Abstract
In 1950, Barbara McClintock published a Classic PNAS article, “The origin and behavior of mutable loci in maize,” which summarized the evidence leading to her discovery of transposition. The article described a number of genome alterations revealed through her studies of the Dissociation locus, the first mobile genetic element she identified. McClintock described the suite of nuclear events, including transposon activation and various chromosome aberrations and rearrangements, that unfolded in the wake of genetic crosses that brought together two broken chromosomes 9. McClintock left future generations with the challenge of understanding how genomes respond to genetic and environmental stresses by mounting adaptive responses that frequently include genome restructuring.
In a 1950 Classic PNAS article, Barbara McClintock summarized and interpreted what had already become a large volume of data on the behavior of loci either harboring or affected by activity of the first transposon family she discovered and studied, called the Activator-Dissociation family (1). Noting that the genetic data were already too voluminous to present in detail, McClintock sought to communicate her most important inferences and insights. McClintock published relatively few papers on transposition in the conventional scientific literature. As required by her employer, the Carnegie Institution of Washington (now the Carnegie Institution for Science), she published summaries of each year’s work in the Carnegie Year Books. The 1950 PNAS article was one of the several efforts she made to communicate her findings on transposition in the wider scientific literature. Back before e-mail and portable document format (PDF) files, colleagues who were interested in an article generally wrote the author or sent a postcard requesting a reprint. McClintock received so few reprint requests for her 1950 PNAS article that she concluded that there was little interest in her work, so she filed her detailed analyses of her observations and went back to publishing just her annual summaries, containing just a fraction of her voluminous primary genetic data, in the Carnegie Year Books (McClintock, personal communication). To understand the lack of interest in what turned out to be a momentous discovery, it is important to reconstruct and understand the context of the time.
Back in the early 1940s, McClintock poked a stick into a sleeping genome’s lair. Out sprang breaking chromosomes, jumping genes, and the reversible mutations we now call epigenetic long before the world was ready to see them. It was in the progeny of self-pollinated plants obtained from crosses between plants each carrying a broken, telomere-less chromosome 9 that she saw a sudden burst of unstable mutations, as well as all kinds of chromosomal rearrangements and aberrations. One of these struck her as especially odd because it caused the chromosome to break repeatedly during development at what seemed to be the same chromosomal location, leading to the simultaneous loss of several genetic markers.
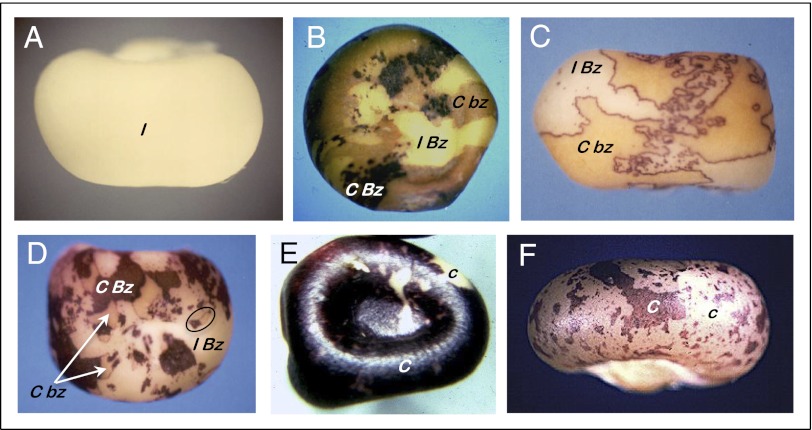
The clues that then led McClintock from the chromosome-breaking locus to transposition are illustrated in Fig. 1 using just two of the several markers affecting kernel phenotypes she routinely used to follow the short arm of chromosome 9. These are the C gene, encoding a transcription factor required for synthesis of the purple anthocyanin pigments in the surface aleurone layer of the kernel, and the Bz gene that encodes a pigment-stabilizing glucosyl transferase in whose absence the aleurone develops the bronze color for which the gene is named (2, 3). McClintock used both a dominant inhibitory allele of the C gene, designated I, and a recessive null allele, designated c, both of which give a colorless aleurone (4). Neglecting the triploidy of the aleurone, kernels that have the genetic constitution I Bz/C bz are colorless (Fig. 1A). Kernels experiencing random chromosome breakage due to what McClintock called the “chromosome-type” of breakage–fusion–bridge cycle exhibit colorless sectors of the original genetic constitution and both pigmented and bronze sectors resulting from progressive loss of the I and Bz alleles during development (Fig. 1B).
Fig. 1.
Phenotypes of kernels that led to McClintock’s discovery of chromosome breakage at Ds. I: dominant inhibitory allele of the C gene; C: full anthocyanin pigmentation when together with the wild-type Bz allele of the Bronze gene; bz: recessive allele of the Bronze gene, bronze pigmentation with C. The chromosome constitution of the kernels shown in A–D is I Bz/C bz (neglecting endosperm triploidy). (A) Colorless I Bz/C bz phenotype. (B) Random breakage of chromosome 9 results in loss of the I allele to reveal fully pigmented C Bz sectors, followed by loss of the Bz allele to reveal the bronze-colored C bz phenotype. (C) Chromosome breakage at a Ds transposon located proximal to both the C and Bz loci results in simultaneous loss of both the I and Bz alleles, giving only the colorless and bronze phenotypes. The colored rims result from complementation between the wild-type C allele in C bz tissue and the wild-type Bz allele in tissue containing the inhibitory I allele of the C gene. (D) Altered phenotype produced by chromosome breakage at Ds after transposition to a new site just proximal to the C gene at the distal end of chromosome 9. The initial chromosome break at Ds eliminates the I allele, revealing pigmented C Bz sectors. The circle highlights a twin sector arising from a dicentric chromatid formed at the cleavage site and subsequent random breakage of the dicentric during cell division, giving rise to adjacent patches of C Bz and C bz tissue. (E) Phenotype resulting from chromosome breakage at the Ds just proximal to the C gene in a kernel having the genetic constitution C Ds/c, where C is the dominant allele (pigmented aleurone) and c is the recessive allele (colorless aleurone). The C → c variegation results from chromosome breakage at Ds and subsequent loss of the C allele. (F) Phenotype of an unstable mutation arising by transposition of Ds into the C gene, causing a mutation to the colorless c allele. The c → C variegation is caused by somatic transposition of Ds out of the gene, restoring the colored phenotype of the C allele.
McClintock’s odyssey began with a kernel showing a startlingly different chromosome breakage phenotype in which both dominant markers were consistently lost together, giving only colorless and bronze sectors, although the colored complementation rims between sectors attest to the presence of the Bz allele in the colorless tissue adjacent to the C bz tissue, in which the pigment is synthesized but not glycosylated (Fig. 1C). Her subsequent analysis of this heritable phenotype led McClintock to designate the site of breakage, located close to the centromere on chromosome 9, the Dissociation (Ds) locus. She soon understood that chromosome breakage at Ds required the presence of another gene, which she designated the Activator (Ac) locus.
Always on the lookout for potentially informative differences, she noticed, grew, and analyzed a kernel that showed chromosome breakage but once again had colored patches (Fig. 1D). Through detailed genetic and cytogenetic studies, she discovered that the chromosome-breaking Ds gene had moved, or transposed, to a location between the I and Bz alleles near the distal end of the chromosome. She also noted many twin sectors of deeply pigmented and bronze tissue, such as the one circled in Fig. 1D, which in time she understood to be attributable to the formation of a dicentric chromatid concomitant with chromosome breakage at Ds. Subsequent random breakage of the dicentric at mitotic divisions could result in the segregation of both newly replicated Bz alleles to one of the daughter cells, giving the adjacent patches of colored tissue bearing both wild-type Bz alleles and bronze tissue containing only the homolog carrying the recessive bz allele of the gene.
McClintock discovered the connection between transposition and unstable mutations in the course of analyzing the behavior of Ac and Ds, once again commencing from an odd kernel that showed an unexpected phenotype. Plants that have the genetic constitution C Ds/c and also carry an Ac give kernels that have colorless patches expressing the recessive c allele, as illustrated in Fig. 1E. This variegation pattern is caused by chromosome breakage at Ds and the subsequent loss of the C allele, exposing the c allele. On such plants, McClintock found a kernel exhibiting an inverse variegation pattern: a colorless c background with sectors of colored C tissue, as illustrated in Fig. 1F. She grew the kernel, made many crosses, and analyzed the progeny. She discovered that the Ds element had moved, or transposed, into the C gene itself, inactivating it. When Ac was present, the Ds element moved out of the locus during development, restoring stable expression of the wild-type C allele. By studying germinal revertants, she understood that Ac-mediated chromosome breakage, the hallmark of Ds, was removed simultaneously. Transposition of Ds before meiosis gives rise to germinally stable revertants of the colorless c allele to the wild-type, deeply pigmented C allele. She found that the site of chromosome breakage had moved to a new chromosomal position in such revertants. McClintock was able to document unequivocally that the unstable mutation was caused by insertion of the chromosome-breaking Ds because she could follow the chromosome-breaking property of Ds, first into and then out of the C gene, first inactivating and then reactivating it.
McClintock’s discovery of transposition was roughly contemporaneous with Watson and Crick’s insights into the structure of DNA in the middle years of the 20th century. Watson and Crick’s landmark contribution, published in 1953, was recognized with the award of a Nobel Prize within a decade. It immediately explained the mechanics of gene inheritance that had by then been under intense study for a half century since the rediscovery of Mendel’s work. Although meticulously documented by a highly respected geneticist, McClintock’s discovery of transposition languished for decades, finally receiving Nobel recognition in 1983 when genomes had begun to spill their secrets and we were awash in jumping genes. Today we know that transposon and retrotransposon sequences constitute an astounding two-thirds of our own genome and 85% of the corn genome (5, 6). We know, too, that the fingerprints of transposable elements and transposition are everywhere in eukaryotic genomes, from the coarsest features of genomic landscapes and how they change through real and evolutionary time to the finest details of gene structure and regulation. So why did transposition remain undiscovered for so long and why, once discovered, did it take decades for transposons to be accepted beyond the maize community as more than a genetic oddity?
I believe that the answers to these questions lie in the epigenetic mechanisms that eukaryotes have elaborated to a much greater extent than prokaryotes (7). Repressive protein complexes, histone methylation, RNAi, and RNA-directed DNA methylation, as well as recombinational regulatory complexes, are among the epigenetic mechanisms that have been discovered in recent years (8–12). These serve a variety of structural and regulatory functions, but perhaps the essential ones for understanding why it took so long to discover transposons are those that regulate transposition and homologous recombination. Epigenetic mechanisms suppress transposon activity, as well as illegitimate and ectopic recombination among homologous sequences in the course of DNA replication and during the repair of DNA breaks. The result is that eukaryotic genomes full of transposons are as stable as genomes with few of them. For this reason, transposons are all but invisible to the geneticist.
It has long been known that heterochromatin, the highly compacted chromosome regions rich in repetitive DNA and transposons, is recombinationally inert (13, 14). Although not all eukaryotes use all of the known epigenetic mechanisms, even lower eukaryotes with relatively small genomes use RNAi to stabilize repetitive DNAs, such as ribosomal RNA genes and centromeric repeats, suppressing illegitimate recombination among repeats (14–16). In fission yeast, noncoding transcripts of repetitive sequences initiate a process that generates small RNAs that, in turn, target further transcripts for degradation and attract protein complexes that induce heterochromatization through histone modification (17, 18). Disruption of the RNAi machinery disturbs the repair of double-strand breaks, stimulating repair by homologous recombination (19).
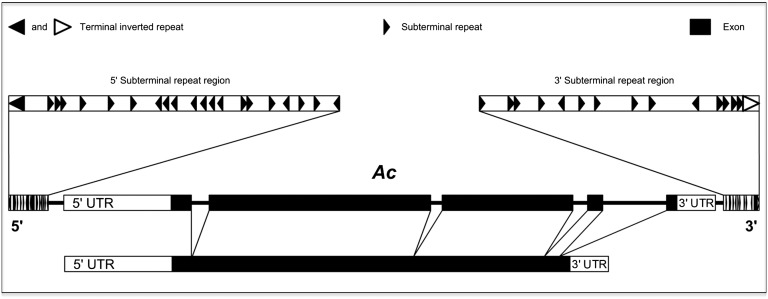
McClintock’s studies on the Ac/Ds transposon family showed that chromosome breaks at the site of insertion of a nonautonomous Ds element could be resolved with attendant duplications, deletions, inversions, and translocations of large chromosomal segments (1, 20). She found that both Ac and Ds were mobile and could cause unstable mutations, speculating that they were made of “the same or similar types of material” (1). Indeed, many years later, when we isolated the first Ac and Ac-derived Ds transposons, it became clear that these Ds transposons were deletion derivatives of Ac (21). By 1950, McClintock knew that not all Ds elements could break chromosomes. What she did not know and what explains the extraordinary chromosome antics she discovered was that the original chromosome-breaking Ds element that led to her ground-breaking discoveries was a very unusual one, comprising one copy of an internally deleted 2-kb derivative of the full 4.6-kb Ac element (Fig. 2) inserted in inverted orientation into the middle of another copy (22, 23). It was this particular element that wreaked the chromosomal havoc that McClintock describes in the 1950 article.
Fig. 2.
Structure of the Activator (Ac) transposon. The 4.6-kb transposon has a single transcription unit with 5 exons, encoding its transposase. The termini are 11-bp inverted repeats, adjacent to which are subterminal repetitive regions containing many copies of sequences homologous or identical to the hexamer AAACGG in both orientations. Figure prepared by T. Peterson and reproduced here with his permission. Note that Fig. 2 was prepared for Peterson T and Zhang J (2013) The mechanism of Ac/Ds transposition. Plant Transposons and Genome Dynamics in Evolution, ed Fedoroff NV (Wiley-Blackwell, Hoboken, NJ), in press.
The underlying explanation, not known for many more years, was that the mechanism that ensures the orderly excision of the transposon’s sequence from just one of the two newly replicated sister chromatids before reinsertion at another site is the correct recognition of the two appropriately hemimethylated copies of the inverted terminal repeats at the elements ends on one chromatid (24). Confusion arises from the presence of the two copies of the transposon in inverted orientation on each newly replicated chromatid. The result is that both sister chromatids are cleaved and rejoined in all possible configurations, forming dicentric chromosomes and conjoined acentric fragments, as well as initiating the other types of chromosomal rearrangements McClintock describes. More recent molecular and genetic analyses of Ac/Ds-associated rearrangements at the P locus identified transposition events that initiate at the 5′ end of one transposon and terminate at the 3′ end of a nearby transposon (25). Termed “alternative transposition,” this can also generate a variety of rearrangements, depending on the relative orientation of the transposon ends, and translocate large segments of intervening DNA.
In the PNAS artice, McClintock goes on to draw parallels between the behavior of these mutable loci in maize and heterochromatin-associated variegation in Drosophila. She also describes her success in using her genetic background to activate a previously quiescent locus called Dotted (Dt) that causes somatic reversion of a long-used stable null allele of the A1 gene in maize, identified some years earlier by her friend and colleague Marcus Rhoades (26). She further notes that Dt is located in a heterochromatic knob, leading her to suspect that Dt’s activity was caused by a modification of the heterochromatin itself. However, the important point here is that McClintock triggered the activation of Dt by crossing in a rearranged chromosome 9 she knew to be capable of introducing a broken chromosome end into a primary endosperm nucleus, causing what McClintock called a genomic “shock” that set off a nuclear earthquake. She deeply appreciated the interconnections among the different manifestations of genomic instability and understood well before the rest of us that genomes have choreographed responses to both unpredictable stresses, such as irradiation, and to predictable stresses, such as heat shock, intended to minimize the impact of the stress. She says in her Nobel Prize lecture (27) that her maize experiments revealed to her
“….how a genome may react to conditions for which it is unprepared, but to which it responds in a totally unexpected manner. Among these is the extraordinary response of the maize genome to entrance of a single ruptured end of a chromosome into a telophase nucleus. It was this event that, basically, was responsible for activations of potentially transposable elements that are carried in a silent state in the maize genome. The mobility of these activated elements allows them to enter different gene loci and to take over control of action of the gene wherever one may enter. Because the broken end of a chromosome entering a telophase nucleus can initiate activations of a number of different potentially transposable elements, the modifications these elements induce in the genome may be explored readily. In addition to modifying gene action, these elements can restructure the genome at various levels, from small changes involving a few nucleotides, to gross modifications involving large segments of chromosomes, such as duplications, deficiencies, inversions, and other more complex reorganizations.”
“A cell capable of repairing a ruptured end of a chromosome must sense the presence of this end in its nucleus. This sensing activates a mechanism that is required for replacing the ruptured end with a functional telomere. That such a mechanism must exist was revealed by a mutant that arose in my stocks. When homozygous, this mutant would not allow the repair mechanism to operate in the cells of the plant. Entrance of a newly ruptured end of a chromosome into the zygote is followed by the chromatid type of breakage-fusion-bridge cycle throughout mitoses in the developing plant. This suggests that the repair mechanism in the maize strains I had been using is repressed in cells producing the male and female gametophytes and also in the endosperm, but is activated in the embryo.”
In the years since McClintock’s pioneering observations more than half a century ago, it has been amply documented that plant transposons are activated in response to a variety of DNA damaging agents and both biotic and abiotic stresses, as well the passage of plant cells through tissue culture (28–33). Other sources of natural chromosomal disturbance are provided by interspecific hybridization and allopolyploidization, both of which trigger the activation of transposons (34–36). This seems to be true, as well, in other eukaryotes, from yeast to flies to humans. Telomerases are relatives of retrotransposon-encoded reverse transcriptases, and transposons either comprise or can fill in for missing telomeres in flies and yeast, respectively (37–40). Telomere loss activates transposons: this is precisely where McClintock’s extraordinary experiments began.
Just as McClintock reported that broken chromosome ends can “heal,” so do transposition bursts subside, sometimes quickly, sometimes after many generations. Some of the Arabidopsis transposons and retrotransposons demethylated and activated in a genetic background devoid of the MET1 DNA methylase are gradually remethylated by RNA-dependent DNA methylation within several generations after reintroduction of a wild-type MET1 gene (41–43). Heat-induced transcription and transposition of the Arabidopsis ONSEN retrotransposon is rapidly silenced, becoming transgenerational only in plants with a compromised RNA-dependent DNA methylation pathway (33). Thus, transcriptional activation by demethylation can also trigger a feedback mechanism that restores methylation and resilences transposons. An example from Drosophila, which lacks DNA methylation, is provided by the recent report that proteins required for the small RNA-based feedback repression of P transposons are mislocalized or degraded in strains undergoing hybrid dysgenesis, a general, sterility-inducing activation of transposons that occurs when P transposons are introduced into a female lacking them by a genetic cross with a male carrying them (44). Females recover their fertility as they age through a complex process that restores the epigenetic repression of transposons.
McClintock concluded her Nobel lecture with the following:
“In the future, attention undoubtedly will be centered on the genome, with greater appreciation of its significance as a highly sensitive organ of the cell that monitors genomic activities and corrects common errors, senses unusual and unexpected events, and responds to them, often by restructuring the genome. We know about the components of genomes that could be made available for such restructuring. We know nothing, however, about how the cell senses danger and instigates responses to it that often are truly remarkable.”
Recent years have seen progress in identifying the components of the restructuring response, but we know little as yet about how cells and organisms perceive and initiate reorganization in response to either genetic disruptions or environmental stressors. It is well known that DNA damage activates transposons (45–47). Recent experiments in Drosophila suggest a link between DNA damage and disruption of the small RNA repression machinery initiated by activation of the checkpoint kinase Chk2, which phosphorylates and destabilizes the Vasa protein involved in the small RNA repression of transposons (44). Conversely, it has been reported in mammalian cells that DNA damage repair depends on small RNA molecules produced by the double-stranded RNA cleaving enzymes involved in epigenetic silencing (48). These observations suggest mutual feedback linkages between the epigenetic regulatory machinery and DNA damage repair, such that disruptions in either trigger a restructuring and resetting of the other. Thus, McClintock’s 21st century challenge is to identify the signaling connections from environmental stress and chromosome damage to the epigenetic/DNA repair nexus, arguably the heart of eukaryotic genome evolvability (7).
Footnotes
References
- 1.McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA. 1950;36(6):344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dooner HK, Nelson OE. Genetic control of UDPglucose:flavonol 3-O-glucosyltransferase in the endosperm of maize. Biochem Genet. 1977;15(5-6):509–519. doi: 10.1007/BF00520194. [DOI] [PubMed] [Google Scholar]
- 3.Dooner HK, Nelson OE. Interaction among C, R, and Vp in the control of the Bz glucosyltransferase during endosperm development in maize. Genetics. 1979;91(2):309–315. doi: 10.1093/genetics/91.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen SM, Coe EH., Jr Control of anthocyanin synthesis by the C locus in maize. Biochem Genet. 1977;15(3–4):333–346. doi: 10.1007/BF00484464. [DOI] [PubMed] [Google Scholar]
- 5.de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12):e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 7.Fedoroff NV. Transposable elements, epigenetics and genome evolution. Science. 2012 doi: 10.1126/science.338.6108.758. 10.1126/science.1228458. [DOI] [PubMed] [Google Scholar]
- 8.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14(2):142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon SA, Meyers BC. Small RNA-mediated epigenetic modifications in plants. Curr Opin Plant Biol. 2011;14(2):148–155. doi: 10.1016/j.pbi.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Moldovan GL, et al. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell. 2012;45(1):75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanderlyn L. The heterochromatin problem in cytogenetics as related to other branches of investigation. Bot Rev. 1949;15:507–582. [Google Scholar]
- 14.Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev. 2008;18(2):204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9(1):25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres-Rosell J, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9(8):923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 17.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303(5658):672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayne EH, et al. Stc1: A critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140(5):666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaratiegui M, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479(7371):135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClintock B. Mutable loci in maize. Carnegie Inst Wash Yr Bk. 1950;49:157–167. [Google Scholar]
- 21.Fedoroff N, Wessler S, Shure M. Isolation of the transposable maize controlling elements Ac and Ds. Cell. 1983;35:243–251. doi: 10.1016/0092-8674(83)90226-x. [DOI] [PubMed] [Google Scholar]
- 22.Döring HP, Pahl I, Durany M. Chromosomal rearrangements caused by the aberrant transposition of double Ds elements are formed by Ds and adjacent non-Ds sequences. Mol Gen Genet. 1990;224(1):40–48. doi: 10.1007/BF00259449. [DOI] [PubMed] [Google Scholar]
- 23.Pohlman RF, Fedoroff NV, Messing J. The nucleotide sequence of the maize controlling element Activator. Cell. 1984;37(2):635–643. doi: 10.1016/0092-8674(84)90395-7. [DOI] [PubMed] [Google Scholar]
- 24.Ros F, Kunze R. Regulation of activator/dissociation transposition by replication and DNA methylation. Genetics. 2001;157(4):1723–1733. doi: 10.1093/genetics/157.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu C, Zhang J, Peterson T. Genome rearrangements in maize induced by alternative transposition of reversed ac/ds termini. Genetics. 2011;188(1):59–67. doi: 10.1534/genetics.111.126847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhoades MM. Effect of the Dt gene on the mutability of the a1 allele in maize. Genetics. 1938;23(4):377–397. doi: 10.1093/genetics/23.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226(4676):792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 28.Grandbastien MA, et al. The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses. Genetica. 1997;100(1-3):241–252. [PubMed] [Google Scholar]
- 29.Sugimoto K, Takeda S, Hirochika H. MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell. 2000;12(12):2511–2528. doi: 10.1105/tpc.12.12.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mhiri C, et al. The promoter of the tobacco Tnt1 retrotransposon is induced by wounding and by abiotic stress. Plant Mol Biol. 1997;33(2):257–266. doi: 10.1023/a:1005727132202. [DOI] [PubMed] [Google Scholar]
- 31.Ki CM, et al. Reprogramming of the activity of the activator/dissociation transposon family during plant regeneration in rice. Mol Cells. 2002;14(2):231–237. [PubMed] [Google Scholar]
- 32.Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA. 1996;93(15):7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito H, et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472(7341):115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- 34.Madlung A, et al. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005;41(2):221–230. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- 35.Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet. 2003;33(1):102–106. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- 36.Kenan-Eichler M, et al. Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics. 2011;188(2):263–272. doi: 10.1534/genetics.111.128348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eickbush TH. Telomerase and retrotransposons: Which came first? Science. 1997;277(5328):911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 38.Pardue ML, Danilevskaya ON, Traverse KL, Lowenhaupt K. Evolutionary links between telomeres and transposable elements. Genetica. 1997;100(1-3):73–84. [PubMed] [Google Scholar]
- 39.Curcio MJ, Belfort M. The beginning of the end: Links between ancient retroelements and modern telomerases. Proc Natl Acad Sci USA. 2007;104(22):9107–9108. doi: 10.1073/pnas.0703224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belfort M, Curcio MJ, Lue NF. Telomerase and retrotransposons: Reverse transcriptases that shaped genomes. Proc Natl Acad Sci USA. 2011;108(51):20304–20310. doi: 10.1073/pnas.1100269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teixeira FK, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323(5921):1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 42.Saze H, Kakutani T. Differentiation of epigenetic modifications between transposons and genes. Curr Opin Plant Biol. 2011;14(1):81–87. doi: 10.1016/j.pbi.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 43.Paszkowski J, Grossniklaus U. Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr Opin Plant Biol. 2011;14(2):195–203. doi: 10.1016/j.pbi.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Khurana JS, et al. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147(7):1551–1563. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradshaw VA, McEntee K. DNA damage activates transcription and transposition of yeast Ty retrotransposons. Mol Gen Genet. 1989;218(3):465–474. doi: 10.1007/BF00332411. [DOI] [PubMed] [Google Scholar]
- 46.Staleva Staleva L, Venkov P. Activation of Ty transposition by mutagens. Mutat Res. 2001;474(1-2):93–103. doi: 10.1016/s0027-5107(00)00165-2. [DOI] [PubMed] [Google Scholar]
- 47.Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Francia S, et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488(7410):231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]