Abstract
Intestinal metaplasia of the stomach, a mucosal change characterized by the conversion of gastric epithelium into an intestinal phenotype, is a precancerous lesion from which intestinal-type gastric adenocarcinoma arises. Chronic infection with Helicobacter pylori is a major cause of gastric intestinal metaplasia, and aberrant induction by H. pylori of the intestine-specific caudal-related homeobox (CDX) transcription factors, CDX1 and CDX2, plays a key role in this metaplastic change. As such, a critical issue arises as to how these factors govern the cell- and tissue-type switching. In this study, we explored genes directly activated by CDX1 in gastric epithelial cells and identified stemness-associated reprogramming factors SALL4 and KLF5. Indeed, SALL4 and KLF5 were aberrantly expressed in the CDX1+ intestinal metaplasia of the stomach in both humans and mice. In cultured gastric epithelial cells, sustained expression of CDX1 gave rise to the induction of early intestinal-stemness markers, followed by the expression of intestinal-differentiation markers. Furthermore, the induction of these markers was suppressed by inhibiting either SALL4 or KLF5 expression, indicating that CDX1-induced SALL4 and KLF5 converted gastric epithelial cells into tissue stem-like progenitor cells, which then transdifferentiated into intestinal epithelial cells. Our study places the stemness-related reprogramming factors as critical components of CDX1-directed transcriptional circuitries that promote intestinal metaplasia. Requirement of a transit through dedifferentiated stem/progenitor-like cells, which share properties in common with cancer stem cells, may underlie predisposition of intestinal metaplasia to neoplastic transformation.
Keywords: CagA, Wnt, β-catenin
Metaplasia is a histological change from one tissue type to another, which is associated with conversion of its respective cell types to the corresponding ones. Metaplastic changes can occur either in a physiological process or a pathological condition, the latter of which predisposes cells to undergo neoplastic transformation via a metaplasia–dysplasia–carcinoma sequence. Intestinal metaplasia, a pathological change of nonintestinal epithelium into an intestinal-like mucosa, is most frequently found in the stomach and esophagus (1, 2). Helicobacter pylori-induced chronic gastritis is a major cause of gastric intestinal metaplasia, from which intestinal-type adenocarcinoma arises (3). Likewise, replacement of the esophageal squamous epithelium by intestinal epithelium known as Barrett’s esophagus substantially increases the risk of esophageal adenocarcinoma (2).
The Drosophila homeobox gene caudal plays a critical role in development of the posterior embryo (4). Caudal has three homologs in vertebrates (CDX1, CDX2, and CDX4 in humans; Cdx1, Cdx2, and Cdx4 in mice; and CdxA, CdxB, and CdxC in chickens) (5, 6). These genes encode Caudal-related homeobox transcription factors (hereafter denoted as CDX family proteins), which play unique roles in axial patterning and gut development by regulating specific genes through binding to an A/T-rich responsive element. The consensus binding sequence for these CDX family proteins is (A/C)TTTAT(A/G), in which TTTAT acts as a conserved core motif (4, 5). In mammals, CDX family members, especially CDX1 and CDX2, are critically involved in development and maintenance of the intestine (6). Indeed, both CDX1/Cdx1 and CDX2/Cdx2 are expressed in the epithelium of the large and small intestines but not in the epithelium of the stomach or esophagus. They are, however, aberrantly expressed in the intestinal metaplastic lesion of the stomach as well as in Barrett’s esophagus (6). CDX1 is transactivated by several distinct signaling mechanisms such as Wnt/β-catenin signal and retinoic acid signal (7). We previously reported that H. pylori CagA, which is delivered into gastric epithelial cells via bacterial type IV secretion, aberrantly stimulates β-catenin signaling and thereby induces Wnt target genes including CDX1 (8). This observation suggested that CagA-mediated Wnt/β-catenin deregulation plays an important role in the ectopic expression of CDX1 in the stomach infected with H. pylori.
Transgenic expression of Cdx1 or Cdx2 in mouse stomach causes intestinal metaplasia (9, 10), indicating a causal and redundant role of ectopically expressed CDX1 and CDX2 in the metaplastic change. Because metaplasia is defined as a switching from one tissue to another, the process may be initiated through changes in cell differentiation status so as to acquire some sort of cell/tissue stemness or multipotency. However, little is known about CDX-governed transcriptional circuitries that give rise to intestinal metaplasia of the stomach. In this work, we investigated genes directly activated by CDX1 in gastric epithelial cells by combining expression microarray and chromatin immunoprecipitation (ChIP)-chip analyses and identified transcription factors, SALL4 and KLF5, both of which are involved in lineage reprogramming and stemness acquisition. We show that CDX1-mediated induction of SALL4 and KLF5 plays an important role in transdifferentiation of gastric epithelial cells into an intestinal phenotype, which underlies intestinal metaplasia of the stomach.
Results
Genes Affected by Ectopic CDX1 Expression in Gastric Epithelial Cells.
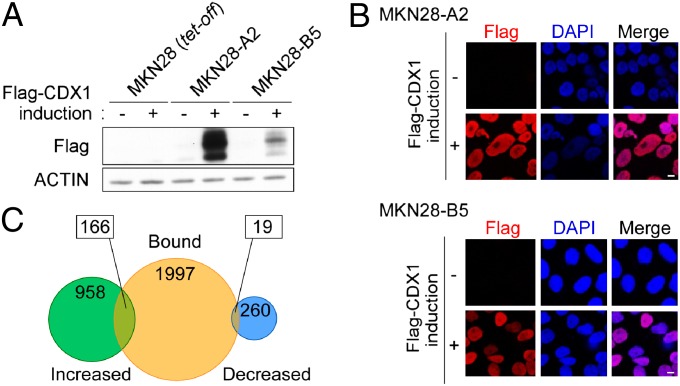
To investigate genes targeted by ectopically expressed CDX1, we established several transfectant clones that inducibly express Flag-tagged CDX1 from MNK28 human gastric epithelial cells using a tetracycline-regulated Tet-Off system (Fig. 1 A and B and Fig. S1A). Among these, MKN28-A2 cells, which showed the highest expression upon depletion of doxycycline (Dox), a water-soluble tetracycline analog, were subjected to expression microarray analysis and ChIP-chip analysis. First, we compared mRNA expression profiles of MKN28-A2 cells cultured in the presence (CDX1 induction −) or absence (CDX1 induction +) of Dox for 24 h through genome-wide expression microarray analysis and identified 958 genes that showed increases in transcript levels of more than twofold after CDX1 induction and 260 genes whose expression levels were decreased to less than half by CDX1 expression (Fig. 1C). Because the removal of Dox had little impact on the mRNA expression profile in parental MKN28 (tet-off) cells (Dataset S1), genes selected in Fig. 1C were due to specific induction of CDX1. We next carried out ChIP-chip analysis using a human promoter array. CDX1 mostly bound to the promoter regions that are localized substantially upstream of the transcription start sites (TSSs) of genes (Fig. S1B). The results of ChIP-chip analysis revealed 1,997 genes to which CDX1 binds at the regulatory regions (Fig. 1C). These identified genes contained known CDX1-target genes (Fig. S1C). By combining data obtained from expression microarray and ChIP-chip analyses, we selected 166 genes that should include genes specifically up-regulated by CDX1 (Fig. 1C and Dataset S2). Unlike a bacterial restriction endonuclease, which strictly recognizes a unique nucleotide sequence, a mammalian transcription factor binds to a range of related sequences (11). Indeed, in the CDX consensus (A/C)TTTAT(A/G), positions 1 and 7 are less stringent compared with the core motif TTTAT (positions 2–6) (5). Given this, we investigated sequences that were enriched in the upstream regions of CDX1-induced genes and found that TTTATT was overrepresented in these regions (Fig. S1D). This result reinforced the importance of the TTTAT core motif for specific DNA binding of CDX family proteins. The result also raised the possibility that, whereas CDX1 can variably interact with a number of sequences related to the CDX consensus (A/C)TTTAT(A/G), it preferentially binds to TTTATT. The slight difference between TTTATT and the CDX consensus may allow CDX1 to regulate genes in a manner that is quantitatively and/or qualitatively different from that by which genes are regulated by other CDX members.
Fig. 1.
Transcriptional targets of CDX1. (A) Lysates prepared from MKN28 (tet-off) cells and the transfectant clones, MKN28-A2 and MKN28-B5 cells, were subjected to immunoblotting with the respective antibodies. Flag-tagged CDX1 was induced by Dox depletion. (B) Anti-Flag immunostaining of MKN28-A2 and MKN28-B5 cells with or without Flag-CDX1 induction. Nuclei were visualized by DAPI. (Scale bars, 10 μm.) (C) Venn diagram showing the overlap between genes to which CDX1 bound and those of which mRNA levels were altered by CDX1 expression.
Transactivation of Reprogramming Factors by Ectopic CDX1.
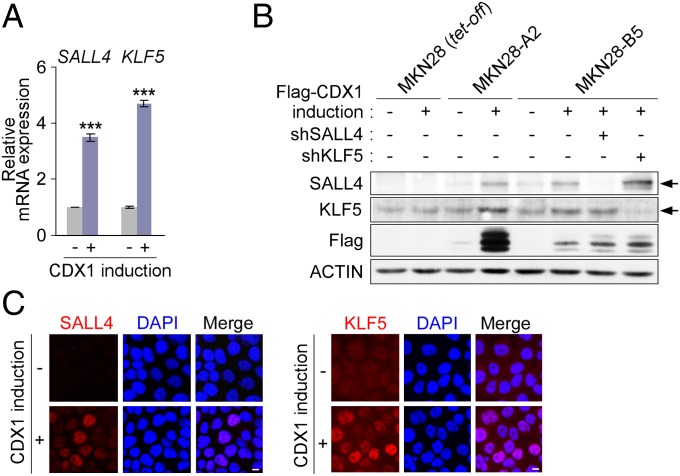
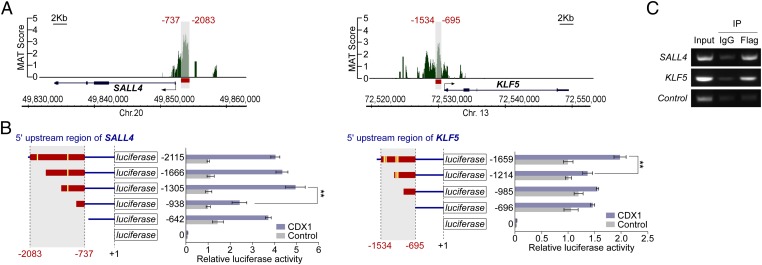
We hypothesized that CDX1 induces stemness-regulating reprogramming factors in gastric epithelial cells that revert cell-differentiation status so that the cells acquire intestinal stem/progenitor-like properties. With this idea, we investigated whether the identified CDX1-target genes included genes that could confer multipotency upon differentiated somatic cells and we found Sal-like 4 (SALL4 in humans and Sall4 in mice), a gene encoding the SALL4/Sall4 transcription factor that is essential for maintaining stemness in embryonic stem (ES) cells (Dataset S2) (12, 13). Of note, Sall4 is not expressed in the adult gastrointestinal tract under physiological conditions (14). The identified CDX1-target genes also included Krüppel-like factor 5 (KLF5 in humans and Klf5 in mice), which encodes the KLF5/Klf5 transcription factor (Dataset S2). Klf5 is capable of replacing Klf4 in generating inducible pluripotent stem (iPS) cells, indicating its role in the acquisition of stemness (15). Whereas Klf5 is predominantly expressed in the small intestine and colon, a small amount of the Klf5 transcript is also detectable in the stomach (16). A reverse–transcription quantitative PCR (RT-qPCR) analysis revealed that both SALL4 and KLF5 mRNAs were induced in MKN28-A2 cells upon ectopic expression of CDX1 (Fig. 2A). Induction of endogenous SALL4 and KLF5 by CDX1 was also demonstrated through immunostaining or immunoblotting (Fig. 2 B and C). ChIP-chip analysis revealed that CDX1 binds to the sequence between −2083 and −737 of the SALL4 promoter, which contains two CDX-binding TTTAT core motifs conserved between the human and mouse sequences (Fig. 3A, Left and Fig. S2). A luciferase reporter assay using a series of deletion mutants of the SALL4 promoter showed that the sequence between −1305 and −938, which contains a single putative CDX-binding sequence, was involved in induction of SALL4 by CDX1 (Fig. 3B, Left). A further deletion of the SALL4 regulatory region from −938 to −642 slightly but significantly restored the reporter activity. This observation was reproduced in the nontransformed human gastric epithelial cell line GES-1 (Fig. S3), indicating the presence of a cis-acting repressor element between −938 and −642. The results of ChIP-chip analysis also showed that CDX1 binds to the sequence between −1534 and −695 of the KLF5 promoter (Fig. 3A, Right and Fig. S2), which contains four CDX1-binding core motifs conserved between the human and mouse sequences. To further elucidate the enhancer element that is used for CDX1-dependent transactivation of KLF5, a luciferase reporter assay was carried out using a series of deletion mutants for the KLF5 promoter region. The results of the experiment revealed that CDX1 transactivates KLF5 via the sequence between −1659 and −1214 that contains two putative CDX1-binding sites (Fig. 3B, Right). In both SALL4 and KLF5 cases, reduction in luciferase activity by deletion of the putative CDX1-binding sites was not robust. In eukaryotes, however, rarely does a single transcription factor govern transcription of the target gene. Instead, different combinations of ubiquitous and cell-type–specific transcription factors act together by binding to the respective binding sites, with each one having a differential functional contribution (17). Hence, despite its partial promoter stimulation, CDX1 may play a pivotal role in passing a certain threshold of the promoter activation that is required for ectopic expression of SALL4 and KLF5. By ChIP experiment, specific binding of CDX1 to the upstream regulatory regions of SALL4 and KLF5 were confirmed (Fig. 3C). Based on these observations, we concluded that SALL4 and KLF5 are direct transcriptional targets of CDX1.
Fig. 2.
Induction of SALL4 and KLF5 by CDX1. (A) SALL4 and KLF5 mRNA levels in MKN28-A2 cells with or without CDX1 induction for 24 h were determined by RT-qPCR. Error bars, ± SD; n = 3. (B) Cells transfected with an expression vector for a SALL4- or KLF5-specific short hairpin RNA (shRNA), pSUPER-shSALL4/1 or pSUPER-shKLF5/1, were induced to express Flag-CDX1 by Dox depletion for 24 h. Lysates prepared were immunoblotted with the respective antibodies. Arrows indicate the positions of SALL4 and KLF5. (C) MKN28-B5 cells were immunostained with the respective antibodies. Flag-CDX1 was induced for 48 h. (Scale bars, 10 μm.)
Fig. 3.
Transactivation of SALL4 and KLF5 by CDX1. (A) ChIP-chip signals at the SALL4 or KLF5 loci. Red lines represent CDX1-binding regions. (B) MKN28 cells transfected with the indicated reporter plasmids together with a CDX1 or control vector were subjected to luciferase reporter assay. Schematic diagrams represent SALL4 (Left) or KLF5 (Right) upstream regions. Numbers indicate the distance from the TSS. Red lines indicate the CDX1-binding regions identified in A and yellow boxes represent putative CDX1-binding sites containing the TTTAT core motif. Error bars, ± SD; n = 3. (C) ChIP-PCR analysis for Flag-CDX1 occupancies of the SALL4 and KLF5 upstream regions in CDX1-induced MKN28-A2 cells.
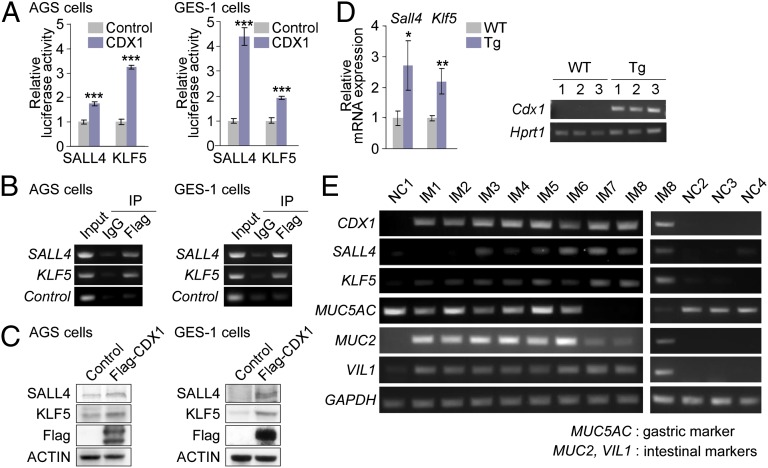
To generalize the above-described observations, we transiently transfected a Flag-tagged CDX1 vector into AGS and GES-1 human gastric epithelial cells. The results of luciferase reporter assays confirmed that CDX1 transactivates SALL4 and KLF5 in both cells (Fig. 4A). A ChIP experiment revealed that CDX1 bound to the upstream regions of SALL4 and KLF5 genes (Fig. 4B). Induction of SALL4 and KLF5 proteins by ectopic expression of CDX1 was also demonstrated in AGS and GES-1 cells (Fig. 4C).
Fig. 4.
Aberrant expression of SALL4 and KLF5 in intestinal metaplasia. (A) Cells transfected with pGL3-SALL4(−1305) or pGL3-KLF5(−1659) reporter plasmid together with a CDX1 or control vector were subjected to luciferase reporter assay. Error bars, ± SD; n = 3. (B) ChIP-PCR analysis for Flag-CDX1 occupancies of the SALL4 and KLF5 upstream regions in cells transiently transfected with a Flag-CDX1 vector. (C) Induction of SALL4 and KLF5 in cells transiently transfected with a Flag-CDX1 vector. (D) Levels of SALL4 and KLF5 mRNAs in the stomach of wild-type (WT) and Cdx1-transgenic (Tg) mice were determined by RT-qPCR. Error bars, ± SD; n = 3 (Left). Transgenic expression of Cdx1 in the stomach of Cdx1-Tg mice was confirmed by RT-PCR (Right). (E) Expression of the indicated mRNAs in human stomachs with (IM1–IM8) or without (NC1–NC4) intestinal metaplasia was determined by semiquantitative RT-PCR.
To investigate the pathophysiological relevance for induction of these reprogramming factors in intestinal metaplasia, expression of Sall4 and Klf5 was investigated in the stomach of Cdx1-transgenic mice using RT-qPCR and found that both of the mRNAs were detectable in the intestinal metaplastic lesions of the mouse stomach (Fig. 4D). The expression of SALL4 and KLF5 was also examined in intestinal metaplasia of the human stomach by semiquantitative RT-PCR (Fig. 4E). In control RNAs obtained from gastric mucosa without intestinal metaplasia [normal control (NC1-NC4)], CDX1, SALL4, or KLF5 was hardly detectable. In contrast, in samples in which CDX1 was expressed [intestinal metaplasia (IM1-IM8)], KLF5 was also detectable and the level of KLF5 expression was in proportion to the level of CDX1 expression. SALL4 was also detected in samples IM3–IM8. However, it was only weakly expressed in samples IM1 and IM2, in which the expression levels of CDX1 were less than those in samples IM3–IM8. Induction of SALL4 may therefore require a higher level of CDX1 expression than that required for KLF5 induction. In samples IM1–IM6, both gastric mucin (MUC5AC) and intestinal mucin (MUC2) were detected, indicating that these samples contained both intestinal metaplastic lesions and normal gastric mucosa. In samples IM7 and IM8, the level of mucin expression, either intestinal or gastric type, was low, whereas that of Villin1 (VIL1) was high. Although VIL1 is known as an enterocyte marker, it is also expressed in gastrointestinal stem/progenitor cells (18). Accordingly, samples IM7 and IM8 may have been derived from lesions that persisted in a less-differentiated state rather than having undergone transdifferentiation. These observations provided in vivo evidence that KLF5 and SALL4 were aberrantly expressed in intestinal metaplasia of the stomach in both humans and mice.
Induction of Intestinal Stem/Progenitor Markers by CDX1.
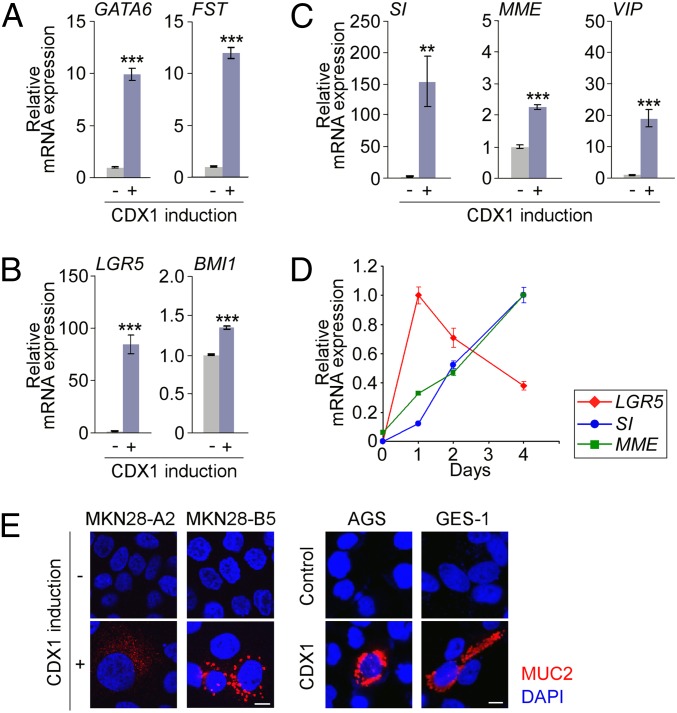
Through microarray analysis, the genes activated by CDX1 in gastric epithelial cells also included genes expressed in intestinal progenitor cells such as GATA binding protein 6 (GATA6) and follistatin (FST). GATA6 is expressed in the intestinal crypt and involved in proliferation of immature cells (19). Likewise, FST, an antagonist of TGF-β superfamily proteins, is expressed in undifferentiated intestinal epithelial cells (20). RT-qPCR analysis exhibited one order-of-magnitude increase in the level of GATA6 or FST upon ectopic CDX1 expression in gastric epithelial cells (Fig. 5A).
Fig. 5.
Induction of intestinal markers by CDX1 in gastric epithelial cells. (A–C) Levels for intestinal-progenitor markers (A), intestinal-stemness markers (B), and intestinal-differentiation markers (C) in MKN28-A2 cells before and after induction of CDX1 for 24 h were determined by RT-qPCR. Error bars, ± SD; n = 3. (D) Kinetic changes in the expression of the respective genes following induction of CDX1 in MKN28-A2 cells were determined by RT-qPCR. Error bars, ± SD; n = 3. (E) Anti-MUC2 immunostaining of cells inducibly expressing Flag-CDX1 (Left) or transiently transfected with a Flag-CDX1 vector (Right) for 4 d. (Scale bars, 10 μm.)
Recent studies have demonstrated that intestinal stem cells are characterized by the expression of intestinal-stemness markers (21). Microarray analysis demonstrated that one of the genes most robustly induced by ectopic CDX1 in gastric epithelial cells was leucine-rich repeat containing G protein-coupled receptor 5 (LGR5), an intestinal-stemness marker (Table S1). Ectopic CDX1 also gave rise to the expression of other intestinal-stemness markers, such as BMI1 polycomb ring finger oncogene (BMI1) (Table S1). RT-qPCR analysis confirmed increased expression of LGR5 and BMI1 upon CDX1 expression in MKN28-A2 cells (Fig. 5B). Although highly reproducible and statistically significant, induction of BMI1 mRNA by CDX1 was relatively weak. This was most probably due to the higher level of CDX1 required for activation of BMI1 than that required for activation of LGR5. From these observations, we concluded that aberrantly expressed CDX1 endowed gastric epithelial cells with an intestinal stem/progenitor-like phenotype.
Up-Regulation of Intestinal-Differentiation Markers by Sustained CDX1 Expression in Gastric Epithelial Cells.
Intestinal metaplasia comprises variably differentiated intestinal epithelial cell lineages such as absorptive enterocytes, goblet cells, enteroendocrine cells, and Paneth cells in nonintestinal epithelium (1, 2). Transgenic expression of Cdx1 has been reported to induce all of those epithelial cell lineages in the mouse stomach (9). Consistently, microarray analysis demonstrated that ectopic expression of CDX1 in gastric epithelial cells induces sucrase-isomaltase (SI) and membrane metallo-endopeptidase (MME), which are physiologically expressed in absorptive enterocytes of the small intestine. Vasoactive intestinal peptide (VIP), a gastrointestinal hormone secreted from enteroendocrine cells in the small intestine, was also induced upon CDX1 expression in gastric epithelial cells. RT-qPCR experiments confirmed up-regulation of these intestine-differentiation markers in gastric epithelial cells by CDX1 (Fig. 5C). The expression levels of these intestinal-differentiation markers increased progressively upon sustained CDX1 expression, whereas that of the intestinal-stemness marker reached a peak within 24 h after CDX1 induction (Fig. 5D). MUC2, a goblet cell marker, was not detected within 24 h after CDX1 induction. However, in all gastric epithelial cells examined (MKN28, AGS, and GES-1), a fraction of cells became MUC2+ after prolonged exposure (∼4 d) to CDX1 (Fig. 5E). The Paneth cell and the enteroendocrine cell markers were negative following sustained CDX1 expression in cultured gastric cells (Fig. S4). Thus, CDX1 on its own may support differentiation into absorptive enterocytes and goblet cells in a cell autonomous fashion. Development of other types of intestinal epithelial cells might require non–cell-autonomous signals in addition to CDX1. Also notably, most of these intestinal-differentiation markers were not likely to be directly transactivated by ectopic CDX1 in gastric epithelial cells (Dataset S2), suggesting that they were induced via de novo formation of CDX1-governed transcriptional circuitries that promote intestinal differentiation.
Requirement of SALL4 and KLF5 in CDX1-Mediated Intestinal Transdifferentiation.
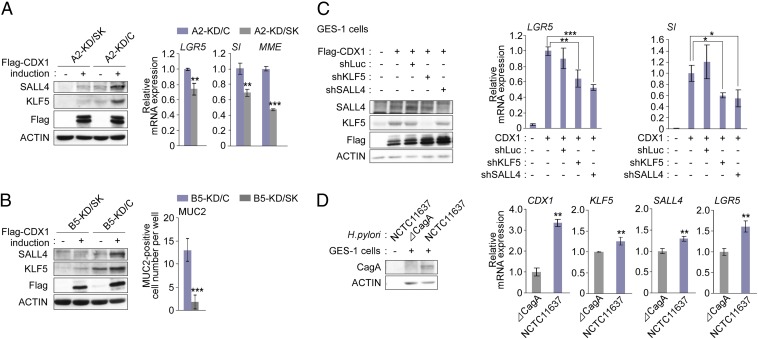
To investigate the role of SALL4 and/or KLF5 induction in the transdifferentiation of gastric epithelial cells by ectopic CDX1, we established two independent CDX1-inducible MKN28 cell lines, A2-KD/SK and B5-KD/SK, in which expression of both SALL4 and KLF5 was suppressed by stable expression of specific shRNA vectors (Fig. 6 A and B, Left). MKN28 cells expressing a luciferase-specific shRNA (A2-KD/C and B5-KD/C) were also established and were used as a control. Increased CDX1 expression or decreased SALL4/KLF5 expression had no relevance to the expression levels of transcription factors such as NF-κB p65/v-rel reticuloendotheliosis viral oncogene homolog A (RelA) and specificity protein 1 (Sp1), which may not be regulated by CDX1, SALL4, or KLF5 (Fig. S5). Induction of the intestinal-stemness marker, LGR5, by ectopic CDX1 was subdued under the condition of suppression of SALL4 and KLF5 (Fig. 6A, Right). Expression of intestinal-differentiation markers, SI and MME, by ectopic CDX1 was much less efficient when the expression of SALL4 and KLF5 was inhibited (Fig. 6A, Right). The number of MUC2+ cells following prolonged exposure to CDX1 was dramatically decreased upon knockdown of SALL4 and KLF5 (Fig. 6B, Right). The degree of induction of intestinal-stemness marker and intestinal-differentiation marker by ectopic CDX1 was also reduced when expression of endogenous SALL4 or KLF5 was transiently inhibited in GES-1 cells (Fig. 6C). Thus, CDX1-mediated induction of SALL4 and KLF5 plays a critical role in the intestinal transdifferentiation of gastric epithelial cells by establishing an intestinal stem/progenitor-like state, from which various intestinal cell types arise.
Fig. 6.
Involvement of SALL4 and KLF5 in CDX1-mediated intestinal transdifferentiation. (A and B) MKN28-derived SALL4/KLF5 double-knockdown (A2-KD/SK or B5-KD/SK) or control-knockdown (A2-KD/C or B5-KD/C) cells were induced to express CDX1 by Dox depletion for 24 h. Cell lysates were subjected to immunoblotting with the respective antibodies (Left). The mRNA levels for intestinal markers were determined by RT-qPCR in CDX1-induced cells (A, Right). The number of MUC2+ cells per well in the eight-well chamber slide was counted in CDX1-induced cells (B, Right). (C) GES-1 cells transiently transfected with a Flag-CDX1 vector together with pSUPER-shLuc, pSUPER-shKLF5/1, or pSUPER-shSALL4/1 were cultured for 24 h. Cell lysates were immunoblotted with the respective antibodies (Left). Levels of the LGR5 and SI mRNAs were determined by RT-qPCR (Right). (D) GES-1 cells infected with H. pylori for 24 h were lysed with 0.1% saponin, which disrupts mammalian cells but not bacterial cells. The lysates were then immunoblotted with the respective antibodies (Left). RNAs isolated from GES-1 cells infected with H. pylori isogenic strains for 96 h were subjected to RT-qPCR analysis for the indicated mRNAs (Right). Error bars, ± SD; n = 3.
We then infected GES-1 cells with a cagA+ or a cagA− H. pylori isogenic strain (Fig. 6D, Left). At 96 h after H. pylori infection, RNAs were isolated from the cells and subjected to RT-qPCR analysis. The results of the experiment revealed that H. pylori infection induced CDX1 in a cagA-dependent manner in GES-1 cells, followed by elevated levels of reprogramming factors, SALL4 and KLF5, and an intestinal stem cell marker, LGR5 (Fig. 6D, Right). These observations provided pathophysiological relevance for the ectopic expression of CDX1 in the induction of intestinal metaplasia in patients infected with H. pylori cagA+ strains.
Discussion
Chronic infection with H. pylori is a major cause of gastric intestinal metaplasia, a precancerous mucosal lesion from which intestinal-type adenocarcinoma arises. CagA, a major virulence factor of H. pylori that is delivered into gastric epithelial cells via type IV secretion, aberrantly activates the Wnt/β-catenin signal and ectopically induces Wnt target genes including CDX1 (8). A causal relationship between ectopic CDX1 and intestinal metaplasia has been provided by the observation that transgenic expression of Cdx1 per se is sufficient to induce intestinal metaplasia in the mouse stomach (9). Persistence of metaplastic changes after removal of triggering agents such as H. pylori indicates that expression of a key inducer of metaplasia must have been maintained in the absence of triggering agents. CDX1 fits this idea in that it establishes an auto-regulatory network to maintain its own expression (22). However, the mechanism by which ectopic CDX1 provokes metaplastic changes has remained poorly understood.
In intestinal epithelial cells, CDX1 acts as a differentiation-promoting factor (6). Assuming that metaplasia requires the conversion of differentiated cells into less-differentiated states, ectopically expressed CDX1 may also activate stemness-regulating reprogramming factors, which allow dedifferentiation of gastric epithelial cells so that they acquire multipotency characteristic of intestinal stem/progenitor-like cells. Consistently, Cdx1 has recently been reported to be a constituent of the transcriptional network that confers pluripotency on ES cells (23). We found that CDX1 directly induces SALL4, a zinc-finger transcription factor playing an important role in maintaining self-renewal and pluripotency (12). Especially, Sall4 positively regulates octamer-binding protein 4 (Oct4), c-Myc, SRY-box containing gene 2 (Sox2), and Klf4, the four defined transcription factors capable of generating iPS cells (13). CDX1 also transactivates KLF5, a gene encoding a member of the KLF family of transcription factors. Importance of KLFs in the acquisition of pluripotency has been highlighted by recent studies showing that depletion of Klf2, Klf4, and Klf5 in mouse ES cells abolishes self-renewal (24). Furthermore, Klf5 can replace Klf4 in generating iPS cells, indicating a redundant role between KLF4 and KLF5 in stemness induction/maintenance (15). In adult tissues, Sall4 and Klf5 are expressed in hepatic stem cells and hair follicle stem cells, respectively, to maintain tissue stemness (25, 26). Identification of the reprogramming factors SALL4 and KLF5 as direct transcriptional targets of CDX1 therefore provides a mechanistic basis underlying intestinal metaplasia of the stomach. On the one hand, CDX1 activates reprogramming transcription factors in gastric epithelial cells and thereby rewires transcriptional circuitries so as to redirect gastric epithelial cells toward a less-differentiated intestinal stem/progenitor-like state. On the other hand, CDX1 creates transcriptional circuitries that direct transdifferentiation of dedifferentiated cells into intestinal epithelial cells. Cancer stem cells have recently been reported to possess properties that are shared in common with tissue stem/progenitor cells (27). The observation indicates that acquisition of stemness traits is linked to cell transformation and suggests that a transition through intestinal stem/progenitor-like states via dedifferentiation predisposes cells to undergo neoplastic changes. This may explain the clinical observation that intestinal metaplasia is a precancerous lesion of the stomach (3).
In intestinal epithelial cells, the two homologous CDX1 and CDX2 (Cdx1 and Cdx2 in mice) transactivate a number of intestine-specific genes (6). Like Cdx1, transgenic expression of Cdx2 in the mouse stomach also causes intestinal metaplasia (10), indicating a redundant role of Cdx1 and Cdx2 in the pathogenic change. Interestingly, genomic binding sites for CDX2 include 5′-flanking regions of SALL4 and KLF5 (28). It is therefore possible that CDX2 provokes intestinal transdifferentiation via direct activation of SALL4 and KLF5, like CDX1. Also of note, Cdx2 is capable of inducing Cdx1 mRNA in the mouse stomach (29). Hence, CDX2-mediated intestinal metaplasia might be at least partly due to CDX2-induced CDX1. Whereas CDX-induced intestinal metaplasia is thought to be a precancerous stomach lesion, the potential role of CDX1 or CDX2 in intestinal carcinogenesis remains unclear. Cdx1 and Cdx2 were originally described as an oncoprotein and tumor suppressor, respectively, in intestinal cells (30, 31). However, recent studies suggested that overexpression of Cdx1 has very limited contribution, if any, to the development of intestinal tumors (32, 33). Hence, CDX1 could promote oncogenesis only when it is ectopic expressed in nonintestinal epithelial cells.
The present work provided evidence that ectopic expression of CDX1 and subsequent induction of stemness-associated reprogramming factors SALL4 and KLF5 by CDX1 are key events underlying gastric intestinal metaplasia. As far as we know, this is a unique demonstration of the involvement of reprogramming factors in metaplastic changes. The finding also gives mechanistic insights into intestinal transdifferentiation; CDX1-directed rewiring of transcriptional circuitries through induction of reprogramming factors converts differentiated gastric epithelial cells to immature intestinal stem/progenitor-like cells, which can transdifferentiate into intestinal cells. Reactivation of such reprogramming factors may broadly contribute to the plasticity of the lineage commitment in both physiological and pathological conditions. Our work therefore provides deeper insights into organogenesis and oncogenesis that can be applied to help the progress of regenerative medicine as well as cancer prevention and treatment.
Materials and Methods
The experiments using human materials were approved by the Research Ethics Committee of the Graduate School of Medicine, The University of Tokyo, and the Ethics Committee of Keio University School of Medicine. Informed consent was obtained from all patients. The experiments using animals were approved by the Committee of Experimental Animal Ethics of Jichi Medical University. Cdx1-transgenic mice have been described previously (9). Luciferase reporter assay, qPCR analysis, and cell-counting assay were evaluated using Student’s t test. P < 0.05 was considered to be statistically significant. For all statistical comparisons in these assays, P < 0.001 was denoted as ***, P < 0.01 as **, and P < 0.05 as *. The Gene Expression Omnibus accession number for expression microarray and ChIP-chip analyses in this study is GSE35369. Details of materials and methods are described in SI Materials and Methods. Primers used in this study are shown in Table S2.
Supplementary Material
Acknowledgments
We thank J. Iovanna, I. Manabe, and R. Nagai for providing plasmids and N. Kamimura, H. Meguro, and D. Sasaya for technical assistance. This work was supported by Grants-in-Aid for the Scientific Research in an Innovative Area from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Footnotes
The authors declare no conflict of interest.
†This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE35369).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208651109/-/DCSupplemental.
See Commentary on page 20173.
References
- 1.Gutiérrez-González L, Wright NA. Biology of intestinal metaplasia in 2008: More than a simple phenotypic alteration. Dig Liver Dis. 2008;40(7):510–522. doi: 10.1016/j.dld.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 2.Reid BJ, Li X, Galipeau PC, Vaughan TL. Barrett’s oesophagus and oesophageal adenocarcinoma: Time for a new synthesis. Nat Rev Cancer. 2010;10(2):87–101. doi: 10.1038/nrc2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Correa P. A human model of gastric carcinogenesis. Cancer Res. 1988;48(13):3554–3560. [PubMed] [Google Scholar]
- 4.Dearolf CR, Topol J, Parker CS. The caudal gene product is a direct activator of fushi tarazu transcription during Drosophila embryogenesis. Nature. 1989;341(6240):340–343. doi: 10.1038/341340a0. [DOI] [PubMed] [Google Scholar]
- 5.Margalit Y, Yarus S, Shapira E, Gruenbaum Y, Fainsod A. Isolation and characterization of target sequences of the chicken CdxA homeobox gene. Nucleic Acids Res. 1993;21(21):4915–4922. doi: 10.1093/nar/21.21.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo RJ, Suh ER, Lynch JP. The role of Cdx proteins in intestinal development and cancer. Cancer Biol Ther. 2004;3(7):593–601. doi: 10.4161/cbt.3.7.913. [DOI] [PubMed] [Google Scholar]
- 7.Prinos P, et al. Multiple pathways governing Cdx1 expression during murine development. Dev Biol. 2001;239(2):257–269. doi: 10.1006/dbio.2001.0446. [DOI] [PubMed] [Google Scholar]
- 8.Murata-Kamiya N, et al. Helicobacter pylori CagA interacts with E-cadherin and deregulates the β-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene. 2007;26(32):4617–4626. doi: 10.1038/sj.onc.1210251. [DOI] [PubMed] [Google Scholar]
- 9.Mutoh H, et al. Cdx1 induced intestinal metaplasia in the transgenic mouse stomach: Comparative study with Cdx2 transgenic mice. Gut. 2004;53(10):1416–1423. doi: 10.1136/gut.2003.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silberg DG, et al. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology. 2002;122(3):689–696. doi: 10.1053/gast.2002.31902. [DOI] [PubMed] [Google Scholar]
- 11.Lapidot M, Mizrahi-Man O, Pilpel Y. Functional characterization of variations on regulatory motifs. PLoS Genet. 2008;4(3):e1000018. doi: 10.1371/journal.pgen.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8(10):1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, et al. Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells. Proc Natl Acad Sci USA. 2008;105(50):19756–19761. doi: 10.1073/pnas.0809321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsubooka N, et al. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells. 2009;14(6):683–694. doi: 10.1111/j.1365-2443.2009.01301.x. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 16.Conkright MD, Wani MA, Anderson KP, Lingrel JB. A gene encoding an intestinal-enriched member of the Krüppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res. 1999;27(5):1263–1270. doi: 10.1093/nar/27.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20(9):1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 18.Qiao XT, et al. Prospective identification of a multilineage progenitor in murine stomach epithelium. Gastroenterology. 2007;133(6):1989–1998. doi: 10.1053/j.gastro.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao X, Sedgwick T, Shi YB, Evans T. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol Cell Biol. 1998;18(5):2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonoyama K, Rutatip S, Kasai T. Gene expression of activin, activin receptors, and follistatin in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2000;278(1):G89–G97. doi: 10.1152/ajpgi.2000.278.1.G89. [DOI] [PubMed] [Google Scholar]
- 21.Lin SA, Barker N. Gastrointestinal stem cells in self-renewal and cancer. J Gastroenterol. 2011;46(9):1039–1055. doi: 10.1007/s00535-011-0424-8. [DOI] [PubMed] [Google Scholar]
- 22.Béland M, et al. Cdx1 autoregulation is governed by a novel Cdx1-LEF1 transcription complex. Mol Cell Biol. 2004;24(11):5028–5038. doi: 10.1128/MCB.24.11.5028-5038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10(3):353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 25.Oikawa T, et al. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology. 2009;136(3):1000–1011. doi: 10.1053/j.gastro.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 26.Morris RJ, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22(4):411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 27.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verzi MP, et al. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc Natl Acad Sci USA. 2010;107(34):15157–15162. doi: 10.1073/pnas.1003822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mutoh H, Hayakawa H, Sakamoto H, Sashikawa M, Sugano K. Transgenic Cdx2 induces endogenous Cdx1 in intestinal metaplasia of Cdx2-transgenic mouse stomach. FEBS J. 2009;276(20):5821–5831. doi: 10.1111/j.1742-4658.2009.07263.x. [DOI] [PubMed] [Google Scholar]
- 30.Chawengsaksophak K, James R, Hammond VE, Köntgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386(6620):84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- 31.Soubeyran P, et al. Homeobox gene Cdx1 regulates Ras, Rho and PI3 kinase pathways leading to transformation and tumorigenesis of intestinal epithelial cells. Oncogene. 2001;20(31):4180–4187. doi: 10.1038/sj.onc.1204551. [DOI] [PubMed] [Google Scholar]
- 32.Bonhomme C, et al. Cdx1, a dispensable homeobox gene for gut development with limited effect in intestinal cancer. Oncogene. 2008;27(32):4497–4502. doi: 10.1038/onc.2008.78. [DOI] [PubMed] [Google Scholar]
- 33.Crissey MA, et al. The homeodomain transcription factor Cdx1 does not behave as an oncogene in normal mouse intestine. Neoplasia. 2008;10(1):8–19. doi: 10.1593/neo.07703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.