Abstract
Secondary metabolites are ubiquitous in bacteria, but by definition, they are thought to be nonessential. Highly toxic secondary metabolites such as patellazoles have been isolated from marine tunicates, where their exceptional potency and abundance implies a role in chemical defense, but their biological source is unknown. Here, we describe the association of the tunicate Lissoclinum patella with a symbiotic α-proteobacterium, Candidatus Endolissoclinum faulkneri, and present chemical and biological evidence that the bacterium synthesizes patellazoles. We sequenced and assembled the complete Ca. E. faulkneri genome, directly from metagenomic DNA obtained from the tunicate, where it accounted for 0.6% of sequence data. We show that the large patellazoles biosynthetic pathway is maintained, whereas the remainder of the genome is undergoing extensive streamlining to eliminate unneeded genes. The preservation of this pathway in streamlined bacteria demonstrates that secondary metabolism is an essential component of the symbiotic interaction.
Keywords: ascidian, trans-acyltransferase, natural products, polyketide, biosynthesis
As symbiotic relationships between animals and bacteria become more exclusive and specific, the symbiont’s genome can undergo a process of reduction, where functions required for independent life (but not life within the host) are lost (1). Those functions that are beneficial to the host are maintained and are clearly discerned through genome sequencing. For example, several insect symbionts are found to contain biosynthetic genes for amino acids essential to the host and not found in the host’s diet (1, 2). Symbiotic bacteria within higher animals are often implicated as the sources of bioactive secondary metabolites (3), particularly in the marine environment. Although some of these compounds have been shown to be protective for the host (4–8), the identity of the producers and the importance of metabolites to the symbiotic relationship mostly remain obscure. The term “secondary metabolite” itself is derived from the idea that these products are nonessential to life (9), yet secondary metabolite pathways are ubiquitous in bacteria, accounting for up to 10% of some genomes (10), and bioactive compounds such as antibiotics and cytotoxics clearly confer a survival advantage.
Lissoclinum patella, a colonial filter-feeding tunicate found throughout the tropical Indo-Pacific (Fig. 1 A and B), is a promising model for studying the importance of secondary metabolites in symbiosis. Like other tunicates, L. patella is a rich source of potentially bioactive drugs and drug leads (11), which in a few cases are known to be produced by symbiotic bacteria (7, 8). Beyond the major symbiont, the cyanobacterium Prochloron didemni, L. patella harbors a complex microbiome (12). Some L. patella samples contain highly potent cytotoxic polyketides, patellazoles (13–15) (IC50 332 pM, KB cells; Fig. 1C), for which the source was unknown. Here, we take a comprehensive approach to identify the patellazole producer, an α-proteobacterium that uniquely maintains a large secondary biosynthetic pathway despite undergoing extensive genome reduction. These results provide definitive evidence for a symbiotic interaction based entirely on essential secondary metabolism.
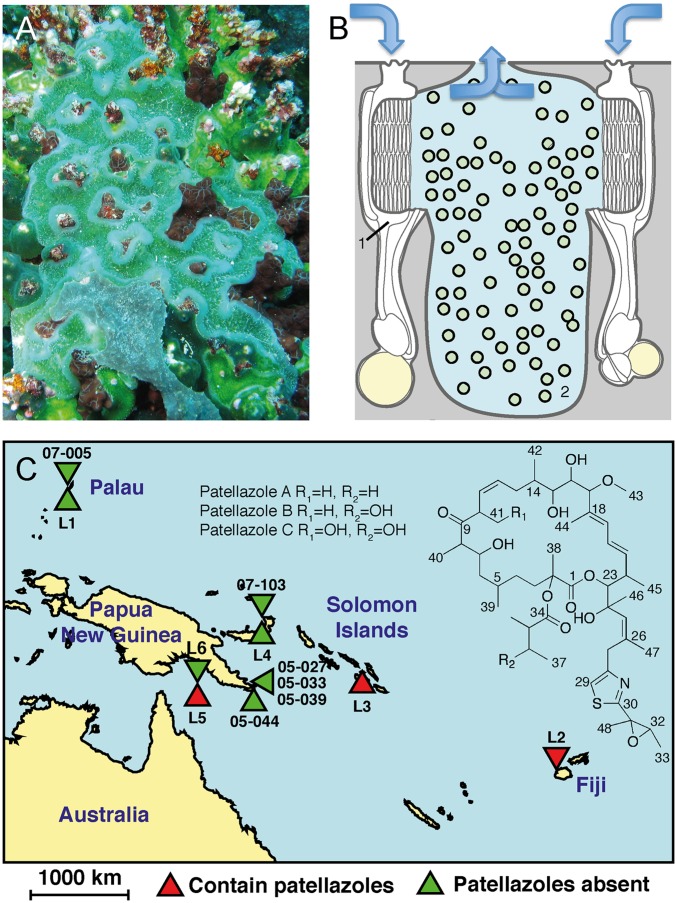
Fig. 1.
Patellazoles are only found sporadically in the tunicate L. patella. (A) Photograph of L. patella sample L3 at collection site (courtesy of C. M. Ireland, University of Utah). The green color is due to photosynthetic P. didemni. (B) L. patella is a colonial ascidian containing individual zooids (1) surrounding shared cloacal-peribranchial cavities (2) that contain P. didemni (green circles). Zooids filter feed, excreting waste into the cloacal cavities (direction shown with blue arrows). (C) Structure of the patellazoles and sites of L. patella collections, showing locations of patellazole-containing samples.
Results
Identification of a Symbiont Associated with the Production of Patellazoles.
L. patella consists of many zooids embedded in a common tunic, surrounding a cloacal-peribranchial cavity (16). P. didemni invariably inhabits the L. patella cloacal cavity, where it makes secondary metabolites (17). However, the presence of patellazoles is highly sporadic (15, 18). We previously sequenced the genome of P. didemni and obtained >4 Gbp of sequence from the tunic and cloacal cavity habitats from both patellazole-containing and patellazole-negative animals (12). In the present study, we made a >30 Gbp DNA clone library from these habitats that we screened for the presence of potential patellazole biosynthetic genes. Our screening strategy was based on the hypothesis that patellazoles would be derived from multiple acetate units and made by a type I polyketide synthase (PKS) pathway (19). The thiazole ring was anticipated to arise from incorporation and subsequent cyclization of a cysteine unit by a nonribosomal peptide synthetase module (20). Surprisingly, exhaustive screening by multiple methods failed to convincingly locate patellazole biosynthetic genes either in P. didemni or in the tunic-cloaca habitats (SI Appendix). Therefore, we turned our attention to the tiny zooids (length ∼1 mm). PCR amplification showed that zooids are dominated by genes consistent with patellazole biosynthesis, whereas other habitats in the tunicate are not (SI Appendix, Figs. S1 and S2). These genes encoded PKS proteins from the trans-acyltransferase (trans-AT) family (19, 21).
Shotgun sequencing of DNA from the zooids fraction (SI Appendix, Table S1), followed by bioinformatics analysis and PCR verification, allowed the assembly of a 1.48-Mbp circular chromosome (Fig. 2). The chromosome contained a complete 86-kbp trans-AT PKS pathway (Fig. 3). Analysis of the genome and phylogenetic analysis of the 16S rRNA gene sequence allowed us to propose a species and genus designation, Candidatus Endolissoclinum faulkneri. The 16S rRNA gene of Ca. E. faulkneri is most closely related (94% identity) to that of the recently described genus Thalassobaculum (22), which are free-living marine α-proteobacteria (Fig. 4A).
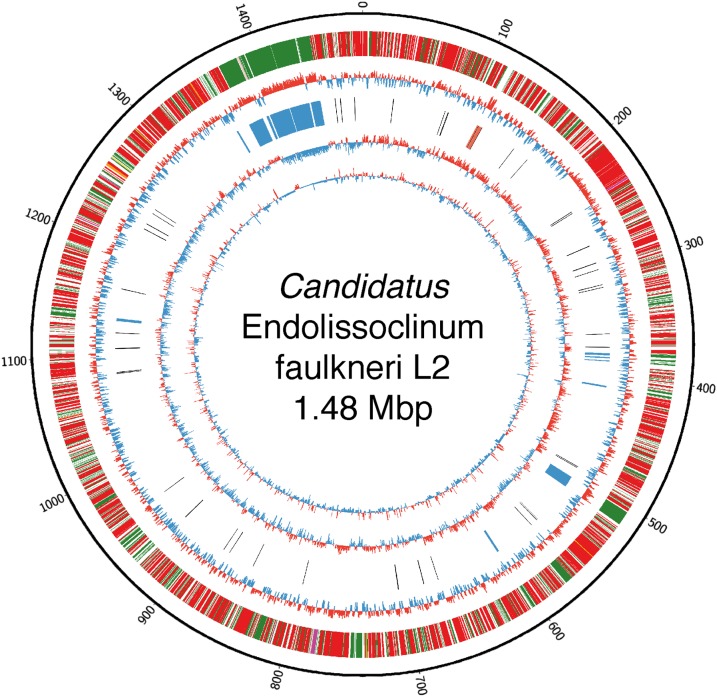
Fig. 2.
Circular map of the genome of Ca. E. faulkneri L2 and genome features. Circles correspond to the following (from outermost, scale is in kbp): (i) protein-coding genes in the genome. Colors indicate results from BLASTP searches in GenBank (red: top hit is BAL199; green, BAL199 not in top 100 hits); (ii) GC% using a 500-bp window size; (iii) Select highlighted features, blue: genes assigned to patellazole biosynthesis, black: tRNAs and other ncRNAs, orange: rRNA genes; (iv) plot of GC skew (G − C)/(G + C) using a 500-bp window size; (v) codon adaptation index (CAI), plotted for each ORF, using nonhypothetical genes to calculate reference RCSU and w values (36).
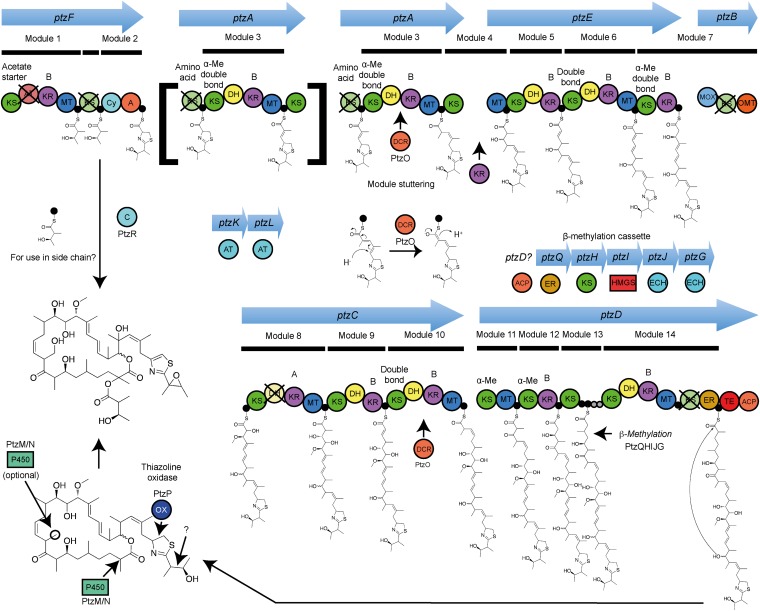
Fig. 3.
Proposed patellazoles biosynthetic pathway. The trans-AT PKS pathway contains 14 modules, with proposed repeated use (stuttering) of module 3 (PtzA) and use of the starting module of PtzF to produce the side chain. Predictions of structures accepted by KSs by phylogenetic analysis (SI Appendix, Fig. S11) are shown above the domains, as are predications for KR stereospecificity (SI Appendix, Fig. S14 and Table S11). A, adenylation; ACP, acylcarrier protein, also denoted by a filled black circle (predicted active) or a filled gray circle (predicted inactive); AT, acyltransferase; C, condensation; Cy, condensation/cyclization; DCR, 2,4-dienoyl-CoA-reductase; DH, dehydratase; ECH, enoyl-CoA-reductase; ER, enoyl-reductase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; KR, ketoreductase; KS, ketosynthase; MOX, flavin monooxygenase; MT, C-methyltransferase; OMT, O-methyltransferase; OX, thiazoline oxidase; P450, cytochrome P450; TE, thioesterase.
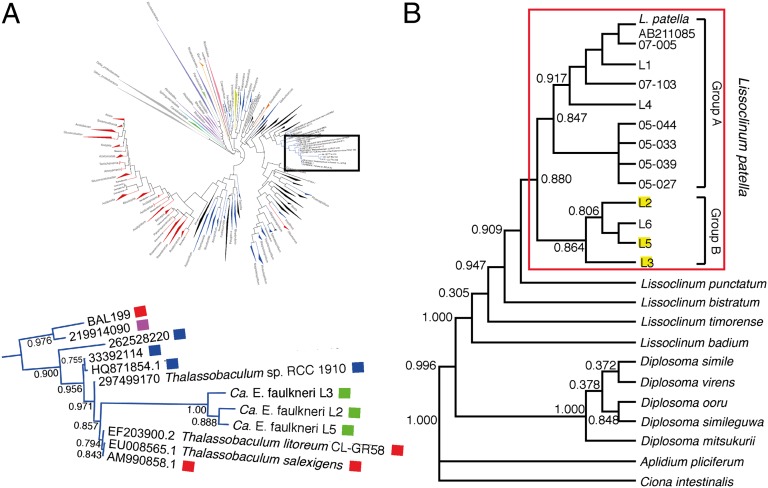
Fig. 4.
Relationship of Ca. E. faulkneri to other α-proteobacteria and to host tunicate phylogeny. (A) Phylogenetic tree based on 16S rRNA gene sequences of α-proteobacteria, showing enlargement of the clade containing Ca. E. faulkneri from three L. patella animals, L2, L3, and L5. Colored squares indicate the source of 16S rRNA gene sequences: red, cultured marine isolate; purple, uncultured terrestrial clone (oil-contaminated soil); blue, uncultured marine clone; green, uncultured marine symbiont clone. The full tree can be found in the SI Appendix, Fig. S15). (B) Phylogenetic tree based on 18S rRNA gene sequences of tunicates referred to in this study along with others from public databases. Patellazole-containing L. patella animals, highlighted in yellow, fall into a distinct clade we refer to here as “group B,” indicating that Ca. E. faulkneri may be specific for this group. The presence of L6 in this group, which does not contain patellazoles or Ca. E. faulkneri indicate that the presence of this symbiont is not necessary for the survival of the tunicate.
The patellazole biosynthetic pathway (ptz) was the only secondary metabolism sequence encoded in the Ca. E. faulkneri genome, and it closely matched the prediction for patellazole biosynthesis (Fig. 3 and SI Appendix, Table S2). To further confirm the association of ptz with patellazoles, we compared 10 L. patella samples from across the tropical Pacific by using multiple chemical and biological approaches (Figs. 1C and 5 and SI Appendix, Figs. S3–S5). Three samples (L2, L3, and L5) contained patellazoles, whereas seven did not. All three patellazole-positive samples were highly enriched both for ptz and the Ca. E. faulkneri 16S rRNA gene, whereas these genes were completely absent from patellazole-negative samples (Fig. 5 A and B and SI Appendix, Table S3).
Fig. 5.
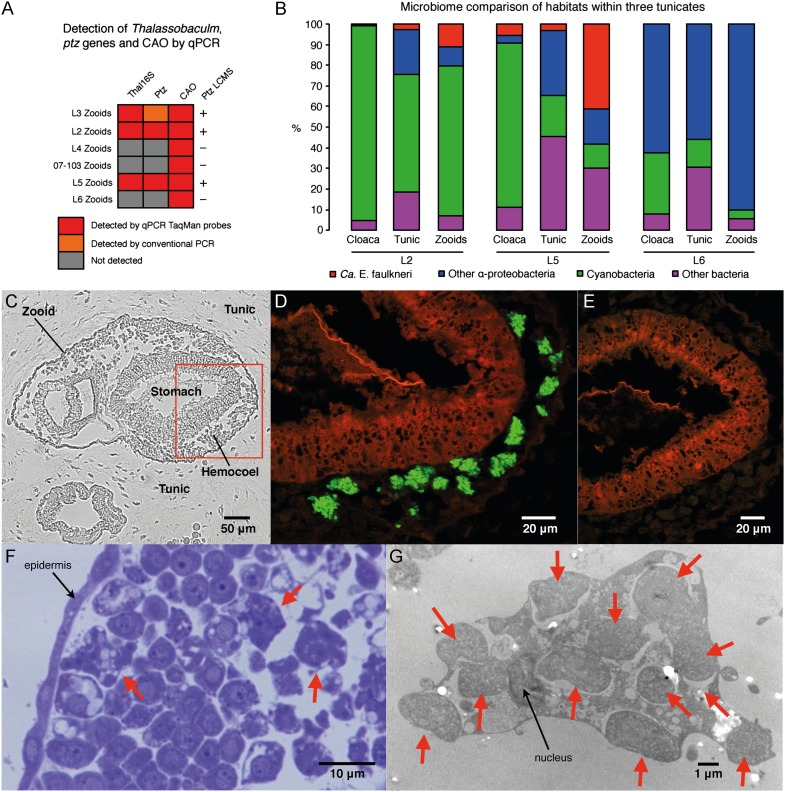
Localization of Ca. E. faulkneri and the patellazoles pathway. (A) Detection of the Ca. E. faulkneri 16S rRNA sequence, the ptz pathway, and the P. didemni CAO gene in zooid fractions of six L. patella animals by quantitative PCR and conventional PCR. (B) Classification of phylogeny of 16S rRNA gene sequences obtained by 454 amplicon sequencing, showing the content of Ca. E. faulkneri 16S rRNA (red, defined as sequences ≥ 98% identity to the Ca. E. faulkneri L2 16S rRNA sequence). (C) Bright-field view of a cross-section of a single zooid (L5) within the tunic, with a red box indicating the area shown in D. (D) Fluorescence in situ hybridization (L5) using a probe specific for Ca. E. faulkneri 16S rRNA (Ptzsymb646-033, green). (E) Negative control (L5) using a mismatch probe (Ptzsymb646NON, green). In D and E, the red channel is tissue autofluorescence. (F) Histological section of L2 stained with toluidin blue showing cells within a zooid, including bacteriocytes shown with red arrows. (G) TEM image (L2) of a single bacteriocyte, showing the cell nucleus and individual bacterial cells shown with red arrows.
We also investigated the microbial diversity within the different tissues of L2, L5, and L6 by 454 pyrosequencing of 16S rRNA genes. In both L2 and L5, Ca. E. faulkneri represents a large portion of the community in the zooids fraction (11.3% and 41.2%, respectively), but we did not find a single instance of the Ca. E. faulkneri 16S rRNA gene sequence in 20,000 reads from the three tissue fractions of L6 (Fig. 5B and SI Appendix, Table S3). Tellingly, samples L5 and L6 were collected within 250 m of each other in the Eastern Fields of Papua New Guinea; L5 contained patellazoles, whereas L6 did not. Both samples are nearly identical by tunicate phylogeny using 18S rRNA (Fig. 4B), by COX1 (SI Appendix, Fig. S6) markers, and by microbial community (SI Appendix, Table S3). Every bacterial strain present in L5 at >1% 16S sequence abundance was also present in L6, with the exception of Ca. E. faulkneri. Because of this extremely close relationship, L6 constitutes a naturally occurring “virtual knockout,” which provides strong evidence of the connection between Ca. E. faulkneri, the patellazoles pathway, and the presence of the compounds. Through microscopic examination using fluorescence in situ hybridization (FISH) with a probe specific to Ca. E. faulkneri, we localized clumps of Ca. E. faulkneri to zooids in patellazole-positive sample L5 (Fig. 5 C–E), whereas Ca. E. faulkneri could not be found in patellazole-negative sample L6. Ca. E. faulkneri was observed in the hemocoel of the zooids, where amorphous bacteria could be visualized by TEM within specialized tunicate bacteriocytes (Fig. 5 F and G).
Genome Streamlined for Secondary Metabolite Synthesis.
The 1.48-Mbp genome of Ca. E. faulkneri (Fig. 2) showed many of the hallmarks of genome reduction, which is thought to be a consequence of very small effective population sizes in host-specific symbionts (1). In these circumstances, purifying selection is very weak, leading to progressive loss of genes through deletion bias and Muller’s ratchet (23). The genome was completely closed and lacked any gaps or stretches of unknown nucleotide sequence, and no possible plasmids were detected (see SI Appendix for details). Predicted proteins in the chromosome were most related to those from the genome of cultivated marine α-proteobacterium BAL199 (ABHC00000000). In fact, nearly all Ca. E. faulkneri proteins with predicted function were ∼60% identical to their homologs in BAL199. Only 63 nonhypothetical genes in the Ca. E. faulkneri genome lacked a BAL199 homolog (SI Appendix, Table S4). Of those genes, 41 are directly explained by the needs of the symbiosis: 18 encompass the ptz pathway, 17 encode transporters, and 6 are related to a fix operon low-oxygen tension cytochrome oxidase system, reflecting low intracellular oxygen tension. The remaining 22 genes encode mostly proteins with predicted domains, but not predicted substrates. Therefore, we obtained the raw draft assembly of BAL199 and analyzed it in parallel with that of Ca. E. faulkneri. The comparison is telling: Whereas the BAL199 genome is 6.1 Mbp with 6,107 predicted genes, the Ca. E. faulkneri genome only encodes a predicted 783 genes (Table 1 and SI Appendix, Table S5). The GC content of Ca. E. faulkneri is much lower than BAL199 (34% versus 65%). This disparity is consistent with a general trend toward higher AT content in obligate intracellular symbionts, which is thought to be a result of (G/C)→(A/T) mutation biases (1). The coding density of the chromosome is also very low at 57% (versus a bacterial average of 85–90%; ref. 24), with a disparity in the GC content between coding (40%) and noncoding (25%) regions. Indeed, many hypothetical ORFs called by automatic annotation were both shorter than the majority of coding sequences and had the GC content of noncoding regions (SI Appendix, Fig. S7) and, therefore, we believe they are nonfunctional. Low coding densities are characteristic of genomes in early stages of genome reduction (24), because a high degree of genomic drift seen in these stages leads to a proliferation of pseudogenes. Indeed, by comparing sections of the ptz pathway in L5 and L2, we found an extremely high evolution rate as measured by the ratio of nonsynonymous to synonymous mutations (1.00; SI Appendix, Fig. S8) (24). Genome reduction appears to be at a much later stage than other low-density genomes, such as the mutualist Sodalis glossinidius, which has a coding density of 51% but a much larger genome than Ca. E. faulkneri (4.2 Mbp vs. 1.48 Mbp) (25). Although S. glossinidius contains many pseudogenes with homology to known proteins (25), Ca. E. faulkneri contains relatively few recognizable pseudogenes (SI Appendix, Table S6).
Table 1.
Comparison of Ca. E. faulkneri and BAL199 genomes
| Ca. E. faulkneri | BAL199* | |
| Length, Mbp | 1.48 | 6.1 |
| % coding (length) | 57.2 | 89.7 |
| GC% (coding) | 40.9 | 65.7 |
| GC% (noncoding) | 25.1 | 59.3 |
| No. of nonhypothetical genes† | 688 | 4,408 |
| No. of hypothetical genes† | 95 | 1,699 |
| % secondary metabolite genes (length) | 10.2 | 0.9 |
Here, the BAL199 genome refers to the raw assembly contigs retrieved from NCBI. For direct comparison, these contigs and Ca. E. faulkneri were annotated by using the same procedure (SI Appendix).
Genes called by automatic annotation that were less than 150 bp in length and less than 30% GC were discounted (SI Appendix, Fig. S7).
Streamlined genomes are found in intracellular pathogens (26) and even free-living bacteria (27, 28). By contrast, certain features distinguish obligate symbionts from bacteria with other lifestyles, principal among which is the loss of control over replication and division accompanied by an exclusively vertical mode of transmission. Much like symbionts with extremely reduced genomes (1), Ca. E. faulkneri lacks both dnaA and ftsZ, which are involved in chromosomal replication and cell division, respectively (29, 30). These genes are maintained even in the free-living bacterium with the smallest genome, Mycoplasma genitalium (27). The genome also completely lacks elements for horizontal transfer, without recognizable viral, plasmid, or transposon sequences, whereas BAL199 contained 285 genes in this category (SI Appendix, Table S5). As with other obligate symbionts, where regulatory mechanisms are typically lost (1), Ca. E. faulkneri has no signal transduction pathways and a low number of transcription factors, compared with 309 genes involved with regulatory functions and signal transduction in BAL199 (SI Appendix, Table S5).
The functions that are maintained in Ca. E. faulkneri are entirely in line with the minimal gene set of obligate symbionts (SI Appendix, Table S7) (1). In addition, functions that are useful to the host are maintained. For example certain insect symbionts (31, 32) harbor biosynthetic pathways for amino acids and other products such as carotenoids (33) not available to the host. In Ca. E. faulkneri, >10% of the coding sequence is devoted to a single secondary metabolite pathway for producing patellazoles. This pathway consists of 18 genes that are almost all related to other proteins in trans-AT PKS pathways (Fig. 3 and SI Appendix, Table S2). The maintenance of such a large pathway indicates its importance to the symbiosis, whereas we were unable to find pathways for other potentially useful products such as the amino acids phenylalanine, tryrosine, tryptophan, valine, isoleucine and leucine, which cannot be synthesized by eukaryotic hosts (34). Based on current models of genome reduction, this result implies that if the ptz pathway were lost, the symbiont would not be maintained within the host.
In some cases, relationships between vertically transmitted symbionts and their animal hosts are thought to be ancient associations where both parties have coevolved over extended periods of time (35). We found evidence of coevolution of tunicate hosts, by examining the phylogenetic relationship of tunicate marker genes (18S rRNA and the mitochondrial cytochrome c oxidase subunit I gene) in patellazole-containing and patellazole-negative animals (Fig. 4B and SI Appendix, Fig. S6). In both trees, all patellazole-containing samples fell into a distinct clade among L. patella that we term “group B”, although only a small number of patellazole-containing animals were available to construct the tree.
Genes in the ptz Pathway Are Not Recent Horizontal Acquisitions.
Only a small subset of proteins present in BAL199 are also found in Ca. E. faulkneri, and the two bacteria are substantially different on the nucleotide level such that alignment of the two genomes was impossible. Analysis of the positions of protein homologs on draft BAL199 genome contigs did not reveal any synteny beyond a few conserved operons (SI Appendix, Fig. S9). ORFs in the two organisms possess different codon use, as measured by codon adaptation index (CAI; SI Appendix, Fig. S10) (36), reflecting the drift toward AT-rich sequence in Ca. E. faulkneri. Importantly, genes of the ptz pathway have typical CAI values compared with the rest of the genome (SI Appendix, Fig. S10), indicating that they are not likely to be recent horizontal acquisitions. In contrast to most bacteria, horizontally acquired genes are not found in obligate symbionts with reduced genomes, which have been shown to maintain near-perfect synteny over vast timescales (37). Whereas late-stage symbiont genomes exhibit fixed synteny, earlier stages of host restriction are characterized by proliferation of mobile elements and widespread genome rearrangements (1). Genes in the ptz pathway are fragmented across seven distinct loci and likely were introduced before early stages in the symbiosis and before gene order became fixed. As nonessential genes were lost (and continue to be lost, e.g., dnaA), ptz remained and retained its function. Coregulation of functional units (38) and ease of horizontal transfer (the “selfish operon” model) (39) are two factors thought to drive selection toward gene clustering. The loss of regulation mechanisms and curtailing of horizontal transfer in early-stage symbionts likely contribute to fragmentation of previously clustered genes.
Model for Patellazoles Biosynthesis.
The putative patellazoles pathway contains five trans-AT PKS proteins (PtzFAEBCD), along with various accessory proteins including two acyltransferases (ATs, PtzKL; SI Appendix, Table S2). This recently described type of PKS protein does not contain an AT domain in each module, as in most cases, but instead ATs on a separate protein load malonate units onto acyl-carrier proteins (ACPs) on the PKS (21). Further accessory proteins are present that would be responsible for β-methylation at position 5 (PtzHIJG), in analogy to the bacillaene pathway (40), and for in trans enoyl reduction (PtzQ), as found in the corallopyronin pathway (40, 41), among others (SI Appendix, Table S2). Despite being distributed in separate loci, the ptz genes clearly belong to the same biosynthetic pathway. The standalone ATs are required for the function of trans-AT PKS proteins, but they are not close to PKS proteins. Likewise, some members of the β-methylation apparatus are found far from any PKS proteins. The ptz genes encode among the only proteins in the chromosome that are not highly related to homologs in BAL199 (Fig. 2 and SI Appendix, Table S4). They are also the only secondary metabolic genes encoded on the genome (SI Appendix, Table S5), and the majority show homology to PKS or accessory proteins from a specific type of pathway containing trans-AT PKSs. Additionally, genes found on separate loci do not form discrete units that could act independently.
Our model for patellazole biosynthesis is shown in Fig. 3, and a detailed description is presented in SI Appendix. In support of the assignment, we applied phylogenetic analysis to the ketosynthase (KS) domains of the PKS proteins (SI Appendix, Fig. S11 and Table S8), following recent work that has shown that in phylogenetic analysis of trans-AT PKS pathways, KS domains group largely according to the structure accepted by each KS (42). These predictions, where possible, are entirely consistent with proposed intermediates. One of the most compelling mysteries about patellazoles biosynthesis is the presence of two β-γ cis-double bonds, which are rare in PKS-produced molecules. In our model, β-γ cis-double bonds would be synthesized from a thioester-conjugated 1,3-diene by the action of PtzO, which lacks similarity to known trans-AT PKS accessory proteins, but is similar to characterized 2,4-dienoyl-CoA-reductases (43). Under this hypothesis, PtzO must act in trans with the PKS to synthesize the cis-double bond during chain elongation. This model is further supported by extensive phylogenetic analysis of dehydratase (DH) domains (SI Appendix, Figs. S12 and S13). Overall, the biosynthetic model is in excellent agreement with the patellazoles structures and strongly supports the assignment. Finally, this analysis allowed prediction of the absolute configuration of four stereocenters, potentially aiding drug development efforts.
Discussion
Symbiotic relationships between bacteria and their hosts exist on a spectrum between casual associations (frequently nonspecific) and mutual codependency, such as the extreme case of organelles in eukaryotic cells (44). Progressive host restriction ultimately reduces the effective population size of the symbiont. In such a situation, purifying selection is weak, allowing fixation of mildly deleterious mutations (23), eventually leading to pseudogene formation and genome reduction due to deletion bias (45). P. didemni is only somewhat host restricted, because it can live in multiple species of didemnid tunicate (16). The genome of P. didemni does not show signs of reduction (12), and although it has never been successfully cultured, genome analysis suggests that it is transmitted horizontally and vertically. P. didemni synthesizes cyclic peptide secondary metabolites known as cyanobactins (8), which could contribute to chemical defense, but there are other potential symbiotic functions for this organism. Other functions are connected to central metabolism: photosynthesis, nitrogen waste recycling, and lipid production (12).
In contrast, Ca. E. faulkneri is in a more advanced state of host restriction, as yet only being found in a subgroup of L. patella. The extent of genome reduction suggests that the bacterium could not live independently of the host and is transmitted vertically, potentially coevolving with L. patella over an extended period. Against the drive of reduction, the patellazoles pathway is maintained, indicating its importance to the symbiotic relationship. The patellazoles are highly toxic to eukaryotic cells and found in high amounts in L. patella (0.1% dry weight) (15), yet we found no evidence of injury or necrosis by microscopic examination. The tunicate may be adapted to tolerating large concentrations of patellazoles, and the compounds probably have a role in chemical defense of the animal. Patellazole B is more toxic to human cells than biosynthetically related compounds, bryostatins (46) and pederins (4), which are produced by bacterial symbionts of bryozoans and insects, respectively, and are both known to have defensive properties.
Here, we describe a symbiotic bacterium in which genome streamlining has eliminated most functions, with the remaining genome focused on the production of a toxic secondary metabolite. This result demonstrates the importance of chemical defense in maintaining this symbiosis, and it provides compelling evidence that secondary metabolism can be essential.
Materials and Methods
Tunicate Collection and Preservation.
Animals were collected at the locations indicated in SI Appendix, Table S9 at 1–5 m depth. Typically part of the animals was preserved in RNAlater at −20 °C, whereas the rest was frozen for later chemical extraction. In addition, L2 was fixed in 2% (wt/wt) glutaraldehyde in artificial seawater for 2–4 h and then transferred to 0.5% glutaraldehyde for storage. For the 2011 collection, which includes L5 and L6, animals were also fixed according to the following protocol. Solid paraformaldehyde (4.0 g) was dissolved in 40 mL of hot (60 °C) deionized water. Solid NaOH was added to aid dissolution, then the solution was chilled on ice. The pH of the solution was corrected to 7.2 using ethanoic acid and then brought up to volume with 10× marine PBS. The resulting fixative was filtered through a 0.2-μm syringe filter and used fresh. Samples were fixed for ∼2−4 h at room temperature, then the fixative was replaced with 70% (vol/vol) EtOH in water.
Other Experiments.
Additional methods, along with supporting tables, figures, and source code are available in SI Appendix.
Supplementary Material
Acknowledgments
Ca. E. faulkneri is named in honor of the late Prof. D. John Faulkner, a pioneer in marine symbiosis and secondary metabolism. We thank the people and governments of Papua New Guinea, Solomon Islands, Fiji, and Palau for permission to conduct this study; C. M. Ireland (University of Utah) for aid with collection; University of Utah Center for High Performance Computing and SHI International for computational resources; J. Ravel, W. F. Fricke, K. Galens, J. Orvis, M. Matalka, C. Arze (all Institute for Genome Sciences, University of Maryland), and B. Milash (University of Utah) for bioinformatics advice; and R. van Wagoner and M. K. Harper (University of Utah) for use of LCMS instrumentation. This work was funded by National Institutes of Health Grant GM092009 and is based on work supported in part by National Science Foundation Grant 0910812 (Future Grid compute grid).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CP003539 and JX033584–JX033588).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213820109/-/DCSupplemental.
References
- 1.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10(1):13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 2.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407(6800):81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 3.Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep. 2004;21(4):519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]
- 4.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA. 2002;99(22):14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudek S, et al. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J Nat Prod. 2007;70(1):67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 6.Lopanik N, Lindquist N, Targett N. Potent cytotoxins produced by a microbial symbiont protect host larvae from predation. Oecologia. 2004;139(1):131–139. doi: 10.1007/s00442-004-1487-5. [DOI] [PubMed] [Google Scholar]
- 7.Rath CM, et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem Biol. 2011;6(11):1244–1256. doi: 10.1021/cb200244t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt EW, et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102(20):7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams DH, Stone MJ, Hauck PR, Rahman SK. Why are secondary metabolites (natural products) biosynthesized? J Nat Prod. 1989;52(6):1189–1208. doi: 10.1021/np50066a001. [DOI] [PubMed] [Google Scholar]
- 10.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26(11):1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt EW, Donia MS, McIntosh JA, Fricke WF, Ravel J. Origin and variation of tunicate secondary metabolites. J Nat Prod. 2012;75(2):295–304. doi: 10.1021/np200665k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donia MS, et al. Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc Natl Acad Sci USA. 2011;108(51):E1423–E1432. doi: 10.1073/pnas.1111712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zabriskie TM, Mayne CL, Ireland CM. Patellazole C: A novel cytotoxic macrolide from Lissoclinum patella. J Am Chem Soc. 1988;110:7919–7920. [Google Scholar]
- 14.Corley DG, Moore RE, Paul VJ. Patellazole B: A novel cytotoxic thiazole-containing macrolide from the marine tunicate Lissoclinum patella. J Am Chem Soc. 1988;110:7920–7922. [Google Scholar]
- 15.Zabriskie TM. 1989. The characterization of cytotoxic metabolites from Fijian marine invertebrates. PhD thesis (Univ of Utah, Salt Lake City)
- 16.Hirose E, et al. 2009. Enigmatic life and evolution of Prochloron and related cyanobacteria inhabiting colonial ascidians. Handbook on Cyanobacteria, eds Gault PM, Marler HJ (Nova Science Publishers, New York) pp 161–189.
- 17.Donia MS, et al. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2(12):729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 18.Richardson AD, Aalbersberg W, Ireland CM. The patellazoles inhibit protein synthesis at nanomolar concentrations in human colon tumor cells. Anticancer Drugs. 2005;16(5):533–541. doi: 10.1097/00001813-200506000-00009. [DOI] [PubMed] [Google Scholar]
- 19.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48(26):4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 20.Strieker M, Tanović A, Marahiel MA. Nonribosomal peptide synthetases: Structures and dynamics. Curr Opin Struct Biol. 2010;20(2):234–240. doi: 10.1016/j.sbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2010;27(7):996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- 22.Zhang GI, Hwang CY, Cho BC. Thalassobaculum litoreum gen. nov., sp. nov., a member of the family Rhodospirillaceae isolated from coastal seawater. Int J Syst Evol Microbiol. 2008;58(Pt 2):479–485. doi: 10.1099/ijs.0.65344-0. [DOI] [PubMed] [Google Scholar]
- 23.Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93(7):2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo C-H, Moran NA, Ochman H. The consequences of genetic drift for bacterial genome complexity. Genome Res. 2009;19(8):1450–1454. doi: 10.1101/gr.091785.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toh H, et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16(2):149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson SGE, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396(6707):133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 27.Fraser CM, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270(5235):397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 28.Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6(2):R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriya S, Kato K, Yoshikawa H, Ogasawara N. Isolation of a dnaA mutant of Bacillus subtilis defective in initiation of replication: Amount of DnaA protein determines cells’ initiation potential. EMBO J. 1990;9(9):2905–2910. doi: 10.1002/j.1460-2075.1990.tb07481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6(11):862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21(16):1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu D, et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4(6):e188. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sloan DB, Moran NA. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol Lett. September 12, 2012 doi: 10.1098/rsbl.2012.0664. 10.1098/rsbl.2012.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guedes RLM, et al. Amino acids biosynthesis and nitrogen assimilation pathways: A great genomic deletion during eukaryotes evolution. BMC Genomics. 2011;12(Suppl 4):S2. doi: 10.1186/1471-2164-12-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104(49):19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharp PM, Li W-H. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Ham RCHJ, et al. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA. 2003;100(2):581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price MN, Huang KH, Arkin AP, Alm EJ. Operon formation is driven by co-regulation and not by horizontal gene transfer. Genome Res. 2005;15(6):809–819. doi: 10.1101/gr.3368805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence JG, Roth JR. Selfish operons: Horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143(4):1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Convergence of isoprene and polyketide biosynthetic machinery: Isoprenyl-S-carrier proteins in the pksX pathway of Bacillus subtilis. Proc Natl Acad Sci USA. 2006;103(24):8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erol Ö, et al. Biosynthesis of the myxobacterial antibiotic corallopyronin A. ChemBioChem. 2010;11(9):1253–1265. doi: 10.1002/cbic.201000085. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen T, et al. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008;26(2):225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 43.Fillgrove KL, Anderson VE. The mechanism of dienoyl-CoA reduction by 2,4-dienoyl-CoA reductase is stepwise: Observation of a dienolate intermediate. Biochemistry. 2001;40(41):12412–12421. doi: 10.1021/bi0111606. [DOI] [PubMed] [Google Scholar]
- 44.Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126(3):453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 45.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17(10):589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 46.Pettit GR, Herald CL, Doubek DL, Herald DL. Isolation and structure of bryostatin 1. J Am Chem Soc. 1982;104(24):6846–6848. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.