Abstract
While overall DNA methylation decreases with age, CpG-rich areas of the genome can become hypermethylated. Hypermethylation near transcription start sites typically decreases gene expression. Klotho (KL) is important in numerous age-associated pathways including insulin/IGF1 and Wnt signaling and naturally decreases with age in brain, heart, and liver across species. Brain tissues from young and old rhesus monkeys were used to determine whether epigenetic modification of the KL promoter underlies age-related decreases in mRNA and protein levels of KL. The KL promoter in genomic DNA from brain white matter did not show evidence of oxidation in vivo but did exhibit an increase in methylation with age. Further analysis identified individual CpG motifs across the region of interest with increased methylation in old animals. In vitro methyl modification of these individual cytosine residues confirmed that methylation of the promoter can decrease gene transcription. These results provide evidence that changes in KL gene expression with age may, at least in part, be the result of epigenetic changes to the 5′ regulatory region.
Keywords: Oxidation, Methylation, Age downregulation, White matter, Pyrosequencing
Introduction
Klotho (KL) is a type I transmembrane protein that is primarily expressed in the brain and kidney (Kuro-o et al. 1997). KL functions both as a transmembrane protein and as a humoral factor (Chen et al. 2007; Kuro-o 2009). KL knockout mice develop abnormalities in multiple-organ systems, many of which are involved in human aging. Animals die from the confluence of these disorders by no later than 4 months of age (Kuro-o et al. 1997). Conversely, KL overexpressing animals live 20–30% longer and are more resistant to oxidative stress (Kurosu et al. 2005; Yamamoto et al. 2005). In the brain of knockout animals, markers of oxidative stress increase prior to the onset of cognitive impairment (Nagai et al. 2003). Rare human cases of profound KL protein alterations result in life-threatening disorders including hypophosphatemic rickets (Brownstein et al. 2008) and tumoral calcinosis (Ichikawa et al. 2007) with decreased and increased KL protein, respectively. As well, KL polymorphisms, resulting in more subtle alterations in KL protein level or function, affect lifespan and the risk of disease development (Kuro-o 2009). Together, research indicates that, both directly and indirectly, KL functions to protect the organism. Our lab observed an age-dependent decrease in KL messenger ribonucleic acid (mRNA) and protein, specifically in white matter of aged rhesus monkey and rodents (Duce et al. 2008). Other groups noted age-related decreases in KL in rodent heart and liver (Nabeshima 2002; Shih and Yen 2007). In addition to decreased expression during normal aging, KL protein decreases in age-related diseases, namely, cancer (Camilli et al. 2010; Chen et al. 2010; Lee et al. 2010; Wolf et al. 2008), and the expression level appears to correlate with disease severity (Camilli et al. 2010; Lee et al. 2010).
Since the rhesus monkey is a long-lived species and a close evolutionary ancestor of humans, it is a stronger model to assess age-related changes than shorter-lived model organisms. Rhesus monkeys are sexually mature by 5 years of age and can live up to 35 years (Tigges et al. 1988) suggesting a 1:3 lifespan ratio to humans (Luebke et al. 2010). While rhesus monkeys do not develop overt neurodegenerative disorders, like aging humans, a subset of animals between the ages of 20−30+ years develop cognitive impairment (Herndon et al. 1997; Hinman and Abraham 2007). While neurons are preserved (Peters et al. 1998), monkeys show dramatic age-related changes in white matter that strongly correlate with cognitive impairment (Hinman and Abraham 2007; Peters 2009; Peters et al. 2000).
The KL promoter is guanine and cytosine (GC)-rich and contains a CpG island that extends into the first exon. Oxidative damage most often occurs to guanine residues, making GC-rich areas of the genome sensitive to oxidative damage. Work in human brain, showed that the promoters of age-downregulated genes are more likely to exhibit evidence of oxidative damage (Lu et al. 2004). Likewise, DNA methyltransferases modify cytosine residues in CpG motifs, and such modification to GC-rich promoters is more likely to occur with age (Christensen et al. 2009; Hernandez et al. 2011). In cervical carcinoma and colon cancer, the promoter of KL can be hypermethylated, particularly, in the late stages of disease and hypermethylation decreases KL expression (Lee et al. 2010; Pan et al. 2011). We hypothesized that age-related decreases in KL mRNA and protein were the result of epigenetic modification (i.e., oxidation and/or methylation) to the KL promoter. Here, we report evidence of increased promoter methylation in the white matter of aged rhesus monkeys. In vitro assessment of methylated cytosine residues confirms that such modification can impact gene transcription. These results provide the first indication of a mechanistic cause for age-related decreases in KL mRNA and protein in the brain.
Methods
Cells, cloning, plasmids
Cell lines were maintained under standard growth conditions and propagated in DMEM (4.5 g/ml glucose) containing 10% FBS (Atlanta Biologicals) and 1% penicillin/streptomycin (100 Units/ml). All cell culture solutions were obtained from Cellgro, unless noted. Human GAPDH and Tau promoter reporter plasmids were a gift of Dr. Bruce Yankner (Harvard Medical School). Commercially available pGL4 (renilla/TK; Promega) was used as a transfection control, as detailed below. All primers used in this study are detailed in the primers section. Human KL promoter [1.8 kb, kidney genomic DNA (Clontech)] and rhesus KL promoter (1.4 kb, young monkey brain) were amplified using BD Advantage polymerase (Clontech). Due to the high concentration of GC residues present in the promoter, addition of 1 M Betaine (Sigma) was required in all PCR reactions. Fragments were cloned into the pCR2.1 vector (Invitrogen) and subcloned into the pGL3 (Promega) using XhoI and HindIII restriction sites. Accuracy of the constructs was confirmed by DNA sequencing.
Monkeys
The monkeys used in this study were Indian-derived Macaca mulatta (rhesus monkey) and were selected from subjects that are part of a larger project investigating cognitive aging. To obtain fresh frozen tissue samples, all subjects were deeply anesthetized and the brain was perfused through the ascending aorta with ice-cold Kreb’s Heinseleit buffer to clear the blood and reduce autolysis. Fresh brain samples were taken from dorsolateral prefrontal cortex (DLPFC) and hippocampus. They were immediately frozen on dry ice and stored at –80°C until used. White and grey matter enriched samples were dissected from a frozen block of DLPFC. Initial assessment of data was conducted to determine whether differences could be detected based on sex or cognitive impairment index. Since no differences were found for either variable, the 22 samples were subsequently analyzed based on age. Seven animals were designated as young and were between the ages of 3.8–15 years old (average of 7.68; four female and three male). Fifteen animals were designated old and were between the ages of 20 and 30.2 years old (average 24.35; eight females and seven male). An additional 11 samples of grey matter were taken from the hippocampus. Assays compared six young animals ranging in age from 4.2 to 12.9 years of age (average 8.6; all male) and five old animals ranging from 20.6 to 30.9 (average 25.3; three male and two female).
Western blotting
Blocks of tissue (50 mg) dissected from the initial five young and eight old animal’s DLPFC and from all 11 hippocampal samples were individually homogenized in a dounce homogenizer on ice in RIPA buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1% Triton X 100, 0.5% deoxycholic acid, 0.1% SDS, fresh protease inhibitors added daily (Roche)). After homogenization, extracts were centrifuged and the supernatant frozen until utilization. BCA protein assay (Pierce) allowed determination of protein concentration and ensured equal protein loading onto 10% tris-glycine polyacrylamide gels. Proteins were transferred to nitrocellulose (Millipore) for western blotting. Nitrocellulose was blocked in 5% nonfat milk prior to overnight incubation in primary antibody (in 1% BSA/TBST). KL was detected using KM2076, a rat monoclonal antibody specific to the KL1 domain of KL (kindly provided from the Antibody Research Laboratories, Kyowa Hakko Kirin, Japan). Brain homogenates from KL knockout, hemizygous, and age-matched wild-type littermates were utilized to ensure specificity of the KM2076 antibody. β-Tubulin (Santa Cruz) antibody was used to normalize protein expression. All washes were conducted in TBST. Relevant secondary antibodies were obtained from KPL. Antibody detection was accomplished using Immobilon (Millipore) or Super Signal Pico West Chemiluminescent (Pierce) reagents. Quantitation of protein bands was determined using Image J software 1.49q.
Oxidation assays
In vitro oxidation
DNA (2 μg) from promoter reporter constructs were incubated with and without 400 μM H2O2 for 1 h at room temperature. Plasmids were diluted in serum free medium and co-transfected into HEK 293 cells with renilla luciferase using Nanofect (Qiagen), per manufacturer’s instructions. After 48 h, cells were lysed and the activity of each luciferase measured using the dual luciferase system (Promega) per manufacturer’s instructions in a Glomax Multi Detection System (Promega). Data were normalized to renilla expression for each well.
FPG assays
HEK 293 cells were incubated 12 h in medium with or without 50 μM H2O2/20 μM FeCl2. Rhesus monkey tissue was dissected as described above. The Fpg (formamidopyrimidine [fapy]-DNA glycosylase; New England Biolabs) assays were performed as described in Lu et al. (2004). Briefly, genomic DNA was isolated from cells or tissue (DNeasy kit; Qiagen). DNA samples were incubated with or without Fpg for 16 h at 37°C. Following enzyme inactivation, 10 ng of genomic DNA was amplified by qPCR using SYBR green iQ supermix (Biorad). The number of cycles to reach threshold was compared to a primer specific standard curve to determine the ng of DNA present in the sample. Results are presented as % intact DNA, calculated as the ng present following Fpg cleavage relative to that in the untreated control.
Methylation assays
SSSI methylase
Reporter plasmid (2 μg) was incubated with and without SSSI methylase and S-adenosylmethionine (New England Biolabs) per manufacturer’s protocol for 1 h. Following heat inactivation at 70°C, an aliquot was used in HpaII (New England Biolabs) methyl-sensitive restriction enzyme digestion to ensure modification of the construct. Samples were separated on ethidium bromide containing 0.8% agarose gels. The remaining reaction was co-transfected, with renilla luciferase, into HEK 293 cells using Nanofect (Qiagen) as above. 48 h after transfection, luciferase activity was measured using the dual luciferase assay system, as above. Data were normalized to renilla expression for each well.
Pyrosequencing
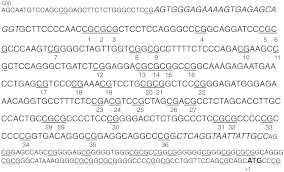
DNA was isolated from white matter of the DLPFC or hippocampus, as above. Unmethylated cytosines were converted to uracils by bisulfite conversion (EZ DNA Methylation Gold Kit, Zymo Research) per manufacturer’s instructions. Methylated cytosines are protected from the reaction and remain as cytosines. Following conversion, a 352 bp section of the rhesus Klotho promoter starting −466 bp from the translation start site was amplified using Hot Start Taq (Qiagen) in two separate PCR reactions. Amplification of appropriately sized PCR products was verified on agarose gels prior to pyrosequencing conducted by EpigenDx (Worcester, MA). Percent methylation was calculated at each CpG residue across the amplified area, 36 total CpGs (Fig. 4).
Fig. 4.
Schematic representation of the Klotho promoter. Shown are the first 500 bp of rhesus KL promoter sequence. Numbering is based on the translation start site ATG that is in bold. This area of the KL promoter is composed of 72% GC residues. Italicized residues are the flanking PCR promoters for pyrosequencing. All CpGs are underlined and those evaluated by pyrosequencing are also numbered
CpG methylation PCR reporter assay
Primers with methyl groups added on selective cytosine residues were used to amplify the rhesus reporter construct as described by Yang et al. (2010). Briefly, forward primers identical, except for the presence of a methyl group to one specific cytosine residue, were used in combination with the common RV4 reverse primer (Promega). PCR amplification resulted in linear constructs of the rhesus KL promoter driving luciferase transcription. PCR products from three independent reactions were gel purified (Stratagene), combined, and co-transfected with renilla luciferase into HEK 293 cells using Nanofect, as described. After 48 h of transfection, luciferase activity was measured using the dual luciferase assay system. Data were normalized to renilla expression for each well. Unmethylated control luciferase activity was defined as 100%.
Transcription factor prediction
Transcription factor binding sites were predicted using MatInspector (Genomatix v 2.1). Binding sites with a matrix similarity greater than 0.8 were deemed likely matches.
Primers
All primers were synthesized by IDT (Des Moines, IA). The human KL promoter was cloned using: Forward GGGAAATGTGATACTCCATGTAGACGTAGC, Reverse GCTGCGCGGGAGCCAGGCTCCGGGGCCCCG. The rhesus KL promoter was cloned using: Forward GGATGCACCTCTGTGAGGAT, Reverse ATGCTGCGCTGGAACCAGGC. Fpg assays were conducted with the following primers for HEK293 cells: GAPDH Forward TGAGCAGTCCGGTGTCACTA, Reverse AAGAAGATGCGGCTGACTGT; Tau Forward GCTTGCTTTAAGCCGATTTG, Reverse CGCTTTCTCCACCTCCTGTA; KL Forward CCAACGCAACCCATAAATCT, Reverse GGACGCTCAGGTTCATTCTC. For rhesus fpg assays: GAPDH Forward ACCTCCAGGCATGCATTTAC, Reverse GACTGTCGAACAGGAGGAGC; Tau Forward TCTCCTGTTGACTCCTGCCT, Reverse TCCCTTGCTCCATATTCCTG; KL Forward GGGGAATCCCTTTCTCTCAG, Reverse GGCAATAATTACCTGAGCCG.
Initial promoter amplification for pyrosequencing was conducted using: Amplicon 1: Forward AGTGGGAGAAAAGTGAGAGTAGG, Reverse 5′biotin labeled AAAAAACACCTATTTCTCCCATCTC; Amplicon 2: Forward GTTTTAGGGAGATGGGAGAAATA, Reverse 5′biotin labeled ACTCCCCTAACAATAATTACCTAAA. Pyrosequencing utilized: Amplicon 1: TGGGAGAAAAGTGAGAGTA and GAAGTAGTTTTAGGGTTGAT. Amplicon 2: GGGAGATGGGAGAAATAG and GTTAGAGAAGTTTTTAGTAT.
In vitro CpG Assays utilized primers synthesized with and without methyl groups. Methyl group sites are identified in bold and underlined: CpG 1 GTGCTTCCCCAACCGCGCGCTCCTCC, CpG 7 CGCGCCCAAGTCGGGGCTAGTTGGTC, CpG 10 CTTTTCTCCCAGACGAAGCCGCTCCAG, CpG 11 CAGACGAAGCCGCTCCAGGGCTG, CpG 12 CAGGGCTGATCTCGGAGGACGCGCGGC, CpG 14 CTCGGAGGACGCGCGGCCGGCAAAG, CpG 15 CTCGGAGGACGCGCGGCCGGCAAAG, CpG 18 CTGAGCGTCCCCGAAACGTCCTG, CpG 19 GTCCCCGAAACGTCCTGCGCGGCTC, CpG 20/21 GAAACGTCCTGCGCGGCTCCCGGGAG, CpG 23 GTGCCTTTCTCCGACGTCCGCTAG, CpG 27 GCTAGCGACGCCTCTAGCAC, CpG 28 CACCTTGCCCACTGCCGCGCCCCTC, CpG 31 CTGGCCCTCCGCGCCCCCGC, CpG 35 CGGTGACAGGGCGGAGGCAGGC. To amplify from the 3′ end of the luciferase construct we used the common reverse primer (RV4; Promega) GACGATAGTCATGCCCCGCG for all reactions.
Statistics
Statistical significance was calculated using Graphpad Prism software version 5.0. Student’s t-test or ANOVA with Bonferroni correction for post hoc multiple comparisons were used where indicated to determine if minimum threshold of at least p < 0.05 had been achieved between groups. Correlation statistics were conducted using Pearson correlation to determine whether the data had achieved statistical significance with a p < 0.05.
Results
KL protein is reduced in white, but not grey matter of aged rhesus monkeys
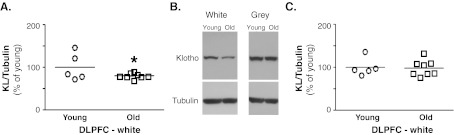
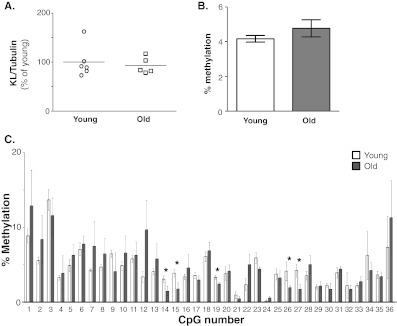
Previous work in our lab described a reduction in KL mRNA and protein in white matter from aged rhesus monkey brain (Duce et al. 2008). To determine whether age-related epigenetic changes were involved in this reduction, we first replicated these findings in a new group of animal tissues. Samples of fresh frozen dorsolateral prefrontal cortex (DLPFC) were removed from storage at −80°C and dissected into blocks of white matter-enriched and grey matter-enriched tissue prior to homogenization. In white matter, quantification of Western blots showed a ~20% decrease in KL protein in old animals (mean age of 24.35 years) compared to young animals (mean age of 7.68 years; Fig. 1a and b). Quantification of DLPFC grey matter from the same animals showed no difference (Fig. 1b and c).
Fig. 1.
Klotho protein decreases in white matter of rhesus monkey. a. Quantification of KL protein in white matter normalized to β-tubulin expression. Old animals show on average a 20% decrease in KL protein (n = 5 young, n = 8 old; Student’s t-test, *p < 0.04). b. Representative blots of KL and β-tubulin expression in white and grey matter from DLPFC. c. Quantification of KL protein in grey matter normalized to β-tubulin expression. No change in KL protein was observed between young and old animals (n = 5 young, n = 8 old; Student’s t-test, *p < 0.05)
Increased sensitivity of the KL promoter to oxidative damage in vitro but not in vivo
The work of Lu et al. (2004) indicates that, in human brains, oxidative damage to promoters is often the cause of age-related gene downregulation. Human GAPDH protein expression is stable as a function of age and does not show evidence of oxidative damage to its promoter with age. Conversely, human Tau protein expression is downregulated with age, and its promoter acquires oxidative damage over time. Since the rhesus promoter is not clearly defined, all promoter numbering is based on the translation start site for both human and rhesus KL. Comparison from −1,500 to +1 of the human and rhesus KL promoters shows 92.8% conservation. The promoters are GC-rich. From −500 to +1, the GC content of the human promoter is 74% and that of rhesus is 72%. Based on these observations, age-related decreases in KL protein could be the result of oxidative damage to the promoter.
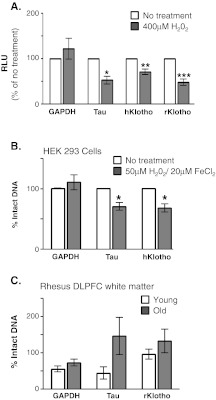
Human GAPDH, Tau, KL, and rhesus KL promoter reporter plasmids were exposed to oxidative conditions prior to transfection. Plasmids were then co-transfected into HEK 293 cells with renilla luciferase and enzyme activity measured 48 h later. As anticipated, luciferase activity after oxidation was stable from the GAPDH reporter and decreased from the Tau reporter construct (Fig. 2a). Oxidation of both the human and rhesus KL promoters resulted in decreased luciferase activity (Fig. 2a). HEK 293 cells were next subjected to oxidizing media conditions followed by Fpg assay to assess the amount of oxidatively damaged promoter DNA (Fig. 2b). As anticipated, statistically significant decreases in the % of intact DNA were observed for tau and KL promoters but not the GAPDH promoter (Fig. 2b). Genomic DNA from young and old rhesus monkey DLPFC was obtained as detailed in methods. Comparison of the ratio of intact PCR product from samples incubated with and without Fpg allowed assessment of the % intact DNA. No significant difference was noted for GAPDH, Tau, or KL promoters in either grey or white matter from the DLPFC in vivo (Fig. 2c white, grey matter data not shown). While in vitro analysis indicates that the KL promoter could be susceptible to age-related downregulation because of oxidative damage, aged monkeys showed no evidence of such damage in vivo.
Fig. 2.
Oxidative damage is not responsible for age-related decrease in Klotho protein in vivo. a. Reporter plasmids were oxidized (400 μM H2O2) prior to cotransfection with renilla luciferase. Unoxidized promoter plasmids (white bars) were processed similarly to oxidized promoter reporter plasmids (grey bars). Following transfection, firefly luciferase was normalized to renilla luciferase in each well. Data in each experiment were compared to relevant unoxidized promoter expression (mean ± S.E.M; n = 4 independent experiments with three replicates per experiment, Student’s t-test, *p < 0.03, **p < 0.003, ***p < 0.0003). b. HEK 293 cells were incubated for 12 h in medium (white bars) or medium containing 50 μM H2O2/20 μM FeCl2 (grey bars) to induce promoter oxidation. Genomic DNA was isolated and incubated with and without Fpg enzyme. GAPDH, Tau, and KL promoters were amplified by PCR and the number of cycles required to cross threshold quantified. Data were normalized to the untreated control (mean ± S.E.M; n = 4 independent experiments with three replicates per experiment; Student’s t-test, *p ≤ 0.002). c. Genomic DNA isolated from the DLPFC white matter of young (white) and old (grey) rhesus monkeys were incubated with and without fpg enzyme. The number of PCR cycles to cross threshold was quantified. Data were normalized to the young animal average (mean ± S.E.M for n = 4 young and n = 8 old animals; Student’s t-test)
Increased methylation of the KL promoter in aged DLPFC white matter
As the KL promoter’s high GC content makes it a candidate for oxidative damage, it likewise makes it a potential target for age-related epigenetic modification via methylation. Hypermethylation of the KL promoter in cervical cancer and colon cancer eliminates KL expression (Lee et al. 2010; Pan et al. 2011). Since increased methylation is observed at the promoters of some age-downregulated genes (Hernandez et al. 2011; Siegmund et al. 2007), promoter methylation with age was assessed in rhesus monkey brain.
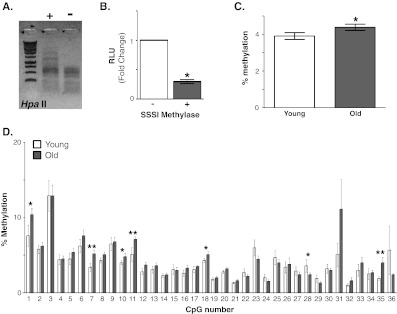
The rhesus KL promoter reporter constructs were incubated with and without SSSI methylase to induce promoter hypermethylation. HpaII methyl-sensitive restriction enzyme digestion confirmed construct modification (Fig. 3a). Following transfection, KL promoter hypermethylation reduced luciferase activity by 70% (Fig. 3b).
Fig. 3.
Methylation of the Klotho promoter in white matter. a. In vitro hypermethylation of the rhesus Klotho promoter. Representative HpaII methyl-sensitive restriction digestion of an aliquot of plasmid treated with and without SSSI methylase to verify modification of the plasmid. Shown is a representative ethidium bromide stained agarose gel. b. HEK293 cells were cotransfected with renilla luciferase and rhesus promoter plasmid treated with and without SSSI methylase. After 48 h, firefly luciferase activity was normalized to renilla luciferase activity in each well. Shown is an average of six experiments, three replicates per experiment (mean ± S.E.M; Student’s t-test, *p ≤ 0.0001). c. To quantify % methylation, white matter isolated from DLPFC of young (n = 7, white) and old (n = 15, grey) rhesus monkey was subjected to pyrosequencing. The % methylation across the promoter region analyzed was determined by comparing the average % methylation across all 36 CpGs for each monkey. An overall 0.4% increase was observed in old monkeys relative to young monkeys (mean ± S.E.M.; Student’s t-test, *p < 0.02). d. The % methylation for each CpG residue was evaluated between young and old animals. Of the 36 residues evaluated, increased methylation in old animals was observed at six sites (CpGs 1, 7, 10, 11, 18, 35). Increased methylation in young animals was observed at one site (CpG 28; mean ± S.E.M.; Student’s t-test, *p < 0.05, **p < 0.02)
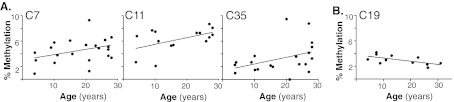
DNA methyltransferases often modify cytosine groups located in CpG motifs (Liu et al. 2009). Fifty eight CpG motifs are found from −500 to +1 of the rhesus KL promoter (Fig. 4, underlined). Pyrosequencing of white matter samples from DLPFC was performed to quantitatively measure the % methylation at CpG residues across the KL promoter. The highly GC-rich rhesus KL promoter sequence made difficult the design of pyrosequencing schemes encompassing the region closest to the translation start site. As such, two PCR reactions, starting −466 bp from the translation start site were used to amplify 352 bp of the rhesus KL promoter (Fig. 4, flanking primer sequence in italics). This allowed assessment of the % methylation at 36 CpGs by pyrosequencing (Fig. 4, numbered). The overall % promoter methylation for each animal was determined by averaging the % methylation of all 36 CpG motifs. A significant increase in methylation of 0.4% was observed in white matter isolated from DLPFC of old animals (Fig. 3c). Increases in methylation were detected at six CpGs (1, 7, 10, 11, 18, 35) in old animals compared to young animals (Fig. 3d). Meanwhile, CpG 28 increased % methylation of DNA in young animals (Fig. 3d). To confirm whether methylation increased with age at these CpGs, the individual CpG % methylation and animal age were correlated. For CpGs 7, 11, and 35, increasing % methylation correlated with increasing age (Fig. 5a).
Fig. 5.
Increased Klotho methylation correlates with age. a. In white matter from DLPFC, of the CpGs that showed an increase in methylation in old animals, CpGs 7, 11, and 35 also showed a significant correlation with age (CpG 7 r2 = 0.14, CpG 11 r2 = 0.26, CpG 35 r2 = 0.15). b. In hippocampus, of the CpGs that showed an increase in methylation in young animals, CpG 19 also showed a significant correlation with age (r2 = 0.4)
Although the changes observed in white matter were statistically significant, they were modest, overall. As such, we sought to determine if an independent area of the brain, enriched in grey matter and with known DNA methylation capabilities would show a similar trend. Since methylation to promoters occurs in hippocampal neurons during learning and memory (Lubin et al. 2008; Penner et al. 2010), rhesus hippocampal samples were evaluated. No changes were detected in KL protein in the hippocampus between groups (Fig. 6a). Following pyrosequencing of samples from hippocampus, the overall percent methylation was assessed for each monkey. Unlike white matter from DLPFC, no change in total promoter methylation was observed in hippocampal samples (Fig. 6b). When individual CpG residues were examined between groups, four CpG residues showed a significant decrease in methylation in old animals (CpGs 14, 15, 19, 27, and 28; Fig. 6c). Only CpG 28 was previously identified in white matter (Fig. 6c). CpG 19 significantly correlated with decreased methylation with age (Fig. 5b).
Fig. 6.
Changes in methylation are specific to white matter. a. Quantification of KL protein expression in hippocampus normalized to β-tubulin (n = 6 young, n = 5 old animals; Student’s t-test). b. The overall level of methylation between young and old was determined by comparing the average % methylation for each monkey. No change was observed in old monkeys relative to young monkeys (mean ± S.E.M.; n = 6 young, n = 5 old animals; Student’s t-test). c. The % methylation for each CpG residue was evaluated between young and old animals. Four of five significant CpGs were hypomethylated in old animals (mean ± S.E.M.; Student’s t-test, *p ≤ 0.05)
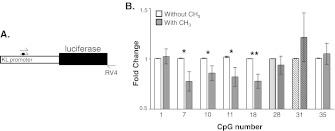
To determine whether CpG methylation affected transcription efficiency, PCR was used to introduce methyl groups to individual C residues (Fig. 7a). The promoter was amplified using forward primers differing only by their incorporation of a methyl group. The reverse primer was common to all PCR reactions amplifying from the 3′ end of the firefly luciferase gene (Fig. 7a). Linear DNA was cotransfected with renilla luciferase into HEK 293 cells. After 48 h, luciferase activity was measured and methylated construct activity compared to its relevant nonmethylated control. In DLPFC white matter, CpGs 1, 7, 10, 11, 18, and 35 were initially identified as sites of increased promoter methylation in old animals (Fig. 3d). In vitro methylation of CpG 7, 10, 11, and 18 caused decreased luciferase activity upon methylation (Fig. 7b, open bars). In vivo, CpG 28 showed disparate results with aging between white and grey matter samples, increasing in methylation in white and decreasing in methylation in grey matter (Figs. 3d and 6c). In vitro methylation of CpG 28 caused no change in reporter activity, indicating that the methylation status of this CpG may not be important (Fig. 7b, vertically stripped bars). While not statistically significant, a striking range in the in vivo % methylation of CpG 31 was observed in white matter. In vitro methylation showed no change in reporter gene activity when C 31 was methylated in vitro (Fig. 7b, diagonally stripped bars). Taken together, % methylation differences between young and old, correlation to age, and in vitro analysis would indicate that CpG 7, 11, 18, and 35 are likely areas of the KL promoter where methylation with age results in a functional consequence.
Fig. 7.
In vitro evaluation of methylation at individual CpG residues. a. Schematic representation of the assay. A region of the rhesus KL promoter and the entire luciferase gene were PCR-amplified. Forward primers differed only in the presence or absence of a methyl group (black circle). b. Linear DNA was cotransfected with renilla luciferase into HEK 293 cells. Luciferase activity was measured 48 h later. Fold change relative to the unmethylated primer are graphed by CpG number. White bars are unmethylated cytosine samples, grey bars are methylated cytosine samples. Bars represent mean ± S.E.M.; n = 4–8, Student’s t-test, *p ≤ 0.05, **p ≤ 0.005
Transcription factor binding sites were predicted in and around the areas where increased methylation was observed in old DLPFC white matter. A total of 114 transcription factors were predicted to bind from −500 to +1 of the KL promoter. Binding sites for SP1 and Egr-1 were identified previously (Choi et al. 2010; Matsumura et al. 1998) and consistent with these results, multiple binding sites for each were predicted by our search. Twenty bases on either side of a CpG motif were searched for consensus binding sites. Eight transcription factors (MASH1, MZF1, SPl1, ZF5, E2F2, CMYB, Max, RREB) were predicted to bind the area around CpG1. CpG 1 is in the core sequence of E2F2 and CMYB. Modification to sites in core sequence would be the most likely to directly impact transcription factor interaction. A number of 20 bp on either side of CpG 7 contains consensus binding sites for Tf2B, XCPE, E2F, CDE, HMX3, and p53. CpG 7 is two bases away from the core sequence for p53 interaction. CpG 10 and 11 are close together, and thus 20-bp upstream of CpG 10 and downstream of CpG 11 were analyzed together. Consensus binding sequences for LYF1, SMAD3, CDE, ZF9, and E2F1 were detected. Of these, CpG 11 is located in the core binding site of ZF9. A number of 20 bp on either side of CpG 18 contains binding sites for WHN, SIX, ZF5, IRF4, and HSF1. CpG 18 is located in the core sequence of both HSF1 and IRF4. Last, 20 bp on either side of CpG 35 contain binding sites for MEIS, Sp1, Egr-1, KLF7, TLX1, HDBP1, p53, and CTCF. CpG 35 is located in the core sequence of SP1 and KLF7.
Discussion
White matter degeneration and KL changes
The rhesus monkey is a model of normal, nonpathological brain aging. Rhesus monkeys do not develop neurodegenerative disorders, but a subset of old animals do develop cognitive impairment (Herndon et al. 1997). As in humans, with age, white matter degeneration is common and widespread in rhesus monkeys. As measured by MRI, only about 2% of overall grey matter is lost, while 11.5% of white matter is lost in aged rhesus monkeys (Wisco et al. 2008). Similarly, with age, astrocytes and microglia become activated (Sloane et al. 1999, 2000) and develop inclusions (Luebke et al. 2010). Myelin proteins are aberrantly expressed or degraded (Hinman and Abraham 2007; Hinman et al. 2006, 2008; Sloane et al. 2003). Protein abnormalities accompany an overall loss of myelinated fibers and an increase in fibers with major myelin deficits like ballooning (Luebke et al. 2010; Peters 2009). The overall number of oligodendrocytes increases with age, but when this increase is coupled to changes in action potential frequency along axons and reorganization of the nodes of Ranvier, it suggests attempts to compensate for widespread dysfunction (Chang et al. 2005; Hinman et al. 2006; Luebke et al. 2010; Peters 2009). This study shows that in the same monkeys where KL protein is decreased, the KL promoter is modified by methylation but not oxidation. Addition of methyl groups to specific cytosine residues in vitro decreased gene transcription and, as such, may explain why less KL protein is detected in the white matter of aged rodents and primates. This interpretation is strengthened by recent reports that hypermethylation of the KL promoter occurs in advanced cervical cancers and results in loss of KL protein expression (Lee et al. 2010). As well, methylation of promoters is an established mechanism behind age-related protein expression changes in the brain (Hernandez et al. 2011; Siegmund et al. 2007).
Many small changes can affect gene expression
In rhesus monkey white matter, we observed a 20% decrease in KL protein. We then show 0.4% increase in overall methylation in old animals. With an average increase of 1.8% at the six individually significant CpGs, the change in % methylation at any of these CpGs is modest. These results are not as profound as those observed in cancer where complete absence of KL is accompanied by hypermethylation across the KL promoter (Lee et al. 2010; Pan et al. 2011). However, in this study, we compared the methylation status of young and old brain during nonpathological aging where effects are not anticipated to be as dramatic as those in a pathological condition. In addition, our samples contained a mix of the different types of cells that make up white matter. Studies in cancer lines or isolated tumors utilize cells from a common precursor and would be expected to show greater effects. While the overall content of 5-methyldeoxycytosines in the genome is reported to decrease with age (Golbus et al. 1990; Wilson et al. 1987), methylation of CpG islands increases with age in the brain (Hernandez et al. 2011; Siegmund et al. 2007). The consequence of increased promoter methylation is most often a decrease in gene expression (Takasugi 2011; Yang et al. 2010; Zschocke et al. 2007). Our findings indicate that KL is among the genes in the brain susceptible to downregulation as a result of age-related promoter methylation. KL is involved in ion homeostasis (Nabeshima 2008) and regulation of signaling pathways (i.e., Insulin/IGF1, FGF23, and Wnt) (Kurosu et al. 2005, 2006; Liu et al. 2007; Utsugi et al. 2000; Yamamoto et al. 2005). Changes in KL gene expression would, over time, impact normal function of all downstream pathways. Evidence of such an effect is observed in the KL polymorphism KL VS (Arking et al. 2002) where slight changes in KL function increases risk of disease (Arking et al. 2002, 2003, 2005; Invidia et al. 2010; Wolf et al. 2010). Of course, the overall contribution of a small change in one gene would not induce a dramatic phenotype in humans as it does in the KL knockout mouse; however, small changes in KL and numerous other genes would add up over time.
Methylation alone is not likely to be the only mechanism resulting in decreased KL mRNA and protein expression (Duce et al. 2008). As well as other epigenetic modifications, some transcription factors are downregulated with age. One example is Egr-1 (Desjardins et al. 1997) which regulates KL transcription (Choi et al. 2010). Decreased expression of Egr-1, as shown by Desjardins et al. (1997), and other relevant transcription factors with age would decrease KL transcription. The compounding effects of a depleted transcription factor pool and epigenetic changes in transcription factor recognition sites would further decrease gene expression. In addition to transcriptional regulatory changes with age, post-transcriptional changes in microRNA (miR) occur with age. Global array profiling of miR show that more miRs are upregulated than downregulated with age (Liang et al. 2009; Maes et al. 2008). Upon binding to their target sequences, miR nearly always decrease protein levels (Liang et al. 2009). As such, an age-related change in the miR milieu of a cell should promote an environment where protein expression decreases. The KL mRNA contains a long 3′UTR indicating it is likely a target for miR regulation. Greater understanding of both the pre and post-transcriptional regulation of KL will further expand our understanding of why it is downregulated with age.
Age and epigenetic differences between species
While in vitro assays with KL promoter constructs showed a clear susceptibility to oxidative damage, such damage was not observed in vivo in rhesus monkeys. As such, while our methylation studies are consistent with human findings (Hernandez et al. 2011), they contrast work examining oxidative damage to human promoters (Lu et al. 2004). Consistent between species, array studies profiling changes in aging brain in humans (Lu et al. 2004) and in rhesus monkeys (Duce et al. 2008) show ~4% of genes examined to be significantly up or downregulated. Of the 4% of age-regulated genes in humans, only a small fraction were the target of oxidative damage to promoter regions. If epigenetic changes at the promoter are a result of age-related dysfunction of DNA repair mechanisms, both oxidative damage and methylation would be expected to contribute, although their contribution to any given gene or in any given tissue may be different. While rhesus monkeys are considered a long-lived species (living up to 35 years), human lifespan is ~ three times greater. It would be interesting to assess whether accumulation of methylated cytosines and oxidative modifications depend on lifespan and whether modifications occur concurrently or sequentially. If methylation and oxidation are a generalized symptom of breakdown in DNA repair integrity (Li et al. 2008), they could occur concurrently and the susceptibility of any given promoter to one or both would likely be a random process with either modification as equally likely to be observed at any given age-downregulated promoter. If, however, accumulation of modifications is a sequential process as DNA repair mechanisms fail over time, our results might suggest more methylation in shorter-lived species (rhesus monkeys) while both persist in longer-lived species (humans). If the process of aging results in a more sequential pattern of DNA modifications, the types of modification observed at age-downregulated genes may serve as a biomarker of overall health.
Why is hypermethylation of the KL promoter only observed in white matter?
Epigenetic modification of the KL promoter may explain why decreased mRNA and protein expression is observed in white matter of aged brains. Since changes in KL mRNA and protein do not change in grey matter, we were not surprised at the difference in CpG methylation between DLPFC white matter and hippocampal grey matter. However, it is interesting to consider whether neurons are specifically protected from age-related promoter modification or rather, if cells that make up the white matter are more vulnerable. Since neurons express both DNA methyltransferases 1 and 3a (Brooks et al. 1996; Siegmund et al. 2007), the lack of modifications in neurons is not the result of an inability to methylate DNA. In fact, transient methylation of genes in hippocampal neurons is critical for learning and memory (Liu et al. 2009; Lubin et al. 2008; Penner et al. 2010). Replication differences exist between aged white and grey matter. Although there is an overall loss of white matter with age in the rhesus monkey brain, oligodendrocyte numbers increase (Luebke et al. 2010). Chu et al. (2007) reported that nonmitotic cells do not show increased CpG methylation with age and suggested that errors in replication are likely the source of age-related methylation increases. DNA repair mechanisms are less efficient with age (Li et al. 2008) and could underscore age-related downregulation as both oxidatively damaged DNA bases and improperly methylated DNA bases are not efficiently detected and removed. If KL protein changes are mostly restricted to glia, mistakes in methylation during the differentiation from progenitor cells would likely explain the white matter restricted decrease in KL protein. However, at this time, the cell types that express KL in the mammalian brain white matter remain to be determined. A third possibility to explain changes in KL expression in white matter only would be altered signaling from neurons. Glutamate transporter (GLT1/EAAT2) is produced by astrocytes to allow uptake of glutamate from the synaptic cleft and prevent exocytotoxic damage to neurons (Rothstein et al. 1996). GLT1 promoter hypermethylation is observed in astrocytes cultured alone (Yang et al. 2010). Meanwhile, low to no methylation of the GLT1 promoter occurs in astrocytes isolated either from neuronal cocultures or from adult mouse brain (Yang et al. 2010). These results suggest that neuronal factors can modify the epigenetic profile of surrounding glia. In aged rhesus monkey white matter, axonal degeneration is apparent (Luebke et al. 2010). It is possible that proper KL expression in white matter relies on currently uncharacterized chemical communication from neurons. In absence of such factors, KL promoter methylation could occur to silence expression.
Summary
This study provides evidence that KL is epigenetically modified by methylation in the white matter of aged rhesus monkey. Methylation at the key residues identified by pyrosequencing can impact gene transcription. These results indicate that KL is among the genes of the CNS that is downregulated with age as a result of epigenetic modification. Understanding the mechanisms that induce age-related gene transcriptional changes is critical in determining the break points between normal aging and development of neurological disorders. Decreased KL protein alone would not be anticipated to result in overt neurodegenerative disease, as none was observed in the knockout animals (Kuro-o et al. 1997). However, KL knockout mice have a short lifespan and most neurodegenerative conditions occur later in life. KL’s absence alone does result in an aged CNS phenotype (Shiozaki et al. 2008), and its function as a homeostatic and neuroprotective (Nagai et al. 2003; Shiozaki et al. 2008; Uchida et al. 2001) molecule indicate that age-related downregulation would participate in creating an environment where white matter damage could accumulate with time.
Acknowledgements
The authors would like to thank Dr. CiDi Chen for cloning of the human KL promoter and for insightful discussions during the early stage of project development. We also thank Dr. J. David Sweatt, Dr. Joerg Kumbrink, Ms. Tracey Tucker, and Mr. Mike Nagle for helpful discussions of the manuscript. This work was funded by the National Institute of Health/National Institute of Aging P01 AG-000001-33 (CRA and DLR), the National Institute of Health/National Institute of Aging 5T32AG000115-24 and NIH/NIA K99/R00 AG034989-01 (GDK), the and National Institute of Health/National Center for Research Resources P51-RR000165 (Yerkes Center).
Contributor Information
Douglas L. Rosene, Email: drosene@bu.edu
Carmela R. Abraham, Phone: +1-617-6384308, FAX: +1-617-6385339, Email: cabraham@bu.edu
References
- Arking DE, Krebsova A, Macek M, Sr, Macek M, Jr, Arking A, Mian IS, Fried L, Hamosh A, Dey S, McIntosh I, Dietz HC. Association of human aging with a functional variant of klotho. Proc Natl Acad Sci U S A. 2002;99(2):856–861. doi: 10.1073/pnas.022484299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Becker DM, Yanek LR, Fallin D, Judge DP, Moy TF, Becker LC, Dietz HC. KLOTHO allele status and the risk of early-onset occult coronary artery disease. Am J Hum Genet. 2003;72(5):1154–61. doi: 10.1086/375035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Atzmon G, Arking A, Barzilai N, Dietz HC. Association between a functional variant of the KLOTHO gene and high-density lipoprotein cholesterol, blood pressure, stroke, and longevity. Circ Res. 2005;96(4):412–8. doi: 10.1161/01.RES.0000157171.04054.30. [DOI] [PubMed] [Google Scholar]
- Brooks PJ, Marietta C, Goldman D. DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci. 1996;16(3):939–945. doi: 10.1523/JNEUROSCI.16-03-00939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein CA, Adler F, Nelson-Williams C, Iijima J, Li P, Imura A, Nabeshima Y, Reyes-Mugica M, Carpenter TO, Lifton RP. A translocation causing increased alpha-klotho level results in hypophosphatemic rickets and hyperparathyroidism. Proc Natl Acad Sci U S A. 2008;105(9):3455–3460. doi: 10.1073/pnas.0712361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli TC, Xu M, O’Connell MP, Chien B, Frank BP, Subaran S, Indig FE, Morin PJ, Hewitt SM, Weeraratna AT. Loss of Klotho during melanoma progression leads to increased filamin cleavage, increased Wnt5A expression, and enhanced melanoma cell motility. Pigment Cell Melanoma Res. 2010;24(1):175–186. doi: 10.1111/j.1755-148X.2010.00792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YM, Rosene DL, Killiany RJ, Mangiamele LA, Luebke JI. Increased action potential firing rates of layer 2/3 pyramidal cells in the prefrontal cortex are significantly related to cognitive performance in aged monkeys. Cereb Cortex. 2005;15(4):409–418. doi: 10.1093/cercor/bhh144. [DOI] [PubMed] [Google Scholar]
- Chen CD, Podvin S, Gillespie E, Leeman SE, Abraham CR. Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc Natl Acad Sci U S A. 2007;104(50):19796–197801. doi: 10.1073/pnas.0709805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang X, Zhao W, Wu J. Klotho inhibits growth and promotes apoptosis in human lung cancer cell line A549. J Exp Clin Cancer Res. 2010;29:99. doi: 10.1186/1756-9966-29-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BH, Kim CG, Lim Y, Lee YH, Shin SY. Transcriptional activation of the human Klotho gene by epidermal growth factor in HEK293 cells; role of Egr-1. Gene. 2010;450(1–2):121–127. doi: 10.1016/j.gene.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu MW, Siegmund KD, Eckstam CL, Kim JY, Yang AS, Kanel GC, Tavare S, Shibata D. Lack of increases in methylation at three CpG-rich genomic loci in non-mitotic adult tissues during aging. BMC Med Genet. 2007;8:50. doi: 10.1186/1471-2350-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins S, Mayo W, Vallee M, Hancock D, Moal M, Simon H, Abrous DN. Effect of aging on the basal expression of c-Fos, c-Jun, and Egr-1 proteins in the hippocampus. Neurobiol Aging. 1997;18(1):37–44. doi: 10.1016/S0197-4580(96)00206-0. [DOI] [PubMed] [Google Scholar]
- Duce JA, Podvin S, Hollander W, Kipling D, Rosene DL, Abraham CR. Gene profile analysis implicates Klotho as an important contributor to aging changes in brain white matter of the rhesus monkey. Glia. 2008;56(1):106–117. doi: 10.1002/glia.20593. [DOI] [PubMed] [Google Scholar]
- Golbus J, Palella TD, Richardson BC. Quantitative changes in T cell DNA methylation occur during differentiation and ageing. Eur J Immunol. 1990;20(8):1869–1872. doi: 10.1002/eji.1830200836. [DOI] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, Singleton AB (2011) Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet [DOI] [PMC free article] [PubMed]
- Herndon JG, Moss MB, Rosene DL, Killiany RJ. Patterns of cognitive decline in aged rhesus monkeys. Behav Brain Res. 1997;87(1):25–34. doi: 10.1016/S0166-4328(96)02256-5. [DOI] [PubMed] [Google Scholar]
- Hinman JD, Abraham CR. What’s behind the decline? The role of white matter in brain aging. Neurochem Res. 2007;32(12):2023–2031. doi: 10.1007/s11064-007-9341-x. [DOI] [PubMed] [Google Scholar]
- Hinman JD, Peters A, Cabral H, Rosene DL, Hollander W, Rasband MN, Abraham CR. Age-related molecular reorganization at the node of Ranvier. J Comp Neurol. 2006;495(4):351–362. doi: 10.1002/cne.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman JD, Chen CD, Oh SY, Hollander W, Abraham CR. Age-dependent accumulation of ubiquitinated 2′,3′-cyclic nucleotide 3′-phosphodiesterase in myelin lipid rafts. Glia. 2008;56(1):118–133. doi: 10.1002/glia.20595. [DOI] [PubMed] [Google Scholar]
- Ichikawa S, Imel EA, Kreiter ML, Yu X, Mackenzie DS, Sorenson AH, Goetz R, Mohammadi M, White KE, Econs MJ. A homozygous missense mutation in human KLOTHO causes severe tumoral calcinosis. J Clin Invest. 2007;117(9):2684–2691. doi: 10.1172/JCI31330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Invidia L, Salvioli S, Altilia S, Pierini M, Panourgia MP, Monti D, Rango F, Passarino G, Franceschi C. The frequency of Klotho KL-VS polymorphism in a large Italian population, from young subjects to centenarians, suggests the presence of specific time windows for its effect. Biogerontology. 2010;11(1):67–73. doi: 10.1007/s10522-009-9229-z. [DOI] [PubMed] [Google Scholar]
- Kuro-o M. Klotho and aging. Biochim Biophys Acta. 2009;1790(10):1049–1058. doi: 10.1016/j.bbagen.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, Iwasaki H, Iida A, Shiraki-Iida T, Nishikawa S, Nagai R, Nabeshima YI. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390(6655):45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, Shimomura I, Takayama Y, Herz J, Kahn CR, Rosenblatt KP, Kuro-o M. Suppression of aging in mice by the hormone Klotho. Science. 2005;309(5742):1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu MC, Moe OW, Kuro-o M. Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem. 2006;281(10):6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Jeong DJ, Kim J, Lee S, Park JH, Chang B, Jung SI, Yi L, Han Y, Yang Y, Kim KI, Lim JS, Yang I, Jeon S, Bae DH, Kim CJ, Lee MS. The anti-aging gene KLOTHO is a novel target for epigenetic silencing in human cervical carcinoma. Mol Cancer. 2010;9(9):109. doi: 10.1186/1476-4598-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Mitchell JR, Hasty P. DNA double-strand breaks: a potential causative factor for mammalian aging? Mech Ageing Dev. 2008;129(7–8):416–424. doi: 10.1016/j.mad.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R, Bates DJ, Wang E. Epigenetic Control of MicroRNA Expression and Aging. Curr Genomics. 2009;10(3):184–193. doi: 10.2174/138920209788185225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J, Malide D, Rovira II, Schimel D, Kuo CJ, Gutkind JS, Hwang PM, Finkel T. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317(5839):803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- Liu L, Groen T, Kadish I, Tollefsbol TO. DNA methylation impacts on learning and memory in aging. Neurobiol Aging. 2009;30(4):549–560. doi: 10.1016/j.neurobiolaging.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429(6994):883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28(42):10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebke J, Barbas H, Peters A. Effects of normal aging on prefrontal area 46 in the rhesus monkey. Brain Res Rev. 2010;62(2):212–232. doi: 10.1016/j.brainresrev.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes OC, An J, Sarojini H, Wang E. Murine microRNAs implicated in liver functions and aging process. Mech Ageing Dev. 2008;129(9):534–541. doi: 10.1016/j.mad.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R, Kuro-o M, Nabeshima Y. Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem Biophys Res Commun. 1998;242(3):626–630. doi: 10.1006/bbrc.1997.8019. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y. Ectopic calcification in Klotho mice. Clin Calcium. 2002;12(8):1114–1117. [PubMed] [Google Scholar]
- Nabeshima Y. The discovery of alpha-Klotho and FGF23 unveiled new insight into calcium and phosphate homeostasis. Cell Mol Life Sci. 2008;65(20):3218–3230. doi: 10.1007/s00018-008-8177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Yamada K, Kim HC, Kim YS, Noda Y, Imura A, Nabeshima Y, Nabeshima T. Cognition impairment in the genetic model of aging klotho gene mutant mice: a role of oxidative stress. FASEB J. 2003;17(1):50–52. doi: 10.1096/fj.02-0448fje. [DOI] [PubMed] [Google Scholar]
- Pan J, Zhong J, Gan LH, Chen SJ, Jin HC, Wang X, Wang LJ (2011) Klotho, an anti-senescence related gene, is frequently inactivated through promoter hypermethylation in colorectal cancer. Tumour Biol. doi:10.1007/s13277-011-0174-5 [DOI] [PubMed]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA (2010) Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging [DOI] [PMC free article] [PubMed]
- Peters A. The effects of normal aging on myelinated nerve fibers in monkey central nervous system. Front Neuroanat. 2009;3:11. doi: 10.3389/neuro.05.011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Sethares C, Moss MB. The effects of aging on layer 1 in area 46 of prefrontal cortex in the rhesus monkey. Cereb Cortex. 1998;8(8):671–684. doi: 10.1093/cercor/8.8.671. [DOI] [PubMed] [Google Scholar]
- Peters A, Moss MB, Sethares C. Effects of aging on myelinated nerve fibers in monkey primary visual cortex. J Comp Neurol. 2000;419(3):364–376. doi: 10.1002/(SICI)1096-9861(20000410)419:3<364::AID-CNE8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Dykes-Hoberg M, Pardo CA, Bristol LA, Jin L, Kuncl RW, Kanai Y, Hediger MA, Wang Y, Schielke JP, Welty DF. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16(3):675–686. doi: 10.1016/S0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Shih PH, Yen GC. Differential expressions of antioxidant status in aging rats: the role of transcriptional factor Nrf2 and MAPK signaling pathway. Biogerontology. 2007;8(2):71–80. doi: 10.1007/s10522-006-9033-y. [DOI] [PubMed] [Google Scholar]
- Shiozaki M, Yoshimura K, Shibata M, Koike M, Matsuura N, Uchiyama Y, Gotow T. Morphological and biochemical signs of age-related neurodegenerative changes in klotho mutant mice. Neuroscience. 2008;152(4):924–941. doi: 10.1016/j.neuroscience.2008.01.032. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2(9):e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloane JA, Hollander W, Moss MB, Rosene DL, Abraham CR. Increased microglial activation and protein nitration in white matter of the aging monkey. Neurobiol Aging. 1999;20(4):395–405. doi: 10.1016/S0197-4580(99)00066-4. [DOI] [PubMed] [Google Scholar]
- Sloane JA, Hollander W, Rosene DL, Moss MB, Kemper T, Abraham CR. Astrocytic hypertrophy and altered GFAP degradation with age in subcortical white matter of the rhesus monkey. Brain Res. 2000;862(1–2):1–10. doi: 10.1016/S0006-8993(00)02059-X. [DOI] [PubMed] [Google Scholar]
- Sloane JA, Hinman JD, Lubonia M, Hollander W, Abraham CR. Age-dependent myelin degeneration and proteolysis of oligodendrocyte proteins is associated with the activation of calpain-1 in the rhesus monkey. J Neurochem. 2003;84(1):157–168. doi: 10.1046/j.1471-4159.2003.01541.x. [DOI] [PubMed] [Google Scholar]
- Takasugi M (2011) Progressive age-dependent DNA methylation changes start before adulthood in mouse tissues. Mech Ageing Dev [DOI] [PubMed]
- Tigges J, Gordon TP, McClure HM, Hall EC, Peters A. Survival rate and life span of rhesus monkeys at Yerkes Regional Primate Research Center. Am J Primatol. 1988;15:263–73. doi: 10.1002/ajp.1350150308. [DOI] [PubMed] [Google Scholar]
- Uchida A, Komiya Y, Tashiro T, Yorifuji H, Kishimoto T, Nabeshima Y, Hisanaga S. Neurofilaments of Klotho, the mutant mouse prematurely displaying symptoms resembling human aging. J Neurosci Res. 2001;64(4):364–370. doi: 10.1002/jnr.1087. [DOI] [PubMed] [Google Scholar]
- Utsugi T, Ohno T, Ohyama Y, Uchiyama T, Saito Y, Matsumura Y, Aizawa H, Itoh H, Kurabayashi M, Kawazu S, Tomono S, Oka Y, Suga T, Kuro-o M, Nabeshima Y, Nagai R. Decreased insulin production and increased insulin sensitivity in the klotho mutant mouse, a novel animal model for human aging. Metabolism. 2000;49(9):1118–1123. doi: 10.1053/meta.2000.8606. [DOI] [PubMed] [Google Scholar]
- Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262(21):9948–51. [PubMed] [Google Scholar]
- Wisco JJ, Killiany RJ, Guttmann CR, Warfield SK, Moss MB, Rosene DL. An MRI study of age-related white and gray matter volume changes in the rhesus monkey. Neurobiol Aging. 2008;29(10):1563–1575. doi: 10.1016/j.neurobiolaging.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf I, Levanon-Cohen S, Bose S, Ligumsky H, Sredni B, Kanety H, Kuro-o M, Karlan B, Kaufman B, Koeffler HP, Rubinek T. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008;27(56):7094–7105. doi: 10.1038/onc.2008.292. [DOI] [PubMed] [Google Scholar]
- Wolf I, Laitman Y, Rubinek T, Abramovitz L, Novikov I, Beeri R, Kuro OM, Koeffler HP, Catane R, Freedman LS, Levy-Lahad E, Karlan BY, Friedman E, Kaufman B. Functional variant of KLOTHO: a breast cancer risk modifier among BRCA1 mutation carriers of Ashkenazi origin. Oncogene. 2010;29(1):26–33. doi: 10.1038/onc.2009.301. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Clark JD, Pastor JV, Gurnani P, Nandi A, Kurosu H, Miyoshi M, Ogawa Y, Castrillon DH, Rosenblatt KP, Kuro-o M. Regulation of oxidative stress by the anti-aging hormone klotho. J Biol Chem. 2005;280(45):38029–38034. doi: 10.1074/jbc.M509039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gozen O, Vidensky S, Robinson MB, Rothstein JD. Epigenetic regulation of neuron-dependent induction of astroglial synaptic protein GLT1. Glia. 2010;58(3):277–286. doi: 10.1002/glia.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschocke J, Allritz C, Engele J, Rein T. DNA methylation dependent silencing of the human glutamate transporter EAAT2 gene in glial cells. Glia. 2007;55(7):663–674. doi: 10.1002/glia.20497. [DOI] [PubMed] [Google Scholar]