Abstract
Microbial communities present in the Gulf of Mexico rapidly responded to the Deepwater Horizon oil spill. In deep water plumes, these communities were initially dominated by members of Oceanospirillales, Colwellia, and Cycloclasticus. None of these groups were abundant in surface oil slick samples, and Colwellia was much more abundant in oil-degrading enrichment cultures incubated at 4 °C than at room temperature, suggesting that the colder temperatures at plume depth favored the development of these communities. These groups decreased in abundance after the well was capped in July, but the addition of hydrocarbons in laboratory incubations of deep waters from the Gulf of Mexico stimulated Colwellia's growth. Colwellia was the primary organism that incorporated 13C from ethane and propane in stable isotope probing experiments, and given its abundance in environmental samples at the time that ethane and propane oxidation rates were high, it is likely that Colwellia was active in ethane and propane oxidation in situ. Colwellia also incorporated 13C benzene, and Colwellia's abundance in crude oil enrichments without natural gas suggests that it has the ability to consume a wide range of hydrocarbon compounds or their degradation products. However, the fact that ethane and propane alone were capable of stimulating the growth of Colwellia, and to a lesser extent, Oceanospirillales, suggests that high natural gas content of this spill may have provided an advantage to these organisms.
Keywords: methane, biodegradation, marine bacteria, archaea, alkane
The explosion and sinking of the Deepwater Horizon in April 2010 caused one of the world's largest marine oil spills, releasing at least 4 million barrels of oil before the well was capped in July 2010 (1). In addition to its size and duration, the Deepwater Horizon spill was different from previous oil spills for two major reasons, both of which are likely to influence the response of the microbial community. First, the spill released not just oil, but large amounts of natural gas (1, 2). The bacteria that typically consume methane, the primary component of natural gas, have limited abilities to consume multicarbon substrates (3), whereas many oil degraders are capable of growth only on larger hydrocarbons (4), and none are known to consume methane (5). The organisms that consume ethane and propane, the other major components of natural gas, remain largely unknown in marine environments, but may include organisms related to methanotrophs or oil-degraders (6). Second, because of the depth of the spill, nearly all of the gas (2, 7, 8) and some components of the oil (9) remained in the deep ocean, forming plumes of dissolved or dispersed hydrocarbons between 1,000 and 1,200 m (10–12). The temperature at these depths was just 4 °C to 6 °C, much colder than surface water. Temperature has a direct effect on microbial physiology and an effect on the physical properties of oil that influence its bioavailability; both factors will influence the response of hydrocarbon-degrading bacteria (13–15).
Crude oil is a complex mixture containing thousands of different hydrocarbon compounds. These compounds differ in solubility and volatility and are degraded at different rates; in general, the smaller, less substituted hydrocarbons are degraded faster than larger ones with more substituted groups (15, 16). Although the ability to degrade hydrocarbons is found in many types of bacteria, the most abundant oil-degraders in marine environments are typically Gammaproteobacteria, particularly organisms such as Alcanivorax, which primarily degrades alkanes, or Cycloclasticus, which specializes in the degradation of aromatic compounds (16). However, most studies of microbial community response to hydrocarbons have been conducted in oil-amended mesocosm experiments with sediment, beach sand, or surface water (16), and little is known about the response to oil inputs in the deep ocean or the impact of natural gas on these communities. An initial report after the Deepwater Horizon spill showed that, in late May, an uncultivated group of Oceanospirillales were dominant in plume samples; 16 other groups of Gammaproteobacteria were also enriched in plume vs. out-of-plume samples (17). In June, plume samples were dominated by two different groups of Gammaproteobacteria, Colwellia and Cycloclasticus (2). The well was capped in mid-July, and by September, these groups were much less abundant and plumes were dominated by methylotrophs (Methylococcaceae, Methylophaga, and Methylophilaceae), Flavobacteria, and Rhodobacterales (18). Although the abundance of these taxa strongly suggests they played a role in hydrocarbon degradation, it is not possible to directly link any individual taxa to the degradation of a particular type of hydrocarbon with only the environmental sequence data. Here we present additional data on microbial communities in environmental samples taken over the course of the spill response, as well as the results of stable isotope probing (SIP) experiments linking Colwellia to the oxidation of ethane, propane, and benzene.
Results and Discussion
16S rRNA Clone Libraries from Environmental Samples.
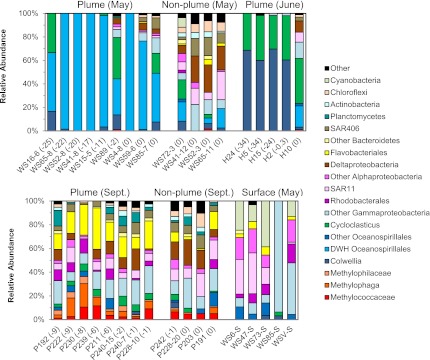
We collected samples on three cruises, referred to as “May” (May 26 to June 5, 2010), “June” (June 11–21, 2010), and “September” (September 7–17, 2010). We sequenced bacterial 16S rRNA genes from 29 deep samples and five surface samples; data from the five June samples and seven of the 12 September samples were previously published (2, 18), but we have since performed additional sequencing on some of these samples, and results from the full set of clone libraries are shown in Fig. 1. Deep water samples (below 800 m) were classified as “plume” or “nonplume” based on the presence or absence of a fluorescence anomaly indicative of aromatic hydrocarbons (11, 18–20). Nonplume samples were taken at plume depth within 5 km of a plume sample or at a plume site, 200 m above or below peak plume depth. Based on their proximity to plume samples, it is likely that these sites were influenced by low concentrations of hydrocarbons not detected fluorometrically and therefore should not be considered true control sites, but as comparison sites with lower exposure to hydrocarbons. Some plume samples also showed decreases in dissolved oxygen concentration indicative of recent respiration pulses. Oxygen anomalies, the difference between measured concentrations and background values (18), expressed in μmol/L, are shown in parentheses after the sample names in Fig. 1.
Fig. 1.
Relative abundances in 16S rRNA gene clone libraries. Plume samples from were taken from 1,000 to 1,300 m at sites with a fluorescence anomaly at that depth. Nonplume samples were from sites with no detectable fluorescence anomaly or from depths above or below the fluorescence anomaly (800–1,300 m). Oxygen anomalies (μmol/L) are shown in parentheses after the sample numbers. The number of sequences in each clone library is shown in Table S1 (average of 67 sequences per sample).
In all nine plume samples from the May cruise, the group of uncultivated Oceanospirillales observed by Hazen et al. (17) (subsequently referred to as DWH Oceanospirillales) accounted for more than 30% of sequences (Fig. 1). DWH Oceanospirillales accounted for more than 95% of sequences in five of those samples, including one sample with no detectable oxygen depletion (WS4-8). The remaining sequences from the May samples with oxygen anomalies more than 2 μmol/L were affiliated with the genus Cycloclasticus or the genus Colwellia. In the plume samples with oxygen anomalies no greater than 2 μmol/L, the remaining sequences were Cycloclasticus, Colwellia, or a mix of other Gammaproteobacteria, Deltaproteobacteria, Alphaproteobacteria, Chloroflexi, and the SAR406 clade similar to those observed in the nonplume samples. Most sequences from the nonplume samples appear to be “typical” deep sea bacteria, nearly all of them closely related to sequences from the deep Arctic Ocean (21). However, three of the four nonplume samples also contained DWH Oceanospirillales, Cycloclasticus, or Colwellia, accounting for 13% to 42% of sequences. Without widespread sampling before the spill, it is difficult to establish true background values for these groups, but given that no sequences closely related to the DWH Oceanospirillales were found in GenBank before the Deepwater Horizon spill, it is highly unlikely that these bacteria are regularly that abundant. Rather, these bacteria were likely responding to low concentrations of hydrocarbons in these samples, or result from extensive mixing of affected waters.
In contrast to the May samples, DWH Oceanospirillales were not detected in four of the five plume samples from June. Instead, Cycloclasticus and Colwellia accounted for more than 95% of sequences. The one sample (H10) in which DWH Oceanospirillales was detected was the closest sample to the wellhead and the one with no detectable oxygen anomaly, suggesting it had experienced the least biodegradation. Based on the observed progression in these samples and the greater abundance of the DWH Oceanospirillales in May, we suggest that the DWH Oceanospirillales bloomed first, followed by Colwellia and Cycloclasticus, although we lack definitive evidence because of the difficulty determining time of exposure. Fluctuations in ocean currents caused water near the wellhead to move away and then return, so the distance from the wellhead does not necessarily correlate with the amount of time that water had been exposed to hydrocarbons (20, 22). The oxygen anomaly is a better indication of the amount of hydrocarbon degradation that has occurred in a given water mass, but it should be noted that oxygen concentrations never reached hypoxic levels, suggesting that, to some extent, mixing was able to replenish oxygen in plume samples (20). Microbial community composition appears to be a much more sensitive indicator of hydrocarbon exposure than oxygen depletion, given the abundance of these putative hydrocarbon degraders in both May and June samples with no detectable oxygen anomaly.
By September, oxygen anomalies could be traced several hundred kilometers to the southwest, and methane, ethane, and propane were lower than background concentrations for the Gulf of Mexico (18). DWH Oceanospirillales and Colwellia were no longer detectable in 11 of the 12 samples, but methanotrophs, other methylotrophs, Flavobacteria (mostly affiliated with the genera Polaribacter or Owenweeksia), and the Alphaproteobacteria order Rhodobacterales were much more abundant than they had been in plume or nonplume samples earlier in the summer. Flavobacteria in particular were more abundant in plume samples than nonplume samples (10–35% of sequences vs. 1–15%). Flavobacteria are abundant in the ocean and are often associated with the degradation of high molecular weight dissolved organic carbon compounds (23), but several methanol-oxidizing strains of Flavobacterium have been isolated (24) and they have often been observed in 13C-labeled DNA from methane SIP studies (25). The Flavobacteria may therefore have been secondary consumers of methane, oil, or cellular decay products. The abundance of Flavobacteria may have been greater than indicated by the clone libraries, as has been observed when measured by techniques (e.g., FISH) that do not require PCR and cloning (26–28). Terminal restriction fragment length polymorphism (T-RFLP) analysis of these samples, which used the same PCR primers, but did not require cloning, showed the Flavobacteria accounted for as much as 56% of total peak area (Fig. S1A). Although the relative abundances of all other groups of bacteria corresponded well between clone libraries and T-RFLP, the Flavobacteria were somewhat underrepresented in clone libraries (Fig. S2), possibly because of biases associated with cloning (29).
We also sequenced archaeal 16S rRNA genes from six deep water samples: plume/nonplume from May (WS41-8/WS41-12), low oxygen anomaly plume/high oxygen anomaly plume from June (H10/H24), and plume/nonplume from September (P203/P230). These samples contained different bacterial communities (WS41-12 and P203 showed relatively little hydrocarbon influence), but the archaeal communities in all six samples were very similar: more than 70% of sequences in each clone library were from one OTU affiliated with the Marine Group II Euryarchaeota (30), with the remainder related to other marine Euryarchaeota or Thaumarchaeota (Fig. S3). The dominance of this OTU in all samples suggests that the presence of hydrocarbons did not have a large impact, although deeper sequencing of these samples could possibly show an impact on less abundant members of the community. Archaea have not been shown to play a significant role in hydrocarbon degradation in aerobic marine environments, and the effect of crude oil on archaea is not well understood. Some evidence suggests that the presence of oil has a strong negative influence on archaea, whereas other studies see little effect (31). That archaea do not bloom in response to a sudden input of oil is consistent with their hypothesized ecology (32).
In addition to deep water plume samples, we analyzed water samples from five sites with surface oiling. The amount of oil at these sites varied: three sites (WS6-S, WS47-S, and WS73-S) had an oil sheen, whereas the other two contained a thicker coating of oil. The three sheen samples were dominated by Cyanobacteria and Alphaproteobacteria (SAR11 clade, Rhodobacterales, and Rhodospirillales), with just 15% of sequences affiliated with possible hydrocarbon degraders from the Alteromonadales and Oceanospirillales (Fig. 1). Gammaproteobacteria were more abundant in the more heavily oiled samples, accounting for 100% of sequences in WS85-S (93% Pseudoalteromonas) and 48% of sequences in WSV-S (Pseudomonas, Vibrio, Acinetobacter, and Alteromonas). No DWH Oceanospirillales were detected in these samples, and Colwellia and Cycloclasticus accounted for less than 5% of sequences in each sample.
Role of Temperature.
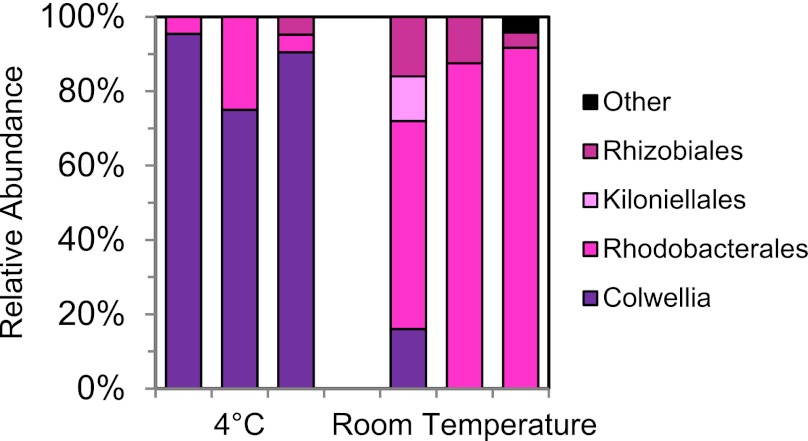
The difference between surface slick and deep water plume communities suggests that temperature may have played a significant role in determining which members of the microbial community responded to the Deepwater Horizon spill. Most cultivated strains of Colwellia are psychrophiles (33), and one of the cultivated organisms most closely related to the DWH Oceanospirillales, Oleispira antarctica, is also psychrophilic (34). Cycloclasticus has been shown to be abundant in oil-degrading microcosms at both 4 °C and 20 °C (14). Although no cultivated strain of Colwellia has been shown to oxidize hydrocarbons and it is not commonly observed in oil degradation studies, several studies have detected Colwellia in oil-contaminated ice cores or sediments incubated at low temperatures (35, 36). To determine the impact of temperature on oil-degrading communities, we added crude oil to seawater collected from a deep water sample on the September cruise and incubated in triplicate at 4 °C and room temperature (∼20 °C) for 10 d. In 16S rRNA gene clone libraries from these samples, Colwellia accounted for 87% of total clones when incubated at 4 °C, but only 5% at room temperature (Fig. 2). Alphaproteobacteria, predominantly Rhodobacteraceae closely related to those abundant in plume samples from September (18), accounted for 93% of clones at room temperature, but 13% at 4 °C. Neither Cycloclasticus nor DWH Oceanospirillales were detected at either temperature.
Fig. 2.
Relative abundances of 16S rRNA genes in clone libraries from crude oil enrichment cultures incubated at 4 °C and room temperature (∼20 °C). n = 20–24 sequences per sample, three samples for each temperature.
SIP.
To identify the bacteria consuming specific hydrocarbon compounds abundant in the deep plume, we used SIP with 13C-labeled methane, ethane, propane, or benzene. Seawater from site P222 from the September cruise was incubated for 10 d at 6 °C, and the 13C-labeled substrates were converted to 13C-labeled dissolved inorganic carbon (DIC; Fig. 3A). 13C-labeled (i.e., “heavy”) and unlabeled (i.e., “light”) DNA were then separated by CsCl density gradient centrifugation. 16S rRNA gene clone libraries from the heavy and light DNA and T-RFLP analysis of the unfractionated DNA and selected gradient fractions showed that the addition of these substrates led to changes in the microbial community (Fig. 3B and Fig. S1B). Colwellia sequences were not detected in the initial sample and were detected in only one of the 11 other September samples, but were the most abundant group in the heavy DNA from the ethane, propane, and benzene incubations (45–72% of sequences) and were also abundant in the unlabeled DNA from all four incubations. The DWH Oceanospirillales were also not detected in any of the September samples, but accounted for 3% to 13% of sequences in the heavy and light DNA from the ethane and propane incubations and light DNA from the methane incubation. DWH Oceanospirillales were not detected in the heavy or light DNA from the benzene incubations, whereas Actinobacteria accounted for 15% of sequences in the benzene heavy DNA clone libraries, but were not detected in any of the others. Heavy benzene fractions and light ethane and propane fractions contained a number of other Oceanospirillales sequences affiliated with the genus Neptunomonas, closely related to the aromatic hydrocarbon degrader Neptunomonas naphthovorans (37) and symbionts of bone-eating Osedax species (38).
Fig. 3.
(A) Conversion of 13C methane, ethane, propane, and benzene to [13C]DIC during SIP incubations of seawater at 6 °C. (B) Relative abundances in 16S rRNA gene clone libraries from heavy (i.e., 13C-labeled) and light (i.e., unlabeled) DNA from SIP incubations with 13C methane, ethane, propane, and benzene, and the initial seawater (sample P222). n = 75 (initial), n = 25 (heavy methane), n = 69 (light methane), n = 44 (heavy methane), n = 81 (light methane), n = 43 (heavy methane), n = 78 (light methane), n = 84 (heavy methane), and n = 76 (light benzene).
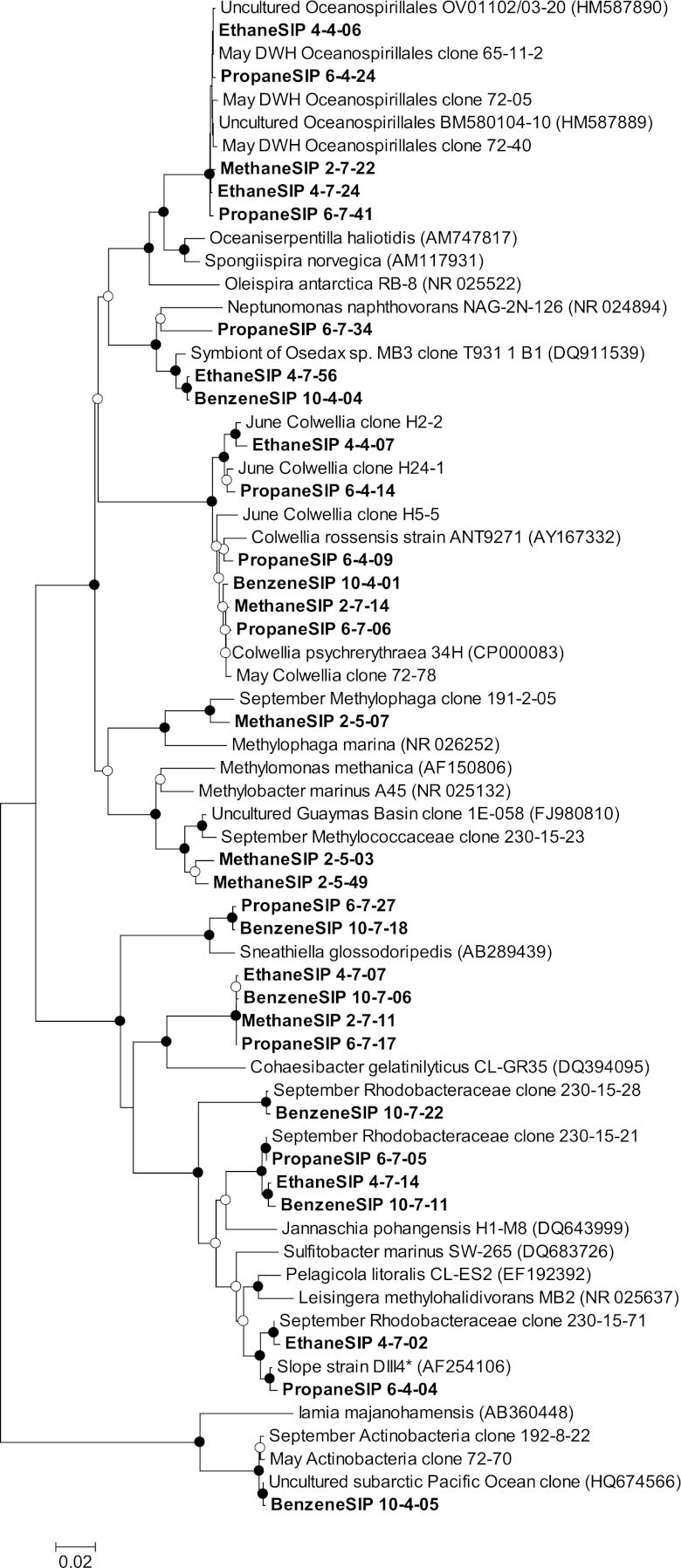
Despite the abundance of methylotrophs in the initial seawater (23% of sequences), methane was converted to [13C]DIC much more slowly than the other substrates (Fig. 3A). This is consistent with the low methane oxidation rates measured in September and supports the hypothesis that the methylotrophs in the September samples were metabolically inactive remnants of a bloom that had peaked earlier in the summer (18). The majority of methylotrophs in the initial sample were affiliated with Methylophaga or Methylophilaceae, which are likely secondary consumers of C1 compounds after Methylococcaceae oxidize methane to methanol. These processes appear to be tightly coupled, but presumably require Methylococcaceae to respond initially (6, 39). Methylococcaceae accounted for only 3% of sequences in the initial sample, and they appear to bloom more slowly in response to natural gas inputs than ethane and propane oxidizers, just as observed earlier in the summer (2). The low DNA concentrations in the heaviest fractions necessitated the use of DNA from a slightly less dense fraction for the heavy DNA clone library, but 84% of sequences in this fraction were affiliated with the Methylococcaceae or Methylophaga, compared with just 7% in the light fraction. These sequences, along with Colwellia, DWH Oceanospirillales, and Actinobacteria, were closely related to the sequences observed in the environmental samples (Fig. 4).
Fig. 4.
Neighbor-joining tree showing phylogenetic relationships between bacterial 16S rRNA gene sequences from the major groups of Gammaproteobacteria, Alphaproteobacteria, and Actinobacteria in SIP samples (bold), environmental samples from May, June, or September, and reference sequences (GenBank accession numbers in parentheses). Filled circles indicate nodes with bootstrap values greater than 90% (1,000 replicates); open circles indicate bootstrap values greater than 50%.
Although SIP experiments typically try to mimic in situ conditions and use low concentrations of substrate to avoid microbial community changes over the course of the incubation, our goal was to replicate the bloom conditions caused by hydrocarbon inputs following the well blowout. Colwellia was able to respond rapidly, becoming abundant in heavy and light DNA. To become so abundant in the heavy DNA, Colwellia had to consume 13C-labeled ethane, propane, or benzene, and then use that 13C in the production of new DNA, a process which is typically associated with cell division (39). The Colwellia sequences in the unlabeled DNA were from organisms that were either consuming the 13C substrate but had not yet synthesized a sufficient amount of DNA with that labeled carbon or from organisms consuming an alternate carbon source. Given that the cultivated strains of Colwellia are capable of degrading many different carbon sources (33, 40), either explanation is possible. It is also important to note that we cannot exclude the possibility of cross-feeding (25, 39), whereby one organism is responsible for the initial oxidation step (e.g., ethane to ethanol) and a different organism then consumes this 13C-labeled intermediate (e.g., ethanol) and incorporates it into 13C biomass. However, given the dominance of Colwellia in the SIP samples and the high rates of ethane and propane oxidation in June, when Colwellia sequences accounted for approximately 70% of sequences in plume samples (2), it is likely that Colwellia was responsible for the bulk of ethane and propane oxidation in situ, although the SIP results show the DWH Oceanospirillales may also have played a role.
Conclusions
The microbial community response to the Deepwater Horizon oil spill was distinct from that observed in previous spills or mesocosm studies. Deep water plume communities were dominated by just three groups of Gammaproteobacteria in May and June, none of which were abundant in surface oil samples. Two of these groups, the DWH Oceanospirillales and Colwellia, are related to known psychrophiles, and Colwellia was much more abundant in crude oil enrichments at 4 °C than at room temperature, suggesting that the temperature played a significant role. The abundance of natural gas in the Deepwater Horizon spill seemingly provided an advantage to Colwellia and the DWH Oceanospirillales, as both these groups became more abundant with the addition of ethane or propane alone. Colwellia was likely responsible for the majority of ethane and propane oxidation, but may also have been involved with the degradation of higher molecular weight hydrocarbons.
Methods
Study Site and Sample Collection.
Samples were collected on three cruises: R/V Walton Smith (May 26 to June 5, 2010), R/V Cape Hatteras (June 11–21, 2010), and National Oceanic and Atmospheric Administration ship Pisces (September 7–17, 2010). Sample locations are shown on the map in Fig. S4 and listed with additional details in Table S1. Dissolved oxygen concentrations were measured with the SBE-43 dissolved oxygen sensors on the ships’ conductivity/temperature/depth (CTD) rosettes and on select samples by Winkler titration, as described previously (2). A 10th-degree polynomial was fit to the dissolved oxygen-depth profiles to determine a background oxygen concentration, as described previously (18). Oxygen anomalies were calculated as the difference between the measured dissolved oxygen concentration and the background concentration for a given depth. For the R/V Walton Smith cruise, oxygen anomalies were calculated with the publically available CTD profile data (41). Fluorescence was measured with a WETLabs ECO FL CDOM fluorometer (excitation of 370 nm, emission of 460 nm) on the R/V Walton Smith and R/V Cape Hatteras and with a Chelsea Technologies UV AquaTracka (excitation of 239 nm, emission of 360 nm) on the Pisces. Deep water samples were collected in Niskin bottles on the ships’ CTD rosettes. One liter of seawater was filtered onto a 0.2-μm Sterivex filter (Millipore) which was stored frozen until extraction. Surface samples were collected only on the May cruise, by using a plastic bucket. An oil/water mixture was collected from the surface of the bucket sample into a 125-mL glass jar and stored frozen for several months. Samples were then thawed and filtered onto 0.2-μm Sterivex filters (Millipore); DNA was extracted from the filters as described later.
16S rRNA Clone Libraries.
DNA was extracted from filters with the FastDNA SPIN kit for soil (MP Biomedicals). Bacterial sequences were amplified with the primers 27F and 1392R, as previously described (6), and archaeal sequences were amplified with 21F and 958R (30). For each sample, duplicate PCR reactions were performed, then pooled and cleaned with the Wizard SV Gel and PCR Clean-Up kit (Promega). PCR products were cloned with the PCR Cloning Kit (Qiagen), and randomly selected clones were selected for sequencing at the University of California, Berkeley, DNA Sequencing Facility. Sequences were edited and assembled with Sequencher (Gene Codes) and screened for chimeras with Bellerophon (42) and Pintail (43). Additional chimeras were detected by comparison with closely related sequences via BLAST searches (44). In plume samples with two or three dominant sequences, as many as one third of sequences were found to be chimeric; all suspected chimeras were omitted from subsequent analysis. Sequences were assigned to operational taxonomic units (OTUs) with MOTHUR (45), and each sequence was also identified using the RDP Classifier tool (46). For bacteria, OTU clustering at the 3% difference threshold corresponded with the RDP taxonomy at the family or genus level, with the exception of the SAR 406 clade. Representative members of relevant OTUs were aligned with ClustalW, and MEGA4 (47) was used to construct neighbor-joining phylogenetic trees (maximum composite likelihood method, 1,000 bootstrap replicates). At least one sequence from each OTU was submitted to GenBank under the accession numbers JN018421 to JN018515 (SIP incubations), JN018516 to JN018646 (archaea), JN018647 to JN018743 (surface samples), and JN018744 to JN019023 (deep water samples).
Enrichment Cultures.
Seawater was collected on the September cruise and stored at 4 °C for 3 mo before starting enrichment cultures. Crude oil (0.5 mL) was added to 100 mL seawater and 50 mL artificial seawater in a 250-mL sterile glass serum bottle, sealed with a Teflon-coated butyl rubber stopper and an aluminum crimp seal. The artificial seawater contained, per liter, 20 g NaCl, 3 g MgCl2·6 H2O, 4 g NaSO4, 0.15 g CaCl2·2 H2O, 0.5 g KCl, 0.2 g NaHCO3, 0.85 g NaNO3, and 20 mL 0.5 M phosphate buffer (pH 7.4). Three bottles were incubated at 4 °C and three at room temperature (∼20 °C) for 10 d. Each bottle was filtered onto a 0.2-μm Sterivex filter (Millipore); DNA was extracted and clone libraries were constructed as described earlier.
SIP.
Immediately after collection, seawater from sample P222 was incubated with 99+% 13C methane, ethane, propane, or benzene (Isotec/Sigma Aldrich), as well as 12C (natural abundance of 13C, ∼1.1%) controls for methane, ethane, and propane (Airgas). For each incubation, 200 mL seawater was added to a 250-mL sterile glass serum bottle, sealed with a Teflon-coated butyl rubber stopper and an aluminum crimp seal. Five milliliters of methane, ethane, or propane, or 70 μmol benzene was added and samples were incubated at 6 °C for 10 d. Ten milliliters of water was removed for [δ13C]DIC analysis, and the remainder was filtered onto a 0.2-μm Sterivex filter (Millipore) and stored frozen until DNA extraction. Replicate bottles for each substrate were also harvested at 2 d and 3.5 d for [δ13C]DIC measurements; DNA from these samples was not analyzed as a result of insufficient 13C uptake. Analysis of [δ13C]DIC was performed as described previously (2), and DNA extraction as described earlier. 12C and 13C DNA were separated by CsCl density gradient centrifugation as described previously (6) and separated into 10 fractions. 16S rRNA gene clone libraries were constructed from DNA from one heavy fraction (1.74 g mL−1 for ethane, propane, and benzene; 1.72 g mL−1 for methane) and one light fraction (1.69 g mL−1) from each 13C substrate. There was insufficient DNA for analysis in the heavy fractions from the 12C controls. Cloning and sequence analysis were performed as described earlier.
T-RFLP.
T-RFLP analysis was performed on selected samples from the SIP experiment and the September cruise. DNA was amplified as described earlier, but with a 6-carboxyfluorescein–labeled forward primer. Cleaned PCR product (100 ng) was digested with MspI, and samples were analyzed with T-REX (48), as described previously (6). Terminal restriction fragments (T-RFs) were identified by in silico digestion of all clone library sequences from each batch of samples (SIP and September samples were analyzed separately). Some T-RFs could be assigned to multiple groups (there was significant overlap between the methanotrophs, methylotrophs, and some other Gammaproteobacteria, and it was not possible to distinguish the Deltaproteobacteria, Actinobacteria, and SAR406 clade from each other, but the Flavobacteria, Alphaproteobacteria, and Gammaproteobacteria were readily differentiated.
Supplementary Material
Acknowledgments
We thank the crews and scientific parties of the R/V Walton Smith, R/V Cape Hatteras, and National Oceanic and Atmospheric Administration (NOAA) ship Pisces, particularly chief scientists Samantha Joye and John Kessler. We also thank Stephanie Mendes for [δ13C]DIC measurements, Mengran Du for oxygen anomaly calculations, and Monica Heintz and Chris Farwell for assistance with sampling. This work was supported by National Science Foundation Awards OCE1042097and OCE 0961725 and US Department of Energy Award DE-NT0005667 (to D.L.V.) and by NOAA through a contract with Consolidated Safety Services.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. JN018421–JN018515 (SIP incubations), JN018516–JN018646 (archaea), JN018647–JN018743 (surface samples), and JN018744–JN019023 (deep water samples)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108756108/-/DCSupplemental.
References
- 1.McNutt M, et al. Assessment of Flow Rate Estimates for the Deepwater Horizon / Macondo Well Oil Spill. Flow Rate Technical Group Report to the National Incident Command. Washington, DC: Department of the Interior; 2011. [Google Scholar]
- 2.Valentine DL, et al. Propane respiration jump-starts microbial response to a deep oil spill. Science. 2010;330:208–211. doi: 10.1126/science.1196830. [DOI] [PubMed] [Google Scholar]
- 3.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yakimov MM, Timmis KN, Golyshin PN. Obligate oil-degrading marine bacteria. Curr Opin Biotechnol. 2007;18:257–266. doi: 10.1016/j.copbio.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Rojo F. Degradation of alkanes by bacteria. Environ Microbiol. 2009;11:2477–2490. doi: 10.1111/j.1462-2920.2009.01948.x. [DOI] [PubMed] [Google Scholar]
- 6.Redmond MC, Valentine DL, Sessions AL. Identification of novel methane-, ethane-, and propane-oxidizing bacteria at marine hydrocarbon seeps by stable isotope probing. Appl Environ Microbiol. 2010;76:6412–6422. doi: 10.1128/AEM.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yvon-Lewis SA, Hu L, Kessler J. Methane flux to the atmosphere from the Deepwater Horizon oil disaster. Geophys Res Lett. 2011;38:L01602. [Google Scholar]
- 8.Ryerson TB, et al. Atmospheric emissions from the Deepwater Horizon spill constrain air-water partitioning, hydrocarbon fate, and leak rate. Geophys Res Lett. 2011;38:L07803. [Google Scholar]
- 9.Reddy CM, et al. Science applications in the Deepwater Horizon oil spill special feature: Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20229–20234. doi: 10.1073/pnas.1101242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camilli R, et al. Tracking hydrocarbon plume transport and biodegradation at Deepwater Horizon. Science. 2010;330:201–204. doi: 10.1126/science.1195223. [DOI] [PubMed] [Google Scholar]
- 11.Diercks AR, et al. Characterization of subsurface polycyclic aromatic hydrocarbons at the Deepwater Horizon site. Geophys Res Lett. 2010;37:L20602. [Google Scholar]
- 12.Socolofsky SA, Adams EE, Sherwood CR. Formation dynamics of subsurface hydrocarbon intrusions following the Deepwater Horizon blowout. Geophys Res Lett. 2011;38:L09602. [Google Scholar]
- 13.Coulon F, McKew BA, Osborn AM, McGenity TJ, Timmis KN. Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ Microbiol. 2007;9:177–186. doi: 10.1111/j.1462-2920.2006.01126.x. [DOI] [PubMed] [Google Scholar]
- 14.Teira E, et al. Dynamics of the hydrocarbon-degrading Cycloclasticus bacteria during mesocosm-simulated oil spills. Environ Microbiol. 2007;9:2551–2562. doi: 10.1111/j.1462-2920.2007.01373.x. [DOI] [PubMed] [Google Scholar]
- 15.Venosa AD, Holder EL. Biodegradability of dispersed crude oil at two different temperatures. Mar Pollut Bull. 2007;54:545–553. doi: 10.1016/j.marpolbul.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Head IM, Jones DM, Röling WF. Marine microorganisms make a meal of oil. Nat Rev Microbiol. 2006;4:173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- 17.Hazen TC, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 18.Kessler JD, et al. A persistent oxygen anomaly reveals the fate of spilled methane in the deep Gulf of Mexico. Science. 2011;331:312–315. doi: 10.1126/science.1199697. [DOI] [PubMed] [Google Scholar]
- 19.Joye SB, MacDonald IR, Leifer I, Asper V. Magnitude and oxidation potential of hydrocarbon gases released from the BP oil well blowout. Nat Geosci. 2011;4:160–164. [Google Scholar]
- 20.Joint Analysis Group Report 2: Review of Preliminary Data to Examine Subsurface Oil In the Vicinity of MC252#1, May 19 to June 19, 2010. 2011. Available at http://service.ncddc.noaa.gov/rdn/www/activities/healthy-oceans/jag/reports/documents/JAG_Data_Report_2_FINAL.pdf. Accessed May 15, 2011.
- 21.Galand PE, Potvin M, Casamayor EO, Lovejoy C. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J. 2010;4:564–576. doi: 10.1038/ismej.2009.134. [DOI] [PubMed] [Google Scholar]
- 22.Valentine DL, et al. Dynamic auto-inoculation and the microbial ecology of a deep water hydrocarbon irruption. Proc Natl Acad Sci USA. 2012;109:20286–20291. doi: 10.1073/pnas.1108820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cottrell MT, Kirchman DL. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boden R, Thomas E, Savani P, Kelly DP, Wood AP. Novel methylotrophic bacteria isolated from the River Thames (London, UK) Environ Microbiol. 2008;10:3225–3236. doi: 10.1111/j.1462-2920.2008.01711.x. [DOI] [PubMed] [Google Scholar]
- 25.Jensen S, Neufeld JD, Birkeland NK, Hovland M, Murrell JC. Methane assimilation and trophic interactions with marine Methylomicrobium in deep-water coral reef sediment off the coast of Norway. FEMS Microbiol Ecol. 2008;66:320–330. doi: 10.1111/j.1574-6941.2008.00575.x. [DOI] [PubMed] [Google Scholar]
- 26.Cottrell MT, Kirchman DL. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol. 2000;66:5116–5122. doi: 10.1128/aem.66.12.5116-5122.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stepanauskas R, Sieracki ME. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc Natl Acad Sci USA. 2007;104:9052–9057. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eilers H, Pernthaler J, Glöckner FO, Amann R. Culturability and In situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. 2000;66:3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Wintzingerode F, Göbel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: Pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 30.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Röling WF, de Brito Couto IR, Swannell RP, Head IM. Response of Archaeal communities in beach sediments to spilled oil and bioremediation. Appl Environ Microbiol. 2004;70:2614–2620. doi: 10.1128/AEM.70.5.2614-2620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valentine DL. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol. 2007;5:316–323. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- 33.Methé BA, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci USA. 2005;102:10913–10918. doi: 10.1073/pnas.0504766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yakimov MM, et al. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int J Syst Evol Microbiol. 2003;53:779–785. doi: 10.1099/ijs.0.02366-0. [DOI] [PubMed] [Google Scholar]
- 35.Brakstad OG, Nonstad I, Faksness LG, Brandvik PJ. Responses of microbial communities in Arctic sea ice after contamination by crude petroleum oil. Microb Ecol. 2008;55:540–552. doi: 10.1007/s00248-007-9299-x. [DOI] [PubMed] [Google Scholar]
- 36.Powell S, Bowman J, Snape I. Degradation of nonane by bacteria from Antarctic marine sediment. Polar Biol. 2004;27:573–578. [Google Scholar]
- 37.Hedlund BP, Geiselbrecht AD, Bair TJ, Staley JT. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov. Appl Environ Microbiol. 1999;65:251–259. doi: 10.1128/aem.65.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goffredi SK, Johnson SB, Vrijenhoek RC. Genetic diversity and potential function of microbial symbionts associated with newly discovered species of Osedax polychaete worms. Appl Environ Microbiol. 2007;73:2314–2323. doi: 10.1128/AEM.01986-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dumont MG, Pommerenke B, Casper P, Conrad R. DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ Microbiol. 2011;13:1153–1167. doi: 10.1111/j.1462-2920.2010.02415.x. [DOI] [PubMed] [Google Scholar]
- 40.Deming J, Somers L, Straube W, Swartz D, MacDonell M. Isolation of an obligately barophilic bacterium and description of a new genus, Colwellia gen. nov. Syst Appl Microbiol. 1988;10:152–160. [Google Scholar]
- 41.National Oceanic and Atmospheric Administration R/V Walton Smith CTD profile data. 2011 Available at http://data.nodc.noaa.gov/DeepwaterHorizon/Ship/Walton_Smith/ORR/Cruise_01/CTD/. Accessed September 20, 2011. [Google Scholar]
- 42.Huber T, Faulkner G, Hugenholtz P. Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 43.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 48.Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH. T-REX: Software for the processing and analysis of T-RFLP data. BMC Bioinformatics. 2009;10:171. doi: 10.1186/1471-2105-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.