days in culture.
FACS
The red fluorescent protein (RFP)+ (tdTomato+) and RFP− SVF cells were isolated from WAT and BAT tissues of aP2-Cre/Rosa26-tdTomato mice using FACS. SVF cells were isolated as described above and filtered through a 30-μm filter before sorting. SVF cells from wild-type mice were used as negative control for gating the RFP+ cells. About 40–50% cells were RFP+, and conservative gating was used to eliminate weak signals and select the bright RFP+ cells (see Fig. 7A, B). After sorting, RFP− and RFP+ cells were collected for extracting RNA directly or seeded in culture plates for differentiation assay. In addition, RFP+ cells from WAT of aP2-Cre/Rosa26-tdTomato were also used for transplantation experiments. To examine subpopulations of progenitor cells in the SVF, we sorted cells based on Lin− (CD45−, CD31−, and TER119−), Sca1+/− (Biolegend, San Diego, CA, USA), and RFP+/−. From the Lin− cells, we isolated 4 subpopulations (RFP+Sca1+, RFP+Sca1−, RFP−Sca1+, and RFP− Sca1−) of SVF cells, then cultured and differentiated them to examine their adipogenic potential in vitro.
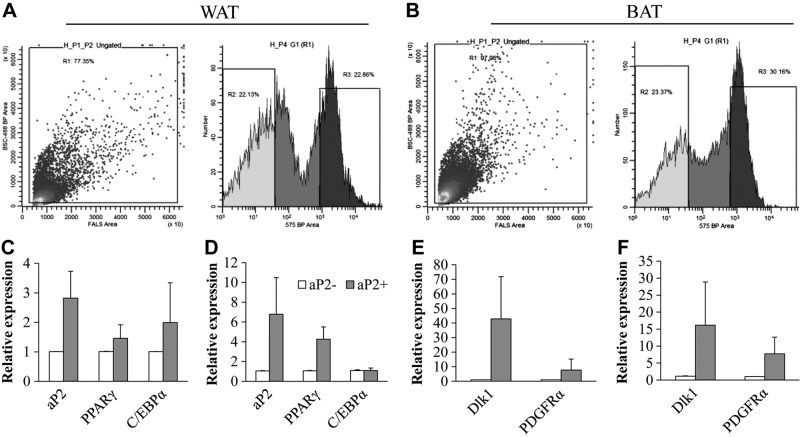
Figure 7.
The aP2-lineage SVF cells express higher levels of adipocyte progenitor cell markers. A, B) FACS of RFP+ (aP2-lineage) and RFP− (non aP2-lineage) SVF cells from the WAT (A) and BAT (B) of aP2-Cre/Rosa26-tdTomato mice. C, D) Relative expression of adipogenic genes in RFP− and RFP+ SVF cells. E, F) Relative expression of adipoprogenitor markers Dlk1 and PDGFRα in RFP− and RFP+ SVF cells. Error bars = se; n = 3.
Oil red O staining
Cultured cells were washed with PBS and fixed with 10% formaldehyde for 15 min at room temperature. Then, the cells were stained using the Oil red O work solutions containing 6 ml of Oil red O stock solution (5 mg/ml in isopropanol) and 4 ml ddH2O for 30 min. After staining, the cells were washed with 60% isopropanol and imaged.
Total RNA extraction, cDNA synthesis, and real-time PCR
Total RNA extraction, cDNA synthesis, and real-time PCR were performed as described previously (19, 20). Briefly, total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. RNA was treated with RNase-free DNase l to remove contaminating genomic DNA. The purity and concentration of total RNA were measured by a spectrophotometer (Nanodrop 3000; Thermo Fisher, Waltham, MA, USA) at 260 and 280 nm. Ratios of absorption (260/280 nm) of all samples were between 1.8 and 2.0. Then 5 μg of total RNA was reverse transcribed using random primers and M-MLV-reverse transcriptase. Real-time PCR was carried out in a Roche LightCycler 480 PCR System (Roche, Basel, Switzerland) with SYBR Green master mix and gene-specific primers. Primer sequences are from published papers (8, 18, 20). The 2−ΔΔCT method was used to analyze the relative changes in each gene's expression normalized against 18S rRNA expression. All measurements were repeated ≥3 times.
Protein extraction and Western blot analysis
Total protein was isolated from cells using RIPA buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS). Protein concentrations were determined using Pierce BCA protein assay reagent (Pierce Biotechnology, Rockford, IL, USA). Proteins were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a PVDF membrane (Millipore Corp., Billerica, MA), and incubated with aP2 antibody (sc-19857, 1:500 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or GAPDH antibody (sc-47778, 1:1000 dilution; Santa Cruz Biotechnology). The secondary antibody (anti-rabbit IgG or anti-mouse IgG, Santa Cruz Biotechnology) was diluted 8000-fold. Immunodetection was performed using enhanced chemiluminescence (ECL) Western blotting substrate (Pierce Biotechnology) and detected with a Gel Logic 2200 imaging system (Carestream, Rochester, NY, USA).
Immunostaining and image acquisition
Immunostaining was performed as described previously (20–22). Briefly, adipose SVF cells were seeded on slide and cultured for 3 h to ensure that the cells were adhered. Cells or tissue sections were fixed with 4% paraformaldehyde, and the fixed cells or sections were blocked with blocking buffer containing 5% goat serum, 2% bovine serum albumin, 0.2% Triton X-100, and 0.1% sodium azide in PBS for 1 h. The samples were incubated with primary antibodies diluted in blocking buffer overnight. After washing with PBS, the samples were incubated with secondary antibodies and Hoechst for 45 min at room temperature. Fluorescent images were captured using Leica DM 6000B fluorescent microscope (Leica Microsystems, Wetzlar, Germany).
Transplantation
FACS-purified RFP+ cells from WAT of aP2-Cre/Rosa26-tdTomato mice were mixed with 1 ng/ml basic fibroblast growth factor (bFGF) and injected subcutaneously into 4-wk-old wild-type mice. After 3 wk, the recipient mice were euthanized, and the adipose tissues near the injection sites were collected and examined for RFP expression using a fluorescent microscope.
Data analysis
All experimental data are presented as means ± se. Comparisons were made by unpaired 2-tailed Student's t tests or 1-way ANOVA, as appropriate. Effects were considered significant at P < 0.05.
RESULTS
The aP2 cell lineage contributes to both mature adipocytes and SVF cells
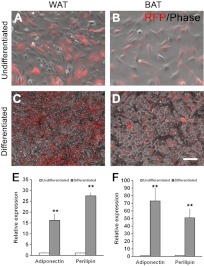
To investigate the progeny of aP2-lineage cells in various tissues, we conducted lineage-tracing experiments using aP2-Cre driver and Rosa26-tdTomato or Rosa26-EYFP reporter mice, in which aP2-lineage cells are labeled by tdTomato, an RFP, or enhanced yellow fluorescent protein (EYFP). We first examined the aP2-Cre/Rosa26-tdTomato mice. As expected, the RFP signals were strongly activated and readily detectable in whole-mount WAT and BAT (Fig. 1A, B). Inspection of WAT and BAT cross sections revealed that aP2 lineage contributes to vast majority of the cells (mature adipocytes) in these tissues (Fig. 1C, D). These results indicate that the aP2-Cre-lineage tracing truthfully marks mature adipocytes.
Figure 1.
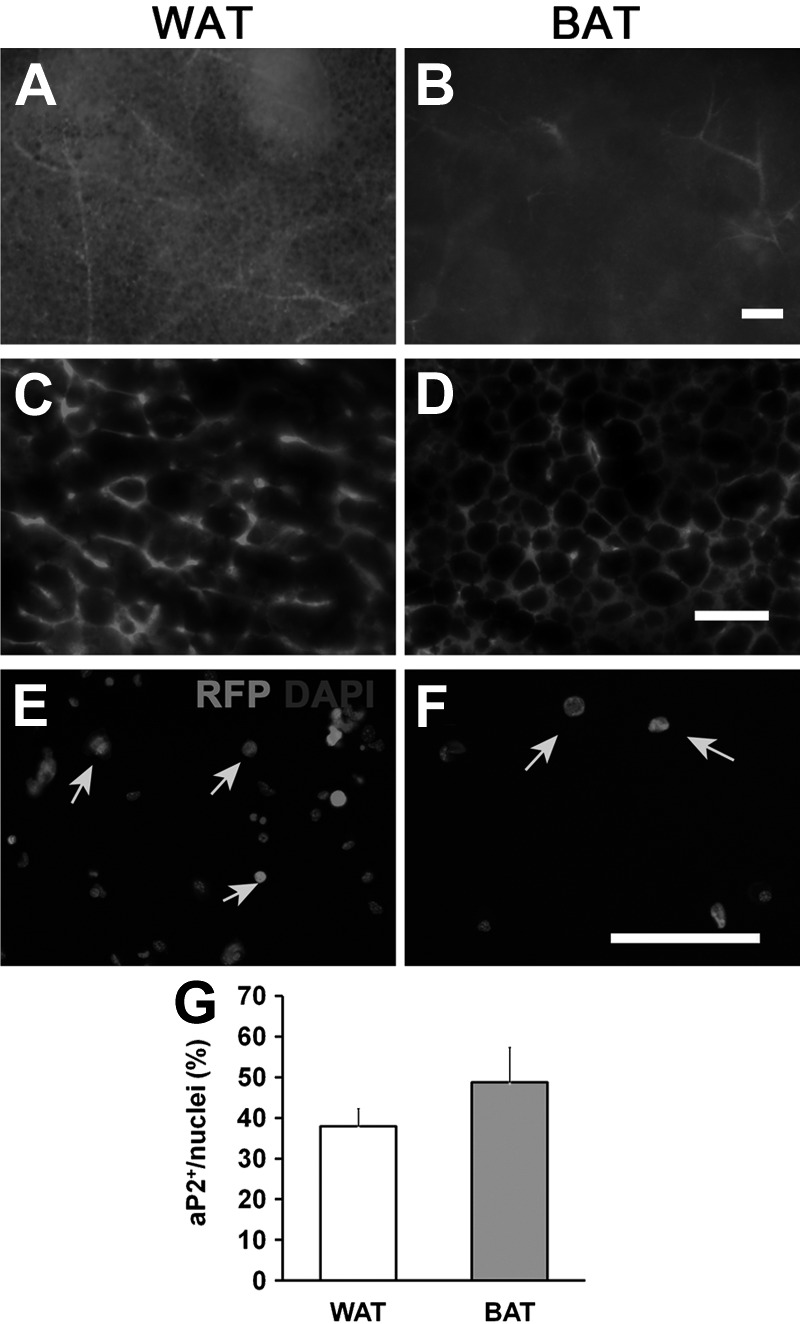
aP2-lineage tracing in adipose tissues and SVF cells. A, B) Whole-mount fresh tissues from WAT (A) and BAT (B) of aP2-Cre/Rosa26-tdTomato mice were used. In this model, all aP2-lineage cells were marked by tdTomato (RFP) expression. Note the intense RFP signal in vascular structures. C, D) Sections of WAT (C) and BAT (D) tissues showing high levels of RFP expression in many cells. E, F) RFP expresses in WAT (E) and BAT (F) SVF cells. Red, RFP; blue, 4′,6-diamidino-2-phenylindole (DAPI). G) Percentage of RFP+ cells in SVFs from WAT and BAT. Arrows indicate RFP positive cells. Error bars = se; n = 5. Scale bars = 200 μm (A, B); 100 μm (C–F).
To examine whether aP2-lineage tracing also labels undifferentiated progenitors and immature preadipocytes in BAT and WAT, we isolated cells from the SVF. After immunostaining the SVF cells, there were many RFP+ cells (Fig. 1E, F). Quantitative analysis indicates that ∼40% cells from the SVF of WAT are RFP+; and 50% cells from the SVF of BAT are RFP+ (Fig. 1G). Using a separate reporter line (aP2-Cre/Rosa26-EYFP), we observed similar results (data not shown), confirming the reproducibility of our models for lineage tracing. Together, these results demonstrate that aP2-lineage cells contribute to both mature and undifferentiated cells within the WAT and BAT.
Concurrent expression of aP2 in the aP2-Cre-lineage-labeled SVF cells
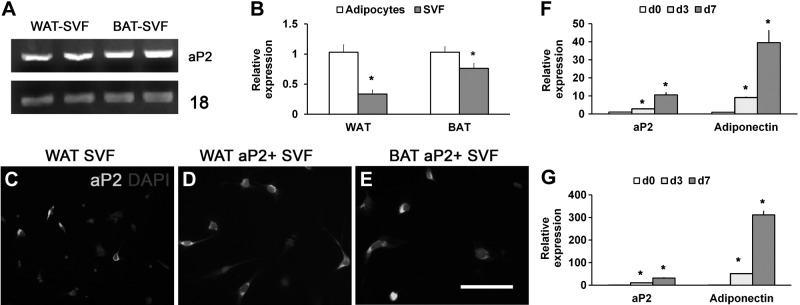
As adipose SVF is enriched with adipogenic progenitors, our result that 40–50% of the SVF cells are marked by aP2-Cre-lineage tracing raises the possibility that some adipoprogenitors concurrently express aP2. Reverse transcriptase (RT)-PCR results showed that aP2 mRNA is indeed expressed in SVF cells from WAT and BAT (Fig. 2A). Immunostaining with aP2 antibody further confirmed that roughly half of the SVF cells are aP2+ (Fig. 2C). Notably, FACS-purified RFP+ SVF cells for the aP2-Cre/Rosa26-tdTomato mice unanimously expressed the aP2 protein (Fig. 2D, E). We next compared the relative expression levels of aP2 in progenitors vs. mature adipocytes, using SVF cells and floating cells as representatives. Real-time quantitative PCR (qPCR) revealed that aP2 expression levels in the SVF cells were ∼30 and 70% of those in the mature adipocytes isolated from WAT or BAT, respectively (Fig. 2B). Thus, adipose SVF cells express the aP2 gene, but at lower levels compared to mature adipocytes. Consistently, the expression levels of aP2 and adiponectin increased in a time-dependent manner during the differentiation course of FACS-purified RFP+ SVF cells (Fig. 2F, G). These results demonstrate that aP2 is concurrently expressed in a population of SVF progenitor cells, and its expression increases during adipogenic differentiation.
Figure 2.
Concurrent expression of endogenous aP2 in SVF cells. A) RT–PCR results showing expression of aP2 in adipose SVF cells. B) Real-time qPCR results of the relative expression of aP2 in mature adipocytes and SVF cells isolated from WAT and BAT. *P < 0.05. C–E) Immunostaining of aP2 expression in unsorted SVF cells (C), and FACS-purified aP2+ SVF cells from WAT (D) and BAT (E). F, G) qPCR results showing the relative expression of aP2 and adiponectin during differentiation of the sorted aP2+ cells. *P < 0.05 vs. d 0. Error bars = se; n = 3.
The aP2-lineage SVF cells express proliferation and adipose stem cell markers
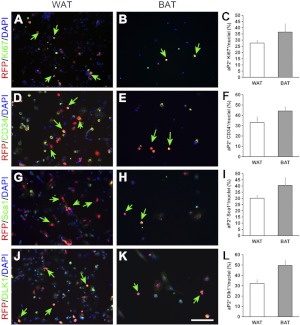
If the SVF cells derived from the aP2 lineage represent a population of progenitor cells, they should be able to proliferate and express established markers for adipose stem cells. Indeed, many aP2-lineage (RFP+) cells from the aP2-Cre/Rosa26-tdTomato mice coexpressed the cell proliferation marker Ki67 in culture (Fig. 3A, B). Overall, ∼25% WAT and 35% BAT SVF cells were RFP and Ki67 double positive (Fig. 3C). Transmembrane glycoprotein CD34 is frequently used as a marker of the adipocyte progenitors (6, 11). We found that >30% SVF cells in WAT and 40% in BAT are double positive for RFP and CD34 (Fig. 3D, F). We also examined the expression of another stem cell marker, Sca1 (6, 8), and found that nearly 30 and 40% of SVF cells in WAT and BAT, respectively, are double positive for Sca1 and RFP (Fig. 3G–I). Furthermore, we examined the expression of Dlk1, an established preadipocyte marker (23) and found that there are ∼30 and 50% of RFP and Dlk1 double-positive cells in SVF of WAT and BAT, respectively (Fig. 3J–L).
Figure 3.
The aP2-lineage (RFP+) SVF cells express proliferation and adipose stem and progenitor cell markers. A–C) Immunostaining results of WAT and BAT SVF cells showing coexpression of RFP (red) and the proliferation marker Ki67 (green) in many cells. D–F) Coexpression of RFP (red) and the progenitor cell marker CD34 (green). G–I) Coexpression of RFP (red) and the stem cell marker Sca1 (green). J–L) Coexpression of RFP (red) and the preadipocyte marker Dlk1 (green). Nuclei are counterstained with DAPI (blue). Green Arrows indicate RFP and progenitor marker double positive cells. Error bars = se; n = 3. Scale bars = 100 μm.
We further compared the relative abundance of CD34+ and Sca1+ cells in RFP+ and RFP− SVF cells. Overall, 83% of aP2+ SVF cells in WAT and BAT were also CD34+, while only 36% and 27% of aP2− cells in WAT and BAT were double positive for CD34 (Supplemental Fig. S1A). Similar profiles were true for the Sca1 expression pattern: 86% and 89% of aP2+ SVF cells in WAT and BAT were also Sca1+, compared to only 25% and 21% of aP2− cells that were Sca1+ (Supplemental Fig. S1B). Conversely, when Lin− (CD31−, CD45−, Ter119−), and Sca1+ SVF cells of WAT and BAT were sorted, we found that >60% of these cells were RFP+ (Supplemental Fig. S1C–E). Further analysis indicates that 60–70% of RFP+ SVF cells in WAT and BAT expressed the proliferation marker Ki67 (data not shown). These results indicated that the majority of RFP+ (aP2-lineage) SVF cells in WAT and BAT express adipogenic stem cell and proliferation markers, and thus represent a population of undifferentiated progenitors.
The aP2-lineage progenitors reside in the adipose stem cell niche
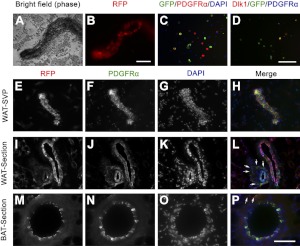
Previous studies have shown that the perivascular niche harbors adipose stem cells in the WAT (6, 7). More recently, it has been shown that BAT and WAT progenitors may also be localized in the vascular endothelium niche (17, 18). We sought to identify the in vivo localization of aP2-lineage progenitor cells. First, we isolated small vessels from the WAT SVP of aP2-Cre/Rosa26-tdTomato mice and found that many cells associated with the vessel were RFP+ (Fig. 4A, B). Immunostaining of cultured SVP cells from aP2-Cre/Rosa26-EYFP mice with PDGFRα, a marker for adipogenic progenitors and vascular mural cells (24, 25), and the preadipocyte marker Dlk1 indicated that a population of vessel-associated aP2-lineage cells coexpress these markers (Fig. 4C, D). Colabeling the SVP vessels from aP2-Cre/Rosa26-tdTomato mice with PDGFRα revealed that almost all RFP+ cells were also PDGFRα+ (Fig. 4E–H). These results indicate that aP2-lineage cells can be found in the perivascular niche on isolated small vessels.
Figure 4.
The aP2-lineage (RFP+) cells are associated with adipose vessels. A, B) Vessel from WAT SVP of aP2-Cre/Rosa26-tdTomato mice photographed under phase-contrast (A) and fluorescent (B) imaging. C) Cells cultured from SVP of aP2-Cre/Rosa26-EYFP mice coexpressed YFP (green) and PDGFRα (red). DAPI (blue) counterstain indicates nuclei. D) Coexpression of YFP (green) and Dlk1 (red). E–H) Whole-mount aP2-Cre/Rosa26-tdTomato SVP vessel was examined for expression of RFP (red) and PDGFRα (green). I–L) Sections of WAT labeled with RFP (red) and PDGFRα (green). M–P) Sections of BAT stained with RFP (red) and PDGFRα (green). Arrows indicate perivascular cells coexpressing RFP and PDGFRα. Scale bars = 100 μm.
Next, we examined sections of aP2-Cre/Rosa26-tdTomato WAT and BAT tissues immunostained with RFP and PDGFRα. Double-positive cells were mostly located in the endothelial layer of all vessels of larger and smaller sizes, in both WAT and BAT (Fig. 4I–P). Only a few RFP/PDGFRα double-positive cells were located in the perivascular niche of large vessels (Fig. 4P, arrows). Interestingly, however, many RFP/PDGFRα double-positive cells were found at the perivascular niche of small-caliber vessels (Fig. 4L, arrows). These results support the notion that aP2-lineage progenitor cells are located in two distinct niches: the endothelial niche and perivascular niche of small capillaries and the endothelial niche of larger vessels.
The aP2-lineage progenitors are necessary for the adipogenic potential of SVF cells
The ability to differentiate into a mature cell type is a defining feature of progenitor cells. To assess the adipogenic potential of aP2-lineage SVF cells, we cultured the SVF cells from aP2-Cre/tdTomato mice and induced them to differentiate. Consistent with our earlier observation (Fig. 1G), ∼40–50% of the cultured SVF cells were RFP+ prior to induced differentiation (Fig. 5A, B). After induced differentiation, there were numerous RFP+ lipid-filled adipocytes (Fig. 5C, D). These mature adipocytes expressed high levels of perilipin and adiponectin, (Fig. 5E, F) two markers for differentiated adipocytes.
Figure 5.
Differentiation of the SVF cells isolated from WAT and BAT of aP2-Cre/Rosa26-tdTomato mice. A, B) WAT (A) and BAT (B) SVF cells before induced differentiation. C, D) WAT (C) and BAT (D) SVF cells after induced differentiation. E, F) Expression of mature adipose markers Adiponectin and Perilipin in adipose SVF cells before and after induced differentiation. Error bars = se; n = 3. Scale bars = 100 μm. **P < 0.01.
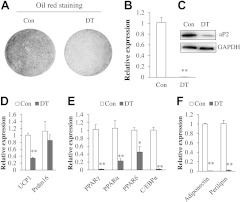
To determine whether the aP2-lineage progenitors are required for the adipogenic differentiation of SVF cells, we established the aP2-Cre/Rosa26-iDTR mouse model. In this model, aP2-Cre induces the expression of DT receptor (DTR; which is normally not expressed by murine cells) and renders the aP2-lineage cells sensitive to DT. Thus, DT treatment will selectively ablate all aP2-lineage cells but not the non-aP2-lineage cells. SVF cells cultured from aP2-Cre/Rosa26-iDTR mice were treated with DT to ablate the aP2-lineage cells, then grown to confluence and induced to undergo adipogenic differentiation. Ablation of aP2-lineage SVF cells nearly eliminated all Oil red staining signals (Fig. 6A), which mark lipid-accumulating mature adipocytes. The mRNA level of aP2 was reduced by >95% (Fig. 6B), and the aP2 protein level was also reduced to a level that is almost undetectable (Fig. 6C). Consistent with the reduced Oil red signal, the brown adipocyte-specific UCP1 expression was also reduced by 70% after ablation of aP2-lineage SVF cells (Fig. 6D). The expression of other adipogenic genes, such as PPARγ, PPARα, PPARδ, and C/EBPα, was also remarkably reduced in the DT-treated cultures (Fig. 6E). Furthermore, expression of mature adipocyte markers adiponectin and perilipin was nearly abolished (Fig. 6F). Similar results were observed in WAT SVF cells after aP2-lineage ablation, although the amplitudes of reduction were less robust in that tissue (Supplemental Fig. S2). These results demonstrate that ablation of aP2-expressing cells severely inhibited the adipogenic potential of SVF cells.
Figure 6.
Ablation of aP2-lineage SVF cells inhibited adipogenic differentiation and gene expression. BAT SVF cells isolated from aP2-Cre/Rosa-iDTR mice were treated with vehicle control or DT that ablates aP2-lineage cells. Cells were grown to confluence and induced to differentiate. A) Oil red O signal of control (Con) and DT-treated cells after differentiation. B, C) mRNA and protein levels of aP2 in control and DT-treated SVF cells. D–F) Expression of adipogenic genes in control and DT-treated SVF cells after induced differentiation. Error bars = se; n = 3.
FACS-purified aP2-lineage cells are adipogenic in vitro and in vivo after transplantation
The much-reduced adipogenic potential of aP2-lineage ablated SVF cells suggests two possibilities. First, the aP2-lineage cells represent the main population of progenitors that gave rise to mature adipocytes. Second, the aP2-lineage cells act as niche cells to support or induce the adipogenic differentiation of aP2− stem and progenitor cells. To distinguish these possibilities, we used FACS to isolate the aP2-lineage cells from the WAT and BAT SVF of aP2-Cre/Rosa26-tdTomato mice (Fig. 7A, B). As expected, sorted RFP+ SVF cells expressed ∼3 times more aP2 mRNA than RFP− cells in both WAT and BAT (Fig. 7C, D). In addition, RFP+ cells expressed higher levels of adipogenic progenitor cell markers Dlk1 and PDGFRα than the RFP− cells in both WAT and BAT (Fig. 7E, F). As macrophages are also known to express aP2 (14, 15), we determined the abundance of macrophage in sorted RFP+ cells. Immunostaining with the macrophage marker CD68 showed that there are 8–9% CD68+ cells in sorted RFP+ cells (Supplemental Fig. S1F–H). Thus, the FACS-purified RFP+ aP2-lineage cells express high levels of progenitor cell markers and contain small numbers of macrophages.
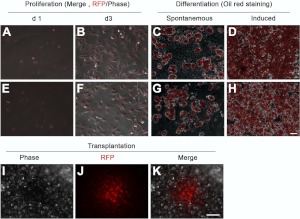
The sorted aP2+ SVF cells from both WAT and BAT proliferated and expanded in culture (Fig. 8A, B, E, F). These cells also differentiated into lipid-filling adipocytes spontaneously or on induction with adipogenic differentiation medium (Fig. 8C, D, G, H). Interestingly, a subpopulation (15–20%) of sorted RFP− cells became RFP+ within 48 h in culture (Supplemental Fig. S3), and subsequently differentiated into mature adipocytes on induction, but less efficiently compared to the RFP+ SVF cells (Supplemental Fig. S3). Specifically, the amount of Oil red signal and expression of adipogenic genes were reduced in RFP− compared to RFP+ SVF after differentiation (Supplemental Fig. S3).
Figure 8.
Proliferation and differentiation of aP2-lineage SVF cells in vitro and in vivo. RFP+ cells isolated by FACS from SVF of BAT and WAT of aP2-Cre/Rosa26-tdTomato were grown in culture and induced to differentiate (A, B, E, F). Oil red O staining indicates adipocytes differentiated from the RFP+ cells (C, D, G, H). I–K) RFP+ cells formed an adipose depot in vivo 21 d after transplantation. Error bars = se; n = 3. Scale bars = 100 μm.
To further examine the adipogenic potential of aP2-lineage SVF cells in vivo, we grafted sorted RFP+ cells into subcutaneous space of wild-type mice (50,000 cells/mouse). At 3 wk after cell transplantation, we detected RFP+ adipose depots in the recipient mice (Fig. 8I–K). These results provide compelling evidence that the aP2-lineage SVF cells represent a population of adipogenic progenitors capable of differentiating into mature adipocytes in culture and in vivo.
Identification of subpopulations of SVF cells with adipogenic and myogenic potentials
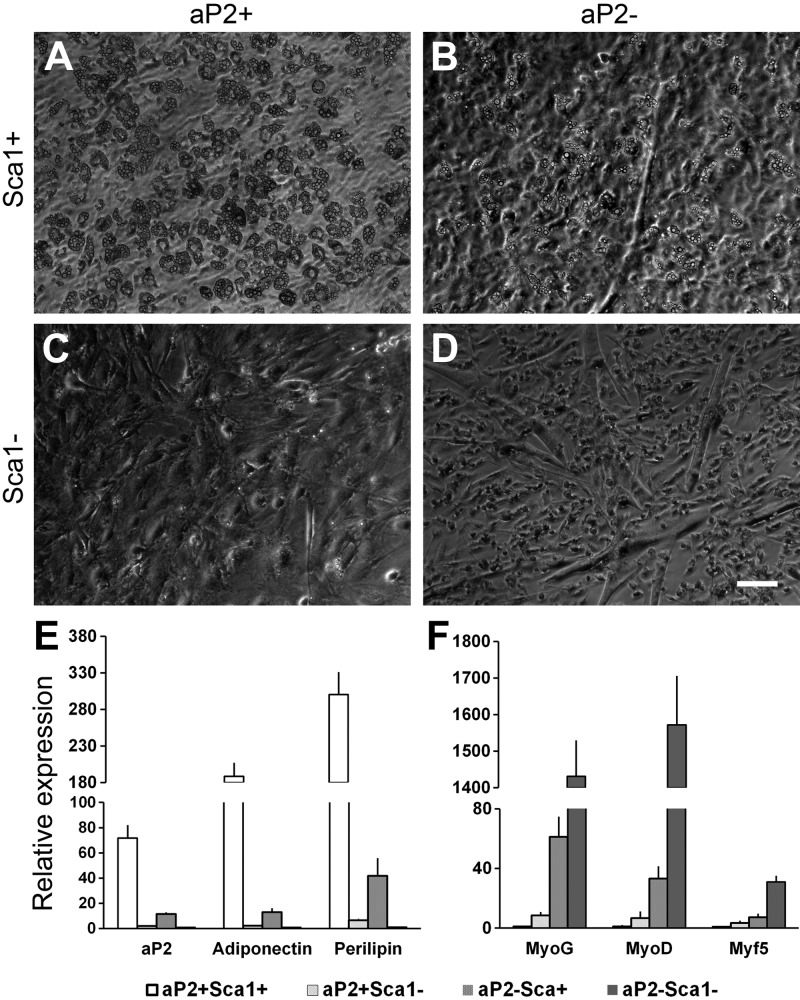
The ability of FACS-purified aP2−-lineage (RFP−) cells to turn on aP2 expression and contribute to adipocyte formation demonstrates the existence of adipogenic progenitors that had never expressed aP2. To further define the adipogenic activity of subpopulations of cells within the aP2+ and aP2− SVF cells, we introduced negative selection for lineage markers (Lin− for CD31−, CD45−, Ter119−) and Sca1+/− selection. On the basis of this, we isolated 4 subpopulations (aP2+Sca1+, aP2+Sca1−, aP2−Sca1+, and aP2−Sca1−) of cells from the Lin− SVF cells of WAT (Fig. 9). When induced to differentiate, the aP2+Sca1+ cells exhibited the highest adipogenic potential (Fig. 9A). Surprisingly, the aP2−Sca1− cells robustly differentiated into myotubes in adipogenic induction medium (Fig. 9D). Interestingly, the aP2−Sca1+ cells differentiated into both adipocytes and myotubes (Fig. 9B), but at a lower efficiency compared to either aP2+Sca1+ or aP2−Sca1− cells. Finally, the aP2+Sca1− cells had neither adipogenic nor myogenic activity (Fig. 9C). These observations were confirmed by qPCR results showing that adipogenic genes aP2, adiponectin, and perilipin are expressed at the highest levels in the differentiated aP2+Sca1+ cells (Fig. 9E), and myogenic genes myogenin, MyoD, and Myf5 are expressed at the highest levels in differentiated aP2−Sca1− cells (Fig. 9F). Both adipogenic and myogenic genes were expressed at modest levels in the aP2−Sca1+ cells, but at very low levels in the aP2+Sca1− cells after induced differentiation (Fig. 9E, F). Similar results were found in the 4 subpopulations of BAT SVF cells (Supplemental Fig. S4). These results suggest that SVF Sca1+aP2− cells represent a population of more primitive bipotential stem cells that can give rise to Sca1+aP2+ adipogenic progenitors and Sca1−aP2− myogenic progenitors.
Figure 9.
Subpopulations of WAT SVF cells and their adipogenic and myogenic potentials. WAT SVF cells were isolated from aP2-Cre/Rosa26-mTmG mice and negatively selected for lineage markers (Lin− for CD31−, CD45−, Ter119−) combined with Sca1+/− and aP2+/− selection. A–D) Lin− SVF cells were sorted into 4 subpopulations of cells: aP2+Sca1+ (A), aP2−Sca1+ (B), aP2+Sca1− (C), and aP2−Sca1− (D). The 4 subpopulations of cells were grown in culture and induced for adipogenic differentiation at confluence. E, F) Relative expression of adipogenic (E) and myogenic (F) genes in the 4 subpopulations of cells after induced differentiation. Error bars = se; n = 3. Scale bars = 100 μm.
DISCUSSION
Our results demonstrated that aP2 is expressed in adipose SVF, and the aP2-expressing cells can proliferate and express the adipose stem cell markers and reside in defined stem cell niche. Furthermore, the aP2+ cells can differentiate to mature adipose in vitro and form adipose depots in vivo. These findings provide compelling evidence that the expression of aP2 is not limited to the mature adipocyte, but also present in a pool of adipogenic progenitors in both WAT and BAT SVFs.
AP2 is a member of the family of FABPs, which plays important roles in the intracellular fatty acid transport and metabolism, especially in maintaining glucose and lipid homeostasis (26). It was originally identified as an adipocyte-specific protein, and is highly expressed in mature adipocytes (27). Thus, aP2 has been used as a marker of mature adipocytes, and its promoter has been widely used in adipocyte-specific recombination in mice (16, 28). AP2 is also expressed in macrophages (29–31) and acts as a key factor connecting vascular and cellular lipid accumulation to inflammation (32). More recent studies indicated that some endothelial cells also express aP2 (26, 33). However, whether aP2 is expressed in adipocyte progenitors is controversial. It was reported that aP2 protein is expressed in some human stromavascular preadipocytes (14, 15), while another study indicated that adipose SVF progenitors are negative for aP2-Cre-lineage labeling, but PPARγ labels a portion of SVF cells (7). The discrepancy between our results and those of Tang et al. (7) may be due to differences of the aP2-Cre-driver mice. We used the widely used transgenic mouse (available from Jackson Laboratory), and Cre expression in our mice is driven by a 5.4-kb aP2 promoter/enhancer fragment. Tang et al. (7) used aP-Cre transgenic mice driven by a similar promoter sequence, but the mice were generated by their own laboratory. The different expression domains may be due to the different insertion sites in the genome between the two lines of transgenic aP2-Cre mice used by us and Tang et al. (7). In the future, a mouse line with Cre directly knocked into the aP2 genomic locus will address such discrepancies. Nevertheless, we confirmed our lineage-tracing results by showing endogenous aP2 mRNA and protein expression in SVF progenitors. Our results are consistent with the observation by Tchoukalova et al. (15). In their study, it was shown that a population of aP2+CD68− (CD68 is a macrophage marker) SVF preadipocytes found in multiple fat depots of humans can proliferate and differentiate into mature adipocytes. Together, our lineage tracing and gene expression results, combined with the earlier study on human adipose tissues, demonstrate that aP2 is, indeed, expressed in a fraction of SVF cells that are adipogenic.
As adipose SVF cells are a mixture of cells, including mesenchymal stem cells, endothelial precursor cells, and adipose progenitors or preadipocytes (13), we double-labeled SVF cells with aP2-reporter and several established adipose stem cell markers. Our immunostaining results showed that many of the aP2-lineage cells also express adipose stem cell markers, such as CD34, Sca1, and PDGFRα (6, 8, 10). Furthermore, >30% of aP2 cells coexpressed the preadipocyte marker Dlk1 (23, 34), whereas in mature adipocytes, there is no expression of Sca1 and PDGFRα. Notably, FACS-purified aP2-expressing cells not only expressed higher levels of aP2 as expected, but also expressed higher levels of progenitor cell markers (Dlk1 and PDGFRα) compared to the aP2− SVF cells. These results indicated that a subset of aP2-expressing cells in SVF meets at least one criterion of stem cells: the expression of adipose stem cell markers.
Another feature of stem cells is their ability to proliferate. Our results showed that >40% of aP2+ cells express the proliferation marker Ki67 (35, 36), which indicated that aP2 cells can proliferate. In addition, FACS-purified aP2-expressing cells (or progeny) readily proliferate and expand in culture. Their proliferating capacity demonstrates that aP2-expressing cells meet another criterion of stem and progenitor cells.
A third feature of stem cells is their ability to differentiate into a mature cell type. We provided multiple lines of strong evidence supporting the differentiation ability of aP2-lineage progenitors in vitro and in vivo. When induced to differentiate, aP2-lineage SVF cells readily accumulate lipid droplets, as revealed by Oil red O staining. In addition, ablation aP2-expressing cells nearly abolished the adipogenic differentiation of BAT SVF cells, and significantly reduced the adipogenesis of WAT SVF cells. Furthermore, FACS-purified aP2-labeled SVF cells spontaneously differentiated into lipid-filling adipocytes and robustly differentiated into large colonies of adipocytes on induced differentiation. Most notably, the sorted aP2-expressing SVF cells gave rise to adipocytes in vivo after transplantation. Together, the capacity to differentiate into mature adipocytes in vitro and in vivo demonstrates that aP2-expressing cells possess an important character of progenitor cells.
Not very surprisingly, we found that FACS-purified aP2-nonexpressing cells can turn on aP2 expression and differentiate into adipocytes. These aP2− cells may represent a population of higher hierarchy cells, for example, mesenchymal stem cells or more primitive adipose stem cells. This notion is supported by the observation that aP2−Sca1+ SVF cells have both adipogenic and myogenic potentials. Interestingly, we also identified the aP2−Sca1− cells as a population of myogenic progenitors that reside in WAT and BAT SVFs. Previous studies show that adipose SVF cells have myogenic potential (37). Our current results provide a molecular signature for the identification of the myogenic progenitors within adipose tissue.
In our aP2-lineage ablation studies, SVF cells depleted of aP2-lineage cells had very little ability to form adipocytes. By contrast, FACS analysis demonstrates that Sca1+aP2− SVF cells can give rise to adipocyte in culture, though at lower efficiency. The discrepancy between the aP2-lineage ablation and cell-sorting results can be due to several reasons. First, in the lineage ablation experiments, SVF cells were continuously treated with DT for 48 h, during which time period any aP2− cells that turned on aP2+ would have been ablated. By contrast, 15–20% sorted aP2− cells became aP2+ within 48 h and subsequently differentiated into adipocytes (Supplemental Fig. S3). Second, the ROSA26-DTR-lineage ablation system may be more sensitive than the ROSA26-TdTomato reporter system. In this scenario, expression of few molecules of the DTR might be sufficient to induce cell death, but many more molecules of TdTomato might be required for its visualization—meaning that many seemingly RFP− adipocytes would have been ablated in the DTR system. Third, it is conceivable that DT-induced massive cell death may trigger a signaling pathway that inhibits the adipogenic differentiation of the aP2− cells that have survived the ablation. Fourth, the aP2+ cells may function to induce the adipogenic differentiation of the aP2− cells. Thus, in the absence of the induction by aP2+ cells after aP2-lineage ablation, the aP2− cells are unable to different into adipocytes. By contrast, some sorted aP2− cells activate aP2 and become aP2+ within 48 h, these aP2+ cells may be sufficient to induce the adipogenic differentiation of other aP2− cells.
Stem cell niche provides functional and anatomical basis for identification of tissue stem cells. The adipose stem cells have been shown to reside in the mural cell compartment of perivascular niche (7). More recent studies also showed that adipocyte progenitors are localized to endothelial layer of BAT and WAT vasculatures (17, 18). However, it is unclear whether there are phenotypical and functional differences between the progenitors occupying the perivascular and endothelial niches, and whether these progenitors represent two hierarchical subpopulations. Our observation that aP2-lineage progenitors are located in the endothelial layer of both large and small vessels, but only in the perivascular niche of small vessels, suggests that different stem cell niches are associated with different vessel types (for example blood vs. lymph vessels). Consistent with this notion that adipose progenitors may also be associated with lymph vessels, it has been shown that lymphatic vasculature dysfunction due to Prox1 haploinsufficiency is associated with adult-onset obesity (38). Our results reconcile the distinct view of endothelial and perivascular niches reported by different studies. More important, identification of aP2+ cells in association with adipose stem cell markers in the stem cell niche further demonstrate that aP2-lineage SVF cells are adipocyte progenitors.
In summary, we provide anatomical, phenotypical, and functional evidence that the expression of aP2 is not limited to the mature adipocyte, but also marks a pool of undifferentiated progenitors associated with the vasculature of adipose tissues. These results enhance our understanding of adipose stem cell biology and further provide insights into the molecular regulation of adipose tissue plasticity and metabolic diseases.
Supplementary Material
Acknowledgments
The authors thank Pengpeng Bi for comments on the manuscript, and Jun Wu for mouse colony support.
The project is partially supported by funding from the Muscular Dystrophy Association, the U.S. National Institutes of Health, and the U.S. Department of Agriculture to S.K.
The authors declare no conflict of interests.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- aP2
- adipocyte protein 2
- BAT
- brown adipose tissue
- DEXA
- dexamethasone
- DAPI
- 4′,6-diamidino-2-phenylindole
- DMEM
- defined minimal essential medium
- DT
- diphtheria toxin
- DTR
- diphtheria toxin receptor
- EYFP
- enhanced yellow fluorescent protein
- FABP4
- fatty acid binding protein 4
- FACS
- fluorescence-activated cell sorting
- FBS
- fetal bovine serum
- IBMX
- 3-isobutyl-methylxanthine
- PAGE
- polyacrylamide gel electrophoresis
- PDGFR
- platelet-derived growth factor receptor
- qPCR
- quantitative PCR
- RFP
- red fluorescent protein
- Sca1
- stem cell antigen 1
- SDS
- sodium dodecyl sulfate
- SVF
- stromal vascular fraction
- SVP
- stromal vascular particulate
- UCP-1
- uncoupling protein 1
- WAT
- white adipose tissue
REFERENCES
- 1. Russell A. P., Crisan M., Leger B., Corselli M., McAinch A. J., O'Brien P. E., Cameron-Smith D., Peault B., Casteilla L., Giacobino J. P. (2012) Brown adipocyte progenitor population is modified in obese and diabetic skeletal muscle. Int. J. Obes. (Lond.) 36, 155–158 [DOI] [PubMed] [Google Scholar]
- 2. Seale P., Conroe H. M., Estall J., Kajimura S., Frontini A., Ishibashi J., Cohen P., Cinti S., Spiegelman B. M. (2011) Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Invest. 121, 96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walden T. B., Hansen I. R., Timmons J. A., Cannon B., Nedergaard J. (2012) Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. Am. J. Physiol. Endocrinol. Metab. 302, E19–E31 [DOI] [PubMed] [Google Scholar]
- 4. Yamamoto Y., Gesta S., Lee K. Y., Tran T. T., Saadatirad P., Kahn C. R. (2010) Adipose depots possess unique developmental gene signatures. Obesity 18, 872–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeve D., Tang W., Graff J. (2009) Fighting fat with fat: the expanding field of adipose stem cells. Cell Stem Cell 5, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodeheffer M. S., Birsoy K., Friedman J. M. (2008) Identification of white adipocyte progenitor cells in vivo. Cell 135, 240–249 [DOI] [PubMed] [Google Scholar]
- 7. Tang W., Zeve D., Suh J. M., Bosnakovski D., Kyba M., Hammer R. E., Tallquist M. D., Graff J. M. (2008) White fat progenitor cells reside in the adipose vasculature. Science 322, 583–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schulz T. J., Huang T. L., Tran T. T., Zhang H., Townsend K. L., Shadrach J. L., Cerletti M., McDougall L. E., Giorgadze N., Tchkonia T., Schrier D., Falb D., Kirkland J. L., Wagers A. J., Tseng Y. H. (2011) Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc. Natl. Acad. Sci. U. S. A. 108, 143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crisan M., Casteilla L., Lehr L., Carmona M., Paoloni-Giacobino A., Yap S., Sun B., Leger B., Logar A., Penicaud L., Schrauwen P., Cameron-Smith D., Russell A. P., Peault B., Giacobino J. P. (2008) A reservoir of brown adipocyte progenitors in human skeletal muscle. Stem Cells 26, 2425–2433 [DOI] [PubMed] [Google Scholar]
- 10. Cawthorn W. P., Scheller E. L., MacDougald O. A. (2012) Adipose tissue stem cells meet preadipocyte commitment: going back to the future. J. Lipid Res. 53, 227–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suga H., Matsumoto D., Eto H., Inoue K., Aoi N., Kato H., Araki J., Yoshimura K. (2009) Functional implications of CD34 expression in human adipose-derived stem/progenitor cells. Stem Cells Dev. 18, 1201–1210 [DOI] [PubMed] [Google Scholar]
- 12. Daquinag A. C., Zhang Y., Amaya-Manzanares F., Simmons P. J., Kolonin M. G. (2011) An isoform of decorin is a resistin receptor on the surface of adipose progenitor cells. Cell Stem Cell 9, 74–86 [DOI] [PubMed] [Google Scholar]
- 13. Poulos S. P., Dodson M. V., Hausman G. J. (2010) Cell line models for differentiation: preadipocytes and adipocytes. Exp. Biol. Med. (Maywood) 235, 1185–1193 [DOI] [PubMed] [Google Scholar]
- 14. Soukas A., Socci N. D., Saatkamp B. D., Novelli S., Friedman J. M. (2001) Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J. Biol. Chem. 276, 34167–34174 [DOI] [PubMed] [Google Scholar]
- 15. Tchoukalova Y. D., Sarr M. G., Jensen M. D. (2004) Measuring committed preadipocytes in human adipose tissue from severely obese patients by using adipocyte fatty acid binding protein. Am. J. Physiol. Regul. Integr. Comp. Physiol. 287, R1132–R1140 [DOI] [PubMed] [Google Scholar]
- 16. Urs S., Harrington A., Liaw L., Small D. (2006) Selective expression of an aP2/fatty acid binding protein 4-Cre transgene in non-adipogenic tissues during embryonic development. Transgenic Res. 15, 647–653 [DOI] [PubMed] [Google Scholar]
- 17. Gupta R. K., Mepani R. J., Kleiner S., Lo J. C., Khandekar M. J., Cohen P., Frontini A., Bhowmick D. C., Ye L., Cinti S., Spiegelman B. M. (2012) Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 15, 230–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tran K. V., Gealekman O., Frontini A., Zingaretti M. C., Morroni M., Giordano A., Smorlesi A., Perugini J., De Matteis R., Sbarbati A., Corvera S., Cinti S. (2012) The vascular endothelium of the adipose tissue gives rise to both white and brown fat cells. Cell Metab. 15, 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shan T., Ren Y., Liu Y., Zhu L., Wang Y. (2010) Breed difference and regulation of the porcine Sirtuin 1 by insulin. J. Anim. Sci. 88, 3909–3917 [DOI] [PubMed] [Google Scholar]
- 20. Liu W., Liu Y., Lai X., Kuang S. (2012) Intramuscular adipose is derived from a non-Pax3 lineage and required for efficient regeneration of skeletal muscles. Dev. Biol. 361, 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuang S., Charge S. B., Seale P., Huh M., Rudnicki M. A. (2006) Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J. Cell Biol. 172, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuang S., Kuroda K., Le Grand F., Rudnicki M. A. (2007) Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 129, 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sul H. S. (2009) Minireview: Pref-1: role in adipogenesis and mesenchymal cell fate. Mol. Endocrinol. 23, 1717–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uezumi A., Fukada S., Yamamoto N., Takeda S., Tsuchida K. (2010) Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 12, 143–152 [DOI] [PubMed] [Google Scholar]
- 25. Hoch R. V., Soriano P. (2003) Roles of PDGF in animal development. Development 130, 4769–4784 [DOI] [PubMed] [Google Scholar]
- 26. Elmasri H., Karaaslan C., Teper Y., Ghelfi E., Weng M., Ince T. A., Kozakewich H., Bischoff J., Cataltepe S. (2009) Fatty acid binding protein 4 is a target of VEGF and a regulator of cell proliferation in endothelial cells. FASEB J. 23, 3865–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spiegelman B. M., Frank M., Green H. (1983) Molecular cloning of mRNA from 3T3 adipocytes. Regulation of mRNA content for glycerophosphate dehydrogenase and other differentiation-dependent proteins during adipocyte development. J. Biol. Chem. 258, 10083–10089 [PubMed] [Google Scholar]
- 28. Barlow C., Schroeder M., Lekstrom-Himes J., Kylefjord H., Deng C. X., Wynshaw-Boris A., Spiegelman B. M., Xanthopoulos K. G. (1997) Targeted expression of Cre recombinase to adipose tissue of transgenic mice directs adipose-specific excision of loxP-flanked gene segments. Nucleic Acids Res. 25, 2543–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Makowski L., Brittingham K. C., Reynolds J. M., Suttles J., Hotamisligil G. S. (2005) The fatty acid-binding protein, aP2, coordinates macrophage cholesterol trafficking and inflammatory activity. Macrophage expression of aP2 impacts peroxisome proliferator-activated receptor gamma and IκB kinase activities. J. Biol. Chem. 280, 12888–12895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maeda K., Cao H., Kono K., Gorgun C. Z., Furuhashi M., Uysal K. T., Cao Q., Atsumi G., Malone H., Krishnan B., Minokoshi Y., Kahn B. B., Parker R. A., Hotamisligil G. S. (2005) Adipocyte/macrophage fatty acid binding proteins control integrated metabolic responses in obesity and diabetes. Cell Metab. 1, 107–119 [DOI] [PubMed] [Google Scholar]
- 31. Hui X., Li H., Zhou Z., Lam K. S., Xiao Y., Wu D., Ding K., Wang Y., Vanhoutte P. M., Xu A. (2010) Adipocyte fatty acid-binding protein modulates inflammatory responses in macrophages through a positive feedback loop involving c-Jun NH2-terminal kinases and activator protein-1. J. Biol. Chem. 285, 10273–10280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Agardh H. E., Folkersen L., Ekstrand J., Marcus D., Swedenborg J., Hedin U., Gabrielsen A., Paulsson-Berne G. (2011) Expression of fatty acid-binding protein 4/aP2 is correlated with plaque instability in carotid atherosclerosis. J. Intern. Med. 269, 200–210 [DOI] [PubMed] [Google Scholar]
- 33. Cataltepe O., Arikan M. C., Ghelfi E., Karaaslan C., Ozsurekci Y., Dresser K., Li Y., Smith T. W., Cataltepe S. (2011) Fatty acid binding protein 4 is expressed in distinct endothelial and non-endothelial cell populations in glioblastoma. Neuropathol Appl Neurobiol 38, 400–410 [DOI] [PubMed] [Google Scholar]
- 34. Lee K., Villena J. A., Moon Y. S., Kim K. H., Lee S., Kang C., Sul H. S. (2003) Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1). J. Clin. Invest. 111, 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Urruticoechea A., Smith I. E., Dowsett M. (2005) Proliferation marker Ki-67 in early breast cancer. J. Clin. Oncol. 23, 7212–7220 [DOI] [PubMed] [Google Scholar]
- 36. Mussig K., Wehrmann T., Dittmann H., Wehrmann M., Ueberberg B., Schulz S., Bares R., Petersenn S. (2012) Expression of the proliferation marker Ki-67 associates with tumour staging and clinical outcome in differentiated thyroid carcinomas. Clin Endocrinol (Oxf.) 77, 139–145 [DOI] [PubMed] [Google Scholar]
- 37. Di Rocco G., Iachininoto M. G., Tritarelli A., Straino S., Zacheo A., Germani A., Crea F., Capogrossi M. C. (2006) Myogenic potential of adipose-tissue-derived cells. J. Cell Sci. 119, 2945–2952 [DOI] [PubMed] [Google Scholar]
- 38. Harvey N. L., Srinivasan R. S., Dillard M. E., Johnson N. C., Witte M. H., Boyd K., Sleeman M. W., Oliver G. (2005) Lymphatic vascular defects promoted by Prox1 haploinsufficiency cause adult-onset obesity. Nat. Genet. 37, 1072–1081 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.