Abstract
The biological consequences of the Deepwater Horizon oil spill are unknown, especially for resident organisms. Here, we report results from a field study tracking the effects of contaminating oil across space and time in resident killifish during the first 4 mo of the spill event. Remote sensing and analytical chemistry identified exposures, which were linked to effects in fish characterized by genome expression and associated gill immunohistochemistry, despite very low concentrations of hydrocarbons remaining in water and tissues. Divergence in genome expression coincides with contaminating oil and is consistent with genome responses that are predictive of exposure to hydrocarbon-like chemicals and indicative of physiological and reproductive impairment. Oil-contaminated waters are also associated with aberrant protein expression in gill tissues of larval and adult fish. These data suggest that heavily weathered crude oil from the spill imparts significant biological impacts in sensitive Louisiana marshes, some of which remain for over 2 mo following initial exposures.
Keywords: ecological genomics, ecotoxicology, microarray, RNA-seq, toxicogenomics
Following the Deepwater Horizon (DWH) drilling disaster on April 20, 2011, in the Gulf of Mexico, acute oiling and the resulting mortality of marine wildlife were evident. In contrast, the sublethal effects, critically important for predicting long-term population-level impacts of oil pollution (1), have not been well described following the DWH disaster. Here, we report the results of a 4-mo field study monitoring the biological effects of oil exposure on fish resident in Gulf of Mexico coastal marsh habitats.
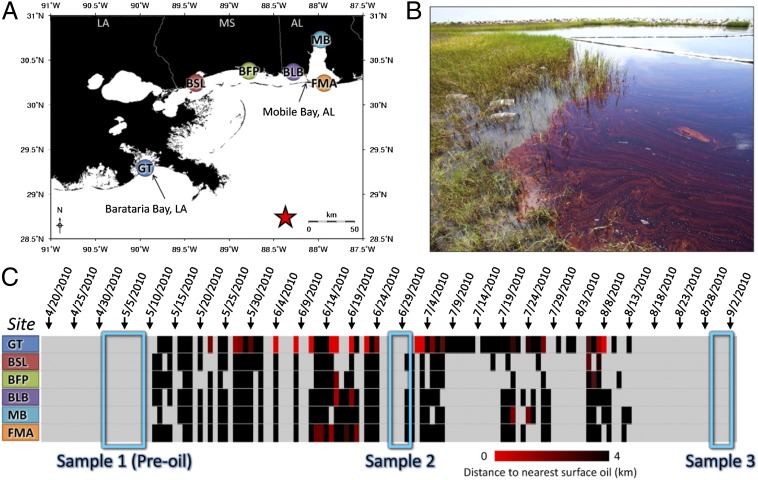
Gulf killifish (Fundulus grandis) were used as our model species because they are among the most abundant vertebrate animals in Gulf of Mexico-exposed marshes (2–4). Furthermore, the Atlantic-distributed sister species to F. grandis (Fundulus heteroclitus) has a narrow home range and high site fidelity, especially during the summer (5, 6), and, among fishes, it is relatively sensitive to the toxic effects of organic pollutants (7). Although home range and toxicology studies are lacking for F. grandis, we infer that F. grandis is also relatively sensitive to pollutants and exhibits high site fidelity, such that the biology of this species is likely affected primarily by the local environment, given the recent shared ancestry of F. grandis with F. heteroclitus (8) and similar physiology, life history, and habitat (9–13). We sampled from populations resident in Gulf of Mexico-exposed marshes before oil landfall (May 1–9, 2010), during the peak of oil landfall (June 28–30, 2010), and after much of the surface oil was no longer apparent 2 mo later (August 30–September 1, 2010) at six field sites from Barataria Bay, Louisiana, east to Mobile Bay, Alabama (Fig. 1 and Dataset S1).
Fig. 1.
Location of field study sites and incidence of oil contamination. (A) Location of field sampling sites, which include Grand Terre (GT), Bay St. Louis (BSL), Belle Fontaine Point (BFP), Bayou La Batre (BLB), Mobile Bay (MB), and Fort Morgan (FMA). Color coding is consistent with other figures. The red star indicates the DWH spill site. (B) Photograph (by A.W.) of the GT field site on June 28, 2010, showing contaminating oil and minnow traps in the marsh. (C) Proximity of nearest surface oil to each field site was determined by SAR, where rows are field sites and columns are days. Light gray represents no data, and black represents the nearest surface oil at a distance of >4 km; the increasing intensity of red indicates closer proximity of oil. Three field sampling trips are highlighted (blue boxes). BSL; BFP; FMA.
Results and Discussion
Remote sensing and analytical chemistry were used to characterize exposure to DWH oil, where remote sensing data are spatially and temporally comprehensive but of low resolution and chemistry data are of high resolution but patchy in space and time. Ocean surface oil was remotely detected through the analysis of images from synthetic aperture radar (SAR) (14). Proximity of the nearest oil slick to each field site (e.g., Fig. S1) was measured for each day that SAR data were available, from May 11 through August 13, 2010, to approximate the location, timing, and duration of coastal oiling (Fig. 1C). Although surface oil came close to many of our field sites in mid-June, only the Grande Terre (GT) site was directly oiled (Fig. 1 B and C). Although the GT site had been clearly contaminated with crude oil for several weeks before our sampling (Fig. 1C and Fig. S2) and retained much oil in sediments (Dataset S2), only trace concentrations of oil components were detected in subsurface water samples collected from the GT site on June 28, 2010, and tissues did not carry abnormally high burdens of oil constituents at any site or time point (Dataset S2). Despite a low chemical signal for oil in the water column and tissues at the time of sampling, we detected significant biological effects associated with the GT site postoil.
We sampled multiple tissues from adult Gulf killifish (average weight of 3.5 g) from each of six field sites for each of three time points [only the first two time points for the Mobile Bay (MB) site] spanning the first 4 mo of the spill event (Fig. 1C). We compared biological responses across time (before, at the peak, and after oiling) and across space (oiled sites and sites not oiled) and integrated responses at the molecular level using genome expression profiling with complimentary protein expression and tissue morphology. Genome expression profiles, using microarrays and RNAseq, were characterized for livers because the organ is internal and integrates xenobiotic effects from multiple routes of entry (gill, intestine, and skin), and because liver is the primary tissue for metabolism of toxic oil constituents. Tissue morphology and expression of CYP1A protein, a common biomarker for exposure to select polycyclic aromatic hydrocarbons (PAHs), was characterized for gills, the organ that provides the greatest surface area in direct contact with the surrounding aquatic environment. In addition, we exposed developing embryos to field-collected water samples to document bioavailability and bioactivity of oil contaminants for this sensitive early life stage.
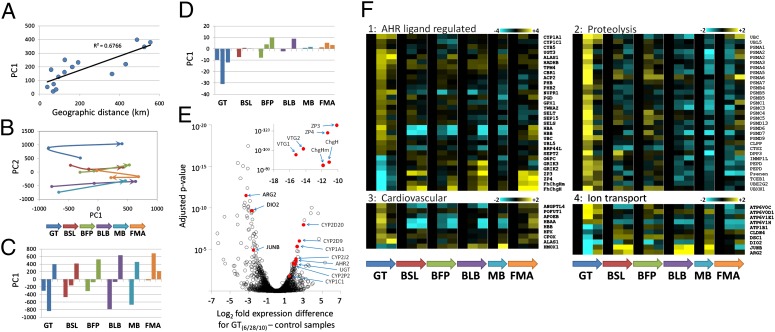
The oiling of the GT site at the end of June 2010 is associated with a clear functional genomic footprint. Of the 3,296 genes included in our analysis, expression of 1,600 and 1,257 genes varied among field sites and throughout the time course, respectively (P < 0.01) (Dataset S3). For the 646 genes that varied in expression only among sites (no significant time effect or site-by-time interaction), site variation followed a pattern of population isolation by distance, which is consistent with neutral evolutionary divergence (Fig. 2A) and population genetic expectations (15). Most importantly, 1,500 genes indicated a pattern of site-dependent time course expression (significant interaction, false discovery rate <0.01), where the trajectory of genome expression through time was divergent at the GT site compared with all other sites (Fig. 2 B and C), particularly at the second time point, which coincides with oil contamination (Fig. 1C).
Fig. 2.
Genome expression between field sites and across time. Field sites include Grand Terre (GT), Bay St. Louis (BSL), Belle Fontaine Point (BFP), Bayou La Batre (BLB), Mobile Bay (MB), and Fort Morgan (FMA). GT was the only site to be directly oiled, which occurred between the first and second sampling times (Fig. 1 and Dataset S2). (A) For genes that vary only among sites (no expression change with time or interaction), pairwise site-specific transcriptome divergence along principal component (PC) 1, as a function of pairwise geographical distance, shows a pattern consistent with isolation by distance. (B) Trajectory of genome expression responses through time for each of six field sites from the preoil sample time (dot at base of arrow) through the peak-oil sample time (middle dot), to the latest postevent sample time (dot at head of arrow) following PC analysis of genes showing statistically significant main effects (site and time) and interaction terms. (C) Divergence along PC1 is isolated, where bars for each site from left to right represent sampling times from the earliest to the latest. (D) Expression divergence along PC1 for the subset of genes that is dose-responsive to PCB exposure (top 10% of PCB-responsive genes). (E) RNAseq data showing genes up- and down-regulated (x axis positive and negative, respectively) in fish from GT sample time 2 (coincident with oil) compared with reference RNA, where select genes are identified. (Inset) Genes are dramatically down-regulated at GT (detailed RNAseq data are presented in Dataset S5). (F) Expression levels for specific genes (rows) and treatments (columns), where cell color indicates up-regulation (yellow) or down-regulation (blue) scaled according to site-specific expression level at the preoil sample time, for genes with divergent expression at the GT site. Genes are grouped into functional categories, and scale bars indicate N-fold up- or down-regulation.
Previous studies have identified genes that are transcriptionally responsive to planar polychlorinated biphenyl (PCB) exposures in killifish (16). Planar PCBs, dioxins, and PAHs (the primary toxic constituents in crude oil) are all mechanistically related insofar as they exert biological effects, in whole or in part, through aryl-hydrocarbon receptor (AHR) signaling pathways; indeed, morpholino knockdown of the AHR is protective of the toxic effects of PAHs and PCBs in killifish (17), and exposures to PCBs and PAHs induce common genome expression responses in flounder (18). Of the genes that were transcriptionally responsive to PCB exposures (16), 380 were included in the current analysis. Expression of this subset of genes is predictive of transcriptional divergence in fish from the GT site coincident with oil contamination compared with other field sites (Fig. S3), especially for the top 10% of PCB-responsive genes (Fig. 2D). Transcriptional activation of these planar PCB-responsive genes in developing killifish embryos is predictive of induction of developmental abnormalities, decreased hatching success, and decreased embryonic and larval survival (16, 19). This set of genes includes members of the canonical battery of genes that are transcriptionally induced by ligand-activated AHR signaling, such as cytochrome P450s, cytochrome B5, and UDP-glucuronosyltransferase (Fig. 2F, set 1), for which increased transcription is particularly diagnostic of exposure to select hydrocarbons (20). Indeed, many genes that are transcriptionally induced or repressed by AHR activators (dioxins, PCBs, and PAHs) show induction or repression at the GT site coincident with crude oil contamination (Fig. 2F, set 1). An independent measure of genome expression, RNAseq, also indicates AHR activation in GT fish from June 28, 2010, compared with reference RNA (e.g., up-regulation of cytochrome P450s, UDP-glucuronosyltransferase (UGT), and AHR itself; Fig. 2E). In parallel, up-regulation of CYP1A protein was detected in gills from GT fish sampled postoil and in early life-stage fish following controlled exposures to GT waters (Figs. 3 and 4). These data appear to be diagnostic of exposure to the toxic constituents in contaminating oil (PAHs) at a sufficient concentration and duration to induce biological responses in resident fish. Sustained activation of the CYP1A gene (Figs. 2F and 3) was predictive of persistent exposure to sublethal concentrations of crude oil components and negative population-level impacts in fish, sea otters, and harlequin ducks following the Exxon Valdez oil spill (reviewed in 1), although PAH toxicity may be mediated through AHR-independent pathways as well (21).
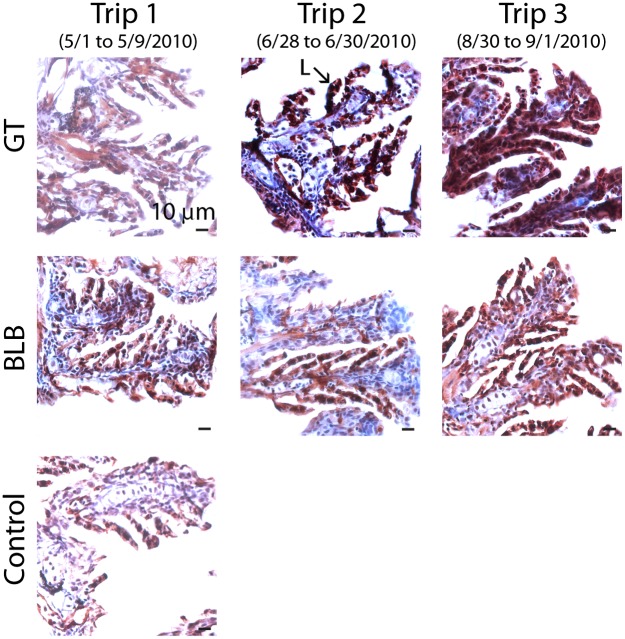
Fig. 3.
CYP1A protein expression (dark red staining) in larval killifish gills (24 d postfertilization) exposed to waters collected from GT (oiled) and BLB (not oiled) during development. (Magnification 40×, scale bars = 10 μm.) CYP1A expression is elevated in the lamellae of larvae exposed during development to waters collected from GT postoil (trips 2 and 3) compared with background levels of CYP1A expression in larvae exposed to GT water preoil (trip 1), compared with CYP1A in fish exposed to waters collected from BLB (which was not directly oiled), and compared with CYP1A in fish reared in laboratory control water. Nuclei were stained using hematoxylin (blue). Analytical chemistry of exposure waters is reported in Dataset S2.
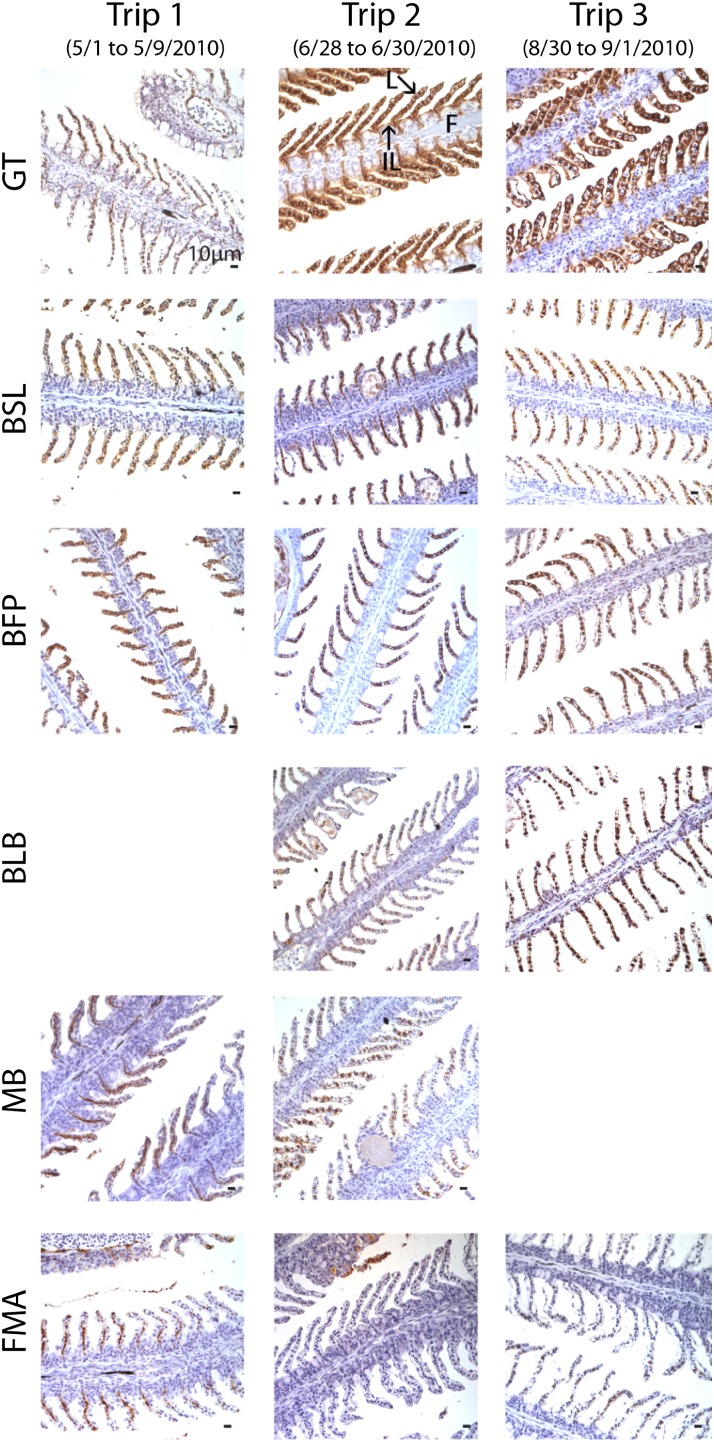
Fig. 4.
CYP1A protein expression in adult killifish gills (dark red staining) sampled in situ from all sampling times (columns) and locations (rows). Locations include Grand Terre (GT), Bay St. Louis (BSL), Belle Fontaine Point (BFP), Bayou La Batre (BLB), Mobile Bay (MB), and Fort Morgan (FMA). (Magnification 40×, scale bars = 10 μm.) The MB site was only sampled on trips 1 and 2, and gills from trip 1 at the BLB site were not available for processing. Fish gills from the GT site during trips 2 and 3 showed high CYP1A expression and an elevated incidence of hyperplasia of the lamellae and interlamellar space on the gill filaments coincident with oil contamination. CYP1A protein was elevated at the GT site postoil (trips 2 and 3) compared with GT preoil (trip 1) as well as with other field sites, none of which were directly oiled. Nuclei were stained using hematoxylin (blue). Exact site locations and sampling dates are reported in Dataset S1.
Transcriptional responses in other sets of coexpressed genes offer insights into the potential biological consequences of contaminating oil exposure at the GT site. Several gene ontology (GO) categories were enriched in the subset of genes that showed GT-specific expression divergence coincident with site- and time-specific oil contamination (Dataset S4). GO enrichment indicates activation of the ubiquitin-proteasome system (Fig. 2F, set 2), which, among diverse functions, is important for cellular responses to stress, cell cycle regulation, regulation of DNA repair, apoptosis, and immune responses (22). The AHR protein itself plays a role as a unique ligand-dependent E3 ubiquitin ligase that targets sex steroid (estrogen and androgen) receptor proteins for proteasomal destruction, thereby impairing normal cellular responses to sex hormones in reproductive tissues, and this response can be activated by planar PAHs (23). Significant down-regulation of transcripts for egg envelope proteins zona pellucida (ZP3 and ZP4) and choriogenin (ChgHm and ChgH) that we detect at the GT site coincident with oil exposure (Fig. 2F, set 1) may be linked to this AHR-dependent proteolytic pathway because their transcription is estrogen-dependent (24, 25) and is down-regulated by exposure to PAHs in fish (25–27). In corroboration, RNAseq detects dramatically down-regulated ZP, ChgH, and vitellogenin transcripts in GT fish (Fig. 2E). Although the transcriptional response that we detect is in male fish, these proteins are synthesized in male livers (reviewed in 25, 27) and down-regulation is consistent with antiestrogenic effects from exposure to PAHs (28). Possible impacts on reproduction merit attention because water only from the GT site induced CYP1A protein in the gills of developing killifish (Fig. 3) at low concentrations of total aromatics and alkanes (Dataset S2) and more than 2 mo after initial oiling, indicating persistent bioavailability of PAHs. Marsh contamination with DWH oil coincided with the spawning season for many marsh animals, including killifish (29), and reproductive effects are predictive of long-term population-level impacts from oil spills (1).
Controlled exposures of developing killifish to water collected from GT on June 28 and August 30, 2010, induced CYP1A protein expression in larval gills relative to fish exposed to GT water preoil and exposed to Bayou La Batre (BLB) site water that was not oiled (Fig. 3). This response is consistent with the location and timing of oil contamination, and it indicates that the remaining oil constituents dissolved at very low concentrations at GT after landfall (Dataset S2) were bioavailable and bioactive to developing fish. Although exposures to PAHs stereotypically induce cardiovascular system abnormalities in developing fish at relatively high concentrations (e.g., 21), none were observed in these animals. However, even very low-concentration exposures during development, insufficient to induce cardiovascular abnormalities in embryos, can impair cardiac performance in adulthood (30). The adult fish sampled in situ from the oil-contaminated GT site showed divergent regulation of several genes involved in blood vessel morphogenesis and heme metabolism coincident with oil contamination (Fig. 2F, set 3). Multigeneration field studies are necessary to confirm cardiovascular effects from DWH oil contamination of marshes that coincided with spawning.
Coastal salt marsh habitats are dynamic and stressful, where changes in environmental parameters, such as temperature, hypoxia, and salinity, can continuously challenge resident wildlife. Regulation of ion transport in fish is particularly important for facilitating homeostasis in response to the salinity fluctuations that are common in estuaries. We found altered regulation of multiple ion transport genes in fish from the GT site coincident with oil contamination (Fig. 2F, set 4). For example, V-type proton ATPases are up-regulated and Na+,K+-ATPase subunits and tight-junction proteins are down-regulated, coincident with oiling at the GT site, in the absence of substantial changes in environmental salinity (Dataset S2). Other genes important for osmotic regulation in killifish (31) are also divergently down-regulated at the GT site, including type II iodothyronine deiodinase (DIO2), transcription factor jun-B (JUNB), and arginase 2 (ARG2). In corroboration, RNAseq data show down-regulation of DIO2, JUNB, and ARG2 in GT fish compared with reference fish (Fig. 2E). Although the physiological consequences of oil exposures are typically studied in isolation, it is reasonable to predict that exposure to oil may compromise the ability of resident organisms to adjust physiologically to natural stressors.
Induction of CYP1A protein expression is a hallmark of AHR signaling pathway activation, making it a sensitive biomarker of exposure to select planar PAHs and other hydrocarbons (20). Although the liver is the key organ for CYP1A-mediated metabolism of these substrates, gill tissues represent the most proximate site of exposure to PAHs. As a result of direct contact with the environment and the nature of the gill as a transport epithelium, the gill may be a more sensitive indicator of exposure to contaminants than the liver (32). CYP1A protein was markedly elevated in GT fish postoil compared with GT fish preoil and compared with fish from other field sites that were not directly oiled (Fig. 4). CYP1A induction was localized predominantly to pillar cells of the gill lamellae and within undifferentiated cells underlying the interlamellar region, which may have contributed to the filamental and lamellar hyperplasia observed during trips 2 and 3, as well as the gross proliferation of the interlamellar region observed during trip 2 in GT fish (Fig. 4). These effects imply a decrease in the effective surface area of the gill, a tissue that supports critical physiological functions, such as ion homeostasis, respiratory gas exchange, systemic acid-base regulation, and nitrogenous waste excretion (33). Currently, the degree to which oil-induced effects may interact with commonly encountered challenges, such as fluctuations in hypoxia and salinity, to compromise physiological resilience is unclear.
By integrating remote sensing and in situ chemical measures of exposure, and linking these with integrated measures of biological effect (genome expression and tissue morphology), we provide evidence that links biological impacts with exposure to contaminating oil from the DWH spill within coastal marsh habitats. Although body burdens of toxins are not high, consistent with reports indicating that seafood from the Gulf of Mexico is safe for consumption (34), this does not mean that negative biological impacts are absent. Our data reveal biologically relevant sublethal exposures causing alterations in genome expression and tissue morphology suggestive of physiological impairment persisting for over 2 mo after initial exposures. Sublethal effects were predictive of deleterious population-level impacts that persisted over long periods of time in aquatic species following the Exxon Valdez spill (1) and must be a focus of long-term research in the Gulf of Mexico, especially because high concentrations of hydrocarbons in sediments (Dataset S2) may provide a persistent source of exposures to organisms resident in Louisiana marshes.
Methods
The locations (latitude and longitude) of our field sampling sites and dates for sampling at each site are summarized in Dataset S1. Gulf killifish (F. grandis) were caught by minnow trap, and tissues were excised immediately. Liver was preserved in RNAlater (Ambion, Inc.) for genome expression (microarray and RNAseq) analysis. Gill tissues were fixed in situ in buffered zinc-based formalin Z-Fix (Anatech LTD). Succinct methods follow, and more detailed methods are available online.
Satellite imagery (SAR) was analyzed to provide estimation of the timing, location, and duration of coastal oil contamination. The calculated distance from each field sampling site to the nearest oil slick was calculated from the “straight-line” distance from the global positioning system position of the station (Dataset S1) to that of the observed oil across any and all intervening geographical barriers (e.g., Fig. S1).
Analytical chemistry of water, tissue, and sediment samples was performed to offer detailed characterization of exposure to contaminating oil (data reported in Dataset S2). Sample dates and locations are summarized in Dataset S1. All sample extracts were analyzed using GC interfaced to an MS detector system. Spectral data were processed by Chemstation Software (Agilent Technologies), and analyte concentrations were calculated based on the internal standard method.
Genome expression across sites and time was characterized using custom oligonucleotide microarrays. Genome expression was measured in liver tissues from five replicate individual male fish per site-time treatment (5 biological replicates) hybridized in a loop design, including a dye swap. Data were lowess-normalized and then mixed model-normalized using linear mixed models to account for fixed (dye) effects and random (array) effects. Normalized data were then analyzed using mixed model ANOVA, with “site” [Grand Terre (GT), Bay St. Louis (BSL), Belle Fontaine Point (BFP), Bayou La Batre (BLB), Mobile Bay (MB), and Fort Morgan (FMA)] and “sampling time” (sampling trips 1, 2, and 3) (Dataset S1) as main effects, including an interaction (site-by-time) term. The false discovery rate was estimated using Q-value (35). Principal components analysis was performed using MeV (36). GO enrichment was tested using DAVID (37).
For RNAseq, transcript abundance was compared between liver mRNA from three replicate fish (RNA was not pooled) from the GT site from June 28, 2010, and mRNA from two control samples. All RNA samples were sequenced on the Illumina Gene Analyzer platform (Expression Analysis, Inc.). Following quality control filtering, quantitative transcript abundance analysis was performed by mapping sequence reads to target sequences (6,810 unique F. heteroclitus target EST sequences, Dataset S5) using the Bowtie short read alignment software (38). A custom Perl script determined the number of fragments mapped to each target sequence. The Bioconductor package DESeq (version 2.8) (39) was used to determine the statistical significance of each differentially expressed target using a negative binomial method with P values adjusted by the Benjamini–Hochberg procedure.
Gill tissues were sampled from all field sites for morphological analysis and immunohistochemical analysis of CYP1A protein expression. Gill tissues from the first and second gill arches were sectioned along the longitudinal axis at a thickness of 4 μm and probed with mAb C10-7 against fish CYP1A (40). Sections were counterprobed using the Vectastain ABC immunoperoxidase system (Vector Laboratories), utilizing the ImmPACT Nova RED peroxidase substrate kit (Vector Laboratories) to visualize the CYP1A protein in red. Tissue sections were counterstained with Vector Hematoxylin QS (Vector Laboratories).
F. grandis embryos obtained from parents not exposed to oil (collected from Cocodrie, LA) were exposed to water samples from the GT and BLB sites collected subsurface on the dates indicated in Dataset S1. Following fertilization, 20 embryos were randomly transferred in triplicate to one of the six field-collected waters (2 field sites × 3 time points) at 3 h postfertilization. Embryos were also exposed to a laboratory control consisting of artificial 17 parts per thousand (ppt) water. Larvae were sampled at 24 d postfertilization and fixed in Z-Fix solution. Sectioning and staining were as described in the previous section.
Supplementary Material
Acknowledgments
K. Carman helped facilitate early field studies. The authors thank R. Brennan, D. Roberts, E. McCulloch, Y. Meng, A. Rivera, C. Elkins, H. Graber, R. Turner, D. Crawford, and M. Oleksiak, for technical assistance. Funding was from the National Science Foundation (Grants DEB-1048206 and DEB-1120512 to A.W., Grant EF-0723771 to A.W. and F.G., and Grant DEB-1048241 to R.B.W.), the National Institutes of Health (R15-ES016905-01 to C.D.R.), and the Gulf of Mexico Research Initiative (A.W., F.G., and N.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data have been deposited to ArrayExpress (accession no. E-MTAB-663).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109545108/-/DCSupplemental.
References
- 1.Peterson CH, et al. Long-term ecosystem response to the Exxon Valdez oil spill. Science. 2003;302:2082–2086. doi: 10.1126/science.1084282. [DOI] [PubMed] [Google Scholar]
- 2.Rozas LP, Reed DJ. Nekton use of marsh-durface habitats in Louisiana (USA) deltaic salt marshes undergoing submergence. Mar Ecol Prog Ser. 1993;96:147–157. [Google Scholar]
- 3.Rozas LP, Zimmerman RJ. Small-scale patterns of nekton use among marsh and adjacent shallow nonvegetated areas of the Galveston Bay Estuary, Texas (USA) Mar Ecol Prog Ser. 2000;193:217–239. [Google Scholar]
- 4.Subrahmanyam CB, Coultas CL. Studies on the animal communities in 2 North Florida salt marshes. 3. Seasonal fluctuations of fish and macroinvertebrates. Bull Mar Sci. 1980;30:790–818. [Google Scholar]
- 5.Lotrich VA. Summer home range and movements of Fundulus heteroclitus (Pisces: Cyprinodontidae) in a tidal creek. Ecology. 1975;56:191–198. [Google Scholar]
- 6.Teo SLH, Able KW. Habitat use and movement of the mummichog (Fundulus heteroclitus) in a restored salt marsh. Estuaries. 2003;26:720–730. [Google Scholar]
- 7.Van Veld PA, Nacci DE. Toxicity resistance. In: Di Giulio RT, Hinton DE, editors. The Toxicology of Fishes. Boca Raton, FL: Taylor and Francis; 2008. pp. 597–641. [Google Scholar]
- 8.Whitehead A. The evolutionary radiation of diverse osmotolerant physiologies in killifish (Fundulus sp.) Evolution. 2010;64:2070–2085. doi: 10.1111/j.1558-5646.2010.00957.x. [DOI] [PubMed] [Google Scholar]
- 9.Able KW, Hata D. Reproductive behavior in the Fundulus heteroclitus-F. grandis complex. Copeia. 1984;(4):820–825. [Google Scholar]
- 10.Kneib RT. The role of tidal marshes in the ecology of estuarine nekton. Oceanography and Marine Biology: An Annual Review. 1997;35:163–220. [Google Scholar]
- 11.Nordlie FG. Physicochemical environments and tolerances of cyprinodontoid fishes found in estuaries and salt marshes of eastern North America. Reviews in Fish Biology and Fisheries. 2006;16:51–106. [Google Scholar]
- 12.Rozas LP, Lasalle MW. A comparison of the diets of Gulf killifish, Fundulus grandis Baird and Girard, entering and leaving a Mississippi brackish marsh. Estuaries. 1990;13:332–336. [Google Scholar]
- 13.Weisberg SB, Lotrich VA. The importance of an infrequently flooded intertidal marsh surface as an energy source for the mummichog Fundulus heteroclitus: An experimental approach. Mar Biol. 1982;66:307–310. [Google Scholar]
- 14.Brekke C, Solberg AHS. Oil spill detection by satellite remote sensing. Remote Sensing of Environment. 2005;95:1–13. [Google Scholar]
- 15.Williams DA, Brown SD, Crawford DL. Contemporary and historical influences on the genetic structure of the estuarine-dependent Gulf killifish Fundulus grandis. Mar Ecol Prog Ser. 2008;373:111–121. [Google Scholar]
- 16.Whitehead A, Pilcher W, Champlin D, Nacci D. Common mechanism underlies repeated evolution of extreme pollution tolerance. Proc R Soc B. 2011 doi: 10.1098/rspb.2011.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark BW, Matson CW, Jung D, Di Giulio RT. AHR2 mediates cardiac teratogenesis of polycyclic aromatic hydrocarbons and PCB-126 in Atlantic killifish (Fundulus heteroclitus) Aquat Toxicol. 2010;99:232–240. doi: 10.1016/j.aquatox.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams TD, et al. Transcriptomic responses of European flounder (Platichthys flesus) to model toxicants. Aquat Toxicol. 2008;90:83–91. doi: 10.1016/j.aquatox.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 19.Whitehead A, Triant DA, Champlin D, Nacci D. Comparative transcriptomics implicates mechanisms of evolved pollution tolerance in a killifish population. Mol Ecol. 2010;19:5186–5203. doi: 10.1111/j.1365-294X.2010.04829.x. [DOI] [PubMed] [Google Scholar]
- 20.Varanasi U. Metabolism of Polycyclic Aromatic Hydrocarbons in the Aquatic Environment. Boca Raton, FL: CRC; 1989. p. 341. [Google Scholar]
- 21.Incardona JP, et al. Aryl hydrocarbon receptor-independent toxicity of weathered crude oil during fish development. Environ Health Perspect. 2005;113:1755–1762. doi: 10.1289/ehp.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 23.Ohtake F, et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446:562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- 24.Modig C, et al. Molecular characterization and expression pattern of zona pellucida proteins in gilthead seabream (Sparus aurata) Biol Reprod. 2006;75:717–725. doi: 10.1095/biolreprod.106.050757. [DOI] [PubMed] [Google Scholar]
- 25.Yu RMK, Wong MML, Kong RYC, Wu RSS, Cheng SH. Induction of hepatic choriogenin mRNA expression in male marine medaka: A highly sensitive biomarker for environmental estrogens. Aquat Toxicol. 2006;77:348–358. doi: 10.1016/j.aquatox.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Holth TF, et al. Differential gene expression and biomarkers in zebrafish (Danio rerio) following exposure to produced water components. Aquat Toxicol. 2008;90:277–291. doi: 10.1016/j.aquatox.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez BC, Carter B, Hammers HR, Sepúlveda MS. Transcriptional response of hepatic largemouth bass (Micropterus salmoides) mRNA upon exposure to environmental contaminants. J Appl Toxicol. 2011;31:108–116. doi: 10.1002/jat.1553. [DOI] [PubMed] [Google Scholar]
- 28.Thomas P. Teleost model for studying the effects of chemicals on female reproductive endocrine function. J Exp Zool Suppl. 1990;4(Suppl 4):126–128. doi: 10.1002/jez.1402560421. [DOI] [PubMed] [Google Scholar]
- 29.Greeley MS, Macgregor R. Annual and semilunar reproductive-cycles of the Gulf killifish, Fundulus grandis, on the Alabama Gulf Coast. Copeia. 1983;(3):711–718. [Google Scholar]
- 30.Hicken CE, et al. Sublethal exposure to crude oil during embryonic development alters cardiac morphology and reduces aerobic capacity in adult fish. Proc Natl Acad Sci USA. 2011;108:7086–7090. doi: 10.1073/pnas.1019031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehead A, Roach JL, Zhang S, Galvez F. Genomic mechanisms of evolved physiological plasticity in killifish distributed along an environmental salinity gradient. Proc Natl Acad Sci USA. 2011;108:6193–6198. doi: 10.1073/pnas.1017542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine SL, Oris JT. CYP1A expression in liver and gill of rainbow trout following waterborne exposure: Implications for biomarker determination. Aquat Toxicol. 1999;46:279–287. [Google Scholar]
- 33.Evans DH, Piermarini PM, Choe KP. The multifunctional fish gill: Dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev. 2005;85:97–177. doi: 10.1152/physrev.00050.2003. [DOI] [PubMed] [Google Scholar]
- 34.State of Louisiana Department of Health and Hospitals . Louisiana Seafood Safety Surveillance Report. Baton Rouge, LA: Louisiana Department of Health and Hospitals; 2011. [Google Scholar]
- 35.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saeed AI, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 37.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 38.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice CD, Schlenk D, Ainsworth J, Goksoyr A. Cross-reactivity of monoclonal antibodies against peptide 277-294 of rainbow trout CYP1A1 with hepatic CYP1A among fish. Mar Environ Res. 1998;46:87–91. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.