Abstract
Contemporary high-throughput technologies permit the rapid identification of transcription factor (TF) target genes on a genome-wide scale, yet the functional significance of TFs requires knowledge of target gene expression patterns, cooperating TFs, and cis-regulatory element (CRE) structures. Here we investigated the myogenic regulatory network downstream of the Drosophila zinc finger TF Lame duck (Lmd) by combining both previously published and newly performed genomic data sets, including ChIP sequencing (ChIP-seq), genome-wide mRNA profiling, cell-specific expression patterns of putative transcriptional targets, analysis of histone mark signatures, studies of TF cooccupancy by additional mesodermal regulators, TF binding site determination using protein binding microarrays (PBMs), and machine learning of candidate CRE motif compositions. Our findings suggest that Lmd orchestrates an extensive myogenic regulatory network, a conclusion supported by the identification of Lmd-dependent genes, histone signatures of Lmd-bound genomic regions, and the relationship of these features to cell-specific gene expression patterns. The heterogeneous cooccupancy of Lmd-bound regions with additional mesodermal regulators revealed that different transcriptional inputs are used to mediate similar myogenic gene expression patterns. Machine learning further demonstrated diverse combinatorial motif patterns within tissue-specific Lmd-bound regions. PBM analysis established the complete spectrum of Lmd DNA binding specificities, and site-directed mutagenesis of Lmd and additional newly discovered motifs in known enhancers demonstrated the critical role of these TF binding sites in supporting full enhancer activity. Collectively, these findings provide insights into the transcriptional codes regulating muscle gene expression and offer a generalizable approach for similar studies in other systems.
The Drosophila larval somatic muscles are multinucleated myotubes, each having unique properties determined by a population of mononucleated myoblasts termed founder cells (FCs). FCs fuse with a group of neighboring muscle cells called fusion-competent myoblasts (FCMs) to form muscle precursors, with the final size of the muscle determined by the number of fusion events (1, 2). The expression patterns of the muscle identity TFs are responsible for the diversity of FC and myotube identities (3). However, FCMs are also a relatively heterogeneous population of myoblasts that possess their own developmental transcriptional regulatory networks (4–8). A central player in the specification of FCMs is a member of the Gli superfamily of zinc finger TFs, Lame duck (Lmd) (5, 7, 8). The absence of multinucleated somatic myotubes in lmd mutants together with the restricted expression of Lmd suggest that the effects of this TF are autonomous to FCMs (5, 8).
A recent study combined expression profiling and ChIP followed by microarray analysis (ChIP-chip) to dissect the regulatory network downstream of Lmd. These experiments used tiling microarrays covering only about half of the genome in large 3-kb fragments and showed significant overlap among Lmd- and Mef2-bound regions (4). Surprisingly, these findings uncovered divergent regulatory effects of Lmd and Mef2, with different genes showing either additive, cooperative, or antagonistic responses to these TFs. However, because the expression patterns of all of the candidate target genes were unknown, it was not possible to assign cell-specific functions of these TFs to each putative target. Furthermore, because the Lmd binding site was unknown, no site-directed mutageneses of Lmd motifs were undertaken, leaving the molecular basis of Lmd regulation unresolved. Such analyses are critical to confirm that direct protein-DNA interactions are necessary for enhancer activity, because ChIP data do not distinguish between this mechanism and TF interactions with other proteins that bind to DNA, and TF binding alone does not always predict gene activity (9).
To address these unanswered questions and to increase the sensitivity and genome-wide coverage of Lmd chromatin occupancy, we used ChIP sequencing (ChIP-seq) to investigate the Lmd binding in purified primary mesodermal cells. Previously published histone mark signatures (10), TF cooccupancy (11), and newly performed machine learning, Lmd binding site determination, and site-directed mutagenesis of Lmd and other key regulatory motifs revealed that Lmd directly orchestrates a diverse network of target genes in FCMs through collaboration with additional mesodermal regulators.
Results and Discussion
An atlas of mesodermal enhancers was described based on in vivo binding of key mesodermal regulators including Twist (Twi, a master regulator of the mesoderm), Tinman (Tin, a regulator of the dorsal mesoderm), and myocyte enhancer factor 2 (Mef2, a regulator of muscle differentiation) [see Fig. S1 for cell types and TF expression patterns (11)]. These TFs function in various combinations to confer both general and subtype-specific (including FC and FCM) mesodermal properties (12). To more thoroughly characterize the FCM gene regulatory network, we first defined a larger collection of FCM enhancers. Because Twi is both necessary and sufficient for formation of all mesodermal derivatives (13), and Mef2 is a myogenic TF with a similar mutant phenotype as Lmd in the somatic mesoderm (5, 7, 8, 14), we reasoned that genomic regions bound by Twi and/or Mef2 and associated with genes expressed in FCMs (Dataset S1) may be enhancers for those genes. To test this hypothesis, we examined 18 such regions for transcriptional activity using transgenic reporter assays (Dataset S1; Fig. S2) (15, 16). Although only 2 of these regions were active in FCMs (Fig. S2), 13 were active in other mesodermal derivatives (Dataset S1). During the course of these studies, the in vivo binding profile of Lmd was published using ChIP-chip tiling arrays containing 3-kb regions encompassing about half the genome (4). Five of the regions we tested were cobound by Lmd, with one being active in FCMs (Dataset S1; Fig. S2). However, because our tested sequences (which were on average 745 bp) are much shorter than the microarray regions and the entire genome was not previously examined, we decided to increase the extent and resolution of Lmd in vivo binding using ChIP-seq. (17).
Genome-Wide Map of Lmd-Bound Genomic Regions.
We previously generated a compendium of gene expression profiles based on known genetic perturbations of myogenesis, used this resource to predict cell-specific mesodermal gene expression patterns, and validated hundreds of these predictions at single cell resolution using whole embryo in situ hybridizations (6, 18, 19). These studies allowed us to functionally divide gene expression in the somatic mesoderm into three categories, which include expression only in FCs (referred to here as FC), only in FCMs (FCM), and in both cell types (FCM+FC) (Dataset S1). Interestingly, the expression of the majority (88%) of FCM and FCM+FC genes coincides with the expression of lmd, with expression beginning at stage 11 when FCMs are specified, continuing throughout myoblast fusion, but not being detected in fully differentiated muscle fibers (20) (Dataset S1). This finding suggests that lmd target genes are first activated before cell fusion and are necessary in the processes preceding terminal myotube differentiation and function. Accordingly, we examined in vivo Lmd binding throughout a 2-h window during which FCMs are first specified but before extensive cell fusion with FCs. Because Lmd-positive cells represent only the minority of the entire embryo (Fig. S1), we chose to increase the sensitivity of detecting site-specific Lmd occupancy by purifying primary mesodermal cells using flow cytometry before performing ChIP-seq (6) (Fig. S3).
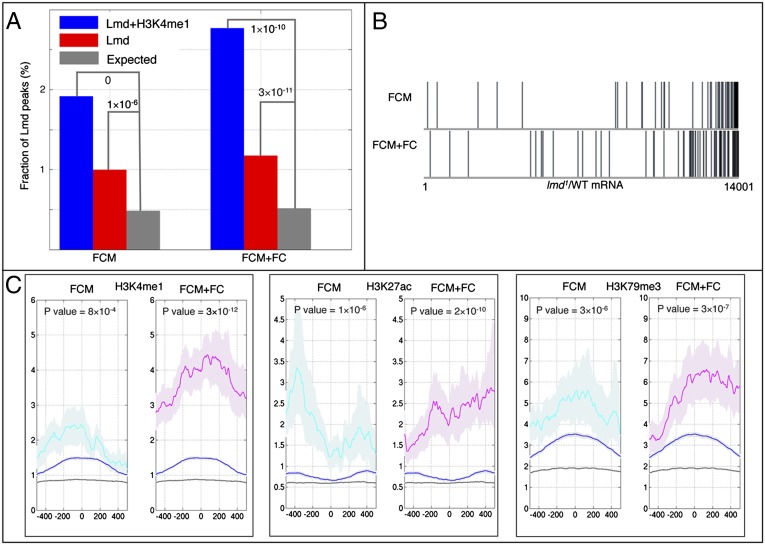
Lmd binding was detected at 5,119 genomic regions, with the majority located in introns and intergenic sequences (Fig. S4A). To provide confirmatory evidence that these regulatory sequences are likely to be functional, we verified that they are enriched for monomethylation of histone H3 on lysine 4 (H3K4me1; Fig. S4B), a histone mark characteristic of enhancers (10, 21). Importantly, Lmd is bound to all eight known enhancers with FCM activity (Fig. S5; Dataset S1), suggesting that Lmd is directly activating these regulatory sequences.
To evaluate the relevance of Lmd binding to the regulation of target genes, we associated significantly bound regions with their nearest neighboring gene and focused on genes that are known to be expressed in FCMs. This analysis revealed that both classes of FCM genes (FCM only and FCM+FC) are enriched for Lmd binding, a result that is significantly more pronounced when Lmd peaks that are also marked with H3K4me1 are considered (Fig. 1A). In addition, the majority of FCM and FCM+FC genes are down-regulated in lmd loss-of-function mutants, suggesting that these genes are activated by Lmd (6) (Fig. 1B). Furthermore, we note a similar enrichment of Lmd binding associated with additional genes that are down-regulated in lmd loss-of-function mutants in our prior genome-wide expression profiling experiment, independent of their assessment by in situ hybridization as being specific to FCMs or FCM+FCs (Fig. S6A). Taken together, these results suggest that Lmd plays a central role in the FCM gene regulatory network, targeting the majority of FCM genes.
Fig. 1.
Lmd-bound CREs associated with FCM and FCM+FC genes. (A) Fraction of Lmd-bound CREs associated with different cell types. Based on gene expression assessed by whole embryo in situ hybridizations, we defined two gene sets: genes up-regulated exclusively in FCMs and genes up-regulated in both FCMs and FCs. We then related Lmd-bound elements with genes such that intergenic sequences are associated with the nearest genes, and intronic sequences are linked to their host gene. The binomial distribution was used to estimate the significance of the observations. (B) mRNA expression profiles of lmd1 mesodermal cells compared with WT mesodermal cells (6). Genes are ranked by Bayesian t-statistic (42) from most likely up-regulated relative to WT (Left) to most likely down-regulated. Responses of previously documented FCM and FCM+FC genes are shown. (C) Enrichment of H3K4me1, H3K27ac, and H3K79me3 signals across Lmd-bound sequences [the vertical axis shows the peak score (10)]. The average signal along control (black line), all Lmd-bound (blue line), FCM Lmd-bound (cyan line), and FCM+FC Lmd-bound (magenta line) sequences. Shading highlights the 25th–75th percentile intervals. The P values are estimated by comparing the average signals along FCM or FCM+FC Lmd-bound sequences with controls using the Wilcoxon rank sum test.
Distinct chromatin modifications are known to be associated with different gene expression states (22). Recently, a series of histone modifications from sorted Drosophila mesodermal nuclei was used to predict mesodermal enhancers (10). In particular, acetylation of histone H3 on lysine 27 (H3K27ac) and trimethylation of histone H3 on lysine 79 (H3K79me3) were associated with active enhancers. Thus, to further evaluate the functionality of Lmd-bound sequences near FCM or FCM+FC genes, we examined these additional aspects of their chromatin state. Indeed, Lmd-bound regions associated with both FCM and FCM+FC genes are enriched for marks of active enhancers, including H3K4me1, H3K27ac, and H3K79me3 (Fig. 1C). Similar results were obtained for additional genes that are down-regulated in lmd loss-of-function mutants, independent of whether they were classified by in situ hybridization as FCM or FCM+FC (Fig. S6B). These results show that the activity state of FCM or FCM+FC genes is mirrored in the associated Lmd-bound candidate cis-regulatory elements (CREs).
Binding Patterns of Other Myogenic TFs Compared with Cell-Specific Lmd-Bound Genomic Regions.
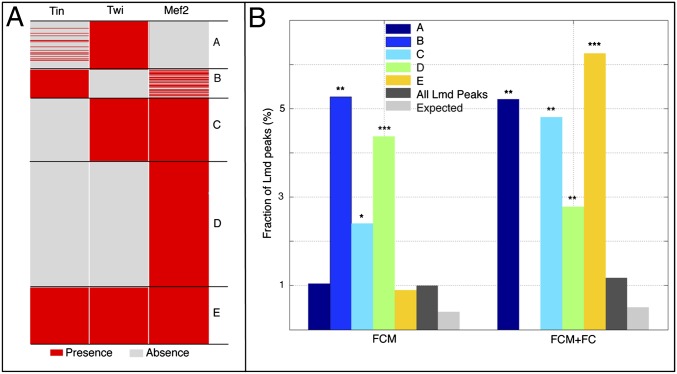
To further dissect the Lmd gene regulatory network, we examined the in vivo cooccupancy of Lmd-bound regions with other key mesodermal regulators, including Twi, Tin, and Mef2 (11). Clustering of Lmd-bound genomic regions occupied by at least one of these TFs (23) revealed five distinct Lmd/TF clusters (Fig. 2), each corresponding to an interaction between Lmd and another mesodermal regulator(s). We then evaluated the enrichment of Lmd/TF clusters associated with FCM genes. This analysis revealed extensive combinatorial regulation of genes expressed in FCMs, as well as similarities and differences in transcriptional input to both FCM and FCM+FC gene sets.
Fig. 2.
Interaction between Lmd and other mesodermal TFs. (A) Clustering Lmd-bound sequences cooccupied by Tin, Twi, and Mef2. K-means clustering based on Euclidean distance was performed according to the presence/absence of the binding of other mesodermal TFs. (B) Distribution of the indicated Lmd peaks associated with FCM or FCM+FC genes. The specificity P value is estimated by comparing the indicated cluster with “all Lmd peaks” using the hypergeometric distribution. (*P < 10−2; **P < 10−3; ***P < 10−4.)
The largest Lmd/TF cluster (39% of all clustered Lmd/TF regions) was cobound by both Lmd and Mef2 but did not include the other two TFs (cluster D; Fig. 2A). Moreover, this cluster was characteristic of both FCM and FCM+FC genes (Fig. 2B). Similarly, an Lmd/TF cluster of regions cobound by Lmd, Mef2, and Twi (cluster C; Fig. 2A) was enriched in both FCM and FCM+FC gene sets.
There were also differences in the combinatorial binding of different TFs to FCM genes. For example, FCM but not FCM+FC genes were cobound by Lmd, Tin, and Mef2, whereas FCM+FC but not FCM genes were cobound by Lmd, Tin, and Twi. The coordinated binding by all four TFs (Lmd, Tin, Twi, and Mef2) predominantly targeted FCM+FC genes (Fig. 2B). The involvement of all four examined TFs in regulating FCM+FC genes likely reflects the broader expression pattern of these genes in both cell types. This result is in agreement with the enrichment of all four TFs with genes down-regulated in lmd loss-of-function mutants (Fig. S7), which is a larger set of genes that includes both FCM and FCM+FC expression patterns. The diversity of transcriptional input, with no single combination of TFs accounting for all aspects of FCM gene expression, supports the heterogeneity of regulatory models available to mediate highly similar but nonidentical gene expression patterns. For example, an enhancer for Mef2 (an FCM+FC gene) that is cobound by all four TFs is indeed active in both cell types (5), whereas the enhancer for blown fuse (blow), an FCM-only gene, is cobound by Twi, Lmd, and Mef2 (4, 24). This heterogeneity of available regulatory models may explain the conservation of expression pattern by nonconserved enhancers, because such enhancers are likely integrating distinct combinations of motifs (25).
These results suggest that Lmd, Twi, and Mef2 are collaborating in various combinations to regulate different subsets of genes that are expressed in FCMs. The cooccupancy of Lmd-bound genomic regions with Mef2 is in agreement with a previous study (4). In addition, the prevalence of cobinding by Twi in these transcriptional codes is interesting, as lmd was discovered as a Twi-dependent gene, and a recently characterized enhancer for lmd is bound by Twi (7, 10, 11). These findings suggest that Twi is directly regulating lmd. Thus, a feedforward loop initiated by Twi may facilitate the regulation of gene expression in somatic FCMs by these two TFs (Fig. S8A) (26). Twi also activates the expression of Tin and Mef2, which often act together to regulate target genes (11, 27), suggesting that numerous feedforward loops are initiated by Twi (28). Similar feedforward loops are initiated by MyoD in initiating and maintaining myogenic differentiation in mammals (29). Interestingly, the binding of Twi is critical for Lmd binding to the blow FCM enhancer (Fig. S8C), suggesting that the combinatorial occupancy of these two TFs may be facilitated by protein-protein interactions that are aided by closely spaced Lmd and Twi binding sites (see Fig. 5A). In summary, these results demonstrate that Lmd is a critical contributor to the somatic myogenic program, with different but overlapping transcriptional codes working together to regulate appropriate spatiotemporal gene expression patterns in FCMs.
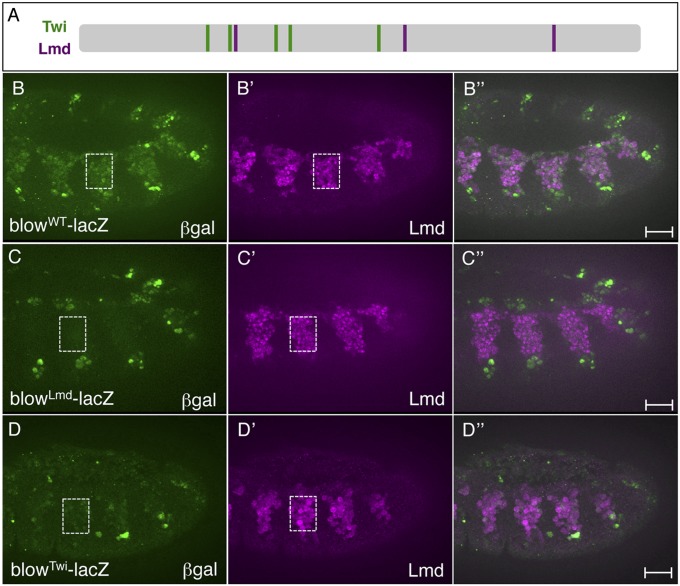
Fig. 5.
Lmd and Twi binding sites are necessary for FCM enhancer activity. (A) Distribution of Twi and Lmd binding sites in the previously characterized blow FCM enhancer (4, 24). Binding sites for Twi were previously published (43), and sequences capable of binding Lmd based on PBM enrichment score are shown. The WT reporter (B) and versions in which Lmd (C) and Twi (D) binding were abolished were designed. The identification of these binding sites and the designs of the mutant versions are described in Dataset S4. (B–D) Stage 11 embryos of the indicated genotypes were stained for β-Gal (B–D) and Lmd (B′–D′) proteins. The β-Gal reporter is extinguished in the somatic and visceral FCMs but not the ectoderm or amnioserosa when Lmd binding sites (blowLmd-lacZ, C) are mutated. The β-Gal reporter is also attenuated in the somatic and visceral FCMs but not the ectoderm when Twi binding sites are mutated (blowTwi-lacZ, D). (Scale bar, 20 μm.)
Machine Learning Identifies Additional Sequence Motifs Enriched Within Cell-Specific Lmd-Bound Genomic Regions.
The preceding analyses suggest similarities and differences in transcriptional input to Lmd-bound CREs, which we hypothesize should involve additional differentially used motifs. To test this idea, we modified a machine learning approach that we previously used to decipher the motifs and enhancers that underlie the gene expression patterns of muscle FCs (16) and the human heart (30). This method identifies specific sequence patterns characteristic of a set of noncoding sequences relying primarily on known TF binding motifs from the literature and available databases. After mapping 1,358 TF binding motifs onto Lmd-bound ChIP-seq and control sequences, we used a support vector machine (SVM) with linear kernels to discriminate Lmd-bound from control sequences based on the occurrence of TF binding motifs (SI Materials and Methods). In a linear SVM where each motif is given a weight and motifs associated with the Lmd-bound sequences receive positive weights, we trained two linear SVMs independently: one for Lmd bound sequences associated with FCM genes and another for Lmd peaks associated with FCM+FC genes. We then selected the top 40 positive motifs with the highest weights in each classifier and grouped these motifs according to the protein families of their respective TFs Most of these TF binding motifs correspond to TFs belonging to a limited number of protein families with a similar binding domain (Fig. 3).
Fig. 3.

Modeling Lmd ChIP-seq peaks reveals motif features associated with FCM gene sets. We trained a linear SVM for each FCM gene set and ranked TF binding motifs according to their linear SVM weights, which reflect the discriminating power of these motifs. For each FCM gene set, we collected the top 40 TF binding motifs and grouped TF binding motifs according to protein families (i.e., binding domain shown in this heatmap) of their respective TFs. A binding domain generally has a set of motifs, and thus corresponds to several rows in this heatmap. The motifs for which TFs are unknown were excluded here (see Dataset S2 for all motifs).
Similar to the heterogeneity of Lmd, Tin, Twi, and Mef2 interactions associated with FCM and FCM+FC genes in vivo, the analysis of Lmd ChIP-seq peaks from FCM and FCM+FC genomic regions revealed differential utilization of previously undescribed TF binding sites derived from the classifiers for these two classes of loci (Fig. 3). We were encouraged that zinc finger motifs, which might represent binding sites for Lmd, were heavily weighted in both FCM and FCM+FC Lmd-bound sequence classes (Fig. 3; Dataset S2). In addition, both FCM classes were enriched for E-box motifs that are bound by members of the basic helix-loop-helix (bHLH) class of TFs and in the mesoderm could include Twi and MyoD (Fig. 3; Dataset S2), although in vivo binding by Twi (Fig. 2) suggests that many of the identified E-boxes may be occupied by this TF. A similar enrichment of zinc finger and E-box motifs was seen among additional genes that are down-regulated in lmd loss-of-function mutants (Dataset S2).
Interestingly, neither FCM- nor FCM+FC-associated Lmd-bound sequences were enriched for Mef2 motifs, despite Mef2 coclustering with Lmd and other TFs near both of the FCM gene sets (Fig. 2). This result is in agreement with a separate study that failed to document an enrichment of Mef2 motifs among Mef2-occupied genomic regions at early developmental stages (31). Thus, the transcriptional response specificity of Mef2 must invoke cooperation with cofactors or other TFs to correctly determine DNA binding site selectivity (32). Alternatively, Mef2 does not directly bind to DNA but is coimmunoprecipitated with other TFs that are bound directly to DNA. In support of the former possibility, Mef2 has been shown to directly regulate gene activity in FCMs (33).
We also note an enrichment of motifs for the Forkhead (Fkh) class of TFs among both FCM+FC and FCM Lmd-bound sequences. We have previously shown that the Fkh class of TFs plays a critical role in directing tissue-specific mesodermal gene expression patterns in the visceral, somatic, and cardiac mesoderm (34). These results imply that Fkh motifs also contribute to the transcriptional response specificity of FCM genes. Other positively weighted motifs suggest candidate TF binding motifs and the corresponding TFs involved in the regulation of FCM and FCM+FC gene expression (Fig. 3; Dataset S2). Collectively, these data show various combinations of motif preferences within Lmd-bound genomic regions that direct somatic mesodermal gene expression patterns, all of which correlate with in vivo binding of TFs and the resulting diversity of cell type–specific gene expression that they confer.
Determination of the Binding Motif for Lmd and Its Enrichment Among Lmd ChIP-seq Peaks.
The enhancer classification showed that motifs from the same TF class as Lmd are enriched among Lmd-bound genomic regions, suggesting direct regulation by Lmd. To evaluate this possibility, we determined the in vitro DNA binding specificities of Lmd using high-resolution universal protein-binding microarrays (PBMs) containing replicates of all possible double-stranded 8-mer DNA sequences and a standardized protocol (35). A position weight matrix (PWM) derived from the bound sequences was generated to visualize the DNA binding preferences of Lmd. Similar to our previous analysis of a mouse homolog of Lmd, Gli-similar 2 (Glis2) (36), a single PWM was unable to capture the entire set of binding preferences for Lmd (Fig. 4A; Dataset S3) We refer to the two separate binding preferences of this TF as Lmd primary (LmdP) and secondary (LmdS) motifs. These results show that the in vitro binding specificities of Lmd are very similar to other Kruppel-like zinc finger proteins, including the closest mouse homolog of Lmd, Gli-similar 3 (37).
Fig. 4.
Determination of the Lmd TF binding site and the locations of this motif in the known Mef2 FCM enhancer. (A) PBM-derived primary and secondary sequence logos for Lmd. (B) Schematic of Fkh and Lmd binding sites in the IED5 Mef2 FCM enhancer. The WT sequence of Lmd and Fkh binding sites within the previously characterized essential 40-bp C/D region of the IED5 enhancer, as well as the WT sequence of two additional Lmd binding sites are shown (5). Additional versions in which those sites are mutated are indicated in purple. The identification of these binding sites and the designs of the mutant versions are described in Dataset S4 (C–E) Stage 13 embryos of the indicated genotypes were stained for GFP (C–E) and Lmd (C′–E′). GFP (green) expression driven by the WT enhancer (IED5WT-GFP) in a subset of FCMs is abrogated when the three Lmd binding sites (D, IED5Lmd-GFP) or the single Fkh binding site (E, IED5Fkh-GFP) is mutated. (Scale bar, 20 μm.)
Having defined the in vitro binding preferences for Lmd, we next asked if these motifs are enriched within the Lmd ChIP-seq peaks by comparing the fraction of Lmd-bound peaks containing a PWM motif match to background controls. These results showed that both LmdP and LmdS motifs were significantly enriched among all Lmd ChIP-seq peaks, and in the vicinity of those associated with genes down-regulated in lmd loss-of-function mutants (Fig. S9). Observing enrichment of both Lmd motifs agrees with previous studies documenting an inability of a single consensus binding site to explain all aspects of in vivo TF binding (4, 38, 39). Despite the enrichment of both motifs among all Lmd-bound genomic regions, only LmdP motifs were significantly enriched among the Lmd peaks associated with FCM-specific gene sets (Fig. S9). Thus, these results suggest that Lmd is directly regulating FCM-specific gene activity through LmdP motifs.
Lmd, Twi, and Fkh Motifs Are Critical Functional Determinants of Individual FCM Enhancers.
A small (170 bp) enhancer called IED5 is associated with the Mef2 gene and drives reporter activity in a subset of somatic FCMs in a lmd-dependent manner (5). A smaller region was identified within this enhancer that is necessary for reporter activity and that is recognized by Lmd in a yeast one-hybrid screen (5) (Fig. 4B). To confirm that Lmd binds to the IED5 enhancer, we scanned it for k-mer matches to the PBM-derived Lmd binding sequence, because individual k-mers provide a much better representation of the sequence preferences than a PWM (Dataset S3). This analysis revealed three sequences that are capable of binding Lmd (Dataset S4). However, none of these motifs fall within the previously characterized region that was proposed to bind this TF (5), although one sequence capable of binding Lmd was found within a nearby region (Fig. 4B; Dataset S4). The latter finding likely explains the identification of Lmd in the yeast one-hybrid screen. To test the functions of the newly identified Lmd motifs in the IED5 enhancer, we mutated these sequences such that their ability to bind Lmd was significantly reduced, as judged by the chosen PBM enrichment score of the mutant sequence. A GFP reporter driven by the WT enhancer (Fig. 4C) is active in a subset of somatic FCMs. However, mutagenesis of the Lmd TF binding sites abrogated activity of the enhancer (Fig. 4D). These functional findings, combined with our in vivo Lmd binding results, strongly suggest that Lmd is directly binding to three separate sites in the IED5 enhancer to activate its activity.
To identity other TFs that may target the IED5 enhancer (5), we used the UniPROBE database of TF binding specificities to search for candidates (40). These results identified a binding site for Foxl1 and Fkh2 (vertebrate and yeast Fkh family TFs, respectively) (Dataset S4). This motif was particularly intriguing because our previous machine learning approach identified Fkh motifs as being positively weighted for classifying Lmd peaks associated with both FCM and FCM+FC gene sets (Dataset S2). Furthermore, a Fkh motif encompasses the two small regions previously found to be necessary for IED5 enhancer activity (Fig. 4B) (5). To test the function of this IED5 Fkh motif, we mutated this sequence such that its ability to bind Fkh TFs was significantly diminished, as judged by its PBM enrichment score. This experiment confirmed that the Fkh motif is indeed essential for full activity of this enhancer (Fig. 4E). In conclusion, our functional studies document the importance of Fkh-like motifs for FCM enhancer activity and help to clarify the identity and locations of the functional Lmd binding sites in the IED5 Mef2 enhancer.
To test the function of Lmd binding sites in another FCM enhancer, we first searched candidates (Dataset S1 for k-mer matches to sequences capable of binding Lmd, as judged by PBM enrichment score, which led to the identification of the previously characterized enhancer associated with the blow FCM gene (Fig. 5A) (4, 24). ChIP results revealed that the blow CRE is bound by Lmd in vivo (Dataset S1; Figs. S5B and S8B). A lacZ reporter driven by the WT blow enhancer is active in the visceral and somatic FCMs, as well as in the amnioserosa and isolated cells of the ectoderm (Fig. 5B). Interestingly, site-directed mutagenesis of the three Lmd binding sites in this enhancer revealed that these sequences are essential for full enhancer activity in both the Lmd-expressing somatic and visceral FCMs. However, as expected, β-Gal reporter activity is not affected in the amnioserosa and ectoderm (Fig. 5C), consistent with Lmd not being expressed in these nonmesodermal cell types (5, 8). These results show that direct Lmd binding to the blow FCM enhancer is required for proper activity in the somatic and visceral FCMs. Given differing opinions on the role of Lmd in the visceral muscle, our results lend support to a role for Lmd in either directly regulating visceral muscle gene expression or at least playing a collaborative role with other TFs necessary for visceral muscle gene activity (5, 8).
To gain a more thorough understanding of FCM gene regulation, we examined additional sequence features associated with the machine learning classification of Lmd peaks. As noted previously, E-box motifs, which could bind Twi, were heavily weighted in the enhancer modeling of FCM gene sets. In addition, Twi cooccupied these Lmd regions in vivo (Fig. 2; Fig. S8B). To test the function of direct Twi binding to a previously characterized FCM enhancer, we mutated the five Twi binding sites in the blow FCM enhancer (Fig. 5A). This analysis showed loss of enhancer activity in both the VM and somatic FCMs. Consistent with the absence of Twi expression in the ectoderm (13), continued activity of the Twi site mutant blow enhancer was maintained in the latter cells (Fig. 5D). Taken together, these results argue that Twi binding directly to FCM enhancers is required to regulate these elements. These findings, combined with our enhancer modeling and in vivo cooccupancy studies, strongly support a regulatory code of Lmd and Twi as being directly required to regulate FCM gene expression (Fig. S8A).
Conclusions
Here we used an integrated genomics approach to define the motif features, in vivo TF combinatorics, and chromatin state associated with Lmd-bound transcriptional regulatory elements. Integrating these data with our prior gene expression atlas identified regulatory combinations of TFs that work together to direct cell-specific patterns of gene expression in the somatic mesoderm. We validated these transcriptional codes by first defining the in vitro binding preferences for Lmd and then using site-directed mutagenesis of overrepresented TF binding sites in known FCM enhancers. In addition, this study suggests the existence of a previously uncharacterized feedforward loop acting between Twi and Lmd in regulating a subset of FCM genes. Furthermore, the cell sorting approach that we used before conducting ChIP-seq studies likely aided in identifying low-level TF occupancy signals that are otherwise difficult to detect for cell-specific TFs. This experimental strategy further focused our analyses of in vivo DNA binding to the cell types of interest, similar to a recent analysis of histone marks associated with sorted mesodermal nuclei (10). We envision that future extension of this integrated approach to additional TFs will further refine the transcriptional codes that direct cell type–specific gene expression patterns in the Drosophila mesoderm, with further generalization of this strategy readily applicable to other model organisms and developmental systems.
Materials and Methods
Details are provided in SI Materials and Methods. ChIP was performed as described (41) on sorted primary mesodermal cells (6). Raw sequencing data were obtained with an Illumina HiSeq-2000 sequencer and deposited in the Gene Expression Omnibus as GSE38402. PBM assays (15), classifier training (16), and analysis of transgenic reporter constructs (15, 16) were performed as described.
Supplementary Material
Acknowledgments
We thank E. Furlong, H. Nguyen, and A. Nose for providing fly strains and antibodies; C. Sonnenbrot for technical assistance; M. Berger for initial PBM analyses of Lmd (Division of Genetics, Brigham and Women’s Hospital); and P. McCoy [Flow Cytometry Core Facility, National Heart Lung and Blood Institute (NHLBI) Division of Intramural Research] for performing cell purifications. This work was funded by the NHLBI Division of Intramural Research (A.M.M.), the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (I.O.), NIH Grant RO1 HG005287 (to M.L.B.), and NIH Grant National Research Service Award F32 GM090645 (to L.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE38402).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210415109/-/DCSupplemental.
References
- 1.Baylies MK, Bate M, Ruiz Gomez M. Myogenesis: A view from Drosophila. Cell. 1998;93(6):921–927. doi: 10.1016/s0092-8674(00)81198-8. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz-Gómez M. Muscle patterning and specification in Drosophila. Int J Dev Biol. 1998;42(3):283–290. [PubMed] [Google Scholar]
- 3.Tixier V, Bataillé L, Jagla K. Diversification of muscle types: Recent insights from Drosophila. Exp Cell Res. 2010;316(18):3019–3027. doi: 10.1016/j.yexcr.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Cunha PM, et al. Combinatorial binding leads to diverse regulatory responses: Lmd is a tissue-specific modulator of Mef2 activity. PLoS Genet. 2010;6(7):e1001014. doi: 10.1371/journal.pgen.1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan H, Skeath JB, Nguyen HT. Drosophila Lame duck, a novel member of the Gli superfamily, acts as a key regulator of myogenesis by controlling fusion-competent myoblast development. Development. 2001;128(22):4489–4500. doi: 10.1242/dev.128.22.4489. [DOI] [PubMed] [Google Scholar]
- 6.Estrada B, et al. An integrated strategy for analyzing the unique developmental programs of different myoblast subtypes. PLoS Genet. 2006;2(2):e16. doi: 10.1371/journal.pgen.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furlong EE, Andersen EC, Null B, White KP, Scott MP. Patterns of gene expression during Drosophila mesoderm development. Science. 2001;293(5535):1629–1633. doi: 10.1126/science.1062660. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz-Gómez M, Coutts N, Suster ML, Landgraf M, Bate M. myoblasts incompetent encodes a zinc finger transcription factor required to specify fusion-competent myoblasts in Drosophila. Development. 2002;129(1):133–141. doi: 10.1242/dev.129.1.133. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonn S, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44(2):148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 11.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462(7269):65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
- 12.Baylies MK, Michelson AM. Invertebrate myogenesis: Looking back to the future of muscle development. Curr Opin Genet Dev. 2001;11(4):431–439. doi: 10.1016/s0959-437x(00)00214-8. [DOI] [PubMed] [Google Scholar]
- 13.Baylies MK, Bate M. twist: a myogenic switch in Drosophila. Science. 1996;272(5267):1481–1484. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- 14.Bour BA, et al. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9(6):730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 15.Busser BW, et al. Molecular mechanism underlying the regulatory specificity of a Drosophila homeodomain protein that specifies myoblast identity. Development. 2012;139(6):1164–1174. doi: 10.1242/dev.077362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busser BW, et al. A machine learning approach for identifying novel cell type-specific transcriptional regulators of myogenesis. PLoS Genet. 2012;8(3):e1002531. doi: 10.1371/journal.pgen.1002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barski A, Zhao K. Genomic location analysis by ChIP-Seq. J Cell Biochem. 2009;107(1):11–18. doi: 10.1002/jcb.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmad SM, et al. Two forkhead transcription factors regulate the division of cardiac progenitor cells by a Polo-dependent pathway. Dev Cell. 2012;23(1):97–111. doi: 10.1016/j.devcel.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmena A, Gisselbrecht S, Harrison J, Jiménez F, Michelson AM. Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev. 1998;12(24):3910–3922. doi: 10.1101/gad.12.24.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson BE, Nowak SJ, Baylies MK. Myoblast fusion in fly and vertebrates: new genes, new processes and new perspectives. Traffic. 2008;9(7):1050–1059. doi: 10.1111/j.1600-0854.2008.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 23.Hamerly G, Elkan C. 2002. Alternatives to the k-means algorithm that find better clusterings. Proceedings of the ACM Conference on Information and Knowledge Management (CIKM) (Association for Computing Machinery, New York, NY), pp 600–607.
- 24.Schröter RH, Buttgereit D, Beck L, Holz A, Renkawitz-Pohl R. Blown fuse regulates stretching and outgrowth but not myoblast fusion of the circular visceral muscles in Drosophila. Differentiation. 2006;74(9–10):608–621. doi: 10.1111/j.1432-0436.2006.00080.x. [DOI] [PubMed] [Google Scholar]
- 25.Weirauch MT, Hughes TR. Conserved expression without conserved regulatory sequence: The more things change, the more they stay the same. Trends Genet. 2010;26(2):66–74. doi: 10.1016/j.tig.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Davidson E. Development and Evolution. Academic, New York; 2006. The regulatory genome: Gene regulatory networks; p. 304. [Google Scholar]
- 27.Sandmann T, et al. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21(4):436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busser BW, Bulyk ML, Michelson AM. Toward a systems-level understanding of developmental regulatory networks. Curr Opin Genet Dev. 2008;18(6):521–529. doi: 10.1016/j.gde.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16(4-5):585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Narlikar L, et al. Genome-wide discovery of human heart enhancers. Genome Res. 2010;20(3):381–392. doi: 10.1101/gr.098657.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandmann T, et al. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10(6):797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity. Molecular Genetics and the Evolution of Animal Design. 2nd Ed. Malden, MA: Blackwell; 2005. [Google Scholar]
- 33.Kelly KK, Meadows SM, Cripps RM. Drosophila MEF2 is a direct regulator of Actin57B transcription in cardiac, skeletal, and visceral muscle lineages. Mech Dev. 2002;110(1-2):39–50. doi: 10.1016/s0925-4773(01)00586-x. [DOI] [PubMed] [Google Scholar]
- 34.Zhu X, et al. Differential regulation of mesodermal gene expression by Drosophila cell type-specific Forkhead transcription factors. Development. 2012;139(8):1457–1466. doi: 10.1242/dev.069005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger MF, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24(11):1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324(5935):1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beak JY, Kang HS, Kim YS, Jetten AM. Functional analysis of the zinc finger and activation domains of Glis3 and mutant Glis3(NDH1) Nucleic Acids Res. 2008;36(5):1690–1702. doi: 10.1093/nar/gkn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji H, Vokes SA, Wong WH. A comparative analysis of genome-wide chromatin immunoprecipitation data for mammalian transcription factors. Nucleic Acids Res. 2006;34(21):e146. doi: 10.1093/nar/gkl803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: Comparison of consensus versus nonconsensus binding sites. Genome Res. 2008;18(11):1763–1777. doi: 10.1101/gr.080622.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robasky K, Bulyk ML. UniPROBE, update 2011: Expanded content and search tools in the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2011;39(Database issue):D124–D128. doi: 10.1093/nar/gkq992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nègre N, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471(7339):527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. 2005. Preferred analysis methods for Affymetric GeneChips revealed by a wholly-defined control dataset. Genome Biol 6(2):R16. [DOI] [PMC free article] [PubMed]
- 43.Philippakis AA, et al. Expression-guided in silico evaluation of candidate cis regulatory codes for Drosophila muscle founder cells. PLOS Comput Biol. 2006;2(5):e53. doi: 10.1371/journal.pcbi.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.