Abstract
Na+-K+-ATPase (NKA) establishes the transmembrane [Na+] gradient in cells. In heart, phospholemman (PLM) inhibits NKA activity by reducing its apparent Na+ affinity, an effect that is relieved by PLM phosphorylation. The NKA crystal structure suggests regions of PLM–NKA interaction, but the sites important for functional effects in live cells are not known. We tested wild type (WT) and CFP–NKA-α1 point mutants (alanine substitution at F956, E960, L964, and F967) for fluorescence resonance energy transfer (FRET) with WT–PLM–YFP in HEK293 cells. NKA–PLM FRET was unaltered with F956A or F967A, reduced with L964A, and nearly abolished with E960A. Mutating the PLM site (F28A) identified by structural analysis to interact with E960-NKA also nearly abolished NKA–PLM FRET. In contrast, NKA–PLM coimmunoprecipitation was only slightly reduced by E960A–NKA or F28A–PLM mutants, consistent with an additional interaction site. FRET titrations indicate that the additional site has higher affinity than that between E960–NKA and F28–PLM. To test whether the FRET-preventing mutations also prevent PLM functional effects, we measured NKA-mediated Na+-transport in intact cells. For WT–NKA, PLM reduced apparent Na+-affinity of NKA and PLM phosphorylation reversed the effect. In contrast, for E960A–NKA the apparent Na+-affinity was unaltered by either PLM or forskolin-induced PLM phosphorylation. We conclude that E960 on NKA and F28 on PLM are critical for PLM effects on both NKA function and NKA–PLM FRET, but also there is at least one additional site that is critical for tethering PLM to NKA.
Intracellular [Na+] ([Na+]i) is critical for electrical excitability and coupled transport. In heart, [Na+]i closely regulates intracellular Ca2+, contraction, and rhythmicity via Na+/Ca2+ exchange (1, 2). Small changes in [Na+]i can have major effects on both [Ca2+]i and intracellular pH (via Na+/H+ exchange) (2). Therefore, [Na+]i regulation is very important for understanding basic ion homeostatic mechanisms.
There are several Na+ entry pathways, whereas the Na+/K+ pump (NKA) is the main Na+ extrusion pathway (2). NKA is a ubiquitous transmembrane protein that establishes and maintains [Na+] and [K+] gradients across the plasma membrane. These gradients ensure osmotic balance, resting membrane potential, and cellular excitability. NKA uses energy derived from hydrolysis of ATP to extrude three Na+ ions in exchange for two K+ ions.
Phospholemman (PLM), a 72-amino acid sarcolemmal protein, is a member of the FXYD protein family, which derives its name from the conserved Phe-X-Tyr-Asp motif in the proximal extracellular domain. FXYDs are tissue-specific NKA regulators that bind to and modulate NKA function by affecting the apparent affinity for internal Na+ or external K+ (3–5). The [Na+]i for half-maximal NKA activation (K0.5) in the heart varies with internal and external ionic conditions and is typically 8–22 mM. This is near the resting [Na+]i in most cells (6). PLM (FXYD1) is highly expressed in heart, brain, and skeletal muscle and we previously showed that PLM associates with and inhibits cardiac NKA, mainly by reducing the apparent affinity for internal Na+ (7, 8). PLM can be phosphorylated on the cytoplasmic domain by protein kinase A (PKA) at Ser-68 and by protein kinase C (PKC) at Ser-68, Ser-63, and Thr-69 (9). PLM phosphorylation at either Ser-63 or Ser-68 is sufficient to relieve NKA inhibition, and it mediates NKA stimulation by PKA and PKC (7, 8, 10, 11). PLM phosphorylation and consequent increase in NKA-mediated Na+ extrusion in cardiac myocytes are an integral part of the sympathetic fight-or-flight response, tempering the rise in both [Na+]i and cellular Ca2+ loading, and perhaps limiting Ca2+ overload-induced arrhythmias (12).
PLM associates with and modulates both NKA-α1 and NKA-α2 isoforms in a comparable but not identical manner (10). Green fluorescent protein-tagged NKA (CFP–NKA) and PLM (PLM–YFP) exhibit robust intermolecular fluorescence resonance energy transfer (FRET). Similar to the PLM effect on the apparent Na+ affinity, FRET is strongly inhibited by PKA and/or PKC phosphorylation of PLM (for both NKA isoforms) (10, 13, 14). Thus, NKA–PLM FRET may reflect the functional state of NKA regulation by PLM.
The site(s) responsible for NKA–PLM interaction are unknown. Cross-linking data (15) and predictions (15–18) based on the crystal structure of the related sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) suggest that the transmembrane domain (TM) of FXYDs could reside in a groove formed by TM2, TM6, and TM9 of the NKA α-subunit. However, crystal structures of shark rectal gland NKA with FXYD10 (19) and pig kidney NKA with FXYD2 (20) show FXYDs in close proximity to NKA TM9 but outside of that groove. X-ray structures clearly show several NKA TM9 sites (F949, E953, L957, and F960, pig kidney NKA numbering) within 5–8 Å of the FXYD transmembrane domain. The corresponding NKA-α1 residues in rat (full transcript numbering) and shark (posttranslational cleavage numbering) are F956, E960, L964, and F967. Mutational analysis and functional studies suggest that some of these sites may be involved in NKA interaction with FXYD2 (18, 21), FXYD4 (18), and FXYD7 (18). However, the relative importance of such sites for NKA–FXYD physical association and functional effects varies among FXYD proteins (18, 21) and no prior analysis has addressed PLM (FXYD1), which is also the only mammalian FXYD regulated by phosphorylation. Here, we combined PLM and NKA mutational analysis, intermolecular FRET, NKA function measurements in live cells, coimmunoprecipitation, and structural modeling to identify key corresponding sites in NKA TM9 and PLM involved in physical association and functional effects. We found that E960 on NKA and F28 on PLM are critical for both interaction and functional regulation of NKA by PLM.
Results
Crystal Structure-Based Screening for NKA–PLM Interaction Sites on NKA.
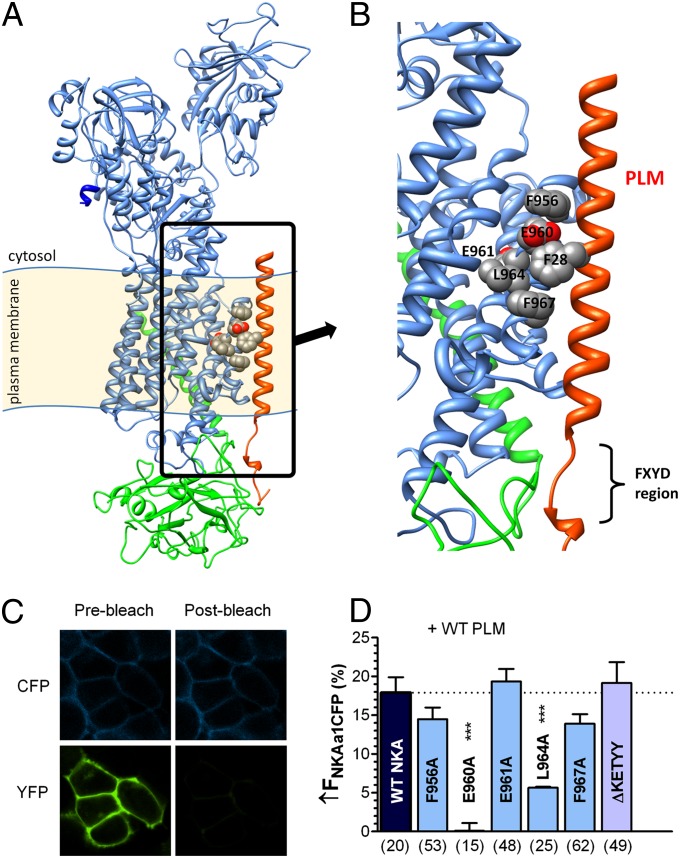
To create a 3D model of the NKA–PLM complex, we used Rosetta software (22–24) and the X-ray structure of shark rectal gland NKA (19) as a template for homology modeling of rat NKA α-subunit and human NKA β-subunit. The NMR structure of human PLM (25) was used for homology modeling of dog PLM. However, this NMR structure, obtained in micellar solutions without NKA, may be different in the cellular environment, as suggested by cross-linking between the FXYD10 C terminus and the NKA A domain (26). Therefore, we omitted the C terminus of PLM in the model and focused on the transmembrane region (Figs. 1 A and B and 2A). Putative NKA–PLM interaction sites on NKA were identified from crystal structures of pig kidney NKA (20) and shark rectal gland NKA (19), where the transmembrane domain of FXYD2 and FXYD10, respectively, is close to the outside of TM9 of the NKA α-subunit. Fig. 1 A and B shows our NKA–PLM model, with the TM9 sites F956, E960, L964, and F967 facing PLM, as previously shown by the crystal structures (19, 20). We mutated these residues, along with the E961 site, on CFP–NKA-α1 and coexpressed the mutated NKAs with WT–PLM–YFP in HEK293 cells. We used NKA–PLM FRET to screen for mutations that affect NKA–PLM interaction. Fig. 1C shows an example with a clear increase of CFP (donor) fluorescence after photobleaching YFP (acceptor). For WT–NKA and WT–PLM, donor fluorescence (FCFP) increased by 17.9 ± 2% (Fig. 1D). Alanine substitution at F956, E961, or F967 did not significantly affect FRET (↑FCFP of 14.4 ± 2%, 19.3 ± 2%, and 13.9 ± 1%, respectively). Truncation of the NKA C terminus (ΔKETYY), thought to be involved in setting the apparent Na+ affinity (27), also failed to alter FRET (↑FCFP was 19.2 ± 3%). However, NKA–PLM FRET was abolished for the E960A mutant (↑FCFP = 0.2 ± 4%) and was significantly reduced for L964A (↑FCFP = 6 ± 1%). We conclude that E960 is crucial for the NKA–PLM interaction that produces FRET.
Fig. 1.
E960 site on NKA is critical for NKA–PLM FRET. (A) Structural model of rat NKA α-subunit (blue), β-subunit (green), and PLM (red). Putative PLM interaction sites on NKA TM9 are highlighted. (B) Magnified picture of highlighted area in Fig. 1A showing putative PLM interaction sites on NKA TM9: F956, E960, E961, L964, and F967. (C) HEK293 cells coexpressing CFP–NKA-α1 and PLM–YFP before and after photobleaching YFP. FRET was measured as the increase in donor (CFP) fluorescence upon acceptor photobleaching. (D) Average FRET (±SEM) between CFP–NKA-α1 and PLM–YFP with indicated NKA mutations (n below the bars, ***P < 0.0001, statistical significance vs. WT–NKA using unpaired t test.
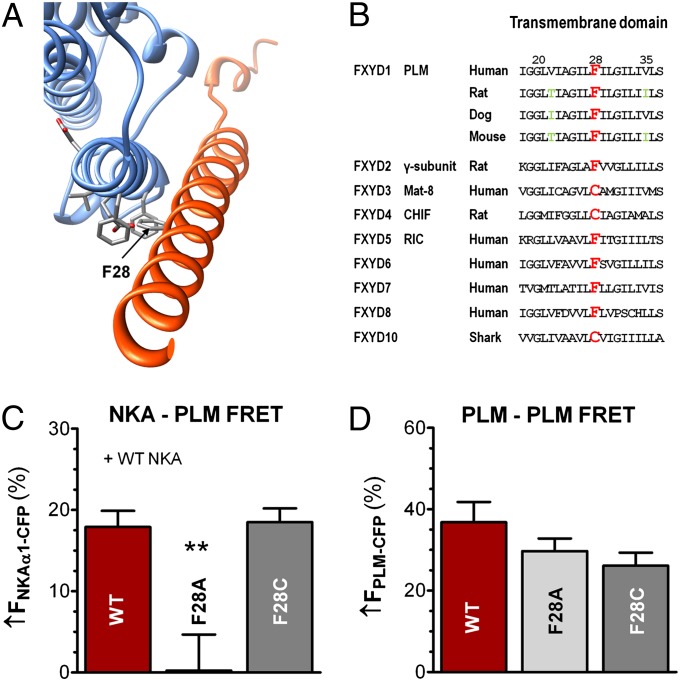
Fig. 2.
Identification and impact of PLM site F28 (opposing site to the NKA–E960 site). (A) NKA structural model with PLM site F28 in stick representation. (B) Transmembrane domain amino acid sequence of FXYDs. F28 is highlighted in red and is naturally cysteine in three FXYDs. Green letters indicate transmembrane variances. (C) NKA–PLM average FRET (±SEM) in HEK293 cells coexpressing WT–NKA and either WT–PLM (n = 20), F28A–PLM (n = 14, P < 0.01), or F28C–PLM (n = 26, P = NS). (D) Mean data of PLM–PLM FRET (±SEM) in control HEK293 cells coexpressing WT–PLM–CFP with WT–PLM–YFP (n = 52), F28A–PLM–YFP (n = 34, P = NS) or F28C–PLM–YFP (n = 24, P = NS).
F28 on PLM and Its Role for NKA–PLM and PLM–PLM Interaction.
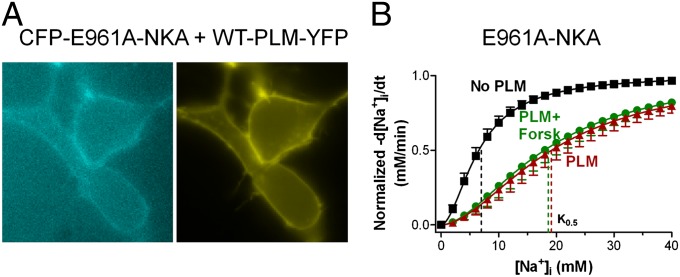
Fig. 2A shows a cytosolic view of our NKA structural model, highlighting the F28 site on PLM, which is in close proximity to TM9 of NKA (distance between F28 and F967 is ∼6 Å; between F28 and E960, ∼4 Å), in good agreement with crystallographic data. We next examined whether alanine substitution at F28–PLM also inhibits NKA–PLM FRET. Known FXYDs have either phenylalanine or cysteine at this position (Fig. 2B, with position 28 in red). Thus, we tested both F28A and F28C mutations. Fig. 2C shows that the F28A mutation was sufficient to prevent detectable FRET upon PLM–YFP photobleach (↑FCFP = 0.2 ± 4%), whereas mutating F28 to cysteine did not affect FRET with WT–NKA (↑FCFP = 18.5 ± 2% vs. 17.9 ± 2%). F28C–PLM has been shown to cross-link with C145 on TM2 of NKA-α1 (15). Thus, we also tested the C145A construct, but found that it was retained in the ER, which prevented further evaluation. PLM is also thought to form PLM–PLM oligomers (14), however, neither F28A nor F28C significantly altered the PLM–PLM FRET (Fig. 2D).
Detailed NKA–PLM FRET Analysis.
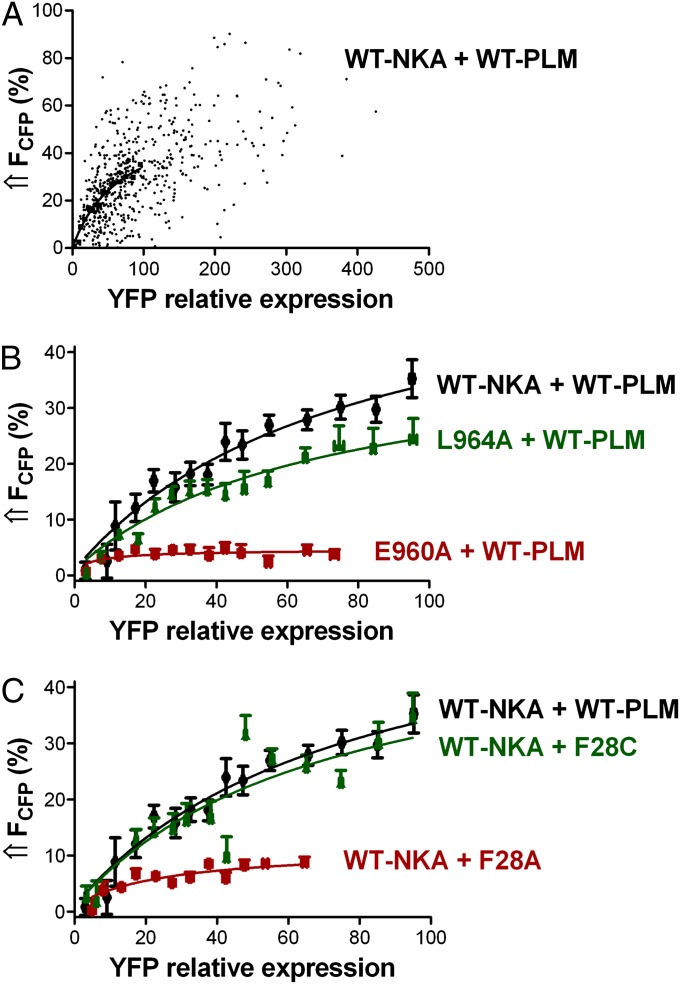
We next explored the PLM–NKA interaction with more detailed FRET analysis, taking advantage of variations in YFP-to-CFP expression ratio. Fig. 3A shows the FRET between CFP–NKA-α1 and PLM–YFP as a function of YFP:CFP ratio (i.e., the relative PLM:NKA expression). The points represent FRET efficiency for each cellular region of interest (n = 587 analysis regions; Fig. S1 and SI Materials and Methods). This is effectively a titration experiment with FRET efficiency fit to FRET = (FRETmax)/(1 + K0.5/[PLM]), where [PLM] is the initial (unbleached) [YFP] in arbitrary units, and FRET is the observed FRET efficiency (14). FRETmax provides structural information related to the proximity of CFP to YFP when PLM is bound to NKA. The half-maximum [YFP] (K0.5) reflects apparent NKA–PLM affinity (in arbitrary units related to surface [YFP]). FRET values in Fig. 3A span nearly the full range from 0 to FRETmax. CFP expression was comparably variable for WT–NKA (Fig. S2A) as well as for NKA mutants L964A and E960A (Fig. S2 B and C). Fig. 3B shows mean FRET efficiency curves for WT–PLM with WT–NKA, L964A–NKA, and E960A–NKA. Mean FRETmax for WT–NKA with WT–PLM was 58 ± 9% and K0.5 was 68.9 ± 20.9. The L964A mutant reduced FRETmax to 38 ± 6% and slightly increased the apparent NKA–PLM interaction affinity (K0.5 = 56.4 ± 16.2). E960A dramatically reduced FRET efficiency (FRETmax = 5 ± 1%), but the residual interaction shows high apparent affinity (K0.5 = 5.1 ± 2.5). Because all NKA constructs targeted effectively to the membrane (Fig. S3), the lower FRETmax for L964A and E960A mutants was not caused by a methodological limitation. Fig. 3C shows mean FRET efficiency curves for WT–NKA with WT–PLM, F28A–PLM, and F28C–PLM. The F28A mutation reduced FRETmax from 58 ± 9% to 11 ± 2% and increased the apparent NKA–PLM interaction affinity (K0.5 = 18.5 ± 7.5). In contrast, F28C mutation had no major effect (FRETmax = 51 ± 12%, K0.5 = 62.9 ± 28.5).
Fig. 3.
Dependence of FRET between CFP–NKA-α1 and PLM–YFP on the relative YFP expression in HEK293 cells. FRET was determined as the increase in donor fluorescence upon acceptor photobleaching. (A) Points represent FRET efficiency plotted region by region for HEK293 cells coexpressing CFP–WT–NKA and WT–PLM–YFP (n = 587 analysis regions). YFP fluorescence is an index of relative [PLM]. (B) FRET efficiency curves from cells coexpressing WT–PLM–YFP with CFP-tagged WT–NKA, L964A, and E960A, respectively (n = 405,344,322). (C) FRET efficiency curves from cells expressing CFP–WT–NKA with YFP-tagged WT–PLM, F28A–PLM, and F28C–PLM, respectively (n = 405,273,301).
Coimmunoprecipitation of NKA with PLM.
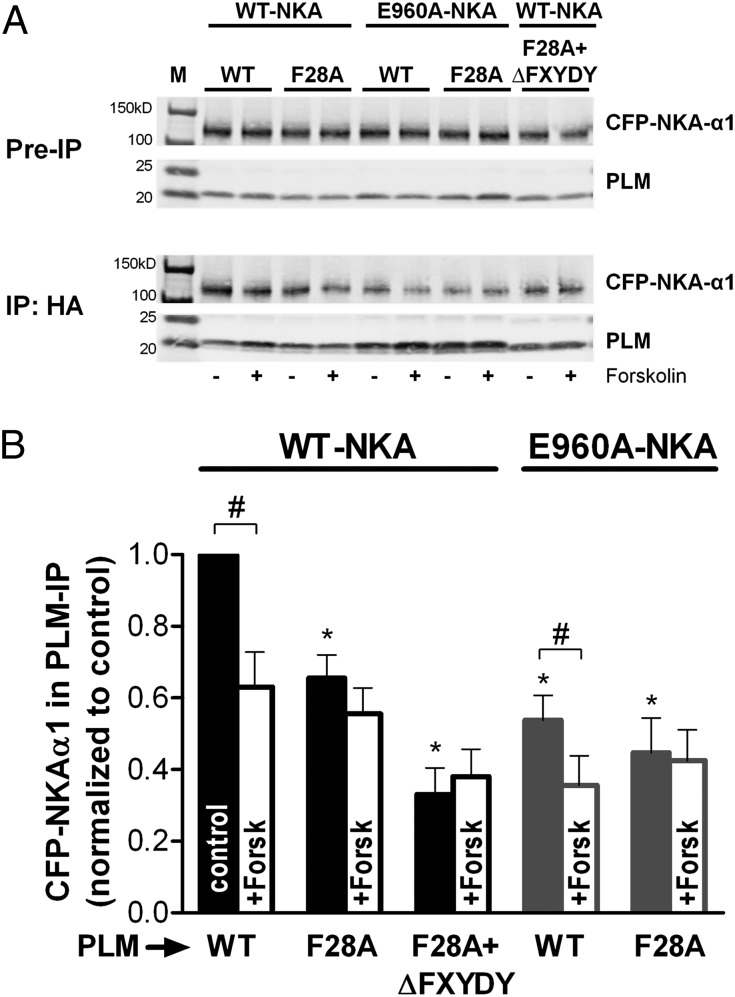
To determine whether the drastic reduction in FRET upon perturbation of the E960–F28 interaction is caused by physical dissociation of NKA and PLM, we performed coimmunoprecipitation (co-IP) experiments. HEK293 cells stably expressing CFP–WT–NKA-α1 or CFP–E960–NKA-α1 were transiently transfected with HA-tagged PLM, either WT or F28A. Before performing the HA-directed co-IP, some cells were exposed to 10 μM forskolin (or vehicle) to induce PLM phosphorylation.
Fig. 4A shows representative co-IP results comparing the amount of NKA-α1 pulled down with PLM for different mutants in the presence and absence of forskolin. A GFP antibody was used to selectively detect CFP-tagged (vs. endogenous) NKA. Fig. 4B shows mean co-IP data normalized to the amount of PLM pulled down. Compared with WT, both E960A–NKA and F28A–PLM significantly reduced the amount of CFP–NKA-α1 pulled down with PLM. Forskolin pretreatment reduced the co-IP of both WT–NKA and E960A–NKA with WT–PLM but not with F28A–PLM. Cells expressing both E960A–NKA and F28A–PLM (±forskolin) were little different from those expressing only one mutant. Thus, either mutant seemed capable of maximally interfering with the interaction, but neither could abolish co-IP, raising the possibility of a second interaction domain. Based on the crystal structure, we hypothesized that the FXYD domain might provide that additional tether, but deletion of this PLM region (ΔFXYDY) along with F28A still did not abolish the co-IP (Fig. 4B).
Fig. 4.
Coimmunoprecipitation of HA-tagged PLM and NKA. (A) Representative immunoblots of the starting material (pre-IP) and the immunoprecipitate (using an agarose-conjugated HA antibody). Recombinant NKA and PLM were detected using GFP and PLM antibodies, respectively. Comparable amounts of PLM were immunoprecipitated for each lane, but variable amounts of NKA were in co-IP, with or without forskolin preincubation. M, molecular weight ladder. (B) Mean data (NKA/PLM signal normalized to control) from different blots (n = 3) show that FRET-abolishing alanine mutations resulted in a reduction, but not an abolition of associated CFP–NKA-α1. *P < 0.01, statistical significance versus control using one-way ANOVA. #P < 0.05, statistical significance between pairs (− vs. + forskolin) using Student’s t test.
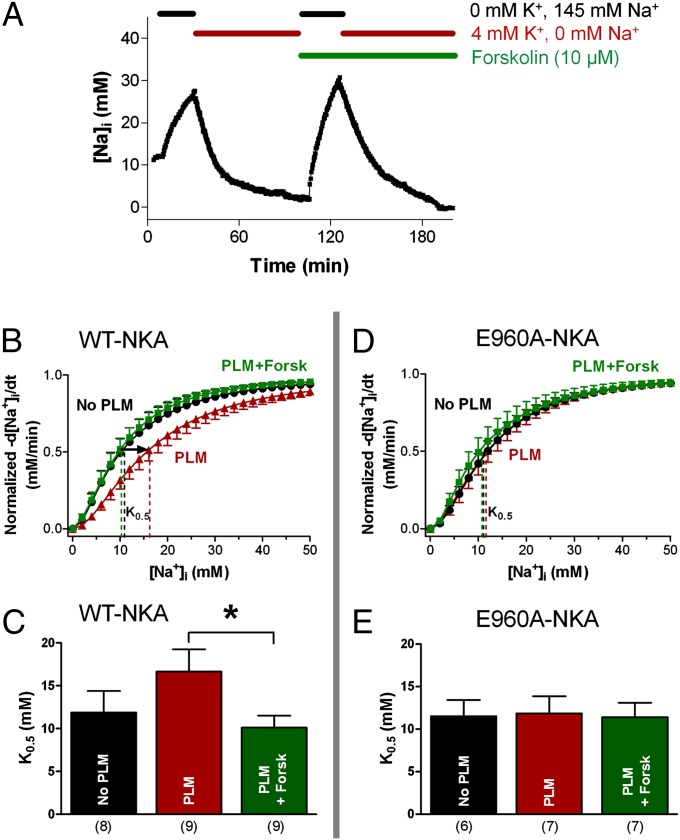
NKA-Mediated Na+ Efflux in HEK293 Cells.
Because E960A–NKA practically abolished FRET with PLM, we tested whether this correlates with a loss of functional effects of PLM on Na+ transport in cells. Using the fluorescent indicator sodium-binding benzofuran isophthalate (SBFI) and dual excitation-single emission fluorescence microscopy (as previously described) (11), we measured [Na+]i decline in HEK293 cells when NKA was reactivated by restoring normal external K+, after a period of Na+-loading with K+-free solution (Fig. 5A). A total of 1 µM ouabain was present, to block endogenous human NKA without inhibiting rat NKA-α1. The procedure was repeated in the presence of 10 µM forskolin (Fig. 5A). The rate of NKA-mediated Na+ extrusion (−d[Na+]i/dt) as a function of [Na+]i indicates the apparent NKA affinity for internal Na+ (7, 10). As expected, WT–PLM reduced the apparent Na+-affinity (raised K0.5) of WT–NKA and the effect was completely reversed by forskolin treatment (Fig. 5 B and C). Vmax was unaltered in any case. However, WT–PLM expression had no effect on the K0.5 for Na+ of E960A–NKA and forskolin failed to stimulate pump-mediated Na+ extrusion (Fig. 5 D and E). Thus, E960A mutation ablated the functional PLM- and PKA-dependent regulation of NKA in HEK293 cells.
Fig. 5.
Functional measurements of NKA-mediated Na+ efflux in HEK293 cells. (A) Experimental protocol: NKA was first inhibited (K+-free external solution) causing cellular [Na+]i loading, then NKA was reactivated by 4 mM K+. One micromole of ouabain was used throughout to inhibit endogenous human NKA. (B–E) NKA-mediated Na+ extrusion curves and mean K0.5 comparing Na+ extrusion in HEK293 cells expressing WT–NKA (B and C) vs. E960A–NKA (D and E). Vmax and K0.5 were derived by fitting the curves with a Hill equation. *P < 0.05. Corresponding n are indicated below bars; error bars indicate SEM.
We also tested the effects of E961A mutation on NKA function. This residue is thought to be a component of the third Na+ binding site (28, 29) but E961A did not alter NKA–PLM FRET (Fig. 1D) despite targeting well to the membrane (Fig. 6A). PLM expression greatly decreased the apparent Na+ affinity of E961A–NKA, however PLM phosphorylation failed to restore the apparent Na+ affinity (Fig. 6B). Thus, whereas E961 is not required for PLM–NKA FRET or PLM’s ability to inhibit NKA function, it is critical for the acute NKA activation by PLM phosphorylation.
Fig. 6.
Expression and functional role of NKA mutant E961A. (A) E961A mutant appears localized at the membrane. (B) Functional experiments with the E961A mutant: Phospholemman reduces the pump’s apparent affinity for intracellular Na+ (increases K0.5), but PLM phosphorylation does not affect the function of the E961A–NKA mutant (error bars indicate SEM; n = 4–5).
Discussion
Our major findings are that: (i) Interaction between E960–NKA and F28–PLM sites is critical for both NKA–PLM FRET and the functional effect of PLM on NKA, but (ii) not for their physical association. (iii) Other sites may contribute to PLM–NKA interaction (e.g., L964) or the ability of PLM phosphorylation to stimulate NKA (E961). A PLM–NKA interaction domain other than E960–NKA/F28–PLM can support the physical association of PLM with NKA.
E960 and F28 Are Essential for NKA–PLM Interaction.
Cross-linking, structural modeling, and crystallographic studies indicate that NKA TM9 is important for NKA interaction with FXYD proteins (15–20). However, the exact sites of interaction seem to vary for different FXYDs. For example, the E960–NKA site is critical for the effect of FXYD2 and FXYD4, but not FXYD7 on the NKA apparent affinity for external K+ (18). The present study is unique in systematically investigating the structural determinants of PLM interaction with NKA. We found that the E960A point mutation abolished both the effect of PLM on NKA apparent affinity for internal Na+ and the NKA–PLM FRET. Moreover, FRET was also prevented by alanine substitution of F28–PLM, the site shown by X-ray crystal structures to be within interaction distance of E960–NKA. Cross-linking experiments (15) also identified the site equivalent to F28–PLM as important for the interaction of PLM, FXYD2, and FXYD4 with NKA. However, in contrast to our findings with PLM, E960A–NKA mutation does not alter regulation of the pump’s apparent Na+ affinity by FXYD2 and FXYD4 (18). One study (21) suggested that the structural association of TM9 with FXYD2 TM domain is not the sole determinant of the FXYD2 effect on the apparent Na+ affinity. The authors pointed out that long-range modulation by the extracellular loop between TMs 7 and 8 is also involved (21). Such fine differences in the FXYD–NKA-α structural interaction may underlie the intrinsic differences in NKA modulation among various FXYDs. We have shown previously that regulation of the cardiac Na+/K+-ATPase by PLM is an integral part of the adrenergic fight-or-flight response (12). A loss of NKA stimulation during sympathetic activation caused by E960A–NKA, E961A–NKA, or F28A–PLM mutations raises the possibility of human NKA or PLM mutations that could prevent this important physiological regulation (in addition to the phosphorylation sites themselves). The failure of normal NKA activation in this setting could cause Na+ and Ca2+ overload and arrhythmias.
Our FRET data suggest that mutation of E960–NKA or F28–PLM sites causes a structural change but does not, by itself, clarify whether a complete dissociation of the complex takes place. Our coimmunoprecipitation study complements the FRET results, by testing whether the detected changes in FRET are paralleled by complete dissociation of PLM from NKA. Disturbing either or both participants in the E960–F28 interaction significantly weakened the NKA–PLM association (reduced amount of NKA pulled down). This fits nicely with our previous results showing dramatically decreased FRET upon PLM phosphorylation, but a smaller effect on co-IP (13). The present co-IP data show that full dissociation of the complex did not occur, even with E960A and F28A mutants combined, suggesting that other sites additionally support NKA–PLM interaction, albeit producing much less FRET (see below).
Second NKA–PLM Interaction Site.
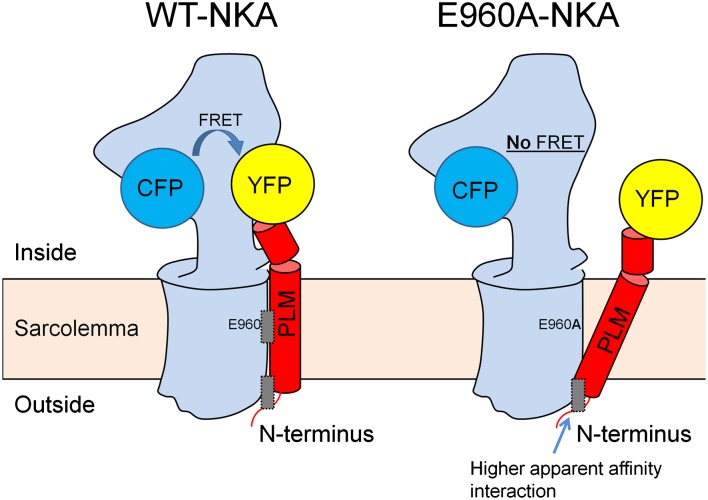
Detailed FRET analysis showed moderate apparent affinity for the interaction between E960–NKA and F28–PLM (K0.5 ∼65). When this interaction and most of the FRET was lost by mutating E960 or F28 to Ala, a higher apparent affinity interaction (K0.5 = 5–18) that exhibited weak FRETmax was unmasked. This suggests that the conformation in the CFP–YFP region is very different for this interaction. Fig. 7 shows a working model for two interacting sites and is consistent with the persistence of co-IP with mutants that nearly abolish FRET. We propose that there is a second NKA–PLM interaction site nearer to the external surface. In the absence of the mid-TM interaction, which draws the PLM C terminus closer to NKA, the CFP–YFP distance is too large to produce FRET. A second higher apparent affinity site also makes teleological sense, in that this nonregulatory interaction may keep the proteins close, allowing rapid functional modulation during acute phosphorylation and rezipping upon dephosphorylation. Indeed, Fig. 7, Right may also represent the phosphorylated state of PLM, in which FRET is drastically reduced but co-IP is still apparent (13, 30).
Fig. 7.
Working model of NKA–PLM complex with attached CFP and YFP. Left shows WT–NKA (blue) and WT–PLM (red). FRET occurs between CFP and YFP and the complex is held together by two binding regions (gray rectangles): E960–F28 functional interaction region and a second region. Right shows a different NKA–PLM conformation in the E960A mutant (or F28A mutant). CFP and YFP are not sufficiently close for appreciable FRET, but the complex is still held together by a higher apparent affinity interaction region near the external membrane surface.
There is crystallographic evidence for a NKA–PLM interaction at the extracellular NKA surface where NKA, FXYD domain, and β-subunit are all in proximity (19). We tested the most obvious possibility, that the FXYD domain itself might be the second PLM anchor point, but the FXYDY deletion coupled with F28A still did not abolish the co-IP. Similarly, the FXYD domain does not seem critical for FXYD7–NKA association (31). Cross-linking revealed that the cytoplasmic domain of FXYD10 interacts with the A domain of shark NKA-α (26), so this could be another possibility. Alternatively, the interaction responsible for NKA–PLM physical association might occur in the TM domain of PLM, as suggested for FXYD3 and FXYD7 (32, 33).
Other NKA Sites Affecting PLM–NKA Interaction.
Of the sites identified in the recent crystal structures of pig NKA (20), which correspond to F956, E960, L964, and F967 in the rat and our study, only E960 and L964 significantly impacted NKA–PLM FRET upon alanine substitution. L964 may play a supporting role in stabilizing the NKA–PLM interaction. The NKA C terminus truncation (ΔKETYY) did not reduce NKA–PLM FRET, suggesting that the C terminus is not critical for the functional NKA–PLM interaction. This is interesting because this truncation seems to reduce apparent Na+ affinity (by affecting Na+ binding at the third site) but not the apparent external K+ affinity (20, 34). Based on functional data in cardiac myocytes, we previously speculated that PLM association may influence binding of intracellular Na+ and extracellular K+ to the common sites. However, PLM phosphorylation may only affect Na+ binding to the third site, with no effect on the two sites common to Na+ and K+ (35). This is further supported by our findings with mutation of the E961 site, which is a component of the third Na+ binding site. Indeed, the E961 mutation preserved NKA–PLM FRET and the PLM-induced decrease in NKA apparent Na+ affinity, but with E961–NKA, PLM phosphorylation did not produce the normal reversal of PLM inhibition. These results suggest that the pair of adjacent glutamates at positions 960 and 961 are central to the complex regulation of NKA by PLM. If PLM were not associated with TM9, the E960 would protrude into the lipid, which would be energetically highly unfavorable for a transmembrane protein (19). In fact, the E960A mutation may weaken the interaction with PLM, but may also simply make that region more energetically favorable in the bilayer. This raises another question regarding whether PLM phosphorylation also moves PLM away from E960 and what the structural consequence of that might be.
In our constructs, CFP was on the NKA N terminus, which is not resolved in recent crystal structures, so we cannot predict its exact position on NKA. There is even evidence that in shark rectal gland, FXYD10 interacts functionally with the NKA N terminus to modulate E1–E2 conformational transitions (36). If the CFP on NKA is very close to the YFP on the C terminus of PLM, then the high degree of FRET we observe would be expected. Moreover, the large changes in FRET upon either PLM phosphorylation or mutation of E960 or F28 suggest that large movements occur in the CFP–YFP region, which are related to PLM-dependent functional NKA regulation.
Lastly, it should be noted that established crystal structures are for one NKA–FXYD conformation (the E2PK2 state). In our study with live cells and functional NKA, we were probably sampling the average behavior of multiple conformations. The interaction of PLM with NKA might be different for different physiological conformations. However, it is reassuring that the E960–NKA and F28–PLM sites that we identified from crystal structures have prominent functional effects in real cells.
Conclusion
By combining mutational analysis, FRET, functional assays in live cells, coimmunoprecipitation, and structural modeling, we demonstrated that the corresponding E960–NKA and F28–PLM sites are critical for NKA–PLM FRET and also for the functional effect of PLM on NKA. An additional PLM–NKA interaction domain contributes to the robust physical association of PLM with NKA.
Materials and Methods
Detailed methods are in SI Materials and Methods.
Constructs, Site-Directed Mutagenesis, Cell Culture, and Transfection.
CFP–WT–NKA-α1 and CFP–NKA-α1 mutants with an Ala substitution at F956, E960, E961, L964, F967, or C145, respectively (rat numbering) or a truncation of the C terminus (ΔKETYY) were constructed. WT–PLM–YFP was generated and PLM–YFP residue F28 was mutated to Ala and Cys, respectively. CFP was on the cytosolic NKA N terminus and YFP on the cytosolic PLM C terminus. CFP–NKA mutants were stably transfected into HEK293 cells (using 10 µM ouabain as selecting agent). Cells were cultured in DMEM with 5% (vol/vol) FBS and penicillin/streptomycin and transiently transfected with cDNAs of PLM–YFP using the transfection reagent Lipofectamine 2000.
Average and Detailed FRET Experiments and Coimmunoprecipitation.
Fluorescence imaging was performed using an inverted microscope and MetaMorph software. FRET efficiency was measured as the increase in CFP (donor) fluorescence upon YFP (acceptor) photobleach. YFP was progressively photobleached as described previously (10, 13). FRET was measured at various PLM–YFP expression levels, plotting FRET efficiency for single regions of interest (Fig. S1) as a function of relative YFP expression. Coimmunoprecipitation experiments were performed as described previously (7, 10).
[Na+]i and NKA-Mediated Na+ Extrusion Measurements and Statistical Analysis.
NKA-mediated Na+ extrusion was measured as a function of [Na+]i in cells expressing NKA alone or in combination with WT–PLM. Cells were loaded for 2 h with the fluorescent indicator SBFI acetoxymethyl ester (SBFI-AM) and dual excitation-single emission fluorescense microscopy was used. Data are expressed as means ± SEM. Statistical discriminations used Student’s t test (paired when appropriate) with P < 0.05 considered significant.
Supplementary Material
Acknowledgments
We thank Khanha Dao, Jeffrey H. Elliott, and Garrick K. Yuen for technical help; Dr. Kenneth S. Ginsburg for comments on the manuscript; and Dr. Seth Robia for valuable intellectual contributions. This work was supported by National Institutes of Health (NIH) Grant R01-HL81562 (to D.M.B.). Molecular graphics images were produced using the chimera package from Resource for Biocomputing, Visualization, and Informatics at the University of California at San Francisco (supported by NIH P41 RR001081).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207866109/-/DCSupplemental.
References
- 1.Bers DM, Despa S. Na+ transport in cardiac myocytes; Implications for excitation-contraction coupling. IUBMB Life. 2009;61(3):215–221. doi: 10.1002/iub.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bers DM, Despa S. Cardiac myocytes Ca2+ and Na+ regulation in normal and failing hearts. J Pharmacol Sci. 2006;100(5):315–322. doi: 10.1254/jphs.cpj06001x. [DOI] [PubMed] [Google Scholar]
- 3.Crambert G, Fuzesi M, Garty H, Karlish S, Geering K. Phospholemman (FXYD1) associates with Na,K-ATPase and regulates its transport properties. Proc Natl Acad Sci USA. 2002;99(17):11476–11481. doi: 10.1073/pnas.182267299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crambert G, Geering K. FXYD proteins: New tissue-specific regulators of the ubiquitous Na,K-ATPase. Sci STKE. 2003;2003(166):RE1. doi: 10.1126/stke.2003.166.re1. [DOI] [PubMed] [Google Scholar]
- 5.Sweadner KJ, Rael E. The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics. 2000;68(1):41–56. doi: 10.1006/geno.2000.6274. [DOI] [PubMed] [Google Scholar]
- 6.Glitsch HG. Electrophysiology of the sodium-potassium-ATPase in cardiac cells. Physiol Rev. 2001;81(4):1791–1826. doi: 10.1152/physrev.2001.81.4.1791. [DOI] [PubMed] [Google Scholar]
- 7.Despa S, et al. Phospholemman-phosphorylation mediates the beta-adrenergic effects on Na/K pump function in cardiac myocytes. Circ Res. 2005;97(3):252–259. doi: 10.1161/01.RES.0000176532.97731.e5. [DOI] [PubMed] [Google Scholar]
- 8.Han F, Bossuyt J, Despa S, Tucker AL, Bers DM. Phospholemman phosphorylation mediates the protein kinase C-dependent effects on Na+/K+ pump function in cardiac myocytes. Circ Res. 2006;99(12):1376–1383. doi: 10.1161/01.RES.0000251667.73461.fb. [DOI] [PubMed] [Google Scholar]
- 9.Fuller W, et al. FXYD1 phosphorylation in vitro and in adult rat cardiac myocytes: Threonine 69 is a novel substrate for protein kinase C. Am J Physiol Cell Physiol. 2009;296(6):C1346–C1355. doi: 10.1152/ajpcell.00523.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossuyt J, et al. Isoform specificity of the Na/K-ATPase association and regulation by phospholemman. J Biol Chem. 2009;284(39):26749–26757. doi: 10.1074/jbc.M109.047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han F, Bossuyt J, Martin JL, Despa S, Bers DM. Role of phospholemman phosphorylation sites in mediating kinase-dependent regulation of the Na+-K+-ATPase. Am J Physiol Cell Physiol. 2010;299(6):C1363–C1369. doi: 10.1152/ajpcell.00027.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Despa S, Tucker AL, Bers DM. Phospholemman-mediated activation of Na/K-ATPase limits [Na]i and inotropic state during beta-adrenergic stimulation in mouse ventricular myocytes. Circulation. 2008;117(14):1849–1855. doi: 10.1161/CIRCULATIONAHA.107.754051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bossuyt J, Despa S, Martin JL, Bers DM. Phospholemman phosphorylation alters its fluorescence resonance energy transfer with the Na/K-ATPase pump. J Biol Chem. 2006;281(43):32765–32773. doi: 10.1074/jbc.M606254200. [DOI] [PubMed] [Google Scholar]
- 14.Song Q, Pallikkuth S, Bossuyt J, Bers DM, Robia SL. Phosphomimetic mutations enhance phospholemman oligomerization and modulate its interaction with the NA/K-ATPase. J Biol Chem. 2011;286(11):9120–9126. doi: 10.1074/jbc.M110.198036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindzen M, Gottschalk KE, Füzesi M, Garty H, Karlish SJ. Structural interactions between FXYD proteins and Na+,K+-ATPase: alpha/beta/FXYD subunit stoichiometry and cross-linking. J Biol Chem. 2006;281(9):5947–5955. doi: 10.1074/jbc.M512063200. [DOI] [PubMed] [Google Scholar]
- 16.Sweadner KJ, Donnet C. Structural similarities of Na,K-ATPase and SERCA, the Ca(2+)-ATPase of the sarcoplasmic reticulum. Biochem J. 2001;356(Pt 3):685–704. doi: 10.1042/0264-6021:3560685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hebert H, Purhonen P, Vorum H, Thomsen K, Maunsbach AB. Three-dimensional structure of renal Na,K-ATPase from cryo-electron microscopy of two-dimensional crystals. J Mol Biol. 2001;314(3):479–494. doi: 10.1006/jmbi.2001.5137. [DOI] [PubMed] [Google Scholar]
- 18.Li C, et al. Structural and functional interaction sites between Na,K-ATPase and FXYD proteins. J Biol Chem. 2004;279(37):38895–38902. doi: 10.1074/jbc.M406697200. [DOI] [PubMed] [Google Scholar]
- 19.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature. 2009;459(7245):446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 20.Morth JP, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450(7172):1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 21.Zouzoulas A, Blostein R. Regions of the catalytic alpha subunit of Na,K-ATPase important for functional interactions with FXYD 2. J Biol Chem. 2006;281(13):8539–8544. doi: 10.1074/jbc.M512700200. [DOI] [PubMed] [Google Scholar]
- 22.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 23.Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62(4):1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barth P, Schonbrun J, Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci USA. 2007;104(40):15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teriete P, Franzin CM, Choi J, Marassi FM. Structure of the Na,K-ATPase regulatory protein FXYD1 in micelles. Biochemistry. 2007;46(23):6774–6783. doi: 10.1021/bi700391b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahmmoud YA, Vorum H, Cornelius F. Interaction of FXYD10 (PLMS) with Na,K-ATPase from shark rectal glands. Close proximity of Cys74 of FXYD10 to Cys254 in the a domain of the alpha-subunit revealed by intermolecular thiol cross-linking. J Biol Chem. 2005;280(30):27776–27782. doi: 10.1074/jbc.M503150200. [DOI] [PubMed] [Google Scholar]
- 27.Yaragatupalli S, Olivera JF, Gatto C, Artigas P. Altered Na+ transport after an intracellular alpha-subunit deletion reveals strict external sequential release of Na+ from the Na/K pump. Proc Natl Acad Sci USA. 2009;106(36):15507–15512. doi: 10.1073/pnas.0903752106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogawa H, Toyoshima C. Homology modeling of the cation binding sites of Na+K+-ATPase. Proc Natl Acad Sci USA. 2002;99(25):15977–15982. doi: 10.1073/pnas.202622299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imagawa T, Yamamoto T, Kaya S, Sakaguchi K, Taniguchi K. Thr-774 (transmembrane segment M5), Val-920 (M8), and Glu-954 (M9) are involved in Na+ transport, and Gln-923 (M8) is essential for Na,K-ATPase activity. J Biol Chem. 2005;280(19):18736–18744. doi: 10.1074/jbc.M500137200. [DOI] [PubMed] [Google Scholar]
- 30.Bossuyt J, Ai X, Moorman JR, Pogwizd SM, Bers DM. Expression and phosphorylation of the na-pump regulatory subunit phospholemman in heart failure. Circ Res. 2005;97(6):558–565. doi: 10.1161/01.RES.0000181172.27931.c3. [DOI] [PubMed] [Google Scholar]
- 31.Crambert G, Li C, Swee LK, Geering K. FXYD7, mapping of functional sites involved in endoplasmic reticulum export, association with and regulation of Na,K-ATPase. J Biol Chem. 2004;279(29):30888–30895. doi: 10.1074/jbc.M313494200. [DOI] [PubMed] [Google Scholar]
- 32.Arimochi J, Ohashi-Kobayashi A, Maeda M. Interaction of Mat-8 (FXYD-3) with Na+/K+-ATPase in colorectal cancer cells. Biol Pharm Bull. 2007;30(4):648–654. doi: 10.1248/bpb.30.648. [DOI] [PubMed] [Google Scholar]
- 33.Li C, et al. Role of the transmembrane domain of FXYD7 in structural and functional interactions with Na,K-ATPase. J Biol Chem. 2005;280(52):42738–42743. doi: 10.1074/jbc.M508451200. [DOI] [PubMed] [Google Scholar]
- 34.Toustrup-Jensen MS, et al. The C terminus of Na+,K+-ATPase controls Na+ affinity on both sides of the membrane through Arg935. J Biol Chem. 2009;284(28):18715–18725. doi: 10.1074/jbc.M109.015099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han F, Tucker AL, Lingrel JB, Despa S, Bers DM. Extracellular potassium dependence of the Na+-K+-ATPase in cardiac myocytes: Isoform specificity and effect of phospholemman. Am J Physiol Cell Physiol. 2009;297(3):C699–C705. doi: 10.1152/ajpcell.00063.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornelius F, Mahmmoud YA, Meischke L, Cramb G. Functional significance of the shark Na,K-ATPase N-terminal domain. Is the structurally variable N-Terminus involved in tissue-specific regulation by FXYD proteins? Biochemistry. 2005;44(39):13051–13062. doi: 10.1021/bi0504456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.