Abstract
Elongation factor-G–catalyzed translocation of mRNA and tRNAs during protein synthesis involves large-scale conformational changes in the ribosome. Formation of hybrid-state intermediates is coupled to counterclockwise (forward) rotation of the body of the 30S subunit. Recent structural studies implicate intrasubunit rotation of the 30S head in translocation. Here, we observe rotation of the head during translocation in real time using ensemble stopped-flow FRET with ribosomes containing fluorescent probes attached to specific positions in the head and body of the 30S subunit. Our results allow ordering of the rates of movement of the 30S subunit body and head during translocation: body forward > head forward > head reverse ≥ body reverse. The rate of quenching of pyrene-labeled mRNA is consistent with coupling of mRNA translocation to head rotation.
Keywords: translocation mechanism, mRNA movement, ribosome dynamics
One of the biggest challenges in the field of protein synthesis is to understand the role of ribosome structural dynamics in the molecular mechanism of translocation, the process by which mRNA and tRNAs are moved through the ribosome in a process catalyzed by elongation factor (EF)-G. After formation of each peptide bond, the newly deacylated tRNA is moved from the P (peptidyl-tRNA) site into the E (exit) site, and the peptidyl-tRNA is moved from the A (aminoacyl-tRNA) site into the P site. In the first step, the acceptor ends of the tRNAs move from the 50S subunit A and P sites to the P and E sites, respectively, from the classical A/A and P/P states to the hybrid A/P and P/E states (1). Productive translocation involves coupled movement of the anticodon stem-loops (ASLs) of the tRNAs and their associated mRNA codons through the 30S subunit, moving the tRNAs from their hybrid A/P and P/E states to the classical P/P and E/E states, advancing the register of the mRNA by one codon. This process is very slow in the absence of EF-G under normal conditions (1–4).
Cryo-EM and crystallographic and solution biophysical studies have shown that translocation is accompanied by large-scale conformational changes in the ribosome, most notably intersubunit rotation (5–8). In cryo-EM reconstructions of ribosomes containing bound EF-G, the 30S subunit was found to be rotated counterclockwise by 5–10° relative to the 50S subunit, as viewed from the solvent surface of the 30S subunit (9, 10). A requirement for intersubunit movement was demonstrated by creation of an intersubunit disulfide bond between proteins S6 and L2, which selectively abolished EF-G–dependent translocation (11). Intersubunit rotation was observed in solution by ensemble FRET experiments in which energy transfer between fluorescent probes attached to the 30S and 50S subunits was found to change upon binding of EF-G (12). Combined use of chemical footprinting and FRET showed that the tRNAs move into hybrid states upon intersubunit rotation (13). The relation between intersubunit rotation and hybrid-state formation was further established by cryo-EM reconstructions of ribosomes trapped in the rotated state containing tRNAs in positions consistent with the P/E and A/P states (14–16) and by a crystal structure of P/E-state tRNA bound to 70S ribosomes cocrystallized with ribosome recycling factor (17). Single-molecule FRET experiments revealed that rotation of the 30S subunit can happen spontaneously and reversibly, driven solely by thermal energy (18); however, these fluctuations do not lead to productive translocation in the absence of EF-G. Recent ensemble FRET studies have shown that mRNA translocation occurs during, but not precisely synchronized with, reverse (clockwise) rotation of the 30S subunit (19), suggesting that translocation may be coupled to some other dynamic event.
Recently, several lines of evidence have led to an increased focus on the dynamics of the head of the 30S subunit, an autonomous structure consisting of the 460-nucleotide 3′-major domain of 16S rRNA (nucleotides ∼930–1390) and eight ribosomal proteins. Crystal structures of 70S ribosomes revealed the existence of a 14Å constriction between the head and platform of the subunit, creating a steric block to passage of the ASL between the P and E sites of the 30S subunit, thus necessitating movement of the head and/or platform during translocation (20). In cryo-EM reconstructions of a eukaryotic 80S eEF-2 sordarin ribosome complex (21), rotation of the 40S subunit head opened the constriction to ∼20 Å, consistent with possible involvement of head rotation in translocation. More recently, multiparticle refinement of cryo-EM data collected from 70S ribosome EF-G complexes trapped with fusidic acid revealed a structure containing a deacylated tRNA bound in a partially translocated state in which the head of the 30S subunit was rotated by about 18° (22). Rotation of the head resulted in movement of the ASL by 8–10 Å toward the E site, resulting in a chimeric state in which it was bound to P-site elements of the head and E-site elements of the platform. In a recent crystal structure of the 70S ribosome bound to release factor RF3, which is structurally related to EF-G, a combined 15° rotation of the head and 6° body rotation resulted in displacement of the P-site elements of the 30S head by 23 Å, matching the distance traveled by the ASL between the P and E sites during translocation (23). These structural observations suggest that rotation of the 30S subunit head in movement of the tRNA ASLs and their associated mRNA codons is a crucial step in the mechanism of translocation. However, the mechanism of translocation of mRNA and tRNA in the 30S subunit remains unclear. To understand how 30S head rotation is related to translocation, it is necessary to observe the events in solution directly in real time.
In this study, we use an ensemble FRET approach (19) to examine movement of the 30S subunit head during translocation in real time. We observe changes in energy transfer between donor and acceptor fluorescent probes attached to specific sites on the head and body of the 30S subunit after rapid mixing of a pretranslocation complex containing labeled ribosomes with EF-G GTP. The rates of forward (counterclockwise) and reverse (clockwise) head rotation are slower than forward rotation of the 30S subunit body, but faster than reverse body rotation. Coupling of mRNA translocation to forward head rotation is consistent with the observed rate of quenching of a pyrene-labeled mRNA. These results provide a basis for understanding the sequence of dynamic structural events that accompany translocation during protein synthesis.
Results
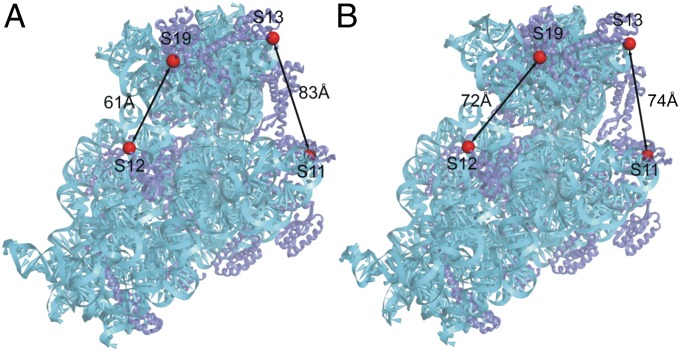
Doubly labeled Escherichia coli 70S ribosomes (constructed as described in Materials and Methods) contained fluorescent Alexa 488 donor and Alexa 568 acceptor probes covalently attached to two different sets of ribosomal proteins at positions based on comparison of crystal structures of ribosomes in which the head is rotated (23) or nonrotated (17) (Fig. 1). In one construct (S11-D/S13-A), the donor probe is attached to the platform at position 101 of protein S11, and the acceptor to the head at position 11 of protein S13. The distance between these two positions decreases by ∼9 Å upon counterclockwise (forward) head rotation (as viewed from the top of the subunit). In the second construct (S12-D/S19-A), the donor probe is attached to position 108 of protein S12 in the 30S body and position 56 of protein S19 in the head. The distance between these two positions increases by ∼11 Å upon forward head rotation. Thus, if EF-G induces head rotation similar to that observed for RF3, we would predict measurable changes in FRET efficiency during translocation, resulting in a decrease for the S11-S13 construct and an increase for the S12-S19 construct during forward head rotation, and conversely for backward rotation.
Fig. 1.
Locations of fluorescent probes in crystal structures of E. coli 70S ribosomes with the head of the 30S subunit trapped in the (A) nonrotated ribosome recycling factor state (17) and (B) rotated RF3 state (23). Distances between the FRET pairs in the doubly labeled S11D-S13A and S12D-S19A constructs for the two conformational states are indicated.
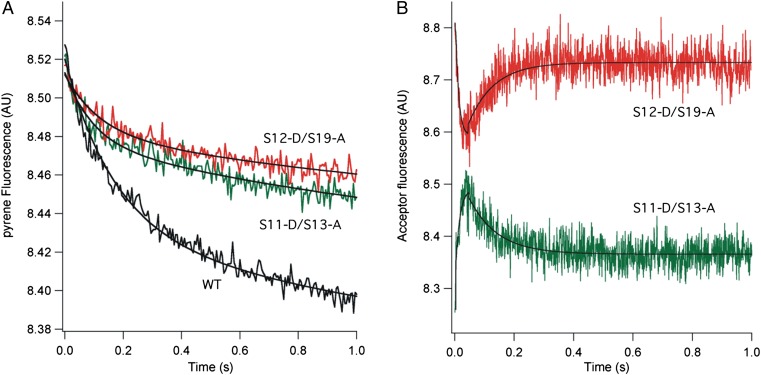
Pretranslocation complexes containing unlabeled or doubly labeled 70S ribosomes were assembled with mv27 mRNA, deacylated tRNAMet (bound to the P site), and N-acetyl-Val-tRNA (bound to the A site), as described in Materials and Methods. Using the assay developed by Joseph and coworkers (24), translocation of mRNA was monitored by quenching of the fluorescence of a pyrene label attached to position +9 of the mRNA after rapid mixing of EF-G GTP with pretranslocation complexes (Fig. 2A). In agreement with previous observations (25–27), the rates of quenching show biphasic behavior. The labeled constructs show apparent translocation initial (k1) and second (k2) rate constants at an initial rate similar to those of WT unlabeled ribosomes (Table 1). Changes in energy transfer between probes in the head and body of the 30S subunit, monitored as changes in acceptor (Alexa 568) emission, revealed rotation of the 30S subunit head. Upon reaction with EF-G GTP, changes in the FRET signal showed EF-G–dependent movement of the 30S subunit head in real time (Fig. 2B). The S11-S13 FRET pair showed a rapid increase in acceptor fluorescence, followed by a slower decrease that reached a plateau at about 0.2 s. Behavior of the S12-S19 FRET pair was anticorrelated with that of the S11-S13 pair, showing a rapid decrease in acceptor emission followed by an increase, also reaching a plateau at around 0.2 s (Fig. 2B). Omission of EF-G, GTP, or N-acetyl-Val-tRNA abolished the changes in FRET signal (Figs. S1 and S2).
Fig. 2.
Rotation of the 30S subunit head during EF-G–catalyzed translocation. (A) mRNA translocation, as followed by quenching of fluorescence of a pyrene dye attached to position +9 of mRNA (24). (B) Rotation of 30S subunit head, followed by changes in FRET efficiency between donor Alexa 488 and acceptor Alexa 568 probes attached to position 101 of S11 and position 11 of S13 for the S11-D/S13-A pair (green) and position 108 of S12 and position 56 of S19 for the S12-D/S19-A pair (red). Pretranslocation 70S ribosome complexes were rapidly mixed with EF-G GTP at time = 0 and pyrene fluorescence (A) or Alexa 568 emission (B) were recorded (19). Each trace represents the average of 10 reactions. AU, arbitrary units; WT, WT unlabeled ribosomes.
Table 1.
Rates of mRNA translocation and 30S head and body rotation during EF-G–catalyzed translocation
| Ribosome construct | mRNA translocation* (s−1) | Forward 30S head rotation (s−1) | Reverse 30S head rotation (s−1) | Forward 30S body rotation (s−1) | Reverse 30S body rotation (s−1) |
| WT | k1 = 6.5 ± 0.5 | — | — | — | — |
| k2 = 0.7 ± 0.1 | |||||
| S11-D/S13-A | k1 = 7.7 ± 1.3 | 80.8 ± 18.5 | 8.8 ± 0.4 | — | — |
| k2 = 0.5 ± 0.1 | |||||
| S12-D/S19-A | k1 = 10.1 ± 1.3 | 76.8 ± 14.2 | 10.8 ± 0.4 | — | — |
| k2 = 0.5 ± 0.1 | |||||
| S6-D/L9-A† | k1 = 8.0 ± 1.3 | — | — | Fast (>200) | 4.5 ± 1.2 |
| k2 = 0.5 ± 0.2 | |||||
| S11-D/L9-A† | ND | — | — | Fast (>200) | 4.1 ± 1.0 |
ND, not determined; S6-D/L9A, construct with donor on S6 and acceptor on L9; S11-D/L9-A, construct with donor on S11 and acceptor on L9; S11-D/S13-A, doubly labeled ribosome construct with donor on protein S11 and acceptor on protein S13; S12-D/S19-A, construct with donor on S12 and acceptor on S19; WT, WT unlabeled ribosomes. Apparent pseudo first-order rate constants for the reaction between pretranslocation. complexes containing deacylated elongator tRNAMet bound to the P site and N-acetyl-Val-tRNA bound to the A site, mixed with EF-G GTP.
*mRNA translocation kinetics obtained from quenching of pyrene-labeled mRNA (24) are biphasic and fitted to two pseudo first-order rate constants.
†Results from ref. 19.
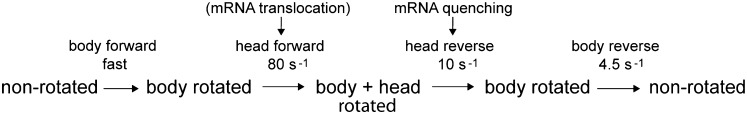
The observation of closely anticorrelated changes in FRET efficiency for the two constructs during translocation (Fig. 2B) validates our interpretation that the changes indeed correspond to rotational movement of the 30S head: the S11-D/S13-A construct shows an initial increase in FRET, followed by a decrease, whereas the S12-D/S19-A construct shows the opposite behavior. Both phases of the reaction can be fit to single-exponential functions, giving pseudo first-order apparent rate constants (Table 1; Fig. S3). The observed changes in energy transfer for the two different FRET pairs give an initial rapid (∼70 s−1) forward (counterclockwise) rotation of the head that is nevertheless slower than the previously observed very fast (>200 s−1) forward intersubunit rotation of the 30S subunit body (19). Reverse (clockwise) rotation of the head to the nonrotated state is slower (∼10 s−1) than forward head rotation, but somewhat faster than reverse rotation of the 30S body (∼5 s−1). The observed apparent rate constants are summarized in Table 1. Thus, together with previously observed rates for 30S body movement (19), we can now order the rates of the main rotational events involving the structural dynamics of the 30S subunit: body forward > head forward > head reverse > body reverse (Fig. 3).
Fig. 3.
Proposed order of dynamic structural events during mRNA translocation. Rates of rotation of the head and body of the 30S subunit, based on FRET experiments and fluorescence quenching of pyrene-labeled mRNA, are from this work (Table 1) and from ref. (19). The nonrotated state is equivalent to the classical state and the body rotated state is equivalent to the hybrid state. Although the rate of mRNA quenching matches the rate of reverse (clockwise) head rotation, translocation of mRNA most likely occurs during forward head rotation, as discussed in the text.
Discussion
The rate of EF-G–catalyzed translocation of mRNA, as measured by k1 of quenching of a 3′-pyrene-labeled mRNA in ref. (24), around 8–10 s−1 at 22° corresponds, within experimental error, to the rate of reverse head rotation (∼9–11 s−1; k2 is very slow, ∼0.5 s−1, and its relationship to the main events of translocation remain unclear). Structural studies have led to the proposal that translocational movement of the ASLs of the tRNAs and their associated mRNA codons is coupled to forward head rotation (22, 23), in apparent contradiction with our findings. However, this can be reconciled by considering the physical events that are responsible for the quenching signal. Quenching is likely due to contact between the pyrene dye attached to position +9 of the mRNA and some part of the ribosome surface at the mRNA entry site to the 30S subunit. Quenching would thus require displacement of the mRNA relative to that part of the ribosome. During forward rotation of the head, both the head and the mRNA rotate together; accordingly, if contact of the pyrene with the head is responsible for the quenching signal, quenching would only be observed upon reverse head rotation. This is consistent with our finding that the quenching rate correlates with the rate of reverse, rather than forward head rotation. The actual rate of initial mRNA movement, relative to the body of the 30S subunit, would thus be about an order of magnitude faster than the rate measured by the quenching assay.
Based on the reasoning presented here and the observed rates of 30S head and body rotation (Table 1), we can propose an order for the structural dynamic events in the ribosome that occur during translocation (Fig. 3). Following peptide bond formation, binding of EF-G GTP triggers a rapid counterclockwise rotation of the 30S subunit body relative to the 50S subunit, accompanied by movement of the tRNAs from their classical A/A and P/P states into A/P and P/E hybrid states. Second is a counterclockwise rotation of the 30S subunit head, which opens the constriction between the 30S P and E sites (20, 21) and moves the tRNA ASLs with their associated mRNA codons into the chimeric ap/P and pe/E hybrid states (22, 23). Next, the head of the 30S subunit relaxes its grip on the ASLs and undergoes reverse (clockwise) rotation, followed by reverse rotation of the 30S body, leaving the ribosome in its nonrotated classical state with mRNA and tRNAs fully translocated into the P/P and E/E states. We note that the rates of reverse head rotation and reverse body rotation differ by only a factor of 2 (Table 1), suggesting that reverse head rotation and reverse body rotation might not be separate, sequential events, but may overlap, such that reverse body rotation begins before completion of head rotation. Indeed, a cryo-EM reconstruction by Spahn and coworkers of an EF-G ribosome complex indicates a state in which the head of the 30S subunit is rotated by 18° but the body by only 4° (22). Together with our findings, their observation raises the possibility that reverse head and body rotation are uncoupled and may not be strictly ordered. Besides the dynamic events involving the 30S subunit, movement of several features of the 50S subunit, including the L1 and L11 stalks and the A-site finger (helix H38 of 23S rRNA), is also linked to translocation. Among the many remaining challenges will be to determine whether and how these and other structural changes are coupled during translocation.
Materials and Methods
Doubly labeled ribosomes were prepared by in vitro reconstitution of 30S ribosomal subunits with donor (Alexa 488) and acceptor (Alexa 568) probes attached to introduced single cysteine residues in proteins S11 or S12 and S19 or S13, respectively, together with the remaining 18 unmodified proteins and 16S rRNA, followed by association with 50S subunits and isolation as 70S ribosomes, as described in detail in SI Materials and Methods. Stopped-flow fluorescence quenching and FRET experiments were carried by rapid mixing of pretranslocation complexes containing a synthetic 24-mer or 27-mer mRNA, respectively, deacylated elongator tRNAMet bound to the ribosomal P site, and N-acetyl-Val-tRNA bound to the P site, with elongation factor EF-G and GTP. Rapid mixing experiments were carried out at 22° using an Applied Photophysics SX-20 stopped-flow apparatus, as previously described (19, 24) and as detailed in SI Materials and Methods. For FRET experiments, the Alexa 488 donor was excited at 490 nm and the Alexa 568 acceptor fluorescence emission was collected using a 590-nm long-pass filter. mRNA translocation rates were obtained using the method of Studer et al. (24), by following quenching of fluorescence of the pyrene residue attached to position +9 of the mRNA. The pyrene dye was excited at 343 nm and fluorescence emission recorded using a 375-nm long-pass filter.
Supplementary Material
Acknowledgments
We thank Dmitri Ermolenko for developing the experimental approach used in these studies and for many helpful discussions, Sarah Gerhardt and Robyn Hickerson for providing mutant protein constructs, and Laura Lancaster and John Paul Donohue for expert advice on experimental methods and computational support, respectively. These studies were supported by National Institutes of Health Grant GM-17129.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218999109/-/DCSupplemental.
References
- 1.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342(6246):142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 2.Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutkevitch NM, Spirin AS. Factor-free (“non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J Mol Biol. 1976;101(4):537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- 3.Southworth DR, Brunelle JL, Green R. EFG-independent translocation of the mRNA:tRNA complex is promoted by modification of the ribosome with thiol-specific reagents. J Mol Biol. 2002;324(4):611–623. doi: 10.1016/s0022-2836(02)01196-8. [DOI] [PubMed] [Google Scholar]
- 4.Semenkov YP, Shapkina TG, Kirillov SV. Puromycin reaction of the A-site bound peptidyl-tRNA. Biochimie. 1992;74(5):411–417. doi: 10.1016/0300-9084(92)90080-x. [DOI] [PubMed] [Google Scholar]
- 5.Noeske J, Cate JH. Structural basis for protein synthesis: Snapshots of the ribosome in motion. Curr Opin Struct Biol. 2012 doi: 10.1016/j.sbi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez CE, Gonzalez RL., Jr In vitro and in vivo single-molecule fluorescence imaging of ribosome-catalyzed protein synthesis. Curr Opin Chem Biol. 2011;15(6):853–863. doi: 10.1016/j.cbpa.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrov A, et al. Dynamics of the translational machinery. Curr Opin Struct Biol. 2011;21(1):137–145. doi: 10.1016/j.sbi.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12(6):674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank J, Agrawal RK. Ratchet-like movements between the two ribosomal subunits: their implications in elongation factor recognition and tRNA translocation. Cold Spring Harb Symp Quant Biol. 2001;66:67–75. doi: 10.1101/sqb.2001.66.67. [DOI] [PubMed] [Google Scholar]
- 10.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114(1):123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 11.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA. 2007;104(12):4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370(3):530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 13.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13(9):1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32(2):190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Julián P, et al. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci USA. 2008;105(44):16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466(7304):329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 17.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30(5):578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18(4):457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 21.Spahn CM, et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23(5):1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468(7324):713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA. 2012;18(2):230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327(2):369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 25.Munro JB, et al. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci USA. 2010;107(2):709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343(5):1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 27.Shi X, Chiu K, Ghosh S, Joseph S. Bases in 16S rRNA important for subunit association, tRNA binding, and translocation. Biochemistry. 2009;48(29):6772–6782. doi: 10.1021/bi900472a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.