Abstract
Telomere extension by telomerase is essential for chromosome stability and cell vitality. Here, we report the identification of a splice variant of mammalian heterogeneous nuclear ribonucleoprotein A2 (hnRNP A2), hnRNP A2*, which binds telomeric DNA and telomerase in vitro. hnRNP A2* colocalizes with telomerase in Cajal bodies and at telomeres. In vitro assays show that hnRNP A2* actively unfolds telomeric G-quadruplex DNA, exposes 5 nt of the 3′ telomere tail and substantially enhances the catalytic activity and processivity of telomerase. The expression level of hnRNP A2* in tissues positively correlates with telomerase activity, and overexpression of hnRNP A2* leads to telomere elongation in vivo. Thus, hnRNP A2* plays a positive role in unfolding telomere G-quadruplexes and in enhancing telomere extension by telomerase.
Human chromosome ends are protected by telomeres, which are composed of TTAGGG DNA repeats and associated proteins. Telomeres shorten with each cell division because of incomplete DNA-end replication (1). Cells compensate for telomere attrition through the action of telomerase, a specialized reverse transcriptase that adds telomeric repeats to the 3′ end of the telomere (2). Normal somatic cells do not express telomerase and undergo replicative senescence. The expression of telomerase is a hallmark of highly proliferative stem cells and most cancer cells. In budding yeast, the recruitment of telomerase to the telomere end is mediated by Cdc13 and Est1. Cdc13 is a single-stranded telomere DNA-binding protein that associates with Est1, a protein that interacts with the RNA component of yeast telomerase (3, 4). Similarly, in Tetrahymena, Teb1 bridges the interaction between telomerase and the telomere, which promotes highly processive telomere extension by telomerase (5). However, the mechanism(s) and factors required to promote a similar interaction between the telomere and telomerase in mammalian cells are poorly understood.
Telomere DNA can adopt a four-stranded G-quadruplex structure (6) that can be either intermolecular and intramolecular. Although an intermolecular G-quadruplex is an excellent substrate for ciliate telomerases (7), an intramolecular G-quadruplex is not. In vertebrates, intramolecular G-quadruplexes (hereinafter referred to as G-quadruplex) preferentially form at the furthest 3′ end of the telomeric DNA (8), rendering it inaccessible to telomerase. As a result, this structure inhibits telomere extension (9–11). Only a few proteins have been identified that can disrupt G-quadruplex. One such protein is protection of telomeres 1 (POT1) (12), a component of the telomere shelterin complex that binds telomere overhangs with high affinity (13). Despite its ability to disrupt G-quadruplex, POT1 actually inhibits telomere extension by binding to the telomere overhang and blocking telomerase access to the overhang (14–16).
Some proteins of the hnRNP family are also able to unfold telomeric G-quadruplex (17, 18). These proteins interact with telomeric ssDNA (19) and telomerase in vitro (20, 21), suggesting that they play a role in telomere biology. However, hnRNPs are highly abundant proteins that are expressed in vast excess compared with telomerase and the number of telomeres in eukaryotic cells (22). hnRNP A2/B1 and A1 alone are represented by at least 107 molecules per cell (23, 24), whereas a typical eukaryotic cell, such as HEK-293, has 20–50 molecules of telomerase (25) and 92 telomere ends. In principle, these proteins should saturate both telomeres and telomerase. Therefore, it is unlikely that they could play a direct role in regulating telomere extension.
Here, we report the discovery of a mammalian protein, hnRNP A2*, that actively unfolds telomeric G-quadruplex and enhances the catalytic activity and processivity of telomerase. hnRNP A2* is an isoform of hnRNP A2 that lacks exons 7–9 of the full-length molecule. Moreover, hnRNP A2* is distinct from the hnRNP A2 and other hnRNP proteins in several respects, including binding specificity, cellular localization, and abundance. We propose that hnRNP A2* plays a critical role in promoting interactions between the telomere and telomerase.
Results
Identification of hnRNP A2*, a Single-Stranded Telomeric DNA-Binding Protein from the Nuclear Matrix.
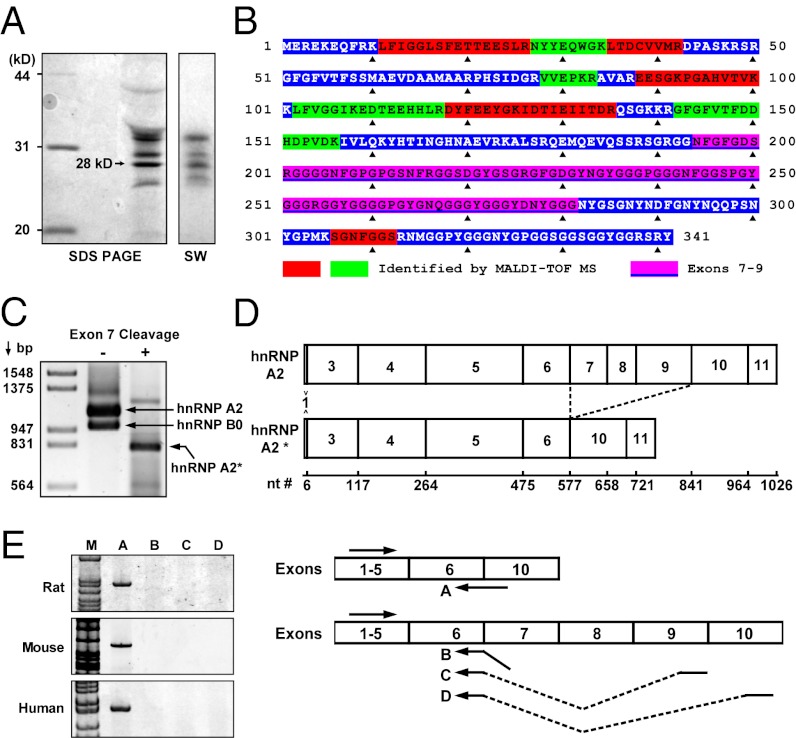
Telomeres attach to the nuclear matrix in mammalian cells (26, 27), and telomere replication is believed to be associated with the nuclear matrix (28). Because telomerase’s action is tightly coupled with telomere replication (29), the nuclear matrix may serve as the site of telomere extension and, thus, may contain telomere accessory proteins. To identify nuclear matrix telomere-binding proteins, nuclear matrix lysates were prepared (30) from rat liver. Telomere binding proteins were then isolated by affinity purification using single-stranded telomere oligonucleotides bound to streptavidin beads. Eluted proteins were analyzed by SDS/PAGE (Fig. 1A, Left), followed by Southwestern blot using a 32P-labeled (TTAGGG)3 probe (Fig. 1A, Right). This experiment identified several telomere-binding proteins. The most abundant protein, with a molecular mass of 28 kDa, was excised from the gel and analyzed by MALDI-TOF mass spectrometry. Nine of the tryptic peptides from this protein mapped to hnRNP A2, with eight peptides being found within the N-terminal half and one near the C terminus (Table S1 and Fig. 1B). Because the peptides span a region of hnRNP A2 greater than 28 kDa and are excluded from the regions encoded by exons 7, 8, and 9, we speculated that the newly isolated telomere-binding protein is a splice variant of hnRNP A2 lacking these exons. To test this, a newly developed “exon-exclusive RT-PCR” (31) was used to specifically amplify splice variants lacking exon 7 (Fig. 1C). Sequence analysis of the PCR product confirmed it as an hnRNP A2 isoform lacking exons 7–9 (Fig. 1D); hereafter, we designate this protein hnRNP A2*. hnRNP A2* is a conserved protein that is expressed in rat, mouse, and human cells (Fig. 1E).
Fig. 1.
Identification of the 28-kDa telomere-/telomerase-interacting protein hnRNP A2*. Nuclear matrix lysate was prepared from rat liver and telomere-binding proteins were isolated by affinity purification using a streptavidin-bound telomere oligonucleotide (TTAGGG)3. (A) SDS/PAGE of affinity-purified proteins (Left) and Southwestern blot (SW) probed with 32P-(TTAGGG)3 (Right). (B) MALDI-TOF mass spectrometric peptide mapping of the 28-kDa protein. Red and green blocks show the tryptic peptides mapped to the hnRNP A2 protein. The pink block shows amino acids encoded by exons 7–9. (C) PCR of hnRNP A2 and hnRNP A2* cDNA without and with prior cleavage at the exon 7 with endonuclease XhoI. The cDNAs bearing exon 7 were annealed with a cDNA at exon 7 and then cut by XhoI to prevent them from being amplified. (D) Exons in hnRNP A2* and hnRNP A2 mRNA. (E) Detection of hnRNP A2* mRNA in rat liver, cultured mouse (MEFs), and human HeLa cells by RT-PCR using junction primers (solid arrows).
hnRNP A2* Uniquely Binds Single-Stranded Telomeric TTAGGG Repeats.
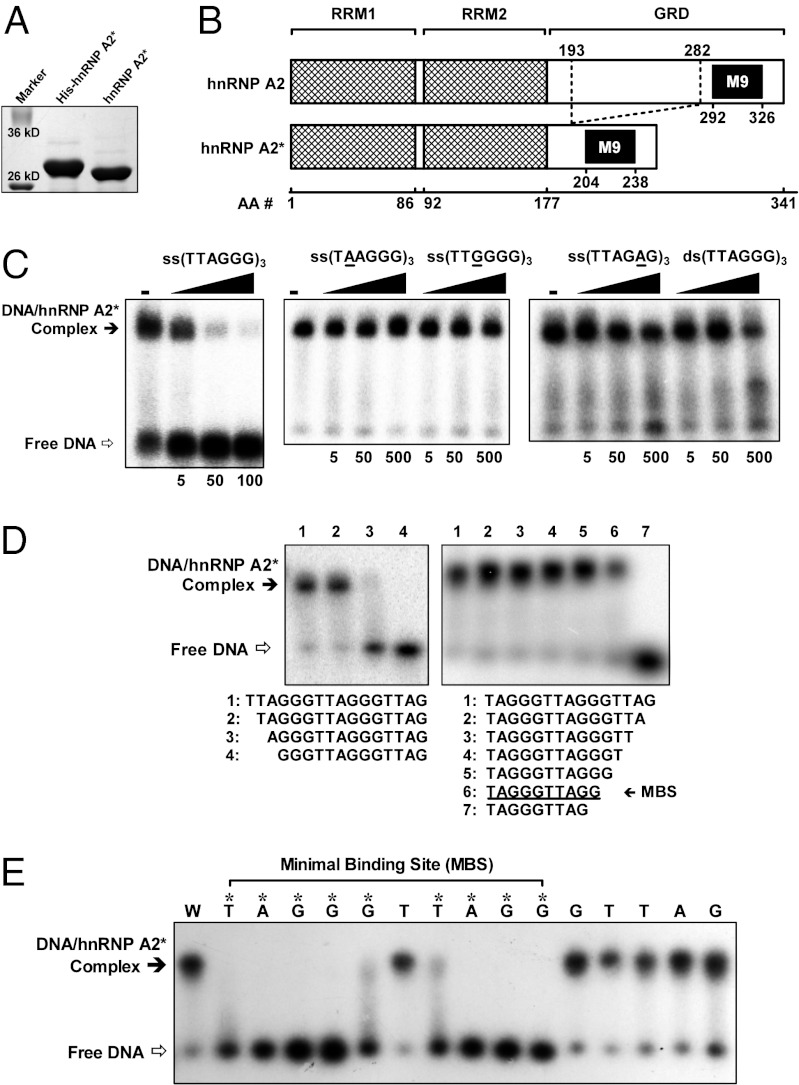
The cDNA encoding hnRNP A2* was cloned and expressed in Escherichia coli, producing a recombinant protein with the expected molecular mass of ∼28 kDa (Fig. 2A). The recombinant hnRNP A2* was then purified, and the binding specificity of hnRNP A2* was examined using an electrophoretic mobility-shift assay (EMSA). The results revealed that hnRNP A2* binds the ssDNA telomeric repeat (TTAGGG)3 with high specificity but has very low or no binding affinity for (TAAGGG)3, (TTGGGG)3, (TTAGAG)3, or double-stranded (TTAGGG)3 (Fig. 2C). This high specificity likely reflects deletion of exons 7–9 of hnRNP A2 within the glycine-rich domain (GRD) (Fig. 2B), which has been shown to be involved in determining binding specificity (32). In contrast to hnRNP A2*, hnRNPs A2/B1, which include exons 7–9, bind DNA sequences that match the consensus sequence N(A,C,T)(C,T)(A,G)G(C,G,T)(A,T)NNN (33), reflecting much broader DNA-binding specificity than hnRNP A2*. The results of additional EMSA experiments revealed a minimal binding site (MBS) of 5′-TAGGGTTAGG-3′ for hnRNP A2* (Fig. 2D). This MBS was verified by constructing 15 mutant variants of the MBS, each carrying a single mutation to cytosine at each nucleotide (Fig. 2E). hnRNP A2* bound the variant carrying a T6 → C mutation, but all other mutations within the MBS completely abolished binding. Mutations outside of the MBS did not affect binding (Fig. 2E). These results further demonstrate the exquisite binding specificity of hnRNP A2*. We note that the putative hnRNP A2* MBS resembles the MBS (5′-TAGGGTTAG-3′) of human POT1, except that it is 1 nt longer at the 3′ end (34).
Fig. 2.
Binding of hnRNP A2* to telomeric DNA analyzed by EMSA. (A) hnRNP A2* and His-tagged hnRNP A2* expressed and purified from E. coli. (B) Functional domains in hnRNP A2* in comparison with hnRNP A2 that contains two RNA-recognition motifs (RRMs), a GRD, and an M9 nuclear localization domain. (C) Binding of hnRNP A2* to (TTAGGG)3 in the presence of competitive DNA of increasing fold concentrations (indicated below images). (D) hnRNP A2* binds a MBS of TAGGGTTAGG. (E) Effect of single-nucleotide mutation on the binding of hnRNP A2* to TAGGGTTAGGGTTAG. DNAs were either wild-type sequence (W) or carrying a mutation to cytosine at the position indicated above the lane. Asterisks indicate mutations that disrupt binding.
hnRNP A2* Actively Unfolds G-Quadruplex, Preferentially Binds to the MBS at the 3′ End of Telomeric DNA, and Exposes a Five-Nucleotide 3′ Tail.
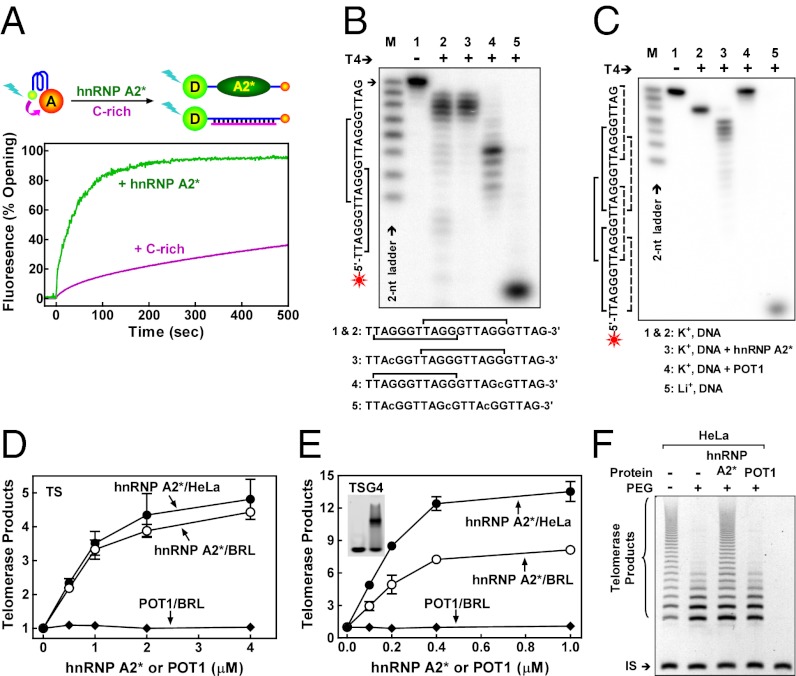
The interaction between hnRNP A2* and telomeric G-quadruplex was analyzed using fluorescence resonance energy transfer (FRET). A G-quadruplex DNA substrate, 5′-(GGGTTA)3GGG-3′, was labeled with fluorescein (FAM) at the 5′ end (the FRET donor) and tetramethylrhodamine (TAMRA) at the 3′ end (the FRET receptor) (Fig. 3A). When this oligomer forms G-quadruplex, the two fluorophores are closely juxtaposed and FRET occurs between the two fluorophores, suppressing the donor fluorescence (35, 36). When G-quadruplex was incubated with hnRNP A2*, donor fluorescence increased rapidly, suggesting that hnRNP A2* promotes G-quadruplex unwinding. We note that the rate of fluorescence increase and G-quadruplex unwinding in the presence of hnRNP A2* was much faster than the slow spontaneous unwinding of G-quadruplex in the presence of excess complementary 5′-CCC(ATTCCC)3-3′; this suggests that hnRNP A2* actively unfolds G-quadruplex. Consistent with the binding specificity of hnRNP A2*, FRET was not altered by hnRNP A2* when an irrelevant linear and hairpin DNA substrate was used (Fig. S1 A and B).
Fig. 3.
hnRNP A2* actively unfolds telomere G-quadruplex, binds to the MBS at the very 3′ end of telomeric DNA, exposes a 5-nt tail, and activates telomerase. (A) FRET analysis showing active opening of G-quadruplex by hnRNP A2* in comparison with the spontaneous opening of G-quadruplex trapped by complementary C-rich DNA. Donor fluorescence monitored in real-time upon addition of hnRNP A2* or excess C-rich DNA. (B) Binding-site preference of hnRNP A2* to telomeric DNA that does not form G-quadruplex. The 5′ 32P-labeled DNA was incubated with hnRNP A2* before digestion with T4 polymerase (T4), which cleaves nucleotides from the 3′ end. Lowercase indicates mutations introduced to manipulate the number and position of MBS (square bracket). (C) Binding-site preference of hnRNP A2* and POT1 to telomeric DNA that forms G-quadruplex. Solid square bracket indicates MBS for hnRNP A2* and dashed for POT1. G-quadruplex forms in K+ but not in Li+ solution. Other conditions are the same as those in B. (D and E) Catalytic activity of telomerase from HeLa or rat cell lysate assayed by the TRAP method using nontelomeric TS (D) or G-quadruplex TSG4 (E) substrate in the presence of increasing amounts of hnRNP A2* or POT1. (E, Inset) Free TSG4 (left lane) and TSG4/hnRNP A2* complex (right lane) in EMSA. (F) Processivity of telomerase from HeLa cell lysate assayed by the modified TRAP method using nontelomeric MTS substrate in the presence of 40% (wt/vol) PEG 200.
The 3′-terminal sequence of most native telomeres is TTAG-3′ (37), which is identical to that of the MBS of POT1. However, as pointed out above, the MBS for hnRNP A2* is 1 nt longer than the MBS for POT1, suggesting that the two proteins might interact differently with telomeric repeats. To explore this idea, the “footprints” of hnRNP A2* and POT1 on a telomeric array were compared by prebinding the protein to its DNA substrate, then exposing the DNA–protein complex to the 3′-exonuclease activity of T4 DNA polymerase. The results show that both hnRNP A2* and POT1 bind to their respective MBS at the very 3′ end when more than one MBS was available (Fig. 3B and Fig. S1C). However, hnRNP A2* failed to protect the terminal GTTAG-3′ from digestion by T4 polymerase (Fig. 3B), whereas POT1 protected the entire DNA substrate from cleavage (Fig. S1C). Importantly, the same 3′-end preference and specificity for each protein was observed when the DNA substrate was telomeric G-quadruplex (Fig. 3C). These results do not reflect secondary structure in the DNA substrate, because they were completely susceptible to digestion by T4 DNA polymerase in the absence of protein (Fig. S1D).
hnRNP A2* Enhances the Catalytic Activity and Processivity of Telomerase in Vitro.
Interestingly, the 3′-terminal sequence left exposed after hnRNP A2* binding, 5′-GTTAG-3′, is complementary to and forms Watson–Crick base pairs with the “alignment sequence,” 5′-CUAAC-3′, in the RNA moiety of vertebrate telomerase (38). It is, thus, conceivable that hnRNP A2* binding could facilitate pairing of telomerase with its telomeric substrate and increase telomerase activity. We tested this possibility in vitro using a conventional telomere repeat amplification protocol (TRAP) assay and found that hnRNP A2* significantly enhanced the activity of human or rat telomerase (rTR) (Fig. 3D and Fig. S2A). Because the TRAP assay uses a nontelomeric TS substrate (39), telomerase must synthesize several telomeric repeats before hnRNP A2* binds and/or G-quadruplex can form. The results suggest that hnRNP A2* may stimulate telomere extension by inhibiting formation of G-quadruplex and leaving an appropriate 3′-terminal sequence available for telomerase binding. In contrast, POT1 does not stimulate telomerase in a TRAP assay (Fig. 3D). This result agrees with a previous study, which reported that POT1 did not stimulate telomerase activity, by a direct, non–PCR-based method using a nontelomeric primer. These data support the proposal that “POT1 may modulate telomerase activity by regulating the access of telomerase to the primer but not during extension” (14).
The ability of hnRNP A2* to unwind telomeric G-quadruplex was also examined by a modified TRAP assay using TSG4 as a substrate in which a thymidine next to the G-tract was mutated into cytosine so that it forms G-quadruplex but is still recognized by hnRNP A2* (Fig. 2E). The TSG4 substrate differs from the sequence added to it by telomerase, allowing PCR amplification using TSG4-specific primers (40). hnRNP A2* stimulates telomerase activity to a greater extent when the TRAP assay is performed with this substrate (Fig. 3E and Fig. S2B) than with the TS substrate (Fig. 3D and Fig. S2A), whereas POT1 does not stimulate telomerase activity with either DNA substrate (Fig. 3 D and E). These data suggest that hnRNP A2* promotes telomerase function by unfolding G-quadruplex during telomerase extension.
Although human telomerase can processively add multiple telomeric repeats to a single primer (41), processive DNA synthesis by telomerase is inhibited by G-quadruplex, because it interferes with telomerase translocation (9, 10, 42). Therefore, we examined the impact of hnRNP A2* on telomerase processivity using a modified TRAP assay (43). In dilute solution, telomerase showed a relatively high processivity (Fig. 3F and Fig. S2C, first lane). We reported previously that in the presence of PEG 200, a crowding agent widely used to mimic the molecularly crowded intracellular environment, the thermal stability of telomere G-quadruplex increases and the processivity of human telomerase decreases (44–46) (Fig. 3F and Fig. S2C, second lane). hnRNP A2* stimulated telomerase processivity in the presence of PEG 200 (Fig. 3F and Fig. S2C, third lane), but POT1 did not (Fig. 3F and Fig. S2C, fourth lane). Because both hnRNP A2* and POT1 can disrupt G-quadruplex, this difference may reflect the fact that hnRNP A2* exposes a free 3′ end when it binds to telomeric DNA, but POT1 does not.
hnRNP A2* Localizes to the Nuclear Matrix and Associates with Telomerase at Telomeres and in Cajal Bodies in Vivo.
The protein region encoded by exons 7–9 in hnRNP A2, which is missing from hnRNP A2*, is entirely hydrophilic (Fig. S3A). As a result, hnRNP A2* is more hydrophobic than hnRNP A2. hnRNP A2 localizes predominantly to the nucleoplasm (27), whereas subcellular fractionation of cells expressing tagged hnRNP A2* shows that hnRNP A2* localizes exclusively to the nuclear matrix (Fig. S3B). This suggests that hnRNP A2 and hnRNP A2* have distinct cellular functions.
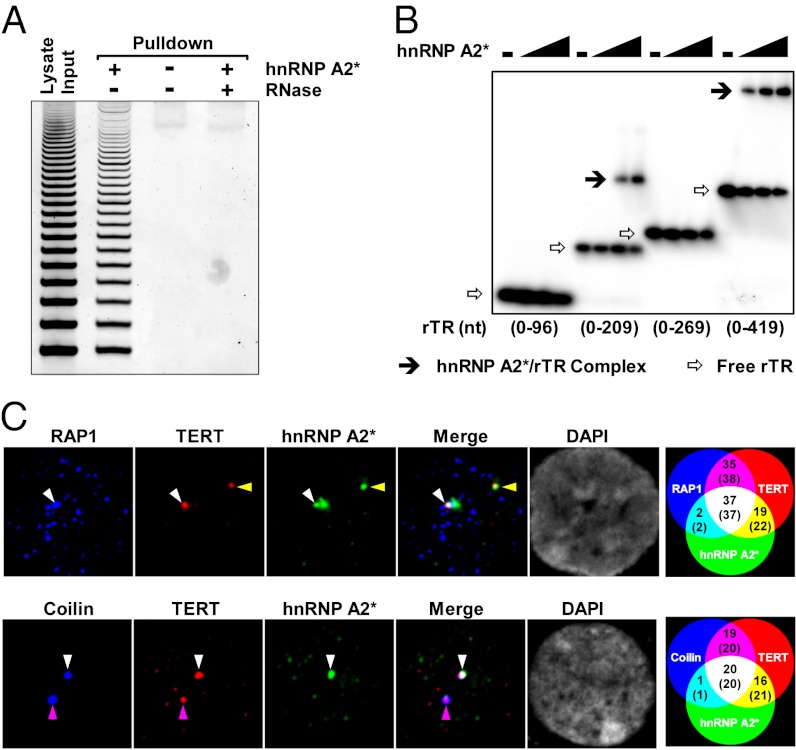
Some proteins from the hnRNP family interact directly with telomerase, as well as with telomeric DNA, in vitro (20, 21). We found that hnRNP A2* could pull-down telomerase activity from cell lysate (Fig. 4A) as did hnRNP A2 (Fig. S4A), demonstrating that hnRNP A2* can physically interact with telomerase. In addition, hnRNP A2* directly binds the RNA component of rTR in vitro (Fig. 4B). This binding is dependent on the size/sequence and possibly the secondary structure of rTR, because hnRNP A2 binds to the 0–269 fragment of rTR (Fig. S4B), whereas hnRNP A2* does not (Fig. 4B).
Fig. 4.
hnRNP A2* interacts with telomerase both in vitro and in vivo. (A) TRAP assay of telomerase activity pulled down from rat cell lysate by His-hnRNP A2* immobilized on nickel beads. (B) Binding to the RNA component of rTR of hnRNP A2* at increasing concentrations (0, 0.25, 0.5, 1 μM) examined by EMSA. (C) Three-color immunofluorescence analysis of colocalization among hnRNP A2*, TERT, and Rap1/Coilin in cultured rat cells. RAP1 and Coilin served as markers for telomere and Cajal body, respectively. Venn diagrams at the farthest right show the number of cells and foci (in parenthesis) that were positive for each combination of markers.
To examine whether hnRNP A2* interacts with telomeres and telomerase in vivo, we expressed HA-tagged hnRNP A2* in rat cells and performed immunofluorescence experiments. The results showed that a fraction of hnRNP A2* colocalized with RAP1, a component of the telomere-associated shelterin complex, and with TERT, the catalytic component of telomerase (Fig. S5A, top and middle images). Control experiments showed that the antibody to HA-hnRNP A2* only stains HA-hnRNP A2*–expressing cells (Fig. S5B) and that suppression of TERT expression by siRNA dramatically reduced the fluorescence in the cells (Fig. S5C). Interestingly, hnRNP A2* can also colocalize with Coilin, a marker of Cajal bodies (Fig. S5A, bottom images). Because Cajal bodies are involved in the processing and positioning of telomerase at telomeres (47), we hypothesized that hnRNP A2* may play a role in these processes.
The interactions between hnRNP A2*, telomerase, telomeres, and Cajal body were further examined using multicolor immunofluorescence. The results can be summarized as follows (Venn diagrams in Fig. 4C, Right): about half of the TERT at telomeres or in Cajal bodies was associated with hnRNP A2*. More importantly, out of the 39 hnRNP A2*/RAP1 foci observed, 37 were also positive for telomerase (hnRNP A2*/RAP1/TERT foci). Similarly, of the 21 hnRNP A2*/Coilin foci observed, 20 were also positive for TERT (Fig. 4C and Fig. S6 A and B). The prevalent colocalization of hnRNP A2* with telomerase at telomeres and in Cajal bodies strongly suggests that hnRNP A2* is a close partner of telomerase. It is possible that hnRNP A2* is assembled into the telomerase holoenzyme at Cajal bodies and delivered to telomeres. Our finding that the binding of hnRNP A2* with rTR (Fig. 4B) involves the 3′ region of rTR (269–419 nt), which contains the Cajal body box (CAB) motif responsible for its mobilization to the Cajal body (38, 48), supports this hypothesis.
hnRNP A2* Expression Correlates with Telomerase Activity, and Overexpression of hnRNP A2* Increases Telomere Length in Vivo.
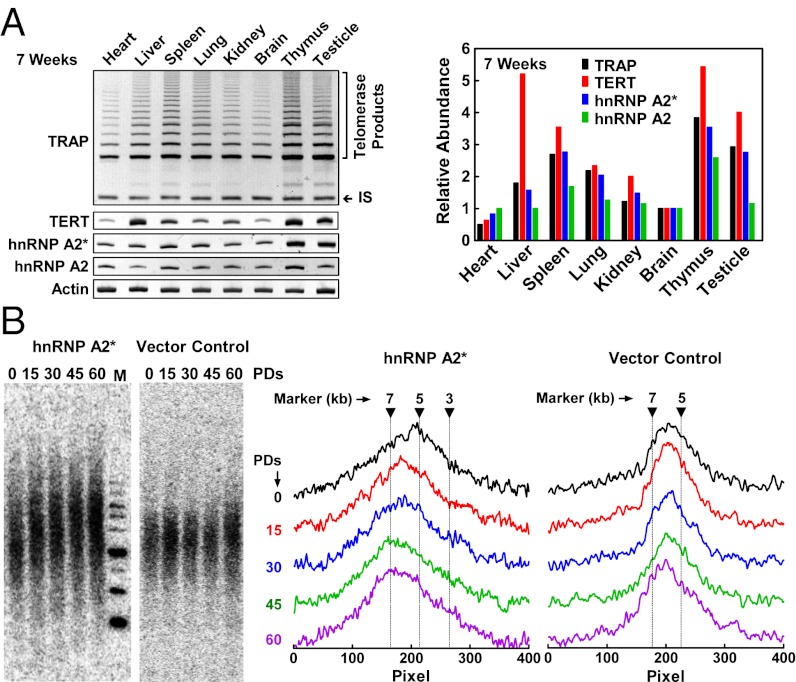
To explore whether hnRNP A2* influences telomerase function in vivo, the amount of hnRNP A2* and telomerase mRNA was determined by RT-PCR and telomerase activity was quantified by TRAP in primary cells from freshly isolated rat tissues (Fig. 5A and Fig. S7A). Interestingly, although telomerase activity did not correlate well with expression of TERT mRNA or hnRNP A2 mRNA, it did correlate well with expression of hnRNP A2*. This correlation implies an important role of hnRNP A2* for telomerase activity in vivo.
Fig. 5.
In vivo correlation of hnRNP A2* expression with telomerase activity and telomere length. (A) Expression of hnRNP A2*, hnRNP A2, TERT, and telomerase activity in 7-wk-old rat tissues. Expression was analyzed by RT-PCR. Telomerase activity was assayed by TRAP. Relative abundance was obtained by normalizing band intensity to actin and then to brain. IS, Internal standard. (B) Overexpression of hnRNP A2* increases telomere length in HeLa cells. Cells were drug-selected and cultured to the indicated population doublings (PDs) after they were transfected with HA-hnRNP A2* or empty control vector. Telomere restriction fragments were detected by Southern blot (Left) and then digitized for quantification (Right). M, marker.
Because hnRNP A2* is expressed at a low level in human and rat cells, and because the abundance of hnRNP A2* correlates with higher telomerase activity in rat tissues (Fig. 5A and Fig. S7A), we predicted that overexpression of hnRNP A2* might correlate with increased telomere length. In fact, when hnRNP A2* was overexpressed in cultured HeLa and rat cells by retrovirus-mediated transfection, telomere length increased relative to control cells carrying empty vector (Fig. 5B and Fig. S7B). Interestingly, the longer telomeres in the rat cells regressed over time, and in this context, subsequent telomere shortening correlated with a gradual decrease in hnRNP A2* expression (Fig. S7B, Lower). This result further supports the conclusion that hnRNP A2* plays an important role in regulating telomerase activity and telomere length.
Discussion
G-Quadruplex Unfolding in Telomere Extension and 3′ Telomere–End Recognition.
Human telomeric G-quadruplex is an extremely stable structure (49), forming within a fraction of a second (50) but taking hours to spontaneously unfold at 37 °C (51). Therefore, if telomeric G-quadruplex forms in vivo, as has been proposed (6, 52–54), cellular mechanisms must exist to promote unfolding of this structure during telomere extension by telomerase. POT1 and proteins from the hnRNP family, like hnRNP A1, are able to disrupt G-quadruplex (55–57). However, these proteins also bind with high affinity to and occlude the 3′ end of the telomere, which could limit access of telomerase to telomere ends and inhibit telomere extension. The MBS of POT1 has the same 3′-terminal nucleotide as a native telomere end, TTAG-3′ (34), and our data show that POT1 binding to a telomere substrate leaves no free 3′ end (Fig. S1C). hnRNP A2/B1 recognizes N(A,C,T)(C,T)(A,G)G(C,G,T)(A,T)NNN, a consensus sequence that encompasses and aligns “in phase” with the telomere repeat TTAGGGTTAG (33), and would also be expected to occlude the telomere end and render it inaccessible to telomerase. This could also be true for other abundant hnRNPs (18, 58–61). Therefore, hnRNP A2* may be unique in its ability to actively unfold telomere G-quadruplex while still leaving a 3′ telomere end available for telomerase binding.
hnRNP A2* Is Functionally Distinct from the hnRNP Proteins and Specific to Telomere Extension.
Although hnRNP A2* is an isoform of hnRNP A2, its low abundance and cellular localization suggest that it is functionally distinct from any other hnRNP. In contrast to previously characterized hnRNPs, the hnRNP A2* transcript is expressed at an extremely low level, making it very difficult to detect by RT-PCR unless other variants are selectively cleaved in the sample before amplification (Fig. 1C). This low abundance makes hnRNP A2* a reasonable candidate for a regulator of telomerase, which is also expressed at very low levels in typical eukaryotic cells (25).
Telomerase is reported to be anchored to the nuclear matrix through its association with chromosome scaffold protein P43 in the ciliated protozoa Euplotes (62). Cajal bodies are also consistently associated with nuclear matrix (63, 64). Data presented here show that hnRNP A2* is localized to the nuclear matrix, consistent with its proposed role in telomere biology. The loss of the GRD in hnRNP A2* selectively targets it to telomeres, and incapacitates its potential role in mRNA metabolism (65). Consistent with this, the level of hnRNP A2*, but not the level of hnRNP A2/B1, correlates with telomerase activity in rat tissues (Fig. 5A and Fig. S7A).
Role of hnRNP A2* in Facilitating Telomerase Action at Telomeres.
The ability of hnRNP A2* to interact with both telomeric DNA and telomerase makes it an ideal protein to recruit telomerase to telomere ends. Telomeric G-quadruplex is not extended by telomerase unless it carries a single-stranded 3′ tail at least 8 nt long (11). During processive extension, hnRNP A2* may intercept newly synthesized telomeric repeats, preventing them from forming G-quadruplex, which would expel telomerase. This would explain the fact that hnRNP A2* stimulates telomerase processivity (Fig. 3F and Fig. S2C), even in the presence of PEG 200, which dramatically stabilizes G-quadruplex (44–46). Interestingly, the function of hnRNP A2* in telomere extension resembles that of Teb1 in Tetrahymena, which bridges the telomere and telomerase, conferring highly processive telomere extension by telomerase (5).
The affinity of hnRNP A2* for telomeric DNA (Kd = 11.05 nM; Fig. S8) is comparable to POT1 (∼10 nM) (13). It is possible that POT1 and hnRNP A2* compete for binding telomeric DNA in vivo (Fig. 6). Under such conditions, the 3′ end of the telomere would be exposed or capped, depending on which protein binds. During most of the cell cycle, the 3′ end of the telomeric overhang invades homologous dsDNA forming a D-loop (66) or is capped by POT1, which protects the single-stranded overhang from triggering a DNA damage response (67). During S phase, hnRNP A2* may unwind G-quadruplex structures, thus releasing the 3′ telomere end to facilitate recruitment of telomerase. This would explain the findings that removal of POT1 (68) or overexpression of hnRNP A2* (Fig. 5B and Fig. S7B) promotes telomere lengthening.
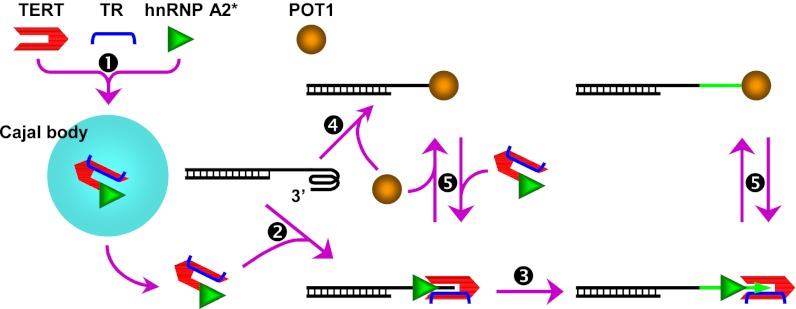
Fig. 6.
Proposed working model of hnRNP A2*. A single-stranded telomere overhang forms intramolecular G-quadruplex. hnRNP A2* assembled in Cajal body with telomerase (1) binds to the telomere end upon unfolding telomere G-quadruplex to facilitate telomere end-telomerase alignment (2) for telomere extension (3). POT1 can also bind to the telomere end (4), making it inaccessible to telomerase. Competition between POT1 and hnRNP A2* (5) determines the accessibility of the telomere end to telomerase and its subsequent extension.
In summary, hnRNP A2* is a telomere- and telomerase-interacting protein that may play an essential role in recruiting telomerase to the telomere during the process of telomerase extension. In this context, hnRNP A2* may play a role similar to TPP1 (69, 70). Intriguingly, although TPP1 is able to recruit telomerase to telomere ends (71), the TPP1-POT1 complex caps telomere ends and blocks access to them by telomerase (72). The mechanistic “switch” that relieves this block and converts inaccessible telomeres to accessible telomeres is not yet understood. However, our findings indicate that hnRNP A2* may play a role in this process. For example, hnRNP A2* and TPP1/POT1 might play opposing but complementary roles, with the former increasing accessibility and the latter decreasing accessibility of telomeric ends to telomerase. Future experiments will be needed to address the role of hnRNP A2* in regulating telomere accessibility, telomerase recruitment, and telomerase processivity in greater detail.
Materials and Methods
Materials and methods are described in SI Materials and Methods, including identification, cloning, expression, and characterization of hnRNP A2*. Animal experiments were approved by the Institutional Animal Care and Use Committee of Wuhan University.
Supplementary Material
Acknowledgments
We thank Dr. Titia de Lange (The Rockefeller University) for providing the rabbit polyclonal anti-hRAP1 antibody, Dr. Joachim Lingner (Swiss Institute for Experimental Cancer Research) for providing the pET-14-POT1 plasmid, and Mary Chaiken and Jason Stewart (University of Cincinnati) for their suggestions and editing of the manuscript. This work was supported by National Natural Science Foundation of China Grants 21072189, 31271472, and 20921062; and Ministry of Science and Technology of China Grants 2012CB720601 and 2010CB945300.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200232109/-/DCSupplemental.
References
- 1.Morin GB. Telomere control of replicative lifespan. Exp Gerontol. 1997;32(4-5):375–382. doi: 10.1016/s0531-5565(96)00164-7. [DOI] [PubMed] [Google Scholar]
- 2.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266(5193):2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 3.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104(3):387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi A, Shore D. How telomerase reaches its end: Mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31(2):153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Min B, Collins K. Multiple mechanisms for elongation processivity within the reconstituted tetrahymena telomerase holoenzyme. J Biol Chem. 2010;285(22):16434–16443. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipps HJ, Rhodes D. G-quadruplex structures: In vivo evidence and function. Trends Cell Biol. 2009;19(8):414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Oganesian L, Moon IK, Bryan TM, Jarstfer MB. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 2006;25(5):1148–1159. doi: 10.1038/sj.emboj.7601006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang J, et al. G-quadruplex preferentially forms at the very 3′ end of vertebrate telomeric DNA. Nucleic Acids Res. 2008;36(4):1200–1208. doi: 10.1093/nar/gkm1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350(6320):718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher TM, Sun D, Salazar M, Hurley LH. Effect of DNA secondary structure on human telomerase activity. Biochemistry. 1998;37(16):5536–5541. doi: 10.1021/bi972681p. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, et al. G-quadruplex formation at the 3′ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011;39(14):6229–6237. doi: 10.1093/nar/gkr164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292(5519):1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 13.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11(12):1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 14.Kelleher C, Kurth I, Lingner J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol Cell Biol. 2005;25(2):808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445(7127):506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 16.Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280(21):20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 17.Nakagama H, et al. Molecular mechanisms for maintenance of G-rich short tandem repeats capable of adopting G4 DNA structures. Mutat Res. 2006;598(1-2):120–131. doi: 10.1016/j.mrfmmm.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Enokizono Y, et al. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J Biol Chem. 2005;280(19):18862–18870. doi: 10.1074/jbc.M411822200. [DOI] [PubMed] [Google Scholar]
- 19.Ford LP, Wright WE, Shay JW. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene. 2002;21(4):580–583. doi: 10.1038/sj.onc.1205086. [DOI] [PubMed] [Google Scholar]
- 20.Ford LP, Suh JM, Wright WE, Shay JW. Heterogeneous nuclear ribonucleoproteins C1 and C2 associate with the RNA component of human telomerase. Mol Cell Biol. 2000;20(23):9084–9091. doi: 10.1128/mcb.20.23.9084-9091.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiset S, Chabot B. hnRNP A1 may interact simultaneously with telomeric DNA and the human telomerase RNA in vitro. Nucleic Acids Res. 2001;29(11):2268–2275. doi: 10.1093/nar/29.11.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattern KA, Humbel BM, Muijsers AO, de Jong L, van Driel R. hnRNP proteins and B23 are the major proteins of the internal nuclear matrix of HeLa S3 cells. J Cell Biochem. 1996;62(2):275–289. doi: 10.1002/(sici)1097-4644(199608)62:2<275::aid-jcb15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 23.Hanamura A, Cáceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4(4):430–444. [PMC free article] [PubMed] [Google Scholar]
- 24.McKay SJ, Cooke H. A protein which specifically binds to single stranded TTAGGGn repeats. Nucleic Acids Res. 1992;20(6):1387–1391. doi: 10.1093/nar/20.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen SB, et al. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315(5820):1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 26.de Lange T. Human telomeres are attached to the nuclear matrix. EMBO J. 1992;11(2):717–724. doi: 10.1002/j.1460-2075.1992.tb05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludérus ME, et al. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996;135(4):867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pardoll DM, Vogelstein B, Coffey DS. A fixed site of DNA replication in eucaryotic cells. Cell. 1980;19(2):527–536. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, et al. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138(3):463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma H, Siegel AJ, Berezney R. Association of chromosome territories with the nuclear matrix. Disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J Cell Biol. 1999;146(3):531–542. doi: 10.1083/jcb.146.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Zhao Y, Hao YH, Tan Z. Identification of low-abundance alternatively spliced mRNA variants by exon exclusive reverse transcriptase polymerase chain reaction. Anal Biochem. 2008;383(2):307–310. doi: 10.1016/j.ab.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Casas-Finet JR, et al. Mammalian heterogeneous ribonucleoprotein A1 and its constituent domains. Nucleic acid interaction, structural stability and self-association. J Mol Biol. 1993;229(4):873–889. doi: 10.1006/jmbi.1993.1093. [DOI] [PubMed] [Google Scholar]
- 33.Moran-Jones K, et al. hnRNP A2, a potential ssDNA/RNA molecular adapter at the telomere. Nucleic Acids Res. 2005;33(2):486–496. doi: 10.1093/nar/gki203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: A nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem. 2004;279(13):13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- 35.Yao Y, Wang Q, Hao YH, Tan Z. An exonuclease I hydrolysis assay for evaluating G-quadruplex stabilization by small molecules. Nucleic Acids Res. 2007;35(9):e68. doi: 10.1093/nar/gkm194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kan ZY, et al. Molecular crowding induces telomere G-quadruplex formation under salt-deficient conditions and enhances its competition with duplex formation. Angew Chem Int Ed Engl. 2006;45(10):1629–1632. doi: 10.1002/anie.200502960. [DOI] [PubMed] [Google Scholar]
- 37.Sfeir AJ, Chai W, Shay JW, Wright WE. Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell. 2005;18(1):131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 38.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100(5):503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 39.Kim NW, Wu F. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP) Nucleic Acids Res. 1997;25(13):2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez D, Mergny JL, Riou JF. Detection of telomerase inhibitors based on g-quadruplex ligands by a modified telomeric repeat amplification protocol assay. Cancer Res. 2002;62(12):3365–3368. [PubMed] [Google Scholar]
- 41.Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003;22(2):304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Cian A, et al. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc Natl Acad Sci USA. 2007;104(44):17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szatmari I, Aradi J. Telomeric repeat amplification, without shortening or lengthening of the telomerase products: A method to analyze the processivity of telomerase enzyme. Nucleic Acids Res. 2001;29(2):E3. doi: 10.1093/nar/29.2.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng KW, Chen Z, Hao YH, Tan Z. Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA. Nucleic Acids Res. 2010;38(1):327–338. doi: 10.1093/nar/gkp898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue Y, et al. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J Am Chem Soc. 2007;129(36):11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 46.Kan ZY, et al. G-quadruplex formation in human telomeric (TTAGGG)4 sequence with complementary strand in close vicinity under molecularly crowded condition. Nucleic Acids Res. 2007;35(11):3646–3653. doi: 10.1093/nar/gkm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cristofari G, et al. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell. 2007;27(6):882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 48.Jády BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol. 2004;164(5):647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36(17):5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36(12):4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Green JJ, Ying L, Klenerman D, Balasubramanian S. Kinetics of unfolding the human telomeric DNA quadruplex using a PNA trap. J Am Chem Soc. 2003;125(13):3763–3767. doi: 10.1021/ja029149w. [DOI] [PubMed] [Google Scholar]
- 52.Schaffitzel C, et al. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci USA. 2001;98(15):8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12(10):847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 54.Chang CC, et al. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal Chem. 2004;76(15):4490–4494. doi: 10.1021/ac049510s. [DOI] [PubMed] [Google Scholar]
- 55.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA. 2005;102(31):10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krüger AC, et al. Interaction of hnRNP A1 with telomere DNA G-quadruplex structures studied at the single molecule level. Eur Biophys J. 2010;39(9):1343–1350. doi: 10.1007/s00249-010-0587-x. [DOI] [PubMed] [Google Scholar]
- 57.Torigoe H, Furukawa A. Tetraplex structure of fission yeast telomeric DNA and unfolding of the tetraplex on the interaction with telomeric DNA binding protein Pot1. J Biochem. 2007;141(1):57–68. doi: 10.1093/jb/mvm011. [DOI] [PubMed] [Google Scholar]
- 58.Dallaire F, Dupuis S, Fiset S, Chabot B. Heterogeneous nuclear ribonucleoprotein A1 and UP1 protect mammalian telomeric repeats and modulate telomere replication in vitro. J Biol Chem. 2000;275(19):14509–14516. doi: 10.1074/jbc.275.19.14509. [DOI] [PubMed] [Google Scholar]
- 59.Huang PR, Tsai ST, Hsieh KH, Wang TC. Heterogeneous nuclear ribonucleoprotein A3 binds single-stranded telomeric DNA and inhibits telomerase extension in vitro. Biochim Biophys Acta. 2008;1783(2):193–202. doi: 10.1016/j.bbamcr.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka E, et al. HnRNP A3 binds to and protects mammalian telomeric repeats in vitro. Biochem Biophys Res Commun. 2007;358(2):608–614. doi: 10.1016/j.bbrc.2007.04.177. [DOI] [PubMed] [Google Scholar]
- 61.Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28(5):773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Möllenbeck M, et al. The telomerase-associated protein p43 is involved in anchoring telomerase in the nucleus. J Cell Sci. 2003;116(Pt 9):1757–1761. doi: 10.1242/jcs.00351. [DOI] [PubMed] [Google Scholar]
- 63.Gedge LJ, Morrison EE, Blair GE, Walker JH. Nuclear actin is partially associated with Cajal bodies in human cells in culture and relocates to the nuclear periphery after infection of cells by adenovirus 5. Exp Cell Res. 2005;303(2):229–239. doi: 10.1016/j.yexcr.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 64.Puvion-Dutilleul F, Besse S, Chan EK, Tan EM, Puvion E. p80-coilin: A component of coiled bodies and interchromatin granule-associated zones. J Cell Sci. 1995;108(Pt 3):1143–1153. doi: 10.1242/jcs.108.3.1143. [DOI] [PubMed] [Google Scholar]
- 65.Mayeda A, Munroe SH, Cáceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13(22):5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 67.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448(7157):1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 68.Churikov D, Price CM. Pot1 and cell cycle progression cooperate in telomere length regulation. Nat Struct Mol Biol. 2008;15(1):79–84. doi: 10.1038/nsmb1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445(7127):559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 70.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24(6):613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong FL, et al. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150(3):481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu D, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6(7):673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.