Thirteen tag SNPs at the CASP8 and CASP10 loci in patients with advanced NSCLC were genotyped in a two-stage analysis consisting of a discovery set and an independent validation set. These SNPs were evaluated for their association with toxicity outcomes with platinum-based chemotherapy.
Keywords: CASP8, CASP10, Polymorphisms, Platinum-based chemotherapy, Toxicity, Non-small cell lung cancer, Association
Abstract
Caspase-8 and caspase-10 play crucial roles in both cancer development and chemotherapy efficacy. In this study, we aimed to comprehensively assess single nucleotide polymorphisms (SNPs) of the caspase-8 (CASP8) and caspase-10 (CASP10) genes in relation to toxicity outcomes with first-line platinum-based chemotherapy in patients with advanced non-small cell lung cancer (NSCLC). We genotyped 13 tag SNPs of CASP8 and CASP10 in 663 patients with advanced NSCLC treated with platinum-based chemotherapy regimens. Associations between SNPs and chemotherapy toxicity outcomes were identified in a discovery set of 279 patients and then validated in an independent set of 384 patients. In both the discovery and validation sets, variant homozygotes of CASP8 rs12990906 and heterozygotes of CASP8 rs3769827 and CASP10 rs11674246 and rs3731714 had a significantly lower risk for severe toxicity overall. However, only the association with the rs12990906 variant was replicated in the validation set for hematological toxicity risk. In a stratified analysis, we found that some other SNPs, including rs3769821, rs3769825, rs7608692, and rs12613347, were significantly associated with severe toxicity risk in some subgroups, such as in nonsmoking patients, patients with adenocarcinoma, and patients treated with cisplatin combinations. Consistent results were also found in haplotype analyses. Our results provide novel evidence that polymorphisms in CASP8 and CASP10 may modulate toxicity outcomes in patients with advanced NSCLC treated with platinum-based chemotherapy. If validated, the findings will facilitate the genotype-based selection of platinum-based chemotherapy regimens.
Introduction
Lung cancer is the leading cause of cancer death worldwide, and the incidence of lung cancer continues to increase in China [1, 2]. Non-small cell lung cancer (NSCLC), including adenocarcinoma, squamous cell carcinoma, and large cell carcinoma, is the major histological type of lung cancer [3], accounting for ∼80% of lung cancer deaths [4], and most cases are diagnosed at advanced stages (III or IV) and cannot be surgically resected.
Platinum (cisplatin or carboplatin) double-agent chemotherapy is the most common first-line treatment for patients with advanced NSCLC at present, and the efficacies of different combinations have been demonstrated to be similar in series of trials in unselected patients, with response rates of 30%–40% [5–7]. Standard treatment using platinum compounds induces both intrastrand and interstrand DNA adducts that result in bulky distortion of DNA and activates apoptosis signaling pathways, leading to cancer cell death [8, 9]. Nonetheless, the cytotoxic effects of platinum drugs may produce severe adverse effects, by damaging normal cells, which may hinder further treatment and impact outcomes. Differences in toxicities experienced and responses in patients who have received the same chemotherapy agent or regimen are commonly observed [10, 11], and these are likely to be a result of polymorphic genetic variation in genes involved in drug metabolism and DNA repair and apoptosis [5, 12]. Recently, numerous clinical studies have elucidated that genes involved in drug transportation, drug metabolism, DNA repair, and apoptosis may modulate platinum-based chemotherapeutic efficacy and drug-related toxicity outcomes [13, 14]. Therefore, the identification of genetic markers may enable us to predict the toxicity outcome with platinum-based chemotherapy and select optimal regimens for personalized therapy.
Caspase proteins are the central components of the apoptotic response [15, 16] and thus are widely recognized to play a crucial role in cancer development and host response to cancer chemotherapy. In the caspase family, caspase-8 and caspase-10 are the initiators and form the death-inducing signaling complex to activate effector caspases (e.g., caspase-3, caspase-6, and caspase-7) by mediating the death signal receptor and/or other signals, including the cytotoxicity of therapeutic regimens [17, 18]. Indeed, several prior basic and clinical studies have suggested that caspases (e.g., caspase-8 and caspase-10) are involved in p53-dependent and p53-independent transcriptional responses in cisplatin-induced apoptosis and are related to the clinical outcomes (including toxicity) of lung cancer patients treated with chemotherapy [19–21]. In addition to previous studies that demonstrated that caspases may play an important role in the etiology of multiple malignancies [22–25], it has also been documented that the gene-expression signatures (including expression levels of the genes encoding caspase-8 [CASP8] and caspase-10 [CASP10]) may be related to the survival rate of patients with stage I NSCLC [26]. In cancer treatment, apoptosis is also the main mechanism of therapeutic effects involving caspases [27]. Downregulation or inhibition of caspase activity often results in resistance of human cancers to chemotherapeutics [28, 29]. Our previous study indicated that three functional candidate single nucleotide polymorphisms (SNPs) (rs3834129, promoter, −652 6 N ins>del; rs13006529, Ile522Leu, A>T; and rs3900115, Ser59Ser, A>G) of CASP8 and CASP10 may be significantly associated with drug response and severe toxicity in subgroups of NSCLC patients (manuscript under review). We believe that it is possible that genetic variants of CASP8 and CASP10 may modulate cell death and affect cancer development and treatment effects or drug-related toxicity.
Currently, there is still a lack of comprehensive studies on the association between CASP8 and CASP10 polymorphisms and chemotherapy toxicity. To expand our previous study, we genotyped 13 tag SNPs at the CASP8 and CASP10 loci in 663 Chinese patients with advanced NSCLC. To minimize type I errors, we performed a two-stage analysis in which the significant associations identified in the first discovery set were further validated in an independent validation set.
Materials and Methods
Study Design and Patient Recruitment
In total, 663 Han Chinese patients with newly histopathologically diagnosed stage IIIA–IV NSCLC in Shanghai in March 2005 to January 2010 were included in this analysis. Of these 663 patients, 279 patients from two participating Hospitals (Zhongshan and Changhai) were used as the discovery set, and an additional 384 patients from another hospital (Shanghai Chest Hospital) were used as the validation set. The study protocol was approved by the Ethical Review Committee of Fudan University and the participating hospitals, and written informed consent was obtained from each individual. The recruitment criteria and clinical data collection were described in our previous report elsewhere [10]. Patient blood samples were collected in ethylenediaminetetraacetic acid tubes and stored at −80°C for later DNA extraction. The research assistants who performed the genotyping assays were blinded to the case–control status and the clinical investigators were blinded to the genotype status of the patients when they scored chemotherapy toxicity, including overall toxicity, gastrointestinal toxicities (nausea and vomiting), and hematologic toxicities (leukocytopenia, neutropenia, amemia, and thrombocytopenia) according to the National Cancer Institution Common Toxicity Criteria version 3.0 [30]. The incidence of grade 3 or 4 toxicity was assessed twice a week during chemotherapy.
Chemotherapy Regimens
All patients enrolled in this study were inoperable and had received first-line platinum-based chemotherapy (definitive chemoradiotherapy was excluded), that is, cisplatin (75 mg/m2) or carboplatin (area under the concentration–time curve of 5), administered on day 1 every 3 weeks, in combination with navelbine (25 mg/m2) on days 1 and 8 every 3 weeks, gemcitabine (1,250 mg/m2) on days 1 and 8 every 3 weeks, paclitaxel (175 mg/m2) on day 1 every 3 weeks, or docetaxel (75 mg/m2) on day 1 every 3 weeks. All chemotherapeutic drugs were administered i.v., and each patient was treated for two to six cycles.
SNP Selection and Genotyping
Common tag SNPs of two apoptotic initiator genes (CASP8 and CASP10) were included in the present study on the basis of biological interactions between these two caspases in the extrinsic apoptosis pathway. The genotyping information of the Han Chinese population was acquired from the HapMap phase II study [31], and tag SNPs with a minor allele frequency (MAF) ≥0.05 and r2 value <0.8 were selected using Haploview software ()[32]. Genomic DNA was extracted using the QIAamp DNA Maxi Kit (Qiagen GmbH, Hilden, Germany). All selected SNPs were genotyped using a customized iSelect HD BeadChip (Illumina, Inc., San Diego, CA). Quality control criteria included a genotyping call rate by SNP >0.95 and a GenCall score >0.2. Each of 13 tag SNPs passed the quality filters. Concordance among the genotyping replicates was >99.9%. Genotyping information is shown in supplemental online Table S1. GenomeStudioV2010.1 (Illumina, Inc., San Diego, CA) and GeneMapper version 4.0 (Applied Biosystems, Foster City, CA) were used to analyze the BeadChip data and generate genotyping reports.
Statistical Analysis
Two-phase screening was performed to investigate associations between CASP8 and CASP10 SNPs and the incidence of severe chemotherapy toxicity for all patients and for subgroups by treatment regimen (platinum plus a DNA-damaging agent and platinum plus a microtubule-targeting agent). Because of the relatively small sample size for each regimen, a dominant model was assumed for the genotypic association test for each SNP. Toxicity outcomes were dichotomized by the presence or absence of (a) any grade 3 or 4 toxicity, (b) any grade 3 or 4 hematologic toxicity (leukocytopenia, neutropenia, anemia, and thrombocytopenia), and (c) any grade 3 or 4 gastrointestinal toxicity (nausea and vomiting). Hardy–Weinberg equilibrium (HWE) was checked using the χ2 goodness-of-fit test. The association between each genetic variant or haplotype and grade 3 or 4 toxicity was estimated by the odds ratio (OR) and its 95% confidence interval (CI), using unconditional logistic regression with adjustment for age (age at diagnosis in cases), sex, performance status, stage, histological type, and treatment regimen. Pairwise linkage disequilibrium (LD) among SNPs was examined using D′ and r2 values, and haplotype blocks were defined using the four-gamete rule with Haploview Software. Individual haplotype frequency was estimated based on the Bayesian algorithm using the PHASE 2.0 program (version 2.0.2) (University of Washington, Seattle, WA) [33]. The haplotype–toxicity association was tested for each LD block using unconditional logistic regression with the same adjustment as mentioned above.
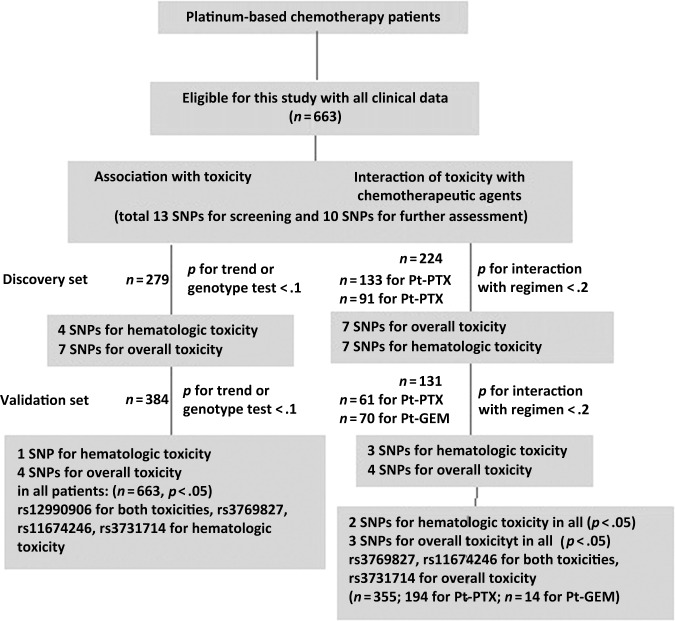
As shown in Figure 1, in the first discovery phase, 13 SNPs were examined for their associations with toxicity outcomes and different levels of interaction with treatment regimen for all 279 and 224 subgroup patients who received a platinum plus a DNA-damaging agent (i.e., paclitaxel) or a platinum plus a microtubule-targeting agent (i.e. gemcitabine), respectively. In the second validation phase, SNPs that had p-values < .10 by the trend test or genotype test for associations with toxicity and p-values < .20 for an interaction with regimen were subjected to confirmation for 384 and 131 subgroup patients who received paclitaxel and gemcitabine, respectively. Finally, SNPs that had p-values < .10 for association with toxicity outcomes and p-values < .20 for an interaction with regimen in patients in the validation set were further subjected to the combined analyses for all 663 and 355 patients.
Figure 1.
Patients and strategy, including selection of eligible cases and a two-phase screening of SNPs associated with toxicity of platinum-based chemotherapy.
Abbreviations: GEM, gemcitabine; Pt, platinum; PTX, paclitaxel; SNP, single nucleotide polymorphism.
Reported p-values are two sided for phase I and one sided for phase II. A p-value < .05 was defined as statistically significant, whereas a p-value < .1 was considered marginal. For multiple statistical testing, the Bonferroni correction was used. Unless otherwise specified, all statistical analyses were performed using SAS software, version 9.2 (SAS Institute, Inc., Cary, NC).
Results
All 663 patients had stage III or IV NSCLC and received two to six cycles of first-line platinum-based chemotherapy. The main patient characteristics and toxicity events are summarized in Table 1 for both the discovery and validation sets. Each of the 13 SNPs met HWE (p > .05). The SNP call rate and sample call rate were >95% and the GenCall score was >0.2. The observed allele frequency was consistent with that previously reported in the HapMap except for three SNPs (two in CASP8 and one in CASP10) that did not meet the quality filter of a MAF ≥0.05, and these were excluded from further analyses (supplemental online Table S1). No significant associations between polymorphisms and patient characteristics were observed (data not shown).
Table 1.
Patient demographics and their association with chemotherapy toxicity outcomes (n = 663)
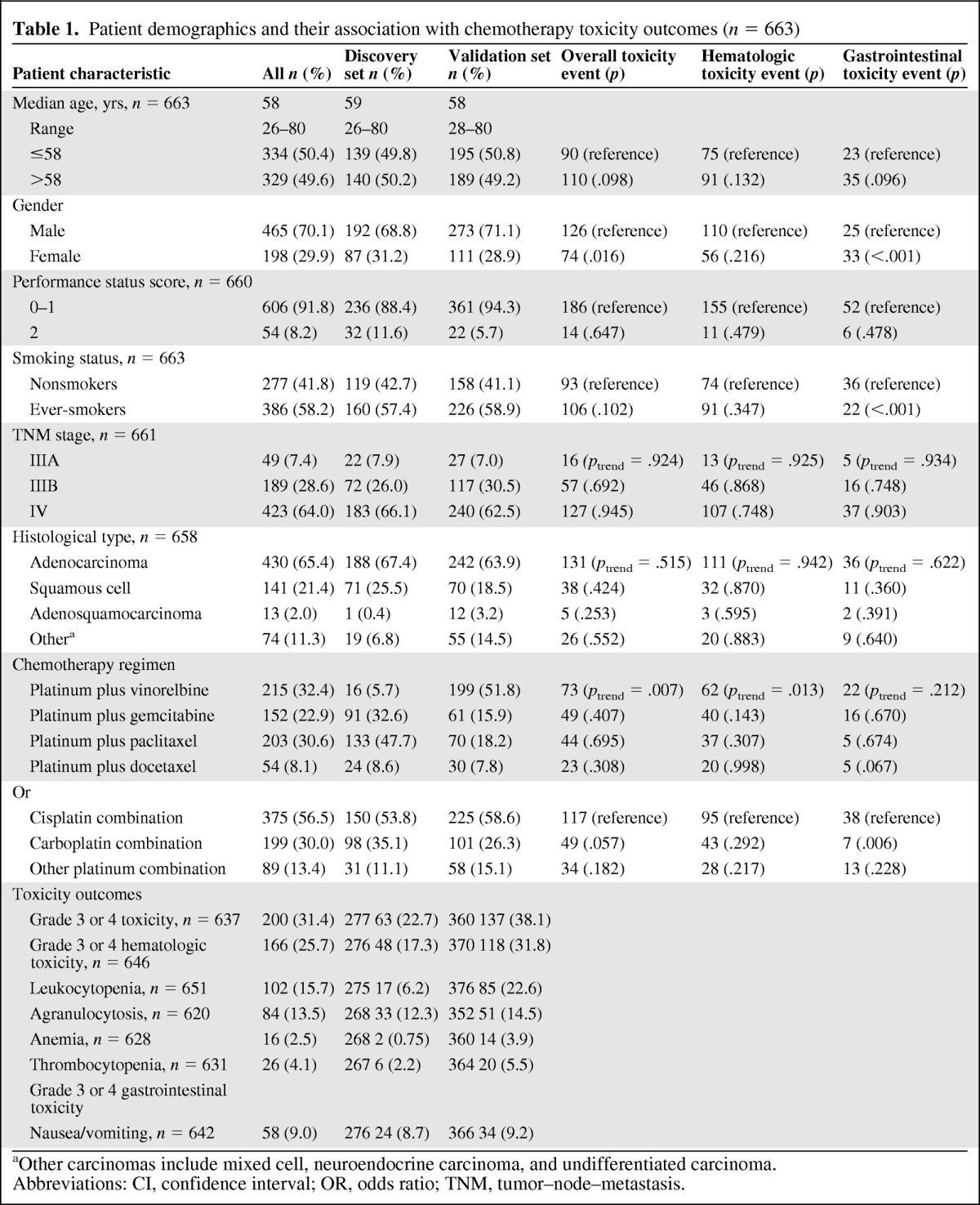
aOther carcinomas include mixed cell, neuroendocrine carcinoma, and undifferentiated carcinoma.
Abbreviations: CI, confidence interval; OR, odds ratio; TNM, tumor–node–metastasis.
Chemotherapy Toxicity by Clinical Characteristics of NSCLC Patients
We stratified patients by age, sex, performance status, stage, smoking status, histopathologic type, and chemotherapy regimen and assessed whether or not clinical variables contributed to chemotherapy toxicity (gastrointestinal toxicity, hematologic toxicity, overall toxicity) using unconditional logistic regression (Table 1). Smoking status and sex were found to be associated with toxicity outcomes. For example, compared with men, women had higher incidences of gastrointestinal toxicity and overall toxicity (p < .001 and p = .016, respectively), and compared with nonsmokers, ever-smokers had a higher incidence of gastrointestinal toxicity (p < .001). Also, patients who were treated with different chemotherapy regimens had varied toxicity outcomes (ptrend = .013 for hematologic toxicity and ptrend = .001 for overall toxicity during platinum-based therapy).
Association Between CASP8 and CASP10 Polymorphisms and Grade 3 or 4 Toxicity
Analysis of Discovery Set
Associations between genetic variants and chemotherapeutic toxicities were assessed using unconditional logistic regression with adjustment for age, sex, performance status, tumor–node–metastasis (TNM) score, histopathologic type, and chemotherapy regimen. In the discovery phase, four SNPs were found to be associated with hematologic toxicity and seven SNPs were associated with overall toxicity (p < .10 by the trend test or genotype test) (Fig. 1, Table 2, and supplemental online Table S2), but none of these SNPs was associated with gastrointestinal toxicity.
Table 2.
Association between CASP8 and CASP10 polymorphisms and grade 3 or 4 chemotherapy toxicity
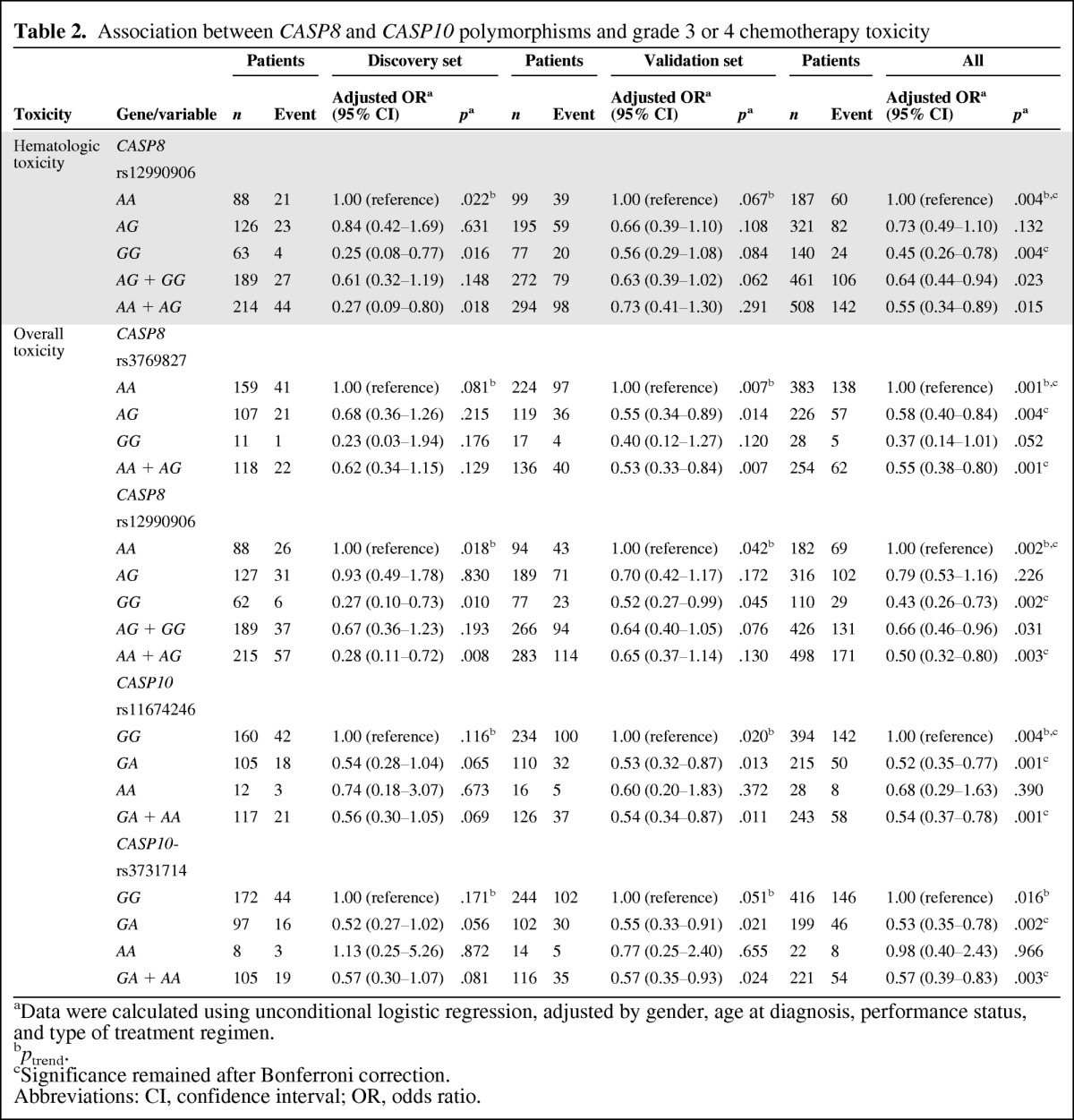
aData were calculated using unconditional logistic regression, adjusted by gender, age at diagnosis, performance status, and type of treatment regimen.
bptrend.
cSignificance remained after Bonferroni correction.
Abbreviations: CI, confidence interval; OR, odds ratio.
Analysis of Validation Set
The associations for one SNP with hematologic toxicity and four SNPs with overall toxicity were reproduced using the aforementioned criteria (p < .10 by the trend test or genotype test) (Fig. 1, Table 2, and supplemental online Table S2) in the remaining 384 patients in the validation set (Table 1). All these five SNPs were associated with the risk for grade 3 or 4 toxicity in the 663 patients overall (p < .01).
In All 663 Patients
For the CASP8 rs12990906 SNP, patients with the variant allele had a lower incidence of severe hematologic toxicity (incidence rate, 24 of 140 versus 60 of 187; adjusted OR, 0.45; 95% CI, 0.26–0.78; p = .004, ptrend = .004). A consistently lower risk for severe toxicity overall was also observed in homozygous variant carriers of rs12990906 (26.4%; p = .002), compared with homozygous carriers of the common allele (37.9%), and the toxicity risk was also lower with higher numbers of minor alleles (p = .002 by the trend test). Heterozygotes of the CASP10 rs11674246 SNP also exhibited a lower incidence of toxicity overall (incidence rate, 50 of 215 versus 142 of 394; adjusted OR, 0.52; 95% CI, 0.35–0.77; p = .001; OR, 0.54; 95% CI, 0.37–0.78; p = .001 under the dominant model; ptrend = .004), and similar results were observed for the CASP8 rs3769827 and CASP10 rs3731714 SNPs (incidence rate, 57 of 226 versus 138 of 384; p= .004 for heterozygotes of rs3769827; incidence rate, 46 of 199 versus 146 of 416; p = .002 for heterozygotes of rs3731714), and the association remained significant after Bonferroni correction (i.e., <α = 0.05/10 = 0.005).
Although carriers of the CASP8 rs3769827 AG and rs3769825 AG genotypes seemed to have a significantly lower risk for severe gastrointestinal toxicity in all 663 patients (p = .028 and p = .013, respectively) (supplemental online Table S2), but they did not show this consistently in both the discovery and validation phases. A similar situation occurred for the CASP8 rs3769825 variant regarding overall toxicity and the CASP8 rs3769827, CASP10 rs11674246, and CASP10 rs3731714 variants regarding hematologic toxicity.
No significant associations (p < .01) were found between CASP8 and CASP10 gene polymorphisms and the risk for severe leukocytopenia or thrombocytopenia (data not shown).
Stratified Analysis of Association Between CASP8 and CASP10 Polymorphisms and Grade 3 or 4 Toxicity
Paclitaxel (203 patients) and gemcitabine (152 patients) were commonly used drugs in the platinum-based regimens of our study among different centers (Table 1). Thus, we looked into differences in toxicity outcomes between these two regimens by genotype.
For overall toxicity, seven of the 10 SNPs met the criteria of p < .20 for an interaction with these two regimens (Fig. 1 and supplemental online Table S3) in the 224 patients in the discovery set with the paclitaxel (133 patients) and gemcitabine (91 patients) regimens. In the validation set, four SNPs reproducibly had a p-value < .20, and in all 355 patients only minor allele carriers of CASP8 rs3769827 still showed a statistically significant protective effect for gemcitabine (incidence rate, 17.6%; OR, 0.29; 95% CI, 0.13–0.69; p = .005 under a dominant model) (Table 3). The association was marginally significant after Bonferroni correction (i.e., <α = 0.05/10 = 0.005).
Table 3.
Stratified analysis of association between CASP8 and CASP10 polymorphisms and grade 3 or 4 chemotherapy toxicity
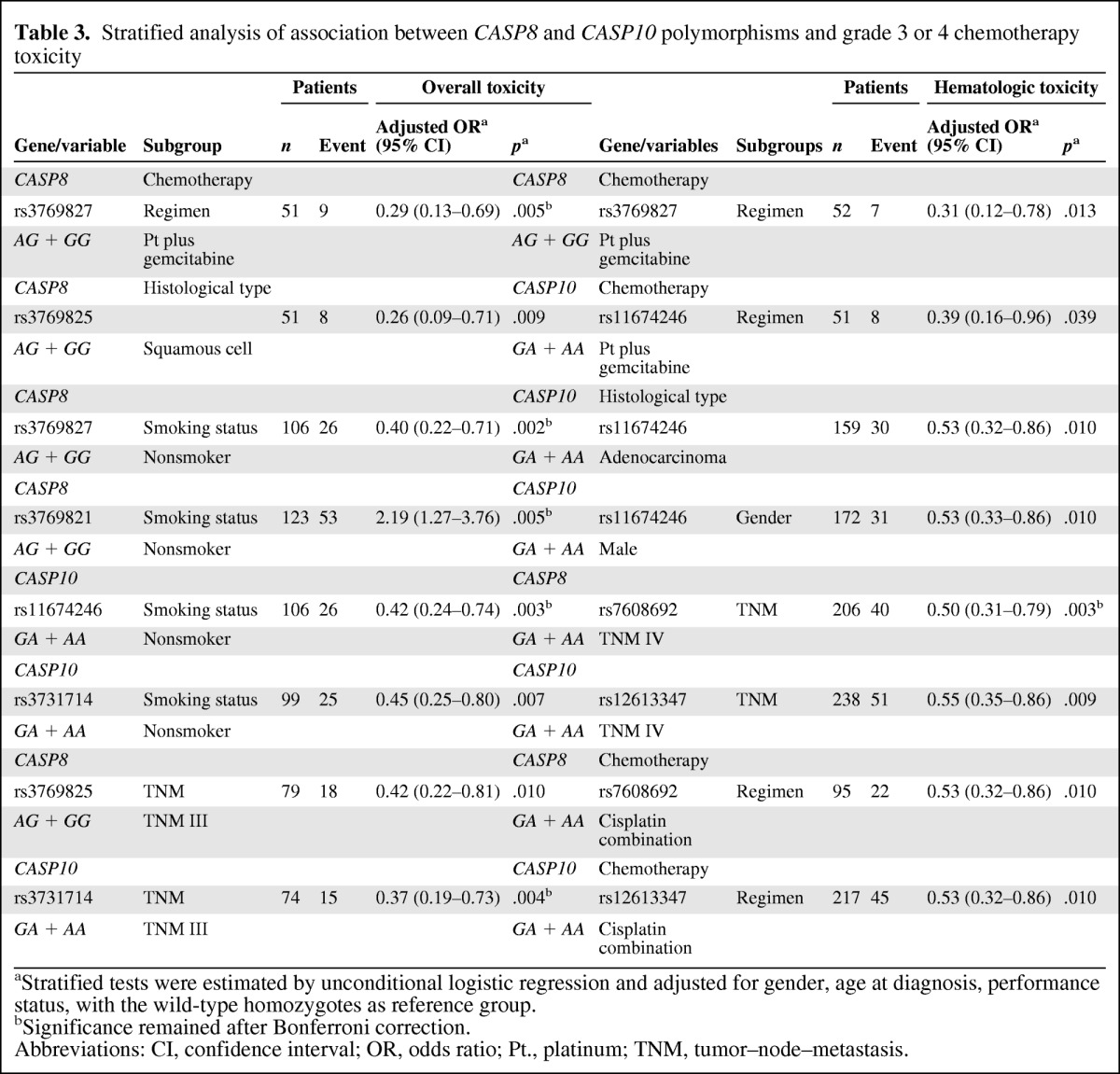
aStratified tests were estimated by unconditional logistic regression and adjusted for gender, age at diagnosis, performance status, with the wild-type homozygotes as reference group.
bSignificance remained after Bonferroni correction.
Abbreviations: CI, confidence interval; OR, odds ratio; Pt., platinum; TNM, tumor–node–metastasis.
For hematologic toxicity, three of seven SNPs remained significant according the criteria p < .20 for interaction after the two phases, and carriers of the minor alleles CASP8 rs3769827 and CASP10 rs11674246 had a lower risk for hematologic toxicity with gemcitabine in all 355 patients (incidence rate, 13.5%; p = .013 and incidence rate, 15.7%; p = 0 .039, respectively).
However, none of the four SNPs discovered in the first phase met our criteria of a p-value < .20 for gastrointestinal toxicity in the end (supplemental online Table S3).
We further performed stratified analyses for chemotherapy toxicity outcomes by other clinical characteristics. The results showed several evident associations between CASP8 and CASP10 polymorphisms and toxicity by sex, smoking status, TNM stage, and histopathologic type (Table 3). For example, minor allele carriers for both the rs11674246 and rs3731714 SNPs had a significant protective effect against overall toxicity (adjusted OR, 0.42; 95% CI, 0.24–0.74; p = .003 and adjusted OR, 0.45; 95% CI, 0.25–0.80; p = .007) in nonsmoking patients, and the same effect against hematological toxicity was found in patients with adenocarcinoma (adjusted OR, 0.53; 95% CI, 0.32–0.86; p = .010 for rs3731714). For rs12613347, minor allele carriers also had a protective effect against hematological toxicity in patients treated with cisplatin combinations (adjusted OR, 0.53; 95% CI, 0.32–0.86; p = .010).
Haplotype Analysis
The reconstructed LD plot of the CASP8 and CASP10 SNPs in 663 patients is shown in supplemental online Figure 1. CASP8 and CASP10 have three blocks defined by the four-gamete rule in the Haploview Software, which were deduced using PHASE 2.1 software. Consistent with the results of the single SNP analysis, we found many evident correlations between haplotypes and severe overall toxicity in all three blocks of CASP8 and CASP10, especially for block 1 of CASP8 (ptrend = 6.86 × 10−4). Haplotypes AA and GG had a protective effect for severe toxicity overall, compared with the most common haplotype AG (OR, 0.73; 95% CI, 0.57–0.93; p = .028 and OR, 0.71; 95% CI, 0.55–0.93; p = 2.74 × 10−4, respectively). Some significant associations were also found between haplotypes of these three blocks and hematological toxicity (Table 4).
Table 4.
CASP8 and 10 haplotypes and grade 3 or 4 chemotherapy toxicity
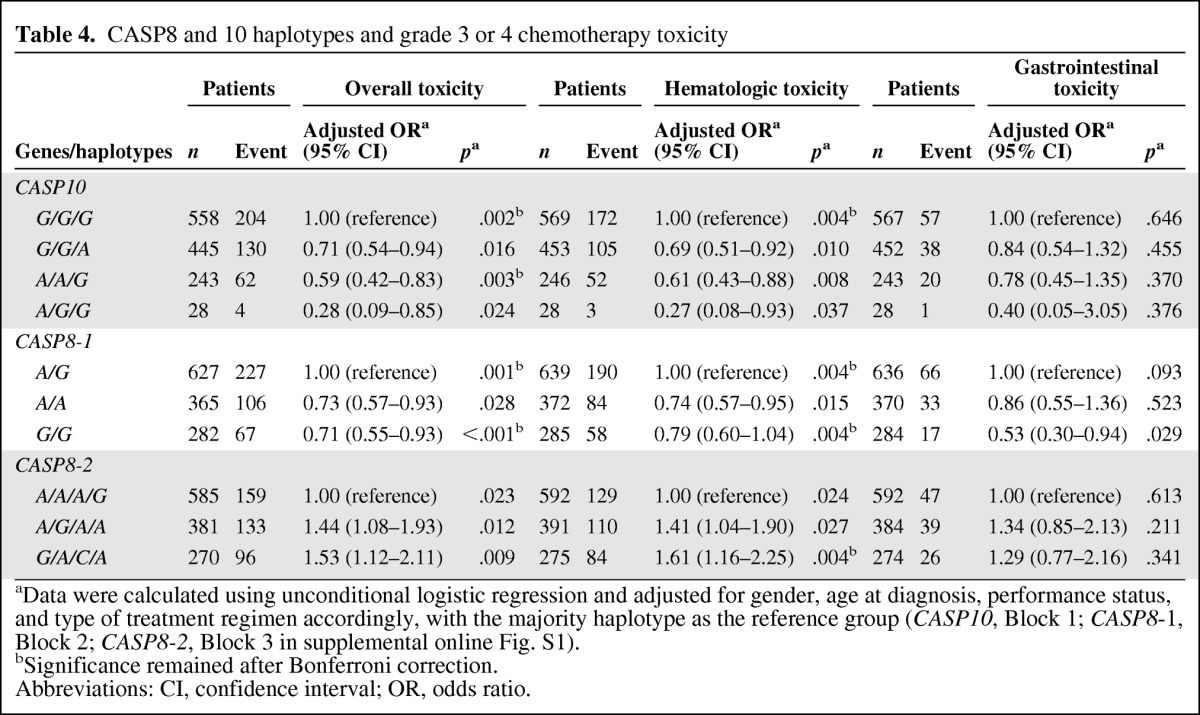
aData were calculated using unconditional logistic regression and adjusted for gender, age at diagnosis, performance status, and type of treatment regimen accordingly, with the majority haplotype as the reference group (CASP10, Block 1; CASP8-1, Block 2; CASP8-2, Block 3 in supplemental online Fig. S1).
bSignificance remained after Bonferroni correction.
Abbreviations: CI, confidence interval; OR, odds ratio.
Discussion
Molecular and cellular studies have provided ample evidence for the roles of the caspase genes in several diseases [34–36]. It is also known that the actions of most cancer chemotherapy regimens are mainly dependent on caspase-mediated apoptosis [27], because the activities of caspase-8 and caspase-10 were found to be required for full induction of cell death during treatment and basal levels of caspase-8 were correlated with treatment sensitivity [37]. However, pharmacogenetic studies on associations between CASP8 and CASP10 polymorphisms and chemotherapy-related cytotoxicity have not been reported. On the basis of these preliminary data, in the current study, we used the tagging SNP approach to comprehensively assess the genetic effects of these two genes on chemotherapy-related toxicity outcomes in lung cancer patients.
Overall, we did find some significant associations between CASP8 and CASP10 tag SNPs and the risk for severe toxicity in NSCLC patients treated with first-line platinum-based chemotherapy. For example, CASP8 rs12990906 was shown to be associated with hematologic and overall toxicities; variants rs3769827, rs11674246, and rs3731714 were associated only with overall toxicity. All associations with these SNPs observed in the discovery set were confirmed in an independent dataset. Furthermore, these SNPs were consistent in their association with a significantly lower risk for toxicity with platinum-based chemotherapy, possibly through the regulation of hematopoietic cell proliferation and survival during chemotherapy.
Previous studies indicated that caspases are not only involved in drug-induced cancer cell death but also play an important role in the development and apoptosis of normal hematological cells [38]. It was reported that caspase-3 is essential in the regulation of normal B-cell homeostasis in a caspase-3 knockout mouse model [39], that caspase-8 regulates T-cell activation and proliferation [40], and that caspase-10 is involved in lymphocyte proliferation [41]. Therefore, it is biologically plausible that SNPs in several genes involved in caspase-related apoptosis pathways could affect the risk for drug- induced toxicity in NSCLC patients treated with first-line platinum-based treatment.
In the present study, we observed that the CASP8 rs12990906 SNP had a significant influence on both hematologic and overall chemotherapy toxicity outcomes with platinum-based treatment, acting in an allele dose–dependent manner. The lower risk associated with rs12990906 was particularly pronounced for overall toxicity (incidence rate, 26.4% with the GG genotype and 37.9% with the AA genotype; p = .002), which could be driven by hematologic toxicity. Because significant SNPs found in our study are mainly tag SNPs, further research is needed to determine the functionality of these SNPs that may be in LD with the identified tag SNPs or molecular mechanisms underlying the identified associations. Nevertheless, several lines of evidence support our findings. Sun et al. [25] reported that T lymphocytes with the deletion variant CASP8 rs3834129, which is in the same LD block as our rs3769827 SNP, had lower caspase-8 activity and activation-induced cell death upon stimulation with cancer cell antigens, leading to lower susceptibility for several cancers. Some SNP–toxicity associations were only observed in heterozygotes (e.g., CASP8 rs11674246), which may be a result of the lower incidence rate of severe toxicity and the relatively small number of homozygous variants (especially in subgroups), if no genetic reason for the selection bias could account for the heterozygotes [42]. However, there may be several other possibilities that could explain this finding, such as loss of heterozygosity in the investigated gene region and the possibility that heterozygotes function in an unknown mode [43].
Our present study also suggested that some genetic variants of CASP8 and CASP10 might differentially affect hematologic and overall toxicity outcomes based on the chemotherapeutic agent administered, including rs3769827 and rs11674246 in the subgroup of patients treated with platinum plus gemcitabine therapy and CASP8 rs7608692 and CASP10 rs12613347 in the subgroup of patients treated with carboplatin. On the other hand, the effects of these SNPs were also apparent in patients with adenocarcinoma and patients in other subgroups, such as males, nonsmokers, and those with stages III or IV disease. However, most of the observed associations were not significant after Bonferroni correction.
Our findings indicate that the predictive role of biomarkers for drug- related toxicity seems to be largely identified by our subgroups, including the specific chemotherapy regimen used, a nonsmoking status, and adenocarcinoma histology. In addition, we found that only the CASP8 rs3769827 variant was weakly associated with response to platinum-based chemotherapy (p = .036). Therefore, genetic factors associated with toxicity are likely to be different from those associated with response and overall survival and progression-free survival outcomes, as also suggested by Shiraishi et al. [5]. Thus, larger studies are needed to validate these subgroup findings, and the underlying mechanism also warrants further investigation.
Residing in tandem order on chromosome 2q33, CASP8 and CASP10 form three LD blocks according to our data, and a consistent correlation was found in block 2 (CASP8–1), which contains rs3769827 and rs7608692. The haplotype GG showed a striking protective effect for severe toxicity overall, suggesting that the lower risk effect of GG was probably driven by rs3769827, especially by its minor allele G. A similar situation was observed for CASP8 rs12990906 and CASP10 rs11674246 and rs3731714, which are located in block 3 (CASP8–2) and block 1 (CASP10), respectively. Although the biologic significance of these tag SNPs and their interactions with chemotherapy agents are unknown at present, it is possible that they may be in strong LD with functionally important but untyped SNPs regulating function and expression of the genes and proteins in these blocks. For instance, a putative microRNA binding site of rs1045487 is completely linked with our SNP rs1045494 (LD r2 = 1.00), according to the SNPINFO Web site [44] (supplemental online Table S4). For CASP10 rs11674246, located in the putative promoter region, this SNP may affect transcription factor binding activity and CASP10 expression at a transcription factor binding site according to prediction using the TFSEARCH [44] and SNPINFO Web sites.
The present study adds to the existing literature on genetic susceptibility to platinum-based chemotherapy toxicity, but a few limitations need to be addressed. First, although our sample size of 663 patients may be one of the largest groups reported to date, the numbers of patients in some subgroups, such as treatment arms and histopathology subtypes (n < 100), were still small. This may result in lower statistical power to detect toxicity associations with mild-effect or low-penetrance SNPs and thus could result in both false-negative and false-positive findings [46]. This problem can only be resolved by studies with larger sample sizes in the future. Additional independent prospective replication is also required to validate our findings. Second, all patients in our present study were treated in three medical centers in Shanghai, which could potentially result in some bias caused by the heterogeneity of the patient populations. However, we used the exact same criteria for patient recruitment and clinical data collection, which may have limited potential confounding effects and reduced the heterogeneity to some extent. In addition, we observed no significant differences in age, sex, or family history in NSCLC patients enrolled from different centers in the study. Third, although we used two independent datasets to validate our findings, this study was performed in a Chinese population, and therefore the results may not be true for other ethnicities. For example, the allele frequencies of all SNPs investigated herein are known to be different among ethnic populations (e.g., rs10192461, whose MAF = 0.00 is lower here than in the HapMap data, MAF = 0.222). Therefore, examination of these tag SNPs in other NSCLC patient populations will help elucidate possible interethnic differences in chemotherapeutic toxicity outcome. We plan to confirm the current findings in our ongoing prospective expansion studies with more stringent conditions and with much larger sample sizes.
Conclusion
In conclusion, the present hospital-based pharmacogenetic study showed some associations between platinum-based chemotherapy toxicity and variants of CASP8 and CASP10 in Chinese NSCLC patients and confirmed that patients homozygous for the CASP8 rs12990906 variant had a significantly lower risk for developing toxicity with platinum-based chemotherapy. Our results support the idea that genetic variations in CASP8 and CASP10 may act as potential predictive biomarkers for platinum-based chemotherapy toxicity in Chinese NSCLC patients. In patients with risk alleles, a cisplatin-based rather than carboplatin-based regimen might be able to reduce severe toxicities. Administration of G-SCF to advanced patients may prevent those patients from experiencing severe hematological toxicities. To date, no genomewide association study (GWAS) has addressed toxicity outcomes with platinum-based chemotherapy in NSCLC patients, although there was a GWAS on the survival outcome of NSCLC patients receiving platinum-based chemotherapy [47] and a GWAS on the efficacy of platinum-based chemotherapy in small cell lung cancer patients [448] that did not identify any genetic association with SNPs from the CASP8 and CASP10 loci. Therefore, further studies or pooled analyses are warranted to validate our findings on the effects on toxicity of CASP8 and CASP10 for potential clinical application.
See www.TheOncologist.com for supplemental material available online.
Editor's Note: See the accompanying editorial on pages 1484–1485 of this issue.
Supplementary Material
Acknowledgments
We greatly acknowledge the collaboration with the participating hospitals and staff.
Ji Qian, Hui-Qi Qu, and Lixin Yang contributed equally.
This work was supported in part by the National Basic Research Program (973 program) of China (2011CB503802), China National High-Tech Research and Development Program (2012AA02A517, 2012AA02A518), Shanghai Science and Technology Research Program (09JC1402200, 10410709100), National Natural Science Foundation of China (30800622, 81001114, 81172093, 30890034), Shanghai Pujiang Program (11PJD005), China Ministry of Health (201002007), Scientific and Technological Support Plans from Jiangsu Province (BE2010715), and Natural Science Foundation of Jilin province (201115215).
Author Contributions
Conception/Design: Ji Qian, Daru Lu, Shaohua Gu, Haijian Wang, Jiucun Wang, Baohui Han, Jin Li, Junjie Wu
Provision of study material or patients: Lixin Yang, Wenqing Chen, Wenting Wu, Xiaoming Tan, Baohui Han
Collection and/or assembly of data: Lixin Yang, Wenqing Chen, Qihan Wu, Xueying Zhao, Wenting Wu, Xiaoming Tan, Baohui Han, Weiwei Fan, Hongyan Chen, Junjie Wu
Data analysis and interpretation: Qingyi Wei, Ji Qian, Hui-Qi Qu, Ming Yin, Xueying Zhao, Xiaoming Tan
Manuscript writing: Qingyi Wei, Ji Qian, Hui-Qi Qu, Ming Yin, Qiming Wang, Junjie Wu
Final approval of manuscript: Daru Lu, Jin Li, Junjie Wu
References
- 1.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Zhao P, Dai M, Chen W, et al. Cancer trends in China. Jpn J Clin Oncol. 2010;40:281–285. doi: 10.1093/jjco/hyp187. [DOI] [PubMed] [Google Scholar]
- 3.Daigo Y, Nakamura Y. From cancer genomics to thoracic oncology: Discovery of new biomarkers and therapeutic targets for lung and esophageal carcinoma. Gen Thorac Cardiovasc Surg. 2008;56:43–53. doi: 10.1007/s11748-007-0211-x. [DOI] [PubMed] [Google Scholar]
- 4.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 5.Shiraishi K, Kohno T, Tanai C, et al. Association of DNA repair gene polymorphisms with response to platinum-based doublet chemotherapy in patients with non-small-cell lung cancer. J Clin Oncol. 2010;28:4945–4952. doi: 10.1200/JCO.2010.30.5334. [DOI] [PubMed] [Google Scholar]
- 6.Azzoli CG, Baker S, Jr, Temin S, et al. American Society of Clinical Oncology Clinical Practice Guideline update on chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol. 2009;27:6251–6266. doi: 10.1200/JCO.2009.23.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 8.Danesi R, de Braud F, Fogli S, et al. Pharmacogenetics of anticancer drug sensitivity in non-small cell lung cancer. Pharmacol Rev. 2003;55:57–103. doi: 10.1124/pr.55.1.4. [DOI] [PubMed] [Google Scholar]
- 9.Siddik ZH. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 10.Wu W, Zhang W, Qiao R, et al. Association of XPD polymorphisms with severe toxicity in non-small cell lung cancer patients in a Chinese population. Clin Cancer Res. 2009;15:3889–3895. doi: 10.1158/1078-0432.CCR-08-2715. [DOI] [PubMed] [Google Scholar]
- 11.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han B, Gao G, Wu W, et al. Association of ABCC2 polymorphisms with platinum-based chemotherapy response and severe toxicity in non-small cell lung cancer patients. Lung Cancer. 2011;72:238–243. doi: 10.1016/j.lungcan.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Braun MS, Richman SD, Thompson L, et al. Association of molecular markers with toxicity outcomes in a randomized trial of chemotherapy for advanced colorectal cancer: The FOCUS trial. J Clin Oncol. 2009;27:5519–5528. doi: 10.1200/JCO.2008.21.6283. [DOI] [PubMed] [Google Scholar]
- 14.Tsunoda A, Nakao K, Watanabe M, et al. Associations of various gene polymorphisms with toxicity in colorectal cancer patients receiving oral uracil and tegafur plus leucovorin: A prospective study. Ann Oncol. 2011;22:355–361. doi: 10.1093/annonc/mdq358. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 16.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 17.Danial NN, Korsmeyer SJ. Cell death: Critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 18.Budihardjo I, Oliver H, Lutter M, et al. Biochemical pathways of caspase activation during apoptosis. Annu Rev Cell Dev Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 19.Konstantakou EG, Voutsinas GE, Karkoulis PK, et al. Human bladder cancer cells undergo cisplatin-induced apoptosis that is associated with p53-dependent and p53-independent responses. Int J Oncol. 2009;35:401–416. [PubMed] [Google Scholar]
- 20.Liu L, Xing D, Chen WR, et al. Calpain-mediated pathway dominates cisplatin-induced apoptosis in human lung adenocarcinoma cells as determined by real-time single cell analysis. Int J Cancer. 2008;122:2210–2222. doi: 10.1002/ijc.23378. [DOI] [PubMed] [Google Scholar]
- 21.Wilson TR, Redmond KM, McLaughlin KM, et al. Procaspase 8 overexpression in non-small-cell lung cancer promotes apoptosis induced by FLIP silencing. Cell Death Differ. 2009;16:1352–1361. doi: 10.1038/cdd.2009.76. [DOI] [PubMed] [Google Scholar]
- 22.Hajra KM, Liu JR. Apoptosome dysfunction in human cancer. Apoptosis. 2004;9:691–704. doi: 10.1023/B:APPT.0000045786.98031.1d. [DOI] [PubMed] [Google Scholar]
- 23.Harada K, Toyooka S, Shivapurkar N, et al. Deregulation of caspase 8 and 10 expression in pediatric tumors and cell lines. Cancer Res. 2002;62:5897–5901. [PubMed] [Google Scholar]
- 24.MacPherson G, Healey CS, Teare MD, et al. Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst. 2004;96:1866–1869. doi: 10.1093/jnci/dji001. [DOI] [PubMed] [Google Scholar]
- 25.Sun T, Gao Y, Tan W, et al. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007;39:605–613. doi: 10.1038/ng2030. [DOI] [PubMed] [Google Scholar]
- 26.Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muguruma K, Nakata B, Yanagawa K, et al. Caspase-1 activity as a possible predictor of apoptosis induced by cisplatin in gastric cancer cells. Int J Mol Med. 2000;6:553–557. doi: 10.3892/ijmm.6.5.553. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Zheng F, Xing H, et al. Resistance to chemotherapy-induced apoptosis via decreased caspase-3 activity and overexpression of antiapoptotic proteins in ovarian cancer. J Cancer Res Clin Oncol. 2004;130:423–428. doi: 10.1007/s00432-004-0556-9. [DOI] [PubMed] [Google Scholar]
- 29.Park SJ, Wu CH, Safa AR. A P-glycoprotein- and MRP1-independent doxorubicin-resistant variant of the MCF-7 breast cancer cell line with defects in caspase-6, -7, -8, -9 and -10 activation pathways. Anticancer Res. 2004;24:123–131. [PubMed] [Google Scholar]
- 30.National Cancer Institute. Cancer Therapy Evaluation Program. Common Toxicity Criteria, version 3.0. [accessed June 10, 2011]. Available at http://ctep.cancer.gov/protocolDevelopment/electronic_applications/ctc.htm.
- 31.International HapMap Project. [accessed August 16, 2011]. Available at http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap28_B36/
- 32.Broad Institute. Haploview. [accessed August 16, 2011]. Available at http://www.broadinstitute.org/haploview.
- 33.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SY, Choi YY, Choi JE, et al. Polymorphisms in the caspase genes and the risk of lung cancer. J Thorac Oncol. 2010;5:1152–1158. doi: 10.1097/JTO.0b013e3181e04543. [DOI] [PubMed] [Google Scholar]
- 35.George GP, Mandal RK, Kesarwani P, et al. Polymorphisms and haplotypes in caspases 8 and 9 genes and risk for prostate cancer: A case-control study in cohort of North India. Urol Oncol. 2011 Mar 9; doi: 10.1016/j.urolonc.2010.08.027. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 36.Yin M, Yan J, Wei S, et al. CASP8 polymorphisms contribute to cancer susceptibility: Evidence from a meta-analysis of 23 publications with 55 individual studies. Carcinogenesis. 2010;31:850–857. doi: 10.1093/carcin/bgq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keuling AM, Felton KE, Parker AA, et al. RNA silencing of Mcl-1 enhances ABT-737-mediated apoptosis in melanoma: Role for a caspase-8-dependent pathway. PLoS One. 2009;4:e6651. doi: 10.1371/journal.pone.0006651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creagh EM, Conroy H, Martin SJ. Caspase-activation pathways in apoptosis and immunity. Immunol Rev. 2003;193:10–21. doi: 10.1034/j.1600-065x.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 39.Woo M, Hakem R, Furlonger C, et al. Caspase-3 regulates cell cycle in B cells: A consequence of substrate specificity. Nat Immunol. 2003;4:1016–1022. doi: 10.1038/ni976. [DOI] [PubMed] [Google Scholar]
- 40.Chun HJ, Zheng L, Ahmad M, et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Zheng L, Lobito A, et al. Inherited human caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 42.Chen D, Jin G, Wang Y, et al. Genetic variants in peroxisome proliferator-activated receptor-gamma gene are associated with risk of lung cancer in a Chinese population. Carcinogenesis. 2008;29:342–350. doi: 10.1093/carcin/bgm285. [DOI] [PubMed] [Google Scholar]
- 43.Ruzzo A, Graziano F, Kawakami K, et al. Pharmacogenetic profiling and clinical outcome of patients with advanced gastric cancer treated with palliative chemotherapy. J Clin Oncol. 2006;24:1883–1891. doi: 10.1200/JCO.2005.04.8322. [DOI] [PubMed] [Google Scholar]
- 44.National Institutes of Health, National Institute of Environmental Health Sciences. SNP Function Prediction (FuncPred) [accessed August 16, 2011]. Available at http://snpinfo.niehs.nih.gov/snpfunc.htm.
- 45.Transcriptional Factor Search. [accessed August 16, 2011]. Available at http://www.cbrc.jp/research/db/TFSEARCH.html.
- 46.Wacholder S, Chanock S, Garcia-Closas M, et al. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, Ye Y, Rosell R, et al. Genome-wide association study of survival in non-small cell lung cancer patients receiving platinum-based chemotherapy. J Natl Cancer Inst. 2011;103:817–825. doi: 10.1093/jnci/djr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu C, Xu B, Yuan P, et al. Genome-wide examination of genetic variants associated with response to platinum-based chemotherapy in patients with small-cell lung cancer. Pharmacogenet Genomics. 2010;20:389–395. doi: 10.1097/FPC.0b013e32833a6890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.