Abstract
Small molecule inhibitors of polyADP-ribose polymerase (PARP) are thought to mediate their antitumor effects as catalytic inhibitors that block repair of DNA single strand breaks. However, the mechanism of action of PARP inhibitors with regard to their effects in cancer cells is not fully understood. In this study we demonstrate that PARP inhibitors trap the PARP1 and PARP2 enzymes at damaged DNA. Trapped PARP-DNA complexes were more cytotoxic than unrepaired single-strand breaks caused by PARP inactivation, arguing that PARP inhibitors act in part as poisons that trap PARP enzyme on DNA. Moreover, the potency in trapping PARP differed markedly among inhibitors with MK-4827 > olaparib (AZD-2281) ≫ veliparib (ABT-888), a pattern not correlated with the catalytic inhibitory properties for each drug. We also analyzed repair pathways for PARP-DNA complexes using 30 genetically altered avian DT40 cell lines with pre-established deletions in specific DNA repair genes. This analysis revealed that, in addition to its function in homologous recombination, PARP also functions in post-replication repair and the Fanconi anemia pathway, and that polymerase β and FEN1 were critical for repairing trapped PARP-DNA complexes. In summary, our study provides a new mechanistic foundation for the rational application of PARP inhibitors in cancer therapy.
Keywords: PARP inhibitor, poly(ADP-ribosyl)ation, PARP-DNA complex, chemotherapy, homologous recombination
Introduction
Poly(ADP-ribose) polymerase 1 (PARP1) is an abundant nuclear protein and the founding member of the PARP family (1–4). It binds damaged DNA through its N-terminal zinc finger motifs, which activates its catalytic C-terminal domain to hydrolyze NAD+ and produce linear and branched PAR chains that can extend over hundreds of ADP-ribose units (1–4) (see Fig. 7B). PARP2, although less abundant and contributing 5% to 10% of the total PARP activity (1, 5), is essential for the viability of PARP1 knockout mice (6). The rapid binding of PARP1 to DNA strand breaks is critical for the resealing of DNA single strand breaks (SSBs) during base excision repair (BER) (7–9) and for the repair of topoisomerase I cleavage complexes (10) and DNA double-strand breaks (DSBs) (11, 12). AutoPARylation of PARP1 and PARP2, and PARylation of chromatin proteins promote the recruitment of DNA repair factors (1, 13, 14). Because of the high negative charge of PAR polymers, extensive autoPARylation of PARP1 and PARP2 leads to their dissociation from DNA, which is required for DNA repair completion (15).
Figure 7. DNA repair pathways for PARP inhibitors.
(A) Sensitivity profiles of olaparib, veliparib and MK-4827 in a panel of thirty DT40 isogenic repair mutant DT40 cell lines (see Table S1 for additional information on the cell lines). The sensitivity in each mutant cell line was determined relative to wild-type cells (see Fig. 1D). Negative and positive scores indicate that a given cell line is hypersensitive or resistant to the drug, respectively. Each bar length reflects the degree of sensitivity or resistance to the drug (log2 units). Each bar is colored according to the category of DNA repair function (see Table S1): black: non-homologous end joining; pink: checkpoint; dark blue: homologous recombination (HR); brown: post replication repair; light blue: PARP1 and PARP1/RAD18; light green; removal of Top1 or Top2 cleavage complexes; magenta; Fanconi anemia (FA) pathway; orange: DNA polymerases; purple; FEN1; red; nucleotide excision repair; green: RecQ helicases. The IC90 for olaparib and MK-4827 and IC50 for veliparib are shown at the bottom (mean ± SD, n = 13). (B and C) Schematic illustration of the dual inhibitory mechanism of PARP inhibitors (poisoning and catalytic inhibition). (B) Molecular interaction map showing the regulatory pathways for the formation and dissociation of PARP-DNA complexes. Symbols are:
 Binding of PARP and DNA with the node corresponding to the PARP-DNA complex;
Binding of PARP and DNA with the node corresponding to the PARP-DNA complex;
 Activation;
Activation;
 Inhibition (50). PARP inhibitors enhance the PARP-DNA complexes by two mechanisms: 1: inhibition of NAD+ binding, and 2: binding of PARP inhibitor in the NAD site activates (allosterically) the binding of PARP to DNA. See discussion for further details. (C) Dual cytotoxic mechanisms of PARP inhibitors. 1: Catalytic inhibition (upper pathway) interferes with the repair of SSBs, leading to replication fork damage that requires homologous recombination (HR) repair. 2: Trapping of PARP-DNA complexes also leads to replication fork damage but utilizes additional repair pathways including Fanconi pathway (FA), template switching (TS), ATM, FEN1 (replicative flap endonuclease) and polymerase β.
Inhibition (50). PARP inhibitors enhance the PARP-DNA complexes by two mechanisms: 1: inhibition of NAD+ binding, and 2: binding of PARP inhibitor in the NAD site activates (allosterically) the binding of PARP to DNA. See discussion for further details. (C) Dual cytotoxic mechanisms of PARP inhibitors. 1: Catalytic inhibition (upper pathway) interferes with the repair of SSBs, leading to replication fork damage that requires homologous recombination (HR) repair. 2: Trapping of PARP-DNA complexes also leads to replication fork damage but utilizes additional repair pathways including Fanconi pathway (FA), template switching (TS), ATM, FEN1 (replicative flap endonuclease) and polymerase β.
A large number of SSBs are generated endogenously as well as BER intermediates (16). When SSBs are encountered by replication forks, they generate DSBs that need to be repaired by homologous recombination (HR) (17, 18). Accordingly, PARP inhibition results in the accumulation of recombinogenic substrates marked by RAD51 and γH2AX nuclear foci (19, 20). This explains why cancer cells deficient in BRCA1 and BRCA2 are selectively hypersensitive to PARP inhibitors (21–23). Recent studies have also suggested that disruptions of Fanconi anemia (FA)- or other HR-related genes also sensitize cells to PARP inhibitors (24, 25), providing a strong rationale for the development of PARP inhibitors for cancer treatment (4, 26, 27).
Olaparib (AZD2281), veliparib (ABT-888) and MK-4827 are PARP inhibitors in advance clinical trials. They are highly efficacious PARP catalytic inhibitors with IC50 (inhibitory concentration 50%) values reaching the low nanomolar range (4, 27–29). Their structure includes a nicotinamide moiety that competes with NAD+ (Fig. S1 and see Fig. 7B). Assuming that the synthetic lethality of PARP inhibitors in the context of HR (BRCA) deficiencies is solely explained by catalytic inhibition, the drugs should have a similar effect as PARP deletion. However, wild-type cells are more sensitive to a PARP inhibitor combined with the alkylating agent MMS than Parp1−/− mouse cells (30–32). Furthermore, PARP inhibition delays SSB repair to a greater extent than PARP depletion (33). To explain these results, a PARP1-trapping model has been proposed (18, 32). This model is based on the idea that PARP1 is trapped on DNA by PARP inhibitors, and PARP1-DNA complexes can interfere with DNA replication. Recently, increased PARP1 association with DNA in alkylation damage by a PARP inhibitor was reported using chromatin immunoprecipitation. However, accumulation of PARP1 was not detected in chromatin bound fraction at the protein level (32).
Our study shows that, in addition to catalytic inhibition, PARP inhibitors induce cytotoxic PARP-DNA complexes and we provide methods to detect such complexes. Secondly, we show that clinically relevant PARP inhibitors differ markedly in their potency to trap PARP-DNA complexes. Finally, we reveal previously unknown repair factors/pathways for the PARP-DNA complexes and we discuss their importance in the context of personalized medicine.
Materials and Methods
Cell lines
The DT40 cell lines used in this study were obtained from the Laboratory of Radiation Genetics Graduate School of Medicine in Kyoto University, Japan. The summary of the DT40 mutant cell lines is described in Table S1. All human cancer cell lines were obtained from the National Cancer Institute Developmental Therapeutics Program (Frederick, USA).
Immunoblotting
To prepare whole cell lysates, cells were lysed with CelLytic™ M lysis reagent (C2978, Sigma-Aldrich, St Louis, MO). After thorough mixing and incubation at 4°C for 30 min, lysate were centrifuged at 15,000 g at 4°C for 10 min, and supernatants were collected. To prepare subcellular fraction of nuclear soluble and chromatin bound fraction, ten million DT40 cells with 10 ml medium in 15 ml tube or semi-confluent human cells with 10 ml medium in 10 cm dish were treated with indicated drugs for 30 min or 4 hours, respectively, and then cells were collected. For the fractionation, we used a Subcellular Protein Fractionation Kit from Thermo Scientific (78840, Rockford, IL, USA) following the manufacturer’s instructions. Immunoblotting was carried out using standard procedures. Densitometric analyses of immunoblots were carried out using Image J software (NIH). Further explanation of fractionation methods is available in Supplemental Experimental Procedures.
Antibodies
Rabbit polyclonal anti-PARP1 antibody (sc-7150), mouse monoclonal anti-PCNA antibody (sc-56), and mouse monoclonal anti-FANCD2 antibody (sc-20022) were from Santa Cruz Biothechnolgy (Santa Cruz, CA, USA). Mouse monoclonal anti-γH2AX antibody (05-636) and rabbit polyclonal anti-histone H3 antibody (07-690) were from Upstate Biotechnology (Lake Placid, NY, USA). Mouse monoclonal anti-actin antibody (A3853) and rabbit anti-γ-tubulin antibody (T3559) were from Sigma (St Louis, MO, USA). Mouse monoclonal anti-Top1 antibody (#556597) was from BD Biosciences (San Diego, CA, USA). Rabbit polyclonal anti-PAR polymer antibody (#4336-BPC-100) was from Trevigen (Gaithersburg, MD, USA). Rabbit polyclonal anti-PARP2 antibody (ab93416) was from Abcam (Cambridge, MA, USA). Secondary antibodies were horseradish peroxidase (HRP)-conjugated antibodies to mouse or rabbit IgG (GE Healthcare, UK).
Immunoassay for PAR substrate
The validated chemiluminescent immunoassay for PAR using commercially available reagents is described previously (34). The detailed lab procedures can be viewed from NCI website at http://dctd.cancer.gov/ResearchResources/biomarkers/PolyAdenosylRibose.htm
Measurement of cellular sensitivity to DNA-damaging agents
To measure the sensitivity of cells to drugs, cells were continuously exposed to various concentrations of the drugs for 72 hours in triplicate. For DT40 cells, 200 cells were seeded into 384-well white plate (#6007680 Perkin Elmer Life Sciences, Waltham, MA) in 40 μl of medium per well. For human cells, 1,500 cells of DU145 or 500 cells of SF295 were seeded in 96-well while plate (#6005680 Perkin Elmer Life Sciences) in 100 μl of medium per well. Cell survival was determined using the ATPlite 1 -step kit (PerkinElmer). Briefly, 20 μl or 50 μl ATPlite solution for 384-well or 96-well plate respectively was added to each well. After 5 min, luminescence was measured by EnVision 2104 Multilabel Reader (PerkinElmer). The ATP level in untreated cells was defined as 100%. Survival of treated cells was defined as ATP treated cells/ATP untreated cells × 100.
Evaluation of the relative cellular sensitivity across a panel of DT40 mutant cells
The cellular sensitivity assay using 384-well plate allowed us to examine 6 different cell lines at the same time, while we always included wild type cells in each plate for normalization. To evaluate the relative cellular sensitivity of each mutant to wild type cells, we drew the single logarithm sensitivity curve by setting the survival of untreated cells as 100%. The concentration that causes 90% reduction of ATP activity (inhibition concentration 90%; IC90) by olaparib or MK-4827, and the concentration that causes 50% reduction (IC50) by veliparib, were determined manually from the sensitivity curves. The IC90 and IC50 of each mutant were divided by the IC90 and IC50 of wild type cells on the same plate, respectively. Finally, the quotient was converted into logarithmic scale (base 2).
siRNA transfection
Gene-specific siRNAs (mix of four sequences) for PARP1 (L-006656-00-0005), PARP2 (L-010127-02-0005) and negative control siRNA (D-001810-10) were products of Dharmacon (Lafayette, CO, USA). Ten nanomolar of each siRNA was transfected to DU145 and SF293 cells with Lipofectamin RNAiMAX Reagent (13778, Invitorogen) according to the manufacturer’s instructions. Culture medium was changed 6–8 hours after the transfection. The transfected cells were trypsinized and seeded on 96-well plate 48 hours after the transfection. Drugs were added 12 hours after the seeding, and cells were incubated for another 72 hours before ATP activity was measured. To confirm the downregulation of each protein, the transfected cells were collected for the western blot analysis 72 hours after the transfection.
Drug preparation
Olaparib, veliparib and MK-4827 were obtained from the Drug Synthesis and Chemistry Branch, National Cancer Institute (Bethesda, MD). 4-amino-1,8-naphthalimide (4-AN) was from Sigma-Aldrich (A0966, St Louis, MO, USA). Drug stock solutions were made in DMSO at 10 μM. The stock solutions were stored at −20°C in dark and diluted in the culture medium immediately before use. 1% or 10% MMS was prepared fresh each time from 99% MMS (129925, Sigma-Aldrich) in PBS, and then diluted in culture medium to final concentration.
Fluorescence anisotropy DNA binding assay
The fluorescence anisotropy (FA) experiment was carried out with a 30 bp duplex labeled with 5′-Alexa Fluor488. The deoxyoligonucleotide (sequence: 5′-Alexa Fluor 488-ACCCTGCTGTGGGCdUGGAGAACAAGGTGAT) was annealed to its complementary DNA strand in buffer containing 50 mM potassium acetate, 20 mM tris-acetate, 10 mM magnesium acetate, and 1 mM dithiothreitol, pH 7.9. All oligonucleotides were purchased from Integrated DNA Technologies. Uracil-DNA glycosylase and APE1 (New England Biolabs) were added to the annealed DNA sample and incubated at 37°C for 1 hour. The resulting DNA construct contains a DNA nick and a 5′-dRP at the nicked site. The completion of digestion is verified by denaturing PAGE. For FA measurements, 250 nM recombinant PARP1 (a kind gift from Dr. Valerie Schreiber, University of Strasbourg, France), 1 nM DNA construct, and increasing concentration of PARP inhibitors were combined and incubated for 30 minutes in buffer containing 50 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 100 mM NaCl, and 100 ng/μL BSA. 1 mM NAD+ was added to the samples to initiate the experiment and the FA values were measured at indicated time using an EnVision 2104 Multilabel Reader equipped with a FITC FP Label (PerkinElmer). The control samples lack NAD+ or PARP inhibitor. The FA values reported here were average of two independent experiments (each experiment was measured in duplicates).
Results
PARP inhibition by olaparib is more cytotoxic than genetic depletion of PARP
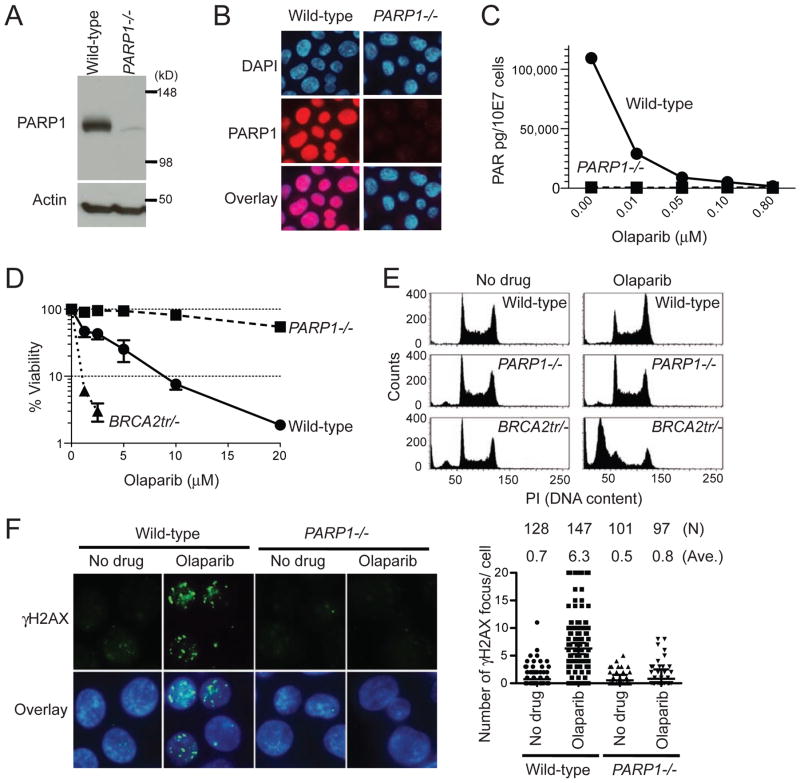
To compare the effect of PARP deletion and PARP inhibition, we took advantage of the fact that avian cells genetically lack PARP2 (35). Therefore, PARP1−/− avian B-lymphoblast DT40 cells are equivalent to PARP1 and PARP2 double knockout (35). Moreover, the catalytic domains and DNA binding zinc finger motifs are highly conserved between human and chicken PARP1 (Fig. S2). Because PARP1 and PAR levels were undetectable in PARP1−/− cells (Fig. 1A–1C) and the PAR level of wild-type cells treated with 0.1 μM olaparib was as low as in PARP1−/− cells (Fig. 1C and 4B), we can compare PARP deletion and catalytic PARP inhibition using PARP1−/− cells and wild-type DT40 cells treated with olaparib. We tested the sensitivity of wild-type, PARP1−/− and BRCA2tr/− (BRCA2 truncated mutant) (36) DT40 cells to olaparib (Fig. 1D). As expected, BRCA2tr/− cells showed greater sensitivity than wild-type. PARP1−/−cells were highly resistant to olaparib, demonstrating the drug selectivity for PARP1. Cell cycle analyses revealed that olaparib had no impact on the cell cycle of PARP1−/− cells, while it induced G2 accumulation in wild-type and massive apoptosis in BRCA2tr/− cells (Fig. 1E). γH2AX levels were also significantly increased by olaparib in wild-type cells but not in PARP1−/− cells (Fig. 1F). These results demonstrate that PARP1 is required for the cytotoxicity of olaparib and that PARP inhibition is not equivalent to PARP deletion.
Figure 1. PARP1 is required for cell killing by olaparib.
(A) Western blot of whole cell lysates prepared from wild-type and PARP1−/− DT40 cells. Blots were probed with the indicated antibodies. (B) Nuclear localization of PARP1 in wild-type DT40 cells and loss of signal in PARP1−/− DT40 cells. Immunofluorescence microscopy was performed with anti-PARP1 antibody and nuclei were stained with DAPI. (C) PAR level in wild-type and PARP1−/− DT40 cells measured by ELISA. Cells were treated with the indicated olaparib concentrations for 2 hours. (D) Survival curves after continuous olaparib treatment for 72 hours. Cellular ATP activity was used to measure cell viability. The survival of untreated cells was set as 100%. Error bars represent standard deviation (SD) (n=3). (E) Cell cycle analyses after olaparib treatment. The indicated cell lines were treated with 10 μM olaparib for 24 hours; after which, cells were analyzed by flow cytometry. The left and right peaks indicate G1 and late S and G2 populations, respectively. (F) DNA damage measured by immunofluorescence microscopy with anti-γH2AX antibody after 2 hours treatments with or without 10 μM olaparib. Left panels show representative images and right plot shows a quantitation of the number ofγH2AX foci per individual cells. The number of cells analyzed (N) and the average number ofγH2AX foci (Ave.) are shown above the plot.
Figure 4. Three clinical PARP inhibitors differ in their potency to poison PARP1 and PARP2 irrespective of their potency to inhibit PARP catalytic activity.
(A) Drug-induced PARylation inhibition. Western blotting of PAR levels in whole cell lysates from wild-type DT40 cells treated as indicated for 30 min. Blots were probed with anti-PAR antibody. Whole cell lysates from untreated PARP1−/− cells were used as a control. Asterisks indicate non-specific bands. (B) Representative PAR ELISA assays in DT40 cells treated with olaparib, veliparib or MK-4827 for 120 min. PAR level of untreated cells was set as 100%. IC50 (inhibitory concentration 50%) of olaparib, veliparib and MK-4827 were 1.2 nM, 10.5 nM and 50.5 nM, respectively. (C–F) Differential cytotoxicity of the three PARP inhibitors. Viability was measured as in Figure 1D. Error bars represent SD (n=3). Survival curves of the wild-type cells (C), PARP1−/− cells (D), wild-type cells treated with a subtoxic MMS concentration (0.00025%; see Fig 2C) (E), and BRCA2-deficient DT40 cells (F). Sensitivity was determined as in Figure 1D. Error bars represent SD (n=3). The viability of PARP1−/− cell to 0.00025% MMS (22%) was shown with a horizontal dashed line with annotation (E). (G) Survival curves of DU145 treated with the indicated PARP inhibitors for 72 hours. (H) Survival curves of DU145 treated with MMS alone or MMS plus 1 μM olaparib or veliparib or MK-4827 for 72 hours.
Olaparib stabilizes toxic PARP1-DNA complexes
The finding that the cytotoxicity of olaparib was selective for PARP1-proficient cells raised the possibility that olaparib might generate toxic DNA lesions mediated by PARP1 at spontaneously occurring base lesions. To examine such possibility, we fractionated wild-type DT40 cell lysates into nuclear-soluble and chromatin-bound fractions. To increase base damage, we combined olaparib with low dose of the classical alkylating agent MMS. Under drug-free condition, most PARP1 was in the nuclear-soluble fraction (Fig 2A. lanes 1 and 7). Olaparib or MMS alone slightly increased PARP1 chromatin binding (Fig 2A. lanes 8 and 9, and 2B). Notably, in the presence of MMS, increasing the concentration of olaparib greatly induced PARP1 accumulation in the chromatin-bound fraction (Fig 2A. lanes 10–12, and 2B). Under these conditions, PAR levels were reduced by olaparib even in the presence of 0.01% MMS (Fig. 2A).
Figure 2. Olaparib stabilizes PARP1-DNA complexes that are more toxic than unrepaired SSBs.
(A) Western blot analysis of nuclear soluble and chromatin bound fractions prepared from wild-type DT40 cells. Cells were treated for 30 min without drug (lane 1 and 7) or as indicated. Blots were probed with indicated antibodies. Histone H3 and topoisomerase I were used as positive markers for chromatin and nuclear soluble fractions, respectively. The asterisk indicates a non-specific band. (B) Quantification of PARP1 levels in chromatin binding fractions 30 min after the indicated drug treatments. Signal intensity was quantified using Image J software (NIH) from 4–7 independent Western blot analyses (mean values ± SD), and normalized to untreated cells set as one.
(C) Survival curves of wild-type (left) and PARP1−/− (right) cells treated with MMS alone or with the indicated olaparib concentrations (indicated beside each curve in micromolar unit). The survival of treated cells without MMS was set as 100%. Data are mean ± SD (n=3).
(D) Cell cycle analysis of wild-type and PARP1−/− DT40 cells after 2 and/or 4 hour continuous treatments. Cells were pulse-labeled with BrdU for 10 min following the drug treatment to measure DNA synthesis.
To examine the cytotoxicity of the olaparib-induced PARP1-DNA complexes, we determined the cytotoxicity of MMS with increasing concentrations of olaparib in wild-type and PARP1−/− cells (Fig. 2C). As expected from the widely accepted concept that PARylation is necessary for BER, PARP1−/− cells were hypersensitive to MMS. Moreover, olaparib had no impact on MMS sensitivity in PARP1−/− cells (Fig. 2C right panel), confirming the lack of “off-target effects” of olaparib. In parallel, olaparib synergistically sensitized wild-type cells to MMS in a concentration-dependent manner (Fig 2C. left panel). Notably, the MMS sensitivity of wild-type cells treated with olaparib was greater than that of PARP1−/− cells (compare left and right panel in Fig. 2C). These results indicate that PARylation inhibition does not simply explain the hypersensitivity of olaparib-treated cells to MMS. Cell cycle responses in wild-type cells (Fig. 2D) also showed that the combination of olaparib with MMS suppressed BrdU incorporation more than either single treatment, and also induced sub-G1 accumulation after 4 hours, indicative of dying cells. By contrast, olaparib had no additional impact on PARP1−/−cells compared to MMS alone. Together, these results demonstrate that olaparib synergistically increases the cytotoxicity of MMS in a PARP1-dependent manner, as it induces the trapping of PARP1-DNA complexes.
Trapping of human PARP1 and PARP2 by olaparib
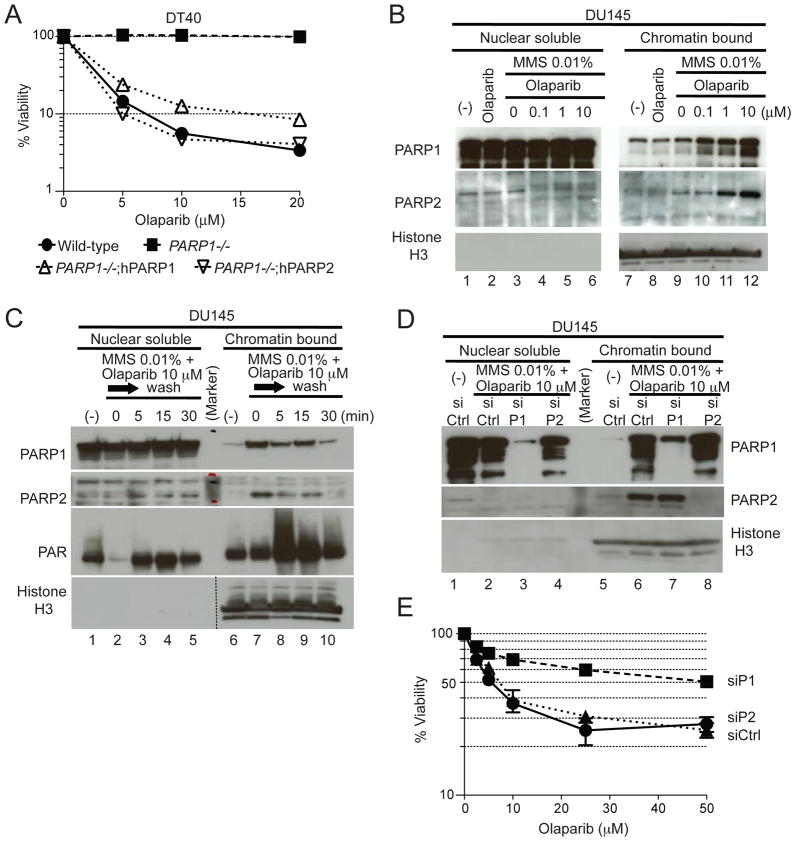
To determine whether both human PARP1 and PARP2 act in the cytotoxicity of olaparib, we compared PARP1−/− DT40 cells complemented either with human PARP1 (PARP1−/−;hPARP1) or PARP2 (PARP1−/−;hPARP2) (35). Complementation with either hPARP1 or hPARP2 restored sensitivity to olaparib (Fig. 3A), demonstrating that both hPARP1 and hPARP2 can be poisoned by olaparib.
Figure 3. Stabilization of toxic PARP1- and PARP2-DNA complexes by olaparib.
(A) Survival curves of wild-type, PARP1−/−, PARP1−/− complemented with human PARP1 (PARP1−/−;hPARP1) and PARP2 (PARP1−/−;hPARP2) DT40 cell lines treated with olaparib. Cells were treated with olaparib as in Figure 1D. Error bars represent SD (n=3). (B–E) Western blotting of nuclear soluble and chromatin bound fractions prepared from human DU145 cells. Blots were probed with the indicated antibodies. (B) Cells were treated for 4 hours without drug (lane 1 and 7), or as indicated. (C) Rapid reduction of PARP1 and PARP2 binding to chromatin after washing out olaparib. Cells were treated for 4 hours without drug or with 10 μM olaparib in the presence of 0.01% MMS. Drugs were removed by washing cells with cold PBS three times, followed by incubation in pre-warmed medium for the indicated times. (D) Control siRNA (siCtrl), PARR1 siRNA (siP1) and PARP2 siRNA (siP2) were transfected into DU145 cells. Three days later, cells were treated for 4 hours with10 μM olaparib in the presence of 0.01% MMS. (E) Reduced cytotoxicity of olaparib in DU145 cells downregulated for PARP1. Cells were transfected with the indicated siRNAs for 60 hours before adding olaparib for an additional 72 hours. Sensitivity was determined as Figure 1D. Error bars represent SD (n=3).
In human DU145 prostate cancer cells, olaparib induced chromatin binding of both PARP1 and PARP2 in the presence of 0.01% MMS (Fig. 3B lanes 10–12). Trapping of PARP1 and PARP2 was rapidly reversible upon drug removal (Fig. 3C) with release of PARP1 and PARP2 from chromatin within 30 minutes. Dual trapping of PARP1 and PARP2 by olaparib was also observed in other human cancer cells: OVCAR4 (ovarian line) and SF295 (CNS) (Fig. S3). Suppression of PARP1 expression level by siRNA reduced the trapping of PARP1 but did not affect the trapping of PARP2 (Fig. 3D). Conversely, siRNA against PARP2 abolished the trapping of PARP2 by olaparib without affecting PARP1 trapping (Fig. 3D). Similar results were obtained in SF295 cells (Fig. S3B), demonstrating that olaparib traps both PARP1- and PARP2-DNA complexes independently of each other.
Next, we tested whether genetic depletion of PARP1 or PARP2 would make human cells tolerant to olaparib, as observed in DT40 cells (see Fig. 1D). As expected, transfection of DU145 cells with siRNA against PARP1 reduced sensitivity to olaparib (Fig. 3E). However, siRNA against PARP2 had minimal impact in spite of the efficiency of the knockdown (see Fig. 3D). These results indicate that the presence of PARP1, whose expression is much higher than PARP2 in human cells (5, 37) is the major mediator of olaparib-induced antiproliferative activity in DU145 cells.
Differential potency of PARP inhibitors to poison PARP1 and PARP2 in cells
Having established the dual effect of olaparib on PARP (catalytic inhibition and PARP trapping), we compared olaparib to two other clinically relevant PARP inhibitors, veliparib and MK-4827 (see structures in Fig. S1) (27). Catalytic PARP inhibition was examined by two methods (Western blotting and ELISA assays) (34, 38). Both assays showed that olaparib was the most potent catalytic PARP inhibitor, followed by veliparib and MK-4827 (Fig. 4A and 4B). For all three drugs, PAR levels were almost undetectable at and above 1 μM drug concentration (Fig. 4A).
In terms of cytotoxicity in wild-type DT40 cells, MK-4827 was the most potent drug while veliparib was the least cytotoxic and olaparib was intermediate (Fig. 4C). The cytotoxicity was mediated by PARP1 as demonstrated by the relative lack of effect of the drugs in PARP1−/− cells (Fig. 4D). Moreover, in the case of veliparib, high drug concentration (50 μM) was required to synergize with MMS beyond the sensitivity of PARP1−/− cells, while for olaparib or MK-4827, 1 μM (50-fold less) was enough to achieve superior synergism (Fig. 4E). These results suggested that veliparib had a lesser potency to poison PARP1 than olaparib and MK-4827. In spite of its minimal cytotoxicity in wild-type cells, veliparib sensitized BRCA2tr/− cells, which is consistent with the general concept of synthetic lethality by PARP inhibitors in BRCA-deficient tumors. Yet, veliparib was approximately 10-fold less cytotoxic than olaparib in the BRCA2tr/− cells, and MK-4827 was even more effective than olaparib as a single agent in the BRCA2tr/− cells (Fig. 4F).
The differential cytotoxicity of the three PARP inhibitors was also observed in human DU145 cells (Fig. 4G and 4H). Single treatment with either olaparib or MK-4827 killed DU145 in a concentration-dependent manner while veliparib had little cytotoxicity (Fig. 4G). Under conditions where 1μM veliparib showed no effect, 1μM olaparib or MK-4827 synergized markedly with MMS (Fig. 4H). Similar results were obtained in the human SF295 cell line (Fig. S4A and S4B). Therefore, the cytotoxicity of three clinically relevant PARP inhibitors shows marked differences across different cell lines with MK-4827 and olaparib being the most cytotoxic and veliparib the least. Moreover, our results demonstrate that the antiproliferative activity of the PARP inhibitors, including in BRCA2tr/− cells, is not directly correlated with their PARP catalytic inhibition potencies.
We next tested whether the differential cytotoxicity of the PARP inhibitors was related to their trapping of PARP1 and PARP2. Even at 50 μM, veliparib induced significantly less trapping of PARP1 and PARP2 on chromatin than olaparib or MK-4827 at 10 μM across different cell types (DT40, human DU145 and SF295 cells; Fig. 5A and S4C). To examine the tightness of PARP-DNA complexes induced by different PARP inhibitors, further fractionation experiments were performed (Fig. 5B and 5C). Most of PARP1 was eluted in fraction B from untreated or veliparib-treated cells, whereas it was mostly eluted in fraction D in olaparib- and MK-4827-teated cells. Histone H3 and monoubiquitinated PCNA were also eluted dominantly in fraction D. These results indicate that PARP1-DNA complexes trapped by olaparib and MK-4827 are tighter than by veliparib. We also compared in our different assays, 4-amino-1,8-naphthalimide (4-AN), a commonly used PARP inhibitor (32, 33, 39). Figure S5 shows that its PARP poisoning potency was stronger than veliparib, but was less than olaparib and MK-4827, while having similar catalytic PARP inhibitory ability as MK-4827. Together, these results demonstrate that PARP inhibitors are not equivalent with respect to trapping PARP-DNA complexes. Veliparib is the least potent poison of PARP1 and PARP2 at concentrations that are effectively blocking PARP catalytic activity.
Figure 5. Differential cellular trapping of PARP1 and PARP2 by clinical PARP inhibitors.
(A) Western blotting of nuclear soluble and chromatin bound fractions prepared from wild-type DT40 cells (upper) and DU145 cells (lower) with or without drug treatment as indicated. Blots were probed with indicated antibodies.
(B,C) Fractionation of PARP-DNA complexes stabilized by PARP inhibitors. DU145 cells were treated for 4 hours without drug (lanes 1–5) or as indicated. (B) Scheme for the preparation of indicated fractions (P1, A–D) using different stringency buffers (Supplementary methods). (C) The eluted proteins in each fraction were blotted with indicated antibodies. Black arrows indicate monoubiqutinated PCNA.
Differential potency of PARP inhibitors to poison PARP1 and PARP2 in biochemical assays
To elucidate whether the differential trapping of PARP-DNA complexes by the three PARP inhibitors could be observed at the molecular level, we set up a PARP1-DNA binding assay based on fluorescence anisotropy. A nicked oligonucleotide duplex DNA with a single 5′-dRP end at the break site was used as fluorescent substrate (Fig. 6A). Its anisotropy was enhanced upon PARP1 binding to the damaged site. PARylation following addition of NAD+ reduced the fluorescence anisotropy signal by freeing the DNA. Figure 6B shows that all three PARP inhibitors enhanced the fluorescence anisotropy signal, which reflects the stabilization of PARP1-DNA complexes. Olaparib and MK-4827 were approximately 10-fold more potent than veliparib. Time-course experiments following NAD+ addition (Fig. 6C) also showed that the PARP inhibitors slowed down the dissociation of PARP1-DNA complexes with olaparib and MK-4827 being significantly more potent than veliparib. These results are in agreement with the cellular data showing greater stabilization of PARP-DNA complexes by olaparib and MK-4827 than by veliparib.
Figure 6. Differential biochemical trapping of PARP1 by clinical PARP inhibitors.
(A) Scheme of the fluorescence anisotropy (FA) binding assay. The star indicates the site labeled on the DNA substrate with Alexa Fluor488. Unbound nicked DNA substrate rotates fast and gives low FA. PARP1 binding to the substrate slows the rotation and gives high FA. Addition of NAD+ leads to PARP1 dissociation from DNA due to autoPARylation. (B) Concentration-dependent PARP1-DNA association using the three different PARP inhibitors. FA was measured 60 min after adding NAD+. (C) Time-course of PARP1-DNA dissociation in the presence of the three different PARP inhibitors (1 μM each). The absence of PARP inhibitors immediately reduces PARP1-DNA complexes (DMSO control). In the absence of NAD+, PARP1-DNA complexes remained stable for at least 120 min (no NAD+).
PARP-DNA complexes are repaired by several repair pathways besides HR
To gain insight into the cellular repair pathways involved in the repair of stabilized PARP1- and PARP2-DNA complexes, we compared the sensitivity of 30 isogenic mutant DT40 cell lines with pre-established genetic DNA repair deficiencies (Fig. 7A and Table S1). An obvious difference between the three PARP inhibitors was the resistance of PARP1−/− cell lines to olaparib and MK-4827 (light blue in Fig. 7A), which is consistent with the requirement of PARP1 for the drugs cytotoxic activities (see above). As expected, the HR deficient cell lines: BRCA2, XRCC3, XRCC2, BRCA1, RAD54 and H2AX (dark blue in Fig. 7A) were hypersensitive to all three PARP inhibitors. The greater differential for veliparib (>16-fold) is probably because of veliparib’s low cytotoxicity in wild-type cells. Notably, deficiencies in check point including ATM and RAD9 (light pink), post replication repair pathways including PCNA ubiquitination and its ubiquitin ligases RAD18 and UBC13 (brown), all FA genes tested (magenta), polymerase β (orange) and FEN1 (purple) conferred greater than ~2-fold hypersensitivity to all three drugs, indicating that DNA lesions induced by PARP inhibitors are not only repaired by classical HR (see discussion). Involvement of the FA pathway was also inferred from enhanced monoubiquitination of FANCD2, a key regulator of the pathway, in response to olaparib (Fig. S6). Interestingly, deletion of PARP1 in RAD18−/− cells (40) restored their resistance to olaparib and MK-4827 (Fig. 7A, compare RAD18 and PARP1/RAD18), suggesting that toxic PARP-DNA complexes but not catalytic inhibition of PARP kills RAD18−/− cells with defective post replication repair.
Discussion
To our knowledge, our study provides the first direct evidence that PARP inhibitors poison both PARP1- and PARP2-DNA complexes. It demonstrates the relevance of this mechanism for the antiproliferative activity (and therefore anticancer activity) of PARP inhibitors and provides novel insights into the molecular pathways that repair PARP-DNA complexes. The clinical relevance of our findings stems from the fact that the concentrations (< 10 μM) required to readily detect PARP1- and PARP2-DNA complexes are below the peak concentration of olaparib (24 μM) in clinical trials (23).
Automodification is a well-established mechanism by which the highly negatively charged PAR polymers dissociates PARP enzymes from DNA (41) (Fig. 7B, step d). In addition, our data demonstrate that PARP inhibitors differ in their potency to stabilize PARP-DNA complexes irrespective of their relative potency to inhibit PARP catalytic activity (see Fig 4, S4 and S5). Indeed, PARP-DNA complexes were detectable under conditions where PAR automodification remained detectable (Fig. 5A, S3 and S4C). Similarly, a recent study showed that nuclear PARylation was observed even after treatment with 10 μM veliparib with MMS (42). Therefore, we propose that PARP inhibitors poison PARP by two mechanisms. One is by inhibition of PARylation, which increases the binding of PARP to DNA (Fig. 7B, annotations 1, c and d) (41, 43); the other is drug binding to the NAD+ site, which allosterically enhances the DNA binding of PARP’s N-terminal zinc finger domain (Fig. 7B, annotations 2 and e). The drug allosteric mechanism fits with the greater potency of the bulky inhibitors, MK-4827 and olaparib to produce PARP-DNA complexes compared to veliparib and 4-AN (see Fig. S1). This hypothetical reverse allosteric mechanism is supported by the fact that tight binding of PARP’s N-terminus zinc domain to damaged DNA (Fig. 7B, step a) is known to allosterically activates the C-terminal catalytic pocket of PARP (1–3, 42, 44) (Fig. 7B, step b). Hence, the PARP inhibitors can be viewed as blocking with the normal allosteric cycle of PARP and as revealing the normal impact of NAD+ binding to PARP. PARP binding to damaged DNA might be normally primed by NAD+ binding. In turn, PARP binding to damaged DNA induces the known allosteric change that activates PARP’s catalytic site (44). The allosteric enhancement of PARP-DNA binding by PARP inhibitors is likely to be more significant for the anticancer activity of PARP inhibitors than catalytic inhibition. Indeed, while PARP1−/− DT40 cells overexpressing catalytic inactive PARP1 are mildly more sensitive to MMS than PARP1−/− cells (45), we find that olaparib, under conditions that induce PARP-DNA complexes, produces a much more drastic hypersensitivity to MMS than PARP−/− cells (Fig. 2C).
Our experiments clearly show that PARP inhibitors that were assumed to be equivalent based on PARP catalytic inhibition, are not equivalent with respect to their potency to poison PARP, suggesting the importance of categorizing PARP inhibitors according to their PARP poisoning potency. Depending on which PARP inhibitor is used, the clinical and experimental results could be very different. Furthermore, this persistent tendency to confuse different PARP inhibitors would make it impossible for the others to recapitulate the results depending on which PARP inhibitor was used. Therefore, our findings are relevant to basic researcher and clinicians who use PARP inhibitors. Specifically, our findings uniquely explain why veliparib has much lower toxicity than olaparib and MK-4827 although the three drugs have similar potency to inhibit PARP catalytic activity.
Our DT40 cell panel data confirm the critical importance of HRs including BRCA1, BRCA2. They are consistent with the lethality of PARP1 siRNAs or shRNAs in BRCA-deficient cells (21, 22) and with the activity of olaparib in BRCA-deficient tumors (23). BRCA2tr/-PARP1−/− double mutant DT40 cells are also unviable (unpublished observations). These lines of evidence strongly support the current anticancer selectivity of PARP inhibitors based on the interference with SSB repair (Fig. 7C upper pathway). On the other hand, in the case of the RAD18−/− cells, their sensitivity to the PARP inhibitors is unambiguously derived from the trapping of toxic PARP-DNA complexes (Fig. 7C lower pathway) rather than from catalytic PARP inhibition since the hypersensitivity of the RAD18−/− cells was rescued in RAD18−/−PARP1−/− double mutant DT40 cells.
Our present data (Fig. 7A) add XRCC2, XRCC3, RAD54 and H2AX to the list of HR genes that confer high sensitivity to PARP inhibitors. The hypersensitivity of check point-deficient cells (ATM−/− and RAD9−/−) and the resistance of non-homologous end joining-deficient cells (KU70−/− and LIGASE IV−/−) extend recent reports (46, 47). The hypersensitivity of FEN1 and polymerase β deficient cells may imply involvement of BER pathway or increase of endogenous DNA damage in these cells. RAD18 and UBC13, which are both responsible for PCNA ubiquitination on lysine 164 (48, 49), were identified in our DT40 panel as novel genes for survival to PARP inhibitors. Consistently, the PCNA mutant K164R also showed hypersensitivity to all three PARP inhibitors. PCNA ubiquitination has been proposed to play a central role in post replication repair that employs translesion synthesis (TLS) and template switching to bypass bulky DNA lesions. For the repair of PARP-DNA lesion, template switching seems more important than TLS because the representative TLS polymerases, POLZ and POLH, mutant cells do not show apparent sensitivity to all drugs. The FA pathway appears critical for the repair of PARP-DNA complexes (see Fig. S6). These results could rationalize the use of PARP inhibitors in tumors with deficiencies in the genes listed above.
In summary, this is the first report showing the toxic PARP-DNA complex by PARP inhibitors, and the greatly different potency to poison PARP among clinical PARP inhibitors irrespective of the potency to inhibit PARP catalytic activity. It also reveals new genetic repair pathways that are critical for the repair of PARP-DNA complexes, and potential synthetic lethality, which can be exploited for additional clinical use of PARP inhibitors.
Supplementary Material
Acknowledgments
We thank Dr. Kouji Hirota and Dr. Hiroyuki Sasanuma (Kyoto University, Japan) for technical advices, Dr. Minoru Takata (Kyoto University, Japan) for sharing FANCC, FANCD2 and FANCG mutant DT40 cell lines, and Dr. Keith Caldecott (University of Sussex, UK) for kindly providing TDP2 mutant DT40 cell line. We also appreciate Dr. Mitsuko Masutani (National Cancer Center Research Institute, Japan) and Dr. Jun Nakamura (University of North Carolina, USA) for a critical discussion.
Grant Support
J.M. is a recipient of a fellowship from the Japan Society for the Promotion of Science (JSPS). This work was supported by the Intramural Program of the National Cancer Institute, Center for Cancer Research and by the JSPS Core-to-Core Program.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–82. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 3.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nature reviews Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, et al. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:17860–8. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 6.Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo J. 2003;22:2255–63. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juarez-Salinas H, Sims JL, Jacobson MK. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979;282:740–1. doi: 10.1038/282740a0. [DOI] [PubMed] [Google Scholar]
- 8.Benjamin RC, Gill DM. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. The Journal of biological chemistry. 1980;255:10493–501. [PubMed] [Google Scholar]
- 9.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–6. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 10.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011;39:3607–20. doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–26. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 12.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 13.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–71. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–33. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–8. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 16.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 17.Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol Cell Biol. 2000;20:3977–87. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Molecular oncology. 2011;5:387–93. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noel G, Godon C, Fernet M, Giocanti N, Megnin-Chanet F, Favaudon V. Radiosensitization by the poly(ADP-ribose) polymerase inhibitor 4-amino-1,8-naphthalimide is specific of the S phase of the cell cycle and involves arrest of DNA synthesis. Molecular cancer therapeutics. 2006;5:564–74. doi: 10.1158/1535-7163.MCT-05-0418. [DOI] [PubMed] [Google Scholar]
- 20.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Molecular and cellular biology. 2005;25:7158–69. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 22.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 23.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. The New England journal of medicine. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 24.D’Andrea AD. Susceptibility pathways in Fanconi’s anemia and breast cancer. The New England journal of medicine. 2010;362:1909–19. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer research. 2006;66:8109–15. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 26.Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr Opin Pharmacol. 2008;8:363–9. doi: 10.1016/j.coph.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 27.Kummar S, Chen A, Parchment RE, Kinders RJ, Ji J, Tomaszewski JE, et al. Advances in using PARP inhibitors to treat cancer. BMC Med. 2012;10:25. doi: 10.1186/1741-7015-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuang HC, Kapuriya N, Kulp SK, Chen CS, Shapiro CL. Differential anti-proliferative activities of poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer cells. Breast Cancer Res Treat. 2012 doi: 10.1007/s10549-012-2106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel AG, Flatten KS, Schneider PA, Dai NT, McDonald JS, Poirier GG, et al. Enhanced Killing of Cancer Cells by Poly(ADP-ribose) Polymerase Inhibitors and Topoisomerase I Inhibitors Reflects Poisoning of Both Enzymes. The Journal of biological chemistry. 2012;287:4198–210. doi: 10.1074/jbc.M111.296475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heacock ML, Stefanick DF, Horton JK, Wilson SH. Alkylation DNA damage in combination with PARP inhibition results in formation of S-phase-dependent double-strand breaks. DNA repair. 2010;9:929–36. doi: 10.1016/j.dnarep.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton JK, Stefanick DF, Naron JM, Kedar PS, Wilson SH. Poly(ADP-ribose) polymerase activity prevents signaling pathways for cell cycle arrest after DNA methylating agent exposure. The Journal of biological chemistry. 2005;280:15773–85. doi: 10.1074/jbc.M413841200. [DOI] [PubMed] [Google Scholar]
- 32.Kedar PS, Stefanick DF, Horton JK, Wilson SH. Increased PARP-1 Association with DNA in Alkylation Damaged, PARP-Inhibited Mouse Fibroblasts. Molecular cancer research : MCR. 2012 doi: 10.1158/1541-7786.MCR-11-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strom CE, Johansson F, Uhlen M, Szigyarto CA, Erixon K, Helleday T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011;39:3166–75. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji J, Kinders RJ, Zhang Y, Rubinstein L, Kummar S, Parchment RE, et al. Modeling pharmacodynamic response to the poly(ADP-Ribose) polymerase inhibitor ABT-888 in human peripheral blood mononuclear cells. PLoS One. 2011;6:e26152. doi: 10.1371/journal.pone.0026152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. The EMBO journal. 2006;25:1305–14. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatanaka A, Yamazoe M, Sale JE, Takata M, Yamamoto K, Kitao H, et al. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Molecular and cellular biology. 2005;25:1124–34. doi: 10.1128/MCB.25.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, et al. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. The Journal of biological chemistry. 2002;277:23028–36. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 38.Kummar S, Chen A, Ji J, Zhang Y, Reid JM, Ames M, et al. Phase I study of PARP inhibitor ABT-888 in combination with topotecan in adults with refractory solid tumors and lymphomas. Cancer Res. 2011;71:5626–34. doi: 10.1158/0008-5472.CAN-11-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nature biotechnology. 2012;30:283–8. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita YM, Okada T, Matsusaka T, Sonoda E, Zhao GY, Araki K, et al. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. The EMBO journal. 2002;21:5558–66. doi: 10.1093/emboj/cdf534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindahl T, Satoh MS, Poirier GG, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends in biochemical sciences. 1995;20:405–11. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 42.Gagné J-P, Rouleau M, Poirier GG. PARP-1 Activation ÄîBringing the Pieces Together. Science. 2012;336:678–9. doi: 10.1126/science.1221870. [DOI] [PubMed] [Google Scholar]
- 43.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–75. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langelier M-F, Planck JL, Roy S, Pascal JM. Structural Basis for DNA Damage-Dependent Poly(ADP-ribosyl)ation by Human PARP-1. Science. 2012;336:728–32. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paddock MN, Bauman AT, Higdon R, Kolker E, Takeda S, Scharenberg AM. Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair. DNA repair. 2011;10:338–43. doi: 10.1016/j.dnarep.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williamson CT, Kubota E, Hamill JD, Klimowicz A, Ye R, Muzik H, et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol Med. 2012;4:515–27. doi: 10.1002/emmm.201200229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3406–11. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W, Qin Z, Zhang X, Xiao W. Roles of sequential ubiquitination of PCNA in DNA-damage tolerance. FEBS Letters. 2011;585:2786–94. doi: 10.1016/j.febslet.2011.04.044. [DOI] [PubMed] [Google Scholar]
- 49.Fox JT, Lee KY, Myung K. Dynamic regulation of PCNA ubiquitylation/deubiquitylation. FEBS Letters. 2011;585:2780–5. doi: 10.1016/j.febslet.2011.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohn KW, Aladjem MI, Kim S, Weinstein JN, Pommier Y. Depicting combinatorial complexity with the molecular interaction map notation. Mol Syst Biol. 2006;2:51. doi: 10.1038/msb4100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.