Abstract
Wheat supplies about 20% of the total food calories consumed worldwide and is a national staple in many countries. Besides being a key source of plant proteins, it is also a major cause of many diet-induced health issues, especially celiac disease. The only effective treatment for this disease is a total gluten-free diet. The present report describes an effort to develop a natural dietary therapy for this disorder by transcriptional suppression of wheat DEMETER (DME) homeologs using RNA interference. DME encodes a 5-methylcytosine DNA glycosylase responsible for transcriptional derepression of gliadins and low-molecular-weight glutenins (LMWgs) by active demethylation of their promoters in the wheat endosperm. Previous research has demonstrated these proteins to be the major source of immunogenic epitopes. In this research, barley and wheat DME genes were cloned and localized on the syntenous chromosomes. Nucleotide diversity among DME homeologs was studied and used for their virtual transcript profiling. Functional conservation of DME enzyme was confirmed by comparing the motif and domain structure within and across the plant kingdom. Presence and absence of CpG islands in prolamin gene sequences was studied as a hallmark of hypo- and hypermethylation, respectively. Finally the epigenetic influence of DME silencing on accumulation of LMWgs and gliadins was studied using 20 transformants expressing hairpin RNA in their endosperm. These transformants showed up to 85.6% suppression in DME transcript abundance and up to 76.4% reduction in the amount of immunogenic prolamins, demonstrating the possibility of developing wheat varieties compatible for the celiac patients.
Keywords: gluten intolerance, autoimmune reaction, prophylactic measure
The highly homologous seed storage proteins of wheat and barley dubbed as prolamins are classified on the basis of their physiochemical properties into alcohol-soluble gliadins and insoluble glutenins. The gliadins, based on their chemical composition and electrophoretic mobilities, are further classified into sulfur-poor ω-gliadins and the sulfur-rich α/β- and γ-gliadins (1). The corresponding proteins in barley are named as C-, γ-, B-, and D-hordeins (1). The complex mixture of these proteins also known as “gluten”, in a single bread wheat variety, is comprised of up to 45 different gliadins, 7–16 low-molecular-weight glutenin (LMWg) subunits and 3–6 high-molecular-weight glutenin (HMWg) subunits (2). These proteins cumulatively represent 80% of proteins stored in wheat endosperm and constitute a major source of plant based-dietary proteins consumed worldwide (3). Despite wheat being the major source of dietary proteins, it is also a key determinant of many diet-induced health issues, especially gluten sensitivity, celiac sprue, schizophrenia, dermatitis herpetiformis, and IgE-mediated allergies including anaphylaxis (3–5). Among these disorders, celiac is one of the most common food-born enteropathies in humans, occurring in various frequencies around the globe (6). In addition to eliciting these clinical responses, wheat proteins are also deficient in the essential amino acids, in particular lysine and threonine (1).
Celiac disease is caused by an autoimmune reaction against epitopes of wheat, barley, and rye grain storage proteins. In HLA DQ2- (or DQ8)-positive individuals, exposure to these gluten proteins can lead to a painful chronic erasure of the microvilli of the epithelium in the intestine and to a permanent intolerance of dietary prolamins (4). The autoimmune response results from the resistance to digestion of proline-/glutamine-rich peptides (epitopes) in the prolamins by gastric, pancreatic, and brush-border membrane proteases. Peptides like PFPQPQLPY are taken up through the intestinal mucosa into the lamina propria and initiate the autoimmune response (7). Celiac disease is commonly detected in congenital cases with severe symptoms in early childhood. In increasing numbers of patients, symptoms arise only later in life as a result of bread and pasta consumption (4). If untreated, celiac disease may cause increased morbidity and mortality. Despite its prevalence in all diagnosed populations comprising 24.4 million registered celiac individuals worldwide, the only effective therapy is strict dietary abstinence from these food grains (8). However, because of the multiple presentations of the disease, many sufferers of this disease have not been formally diagnosed.
Because celiac patients are sensitive to the epitopes of different LMWgs and/or gliadins, a general therapy requires elimination of all immunoreactive prolamins (4). The results of two studies conducted in wheat and barley indicated that it is practically possible to engineer wheat varieties lacking immunoreactive prolamins without having major influence on their baking properties (9, 10). The first study demonstrated that gliadins and LMWg subunits are superfluous for baking by mixing recombinant HMWg subunits (HMWDx5 and HMWDy10) with the washed-out wheat flour residues containing starch, soluble protein, fat, fibers, and minerals. The dough kneaded from the mixture showed excellent elasticity and after baking resulted in bread rolls with desired volume and internal structure, leading to the conclusion that the HMWgs are essential for baking quality (9, 11) (SI Text). The second study demonstrated that accumulation of gliadins and LMWgs in barley grains is determined by demethylation of their promoters in the endosperm (10). The author showed by ligation-mediated PCR and sequencing that cytosine of 10 CpGs in the promoters and 4 CpGs in the adjacent coding regions of the B-hordein genes are hypomethylated in the endosperm but are fully methylated in the leaves allowing their specific accumulation in the developing endosperm. It was subsequently confirmed by gel-blot analysis using methylation-sensitive and -insensitive restriction enzymes and mRNA expression (12). In the developing endosperm of high lysine barley mutant Risø1508 (lys3a), demethylation of the B-hordein promoters does not take place, which results in transcriptional suppression of these proteins with compensatory increase in the amount of lysine-rich proteins responsible for the high lysine content of the mutant grains (10). Later genomic sequencing of the D-hordein gene promoter using bisulfite treated DNA showed presence of a CpG island (CpGi) and its role in keeping the promoter unmethylated in the leaf, as well as endosperm (13). These results suggest that HMWgs are differentially regulated, and their expression in endosperm solely depends on the removal of repressor(s) and/or induction of tissue-specific transcription factors. This differential regulation of HMWgs became the rationale of the present study for selective elimination of undesirable prolamins.
This form of transcriptional control by DNA methylation and demethylation was later rediscovered in Arabidopsis in connection with gene imprinting (14). Additionally, the gene, named DEMETER (DME), responsible for active demethylation of the imprinted genes was cloned. DME encodes a bifunctional DNA glycosylase of HhH-GPD superfamily, named after its hallmark helix–hairpin–helix and Gly/Pro-rich loop followed by a conserved aspartate. The glycosylase domain of DME serves as the catalytic center for 5-methylcytosine excision reaction that results in hypomethylation of the imprinted genes (15, 16).
In view of the functional similarity between barley regulatory gene Lys3 and DME, we undertook in this study molecular cloning of barley and wheat DME homologs using tool and techniques of translational genomics: i.e., performed cytogenetic localization of the DME homeologs to specific chromosome arms and studied epigenetic effect of DME suppression on the accumulation of gliadins and LMWgs in wheat endosperm using RNA interference (RNAi). Finally, the potential of this approach for the production of celiac-compatible wheat cultivars is discussed.
Results
CpGis in the Wheat Prolamin Genes.
High homology between the promoters (85–89%) and transcribed (71–79%) regions of wheat HMWg and barley D-hordein genes, and the presence of CpGis bracketing the transcription start site of the D-hordein genes preventing their methylation and subsequent silencing, instigated us to examine wheat and barley prolamin sequences for the presence of CpGis (Dataset S1). Of 47 HMWg sequences derived from different wheat cultivars and representing various HMWg alleles, CpGis could be predicted in 38 cases, leaving 9 cases where no island was detected (SI Text). In contrast, when LMWg and gliadin sequences were analyzed, no CpGi could be detected in any case, except for two α-gliadin sequences (Dataset S1).
Cloning and Cytogenetic Mapping of DME Genes.
A total of 38.9 Mb of high-quality sequence was obtained for three BAC clones, namely 1946D08, 2106P11, and 2159B03, selected after partial sequencing of seven BAC clones identified from the macroarray hybridization of the “Chinese Spring” (CS) BAC library (SI Materials and Methods). Ninety-four percent of 13.4-Mb sequence obtained for 1946D08 was assembled into 10 large contigs covering 147 kb of the BAC insert. Similarly, 88% of 14.5 Mb of the sequence obtained for 2106P11 and 87% of 11 Mb of the sequence obtained for 2159B03 were assembled into five and six large contigs, respectively, covering 157 and 102 kb of the BAC inserts. Analysis of sequences obtained from the above three BAC clones revealed that each of them harbors a full-length DME sequence. Similarly, a 26.64-kb contig encompassing the full-length DME gene was assembled from the sequence of barley BAC clone 27314i4 (SI Materials and Methods).
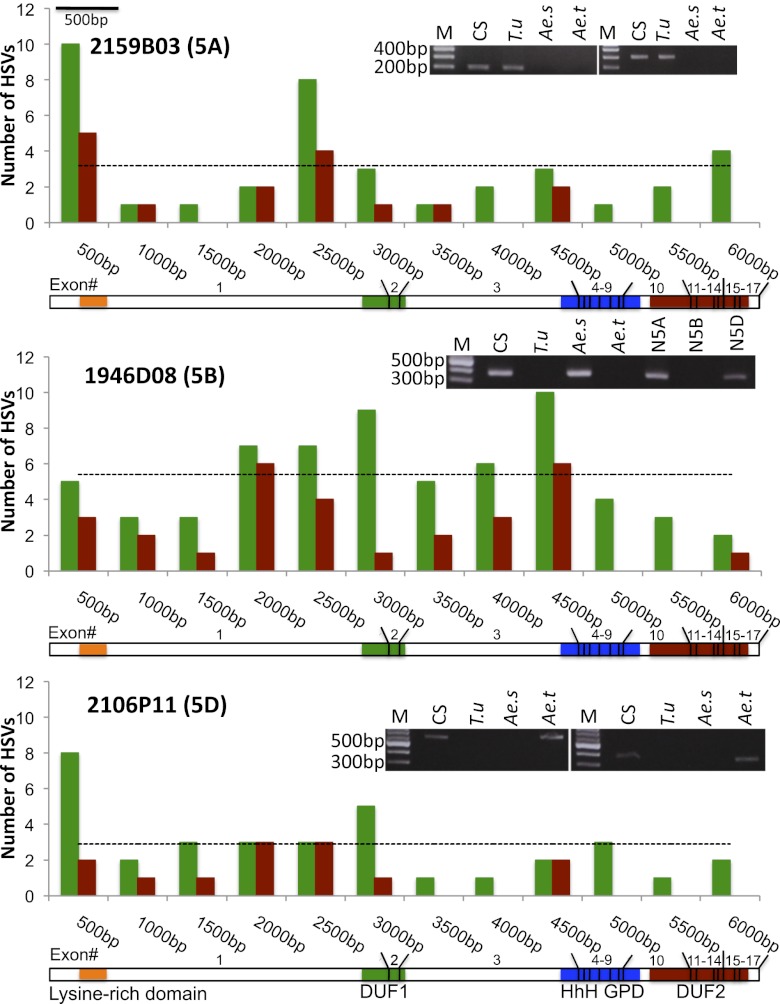
Comparison of the full-length wheat DME sequences with all available wheat ESTs in the public domain revealed that the three homeologs are transcriptionally active (SI Text and Table S1). In addition, comparison of wheat DME homeologs with mapped wheat ESTs showed high homology (e value, 0.0; score, 731) with BE471039 and allowed their assignment to the long arm of wheat group 5 chromosomes (5AL, 5BL, and 5DL; Fig. 1). Assignment of DME homeologs to the specific subgenomes of bread wheat was performed using homeolog-specific primers, designed by tagging their 3′ ends at homeologous sequence variants (HSVs) (Fig. S1 and Table S2). Each of the homeolog-specific primer pairs was used on the genomic DNA of diploid wheat progenitors [Triticum urartu (AA), Aegilops speltoides (BB), and Aegilops tauschii (DD)] with CS [Triticum aestivum (AABBDD)]. These primers allowed unambiguous assignment of 2159B03 to A subgenome, 1946D08 to B subgenome, and 2106P11 to D subgenome of common wheat (Fig. 2). Additionally, the chromosomal and subgenome assignment of DME homeologs was further validated by the use of wheat group 5 nullitetrasomic lines (Fig. 1). Whereas, subchromosomal location of 5B DME homeolog to the subcentromeric bin bracketed by the deletion breakpoints of 5BL-12 [fraction length (FL), 0.08] and 5BL-2 (FL, 0.26), encompassing 98.78 Mb of genomic DNA on 5BL, was determined using 5B-specific terminal and interstitial deletion lines (Fig. 1).
Fig. 1.
Cytogenetic deletion map of wheat and barley DME homologs on wheat group 5 (5A, 5B, and 5D) and barley 5H chromosomes. The green bars show gross chromosomal locations (based on homology with mapped wheat ESTs and/or synteny and colinearity in case of barley), and the red bar shows precise location (based on wheat nullitetrasomic and deletion lines). Subchromosomal localization of wheat DME homeolog on 5B was determined using subgenome specific primers 5B_1946_2 forward (F) and reverse (R) (Table S2) on the genomic DNA of wheat cultivar CS (control), nullitetrasomic lines for group 5 chromosomes, deletion lines for long and short arms of chromosome 5B, and an interstitial deletion line ph1b. TaDME-5B localizes to the subcentromeric bin of the long arm of chromosome 5B (5BL). Specific product indicated by arrow head. M, 100-bp ladder.
Fig. 2.
Bar diagrams showing observed and expected (dotted line) frequencies of HSVs (synonymous changes in green and nonsynonymous changes in red) across the length of wheat DME homeologs. Location of different exons and four functional domains were shown below each bar diagram. PCR amplification profiles obtained using subgenome specific primers on the genomic DNAs extracted from diploid wheat progenitors (T.u, T. urartu “A” subgenome donor; Ae.s, Ae. speltoides most likely “B” subgenome donor; and Ae.t, Ae. tauschii “D” subgenome donor) and CS (AABBDD) was shown at the top of each bar diagram. To confirm assignment to wheat DME homeologs two primer pairs each from BAC clones 2159B03, 1946D08, and 2160P11 were designed and tested (Table S2). The primers for 1946D08 were additionally validated on wheat nullitetrasomic lines for group 5 chromosomes.
Genomic location of the barley DME homolog (HvDME) was determined using rice as a proxy, where wheat ESTs mapped to specific chromosomal bins were used to identify syntenous region on rice chromosome 9, and rice BAC/PAC sequences were searched against mapped barley ESTs. The analysis allowed localization of HvDME to chromosome 5H between centromere (C) and 5HL-15 (Fig. 1).
Nucleotide Diversity Among Wheat DME Homeologs.
The three DME sequences differ in length from each other ranging from 12.27 kb for 2106P11 to 12.63 kb for 1946D08. The observed differences in the length of DME homeologs are mostly attributable to insertions and deletions (InDels) in the introns. A large number of point mutations and small InDels (collectively referred as HSVs) between DME homeologs also exist in exons and contribute to the observed diversity in the protein sequences. A total of 135 HSVs giving a frequency of 22.7 HSVs/kb in exons and 584 HSVs giving a frequency of 90 HSVs/kb in introns was observed. Of 135 HSVs, 60 (44.44%) contribute to amino acid substitutions in at least one of the three DME homeologs (Tables S3 and S4). When expected and observed HSV frequencies were plotted against the distance in nucleotides for three DME homeologs, interestingly, the highest level of nucleotide diversity was observed in TaDME-5B, followed by TaDME-5A and TaDME-5D (Fig. 2; see SI Text for details).
Phylogenetic Analysis of DME.
Comparison of amino acid sequences of DME revealed high levels of similarity within the grass lineage, as well as with Arabidopsis (Fig. 3A). The sizes of five functional domains and the linker regions connecting various domains were kept highly conserved between plants belonging to very distant taxonomic groups. Specifically, sizes of domain of unknown function (DUF)1 ranged from 108 to 111 aa, glycosylase domain [including HhH-GPD and iron-sulfur cluster (FES) motifs] ranged from 191 to 214 aa, linker region 3 and DUF2 ranged from 41 to 45 aa and 245 to 274 aa, respectively, among different plant species (Fig. 3A and Table S5). The HhH-GPD domain showed conservation across kingdoms, suggesting the functional conservation of these proteins in short-patch DNA base excision repair (BER) pathway (Fig. 3 B and C).
Fig. 3.
(A) Phylogenetic analysis of DME homologs from Arabidopsis (REFSEQ accession no. NP_001078527.1), rice (GenBank accession no. BAF04322.1), sorghum (GenBank accession no. JF683319), barley (GenBank accession no. CAQ58412.1), and wheat (GenBank accession nos. JF683316–JF683318) showing high level of conservation at sequence, as well as structural, levels. (B) Diagrammatic representation of DME protein showing four conserved domains with a magnified view of helix–hairpin–helix domain and iron sulfur cluster underlying the active site of the enzyme showing homology among 5-methylcytosine DNA glycosylases obtained from different organisms. (C) Three-dimensional structural model of the glycosylase domain of EndoIII in complex with DNA to show close functional conservation among different glycosylases.
Transcriptional Suppression of DME Homeologs and Its Influence on Accumulation of Prolamins.
Of 56 putative T0 transformants produced using the DME silencing hairpin construct (p728), integration of transgene has been confirmed in 21 (37.5%) cases using a construct-specific primer pair (Table S2 and Fig. S2). Of these 21 transformants, 1 plant died before maturity; thus, further analysis is based on the remaining 20 plants. Immature spikes from these 20 T0 plants were collected for RNA extraction and quantitative (q)RT-PCR analysis to study DME transcript level. The results of qRT-PCR showed suppression in DME transcript abundance ranging from 3.0 to 85.2% in different transformants (Table S6). In view of the chimerism problem associated with the biolistic approach, 10% of the bulk harvested T1 grains from individual T0 transformants were planted in glasshouse, where 4.5–71.7% of progeny plants belonging to different transformants showed faithful inheritance of the transgene (Table S6 and Fig. S2). A wide range of plants showing transgene inheritance in the T1 progeny confirmed chimeric origin of these plants. Immature spikes from the T1 plants showing faithful inheritance of transgene were collected for RNA extraction and qRT-PCR analysis (SI Materials and Methods). The level of suppression observed for DME using qRT-PCR performed on the cDNAs prepared from the immature T2 grains belonging to different transformant families ranged from 3.5 to 85.6% (Table S6 and Dataset S2).
The selected transformants showing suppression in DME transcript abundance close to or more than 50% were examined for the accumulation of immunogenic prolamins in their endosperm by sequential extraction of seed storage proteins, followed by SDS/PAGE and RP-HPLC (SI Materials and Methods). The PAGE and HPLC results revealed that different transformants show elimination of different prolamin groups (Fig. 4 and Figs. S3–S5), for instance, P32F2 and P31D12, progeny of T0 transformant 10-728, showed reduced accumulation of α-gliadins, respectively, by 56.2% and 37.6% and γ-gliadins by 21.8% and 20.7%. These lines also show reduced accumulation for LMWgs, which together with gliadins account for ∼67% reduction in the amount of total immunogenic prolamins (Dataset S2). However, a slight increase in the amount of ω-gliadins and a significant increase in the amount of HMWgs was also observed in these lines, which will be beneficial in maintaining the rheological properties of these lines (Fig. 4). Not only reduction in amount of immunogenic prolamins but also total elimination of specific gliadins and/or LMWgs was also observed in different transformants (Fig. S3).
Fig. 4.
RP-HPLC and SDS/PAGE of gliadins and glutenin fractions extracted from the T2 grains of the two progeny plants (P31D12 and P32F2) of transformant 10-728 (Table S6) expressing DME silencing hpRNA and the wild-type “Brundage 96” (B96). A random sample of T2 grains with their respective thousand kernel weights (TKWs) is shown at right.
The level of reduction for immunogenic prolamins (LMWgs and gliadins) calculated using the area covered by specific prolamin groups on the HPLC chromatograms ranged from 45.2 to 76.4% and was used as an independent variable to regress on the values obtained for DME transcript level expressed as percentage of suppression (Table S6 and Dataset S2). The two variables regressed very well on each other, with a r2 value of 0.877, suggesting 87.7% correspondence among the two variables and indicating that level of DME transcript determines the amount and type of immunogenic prolamins accumulated in the wheat grain (Fig. S6).
Discussion
Celiac disease is unique among other autoimmune disorders as the elicitor or the environmental factor responsible for the disease onset in the genetically predisposed individuals is well known, and elimination of which has shown to remit disease symptoms (4). Complete gluten abstinence for life has, so far, been the only treatment for the disease but is quite difficult to comply with. In addition, following a gluten-free diet has adverse influence on the gut microbiota, deteriorating gut health and also the ability of fecal residues to stimulate the immune system (17). Thus, finding alternative therapy is always a thrust area of research. In this direction, several attempts and suggestions were made where efforts to identify wheat cultivars or close relatives naturally deficient in immunogenic prolamins have always been a very appealing approach (4). Despite of several efforts to screen for wheat and barley accessions including old landraces, cultivars, and deletion lines, none of the tested materials was completely nontoxic for celiac patients and, thus, could not be recommended for general consumption by all celiac patients (refs. 18–21; consult SI Text). Later efforts have also been made to develop wheat and barley lines expressing RNAi constructs targeting specific gliadin and hordein groups, respectively, with some success (22–25). Another approach followed in this direction made use of various “glutenases” of plant, bacterial, and fungal origin for oral enzyme therapy (cf. ref. 26). In view of the practicality issues, an epigenetic approach was adopted and reported in the present study; in the light of current results, it is likely to identify wheat transformants showing elimination of all gliadins and LMWgs. In addition, this approach is appropriate for almost all celiac patients except for very few cases that are registered to show sensitivity against HMWgs.
Cloning and characterization of 5-methylcytosine DNA glycosylases from wheat and barley is the first effort to explore the active demethylation pathway in the Triticeae cereals. Understanding this pathway is not only important to unravel the mechanism of gene imprinting or “parent-of-origin” effect in the developing endosperm (14) but also for epigenetic elimination of immunoreactive prolamins from the developing grains (8). Cloning of the DME gene from barley using the structural and functional information existing for Arabidopsis DME gene is a perfect example of “translational genomics”, where information developed in a model species was translated in an agronomically relevant plant species. Later, the information that became available from barley was used to clone and characterize DME homeologs from bread wheat. Additionally, the genomic and cytogenetic resources existing for wheat, barley, and rice were deployed for subgenomic, chromosomal, and subchromosomal localization of the genes.
As expected on the basis of functional similarities between barley regulatory gene Lys3 and DME, silencing of wheat DME homeologs using a hairpin (inverted-repeat) construct derived from the conserved N-terminal region of DME enzyme resulted in significant reduction in the amount of gliadins and glutenins accumulated in developing endosperm. Interestingly, different transformants showed eliminations and/or reductions in the amounts of specific gliadin and/or glutenin family members, which suggests the complex transcriptional regulation of different gliadins and LMWgs by the DME homeologs. This might be a consequence of subfunctionalization following polyploidization events that occurred during the evolution of the common wheat from its progenitors. Pseudogenization, concerted evolution, subfunctionalization, or neofunctionalization of ortho(homeo)/paralogous copies of genes in a gene family are commonly reported mechanisms of evolution in polyploids (27–30).
In summary, these results suggest differential regulation of specific prolamin groups by the different DME homeologs, the expression level of which varies in the individual transformants and also within their progenies, which depend upon the their level of suppression determined by the number and type of siRNAs generated during the processing of double-stranded hairpin (hp)RNA (compare with SI Text). In addition, another layer of complexity arises from the variation in the number and site of integrations and their segregation pattern. This intricate pattern of regulation made it clear that it is important to obtain plants showing complete silence (knockout) of DME homeologs to obtain wheat cultivars compatible for general use by celiac patients.
In perspective, the transformants described in the present report already have lines deficient in α/β- and γ-gliadins and LMWgs, which are the known sources of most immunogenic epitopes (31, 32). Thus, these lines themselves hold good potential to be tested for immunotoxicity by T-cell assays (33), followed by feeding trials on model organisms, including DQ2/DQ8+ transgenic mice, gluten sensitive apes, and/or celiac patients showing sensitivities against epitopes derived from α/β- and/or γ-gliadins. Once tested, these lines could be released with proper labeling for consumption by specific celiac patients. To obtain the wheat cultivars for general use by all celiac patients, selected transformants could be converted to doubled haploids by microspore culture and crosses could be made between different transformants showing elimination of specific prolamins to obtain plants showing complete or near complete elimination of all immunogenic prolamins. Site-directed mutagenesis of DME homeologs using designer transcription activator like effector nuclease (dTALEN) is also a viable option to obtain DME knockout lines.
Materials and Methods
Plasmid Construction.
A 938-bp hairpin including a 185-bp stem derived from the first exon of DME homeologs (Fig. S7) and a 568-bp loop derived from second intron of wheat TAK14 (AF325198) gene was synthesized from GeneScript. The fragment was ligated between a wheat endosperm–specific 1Dy HMWg promoter and the nopaline synthase (Nos) terminator in modified pGEM.HMWGlut.nos vector (R. J. Henry, Southern Cross University, Lismore, Australia).
Transformation Procedure.
The hpRNA (p728; HMWGlut.hpRNA.nos; Fig. S8) and BAR (pDPG165; ref. 34) plasmids were used in 2:1 molar ratio to cotransform scutellar calli of wheat variety “Brundage 96” by microprojectile bombardment according to Okubara et al. (35) with the following modifications. Both shoot and root regeneration media are supplemented with 1.5 mg/L bialaphos for initial selection of transformants. The survivors were later transferred to the selective media with increased quantity of herbicide (up to 5 μg/mL) to reduce the number of false positives. Integration of cassettes expressing DME-silencing hpRNA was confirmed by PCR using construct-specific primers followed by sequencing of the PCR product (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank P. Reisenauer and E. Mackenzie and Drs. N. Ankrah, J. L. Ullman, and R. Brueggeman for field and laboratory assistance. This work was supported by National Institutes of Health Grants GM080749-01A2 and 2R42DK072721-02, Life Sciences Discovery Fund Grant 3143956, a Mercator Professorship from the German Research Foundation (to D.v.W.), and Programme of Introducing Talents of Discipline to Universities Project B07017 to Key Laboratory of Vegetation Ecology, Ministry of Education, People's Republic of China.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FM164415.1, JF683316, JF683317, and JF683318).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217927109/-/DCSupplemental.
References
- 1.Shewry PR, Halford NG. Cereal seed storage proteins: Structures, properties and role in grain utilization. J Exp Bot. 2002;53(370):947–958. doi: 10.1093/jexbot/53.370.947. [DOI] [PubMed] [Google Scholar]
- 2.Payne PI. Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Annu Rev Plant Physiol. 1987;38:141–153. [Google Scholar]
- 3.Tatham AS, Shewry PR. Allergens to wheat and related cereals. Clin Exp Allergy. 2008;38(11):1712–1726. doi: 10.1111/j.1365-2222.2008.03101.x. [DOI] [PubMed] [Google Scholar]
- 4.Osorio C, et al. Targeted modification of wheat grain protein to reduce the content of celiac causing epitopes. Funct Integr Genomics. 2012;12(3):417–438. doi: 10.1007/s10142-012-0287-y. [DOI] [PubMed] [Google Scholar]
- 5.Samaroo D, et al. Novel immune response to gluten in individuals with schizophrenia. Schizophr Res. 2010;118(1-3):248–255. doi: 10.1016/j.schres.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abadie V, Sollid LM, Barreiro LB, Jabri B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol. 2011;29:493–525. doi: 10.1146/annurev-immunol-040210-092915. [DOI] [PubMed] [Google Scholar]
- 7.Shan L, et al. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297(5590):2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 8.von Wettstein D. Mutants pave the way to wheat and barley for celiac patients and dietary health. In: Shu QY, editor. Induced Plant Mutations in the Genomics Era. Rome: Food and Agriculture Organization of the United Nations; 2009. pp. 187–190. [Google Scholar]
- 9.Bauer I. 2006. Produktion funktioneller Weizen-speicherproteine in transgenen Stämmen der Hefe Saccharomyces [Production of functional wheat storage proteins in transgenic strains of the yeast Saccharomyces]. PhD dissertation (Technical Univ. Berlin, Berlin, Germany), 1–178 pp. German.
- 10.Sørensen MB. Methylation of B-hordein genes in barley endosperm is inversely correlated with gene activity and affected by the regulatory gene Lys3. Proc Natl Acad Sci USA. 1992;89(9):4119–4123. doi: 10.1073/pnas.89.9.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieffer R, Wieser H, Bauer I, Hoffmann R, Meuser F. Functionality of glutenin subunits produced by transgenic yeast. In: Lookhart GL, Ng PKW, editors. Gluten Proteins 2006. St. Paul: American Association of Cereal Chemists, Inc.; 2007. pp. 54–57. [Google Scholar]
- 12.Radchuk VV, Sreenivasulu N, Radchuk RI, Wobus U, Weschke W. The methylation cycle and its possible functions in barley endosperm development. Plant Mol Biol. 2005;59(2):289–307. doi: 10.1007/s11103-005-8881-1. [DOI] [PubMed] [Google Scholar]
- 13.Sørensen MB, Müller M, Skerritt J, Simpson D. Hordein promoter methylation and transcriptional activity in wild-type and mutant barley endosperm. Mol Gen Genet. 1996;250(6):750–760. doi: 10.1007/BF02172987. [DOI] [PubMed] [Google Scholar]
- 14.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110(1):33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J-K. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mok YG, et al. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc Natl Acad Sci USA. 2010;107(45):19225–19230. doi: 10.1073/pnas.1014348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Palma G, Nadal I, Collado MC, Sanz Y. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. Br J Nutr. 2009;102(8):1154–1160. doi: 10.1017/S0007114509371767. [DOI] [PubMed] [Google Scholar]
- 18.Spaenij-Dekking L, et al. Natural variation in toxicity of wheat: Potential for selection of nontoxic varieties for celiac disease patients. Gastroenterology. 2005;129(3):797–806. doi: 10.1053/j.gastro.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 19.van den Broeck HC, et al. Removing celiac disease-related gluten proteins from bread wheat while retaining technological properties: A study with Chinese Spring deletion lines. BMC Plant Biol. 2009;9:41. doi: 10.1186/1471-2229-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Broeck HC, et al. Presence of celiac disease epitopes in modern and old hexaploid wheat varieties: Wheat breeding may have contributed to increased prevalence of celiac disease. Theor Appl Genet. 2010;121(8):1527–1539. doi: 10.1007/s00122-010-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comino I, et al. Significant differences in coeliac immunotoxicity of barley varieties. Mol Nutr Food Res. 2012;56(11):1697–1707. doi: 10.1002/mnfr.201200358. [DOI] [PubMed] [Google Scholar]
- 22.Becker D, Folck A, Knies P, Lörz H, Wieser H. Silencing the α-gliadins in hexaploid bread wheat. In: Lookhart GL, Ng PKW, editors. Gluten Proteins 2006. St. Paul: American Association of Cereal Chemists, Inc.; 2007. pp. 86–89. [Google Scholar]
- 23.Hansen M, et al. Antisense-mediated suppression of C-hordein biosynthesis in the barley grain results in correlated changes in the transcriptome, protein profile, and amino acid composition. J Exp Bot. 2007;58(14):3987–3995. doi: 10.1093/jxb/erm254. [DOI] [PubMed] [Google Scholar]
- 24.Gil-Humanes J, et al. Silencing of γ-gliadins by RNA interference (RNAi) in bread wheat. J Cereal Sci. 2008;48(3):565–568. [Google Scholar]
- 25.Gil-Humanes J, Pistón F, Tollefsen S, Sollid LM, Barro F. Effective shutdown in the expression of celiac disease-related wheat gliadin T-cell epitopes by RNA interference. Proc Natl Acad Sci USA. 2010;107(39):17023–17028. doi: 10.1073/pnas.1007773107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bethune MT, Khosla C. Oral enzyme therapy for celiac sprue. Methods Enzymol. 2012;502:241–271. doi: 10.1016/B978-0-12-416039-2.00013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doyle JJ, et al. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- 28.Wang HW, et al. Expressional diversity of wheat nsLTP genes: Evidence of subfunctionalization via cis-regulatory divergence. Genetica. 2010;138(8):843–852. doi: 10.1007/s10709-010-9467-7. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, et al. Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc Natl Acad Sci USA. 2011;108(46):18737–18742. doi: 10.1073/pnas.1110552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pont C, Murat F, Confolent C, Balzergue S, Salse J. RNA-seq in grain unveils fate of neo- and paleopolyploidization events in bread wheat (Triticum aestivum L.) Genome Biol. 2011;12(12):R119. doi: 10.1186/gb-2011-12-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitea C, et al. A universal approach to eliminate antigenic properties of α-gliadin peptides in celiac disease. PLoS ONE. 2010;5(12):e15637. doi: 10.1371/journal.pone.0015637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi PF, et al. The gamma-gliadin multigene family in common wheat (Triticum aestivum) and its closely related species. BMC Genomics. 2009;10:168. doi: 10.1186/1471-2164-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molberg Ø, McAdam SN, Lundin KEA, Sollid LM. Studies of gliadin-specific T-cells in celiac disease. Methods Mol Med. 2000;41:105–124. doi: 10.1385/1-59259-082-9:105. [DOI] [PubMed] [Google Scholar]
- 34.Gordon-Kamm WJ, et al. Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell. 1990;2(7):603–618. doi: 10.1105/tpc.2.7.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okubara PA, et al. Engineering deoxynivalenol metabolism in wheat through the expression of a fungal trichothecene acetyltransferase gene. Theor Appl Genet. 2002;106(1):74–83. doi: 10.1007/s00122-002-1066-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.