Abstract
Cellulose-degrading enzyme systems are of significant interest from both a scientific and technological perspective due to the diversity of cellulase families, their unique assembly and substrate binding mechanisms, and their potential applications in several key industrial sectors, notably cellulose hydrolysis for second-generation biofuel production. Particularly fascinating are cellulosomes, the multimodular extracellular complexes produced by numerous anaerobic bacteria. Using single-molecule force spectroscopy, we analyzed the mechanical stability of the intermolecular interfaces between the cohesin and the dockerin modules responsible for self-assembly of the cellulosomal components into the multienzyme complex. The observed cohesin–dockerin rupture forces (>120 pN) are among the highest reported for a receptor–ligand system to date. Using an atomic force microscope protocol that quantified single-molecule binding activity, we observed force-induced dissociation of calcium ions from the duplicated loop–helix F-hand motif located within the dockerin module, which in the presence of EDTA resulted in loss of affinity to the cohesin partner. A cohesin amino acid mutation (D39A) that eliminated hydrogen bonding with the dockerin’s critically conserved serine residues reduced the observed rupture forces. Consequently, no calcium loss occurred and dockerin activity was maintained throughout multiple forced dissociation events. These results offer insights at the single-molecule level into the stability and folding of an exquisite class of high-affinity protein–protein interactions that dictate fabrication and architecture of cellulose-degrading molecular machines.
Keywords: molecular recognition, protein unfolding
Through the course of evolution, as plants developed dense cross-linked networks of structural cell wall components to provide them with strength and support, simultaneous coevolution by microorganisms produced a variety of intricate enzyme systems for harvesting the abundant lignocellulosic carbon sources found in nature. These included secreted free cellulases, individual surface-bound cellulases, and an exquisite class of multimodular protein assemblies, the cellulosomes (1). Cellulosomes are nanomachines honed through nature to self-organize on bacterial and fungal cell surfaces, adhere to plant materials, and deconstruct plant cell wall lignocellulose. Anaerobic bacteria such as Clostridium thermocellum express the various cellulosome components, which are transported to the cell surface and assembled into a large (>2 MDa) extracellular macromolecular complex ∼100 nm in size.
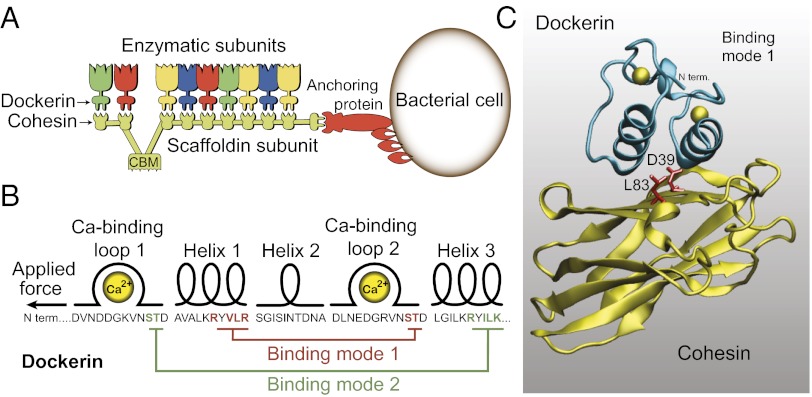
Cellulosomes contain an assortment of enzymatic subunits, each designed for degrading specific components of the substrate. The enzymes are organized along a single protein scaffold, the “scaffoldin,” which itself is not catalytically active but serves to organize the catalytic enzymes at high density and target the entire complex to the plant material via the cellulose-binding module (CBM), as shown in Fig. 1A. To integrate the enzymes into the cellulosome, nature evolved the high-affinity cohesin–dockerin interaction. The dockerin module comprises a highly conserved ∼70-aa sequence borne by each of the cellulosome-destined enzymes. These small modules direct assembly onto the scaffoldin, which bears multiple copies of the conserved cohesin modules that serve as docking sites for the dockerin-bearing enzymes.
Fig. 1.
Schematics of the C. thermocellum cellulosome. (A) Organization of the CipA scaffoldin of C. thermocellum, with dockerin-containing enzymes and anchoring protein. The enzymatic subunits are organized at high density along the scaffoldin subunit, mediated by high-affinity type I cohesin–dockerin interactions. (B) Primary sequence and secondary structural elements of the dockerin module. Key residues involved in binding modes 1 and 2 are highlighted in red and green, respectively. (C) Equilibrated structural model of the cohesin–dockerin complex used in this work. The alanine mutation sites D39 and L83 on the cohesin are shown in red.
The cohesin–dockerin interaction is among the highest affinity protein–protein interactions known, with a dissociation constant of <10−11 M. Dockerins form their binding interface to the cohesin through a duplicated 22-residue calcium binding loop–helix F-hand motif. The dockerin modules are believed to bind to cohesins in two different configurations, a phenomenon referred to as the dual binding mode (2), shown in Fig. 1B. The two binding modes are thought to have evolved as a way to increase the conformational space for enzymes bound to the extracellular scaffoldin, and provide alternative modes of interaction between the enzymes and substrate (3). Prior studies demonstrated that a dockerin truncated at the N terminus to eliminate one of the binding modes still exhibited high affinity due to the presence of the alternative binding mode (4). Additionally, an S45A/T46A double alanine dockerin mutant was shown to exhibit an alternative binding mode compared with the wild-type (WT) complex (2). It would be of significant scientific interest to determine whether one of the binding modes is preferred in nature and how they are populated. Bulk assays have routinely been used to analyze cohesin–dockerin binding and species specificity. However, accurate measurement of the off-rate has proven problematic due to the ultralow off-rate of the complex. Bulk assays can neither provide details regarding unbinding pathways nor resolve differences due to the presence of two binding modes. Therefore, a single-molecule approach was used here to resolve the cohesin–dockerin complex and its dissociation pathways in unprecedented detail.
Using atomic force microscope (AFM)-based single-molecule force spectroscopy, we measured rupture forces that were among the highest receptor–ligand interaction strengths reported to date. Using barrier position analysis, we identified characteristic unfolding fingerprints of the xylanase and CBM fusion partners, and confirmed the contour lengths predicted by their amino acid sequences. In a series of calcium dependency experiments, we characterized how application of force to the cohesin–dockerin complex in the presence of the divalent metal ion chelator EDTA resulted in dissociation of calcium from the dockerin module and loss of binding activity. A cohesin amino acid mutation (D39A) that destabilized the interaction with the dockerin’s calcium-binding loop maintained activity during repeated forced dissociation in the presence of EDTA. This suggested that the high interaction strength between the calcium binding loop and the WT cohesin was responsible for calcium dissociation. Finally, we describe a unique double rupture event and discuss how the double event is likely a consequence of the dockerin’s dual mode of binding. This single-molecule investigation into forced dissociation of cohesins and dockerins represents a step forward in cellulosome research and suggests significant future opportunities for elucidating the structural and functional properties of these refined molecular systems.
Results
Cohesin and Dockerin Fusion Constructs.
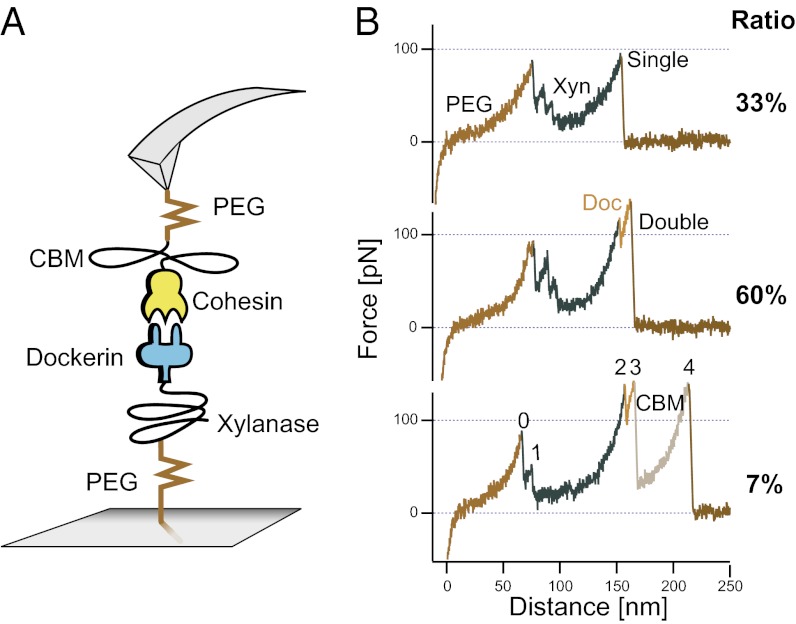
The dockerin under investigation comprised the WT Cel48S dockerin module from C. thermocellum. This protein was expressed in Escherichia coli as a C-terminal fusion to the xylanase T6 enzyme from Geobacillus stearothermophilus to increase stability and expression levels as described previously (5). The xylanase T6 enzyme was modified with an internal T129C mutation to introduce a cysteine residue at a position spatially removed from the C-terminally fused dockerin. This cysteine was used in conjunction with maleimide chemistry to site-specifically immobilize the protein on the AFM cantilever or sample surface, as shown in Fig. 2A. The xylanase module crystal structure showing the position of the internal cysteine residue is shown in Fig. S1A. The construct is denoted xylanase–dockerin, and a version without the T129C mutation had been produced and characterized in previous works by our group (4, 6). Protein amino acid sequences are provided in Dataset S1.
Fig. 2.
Force spectroscopy of the type I cohesin–dockerin interaction. (A) Schematic of the pulling geometry. (B) Typical unfolding patterns of the CBM–cohesin:xylanase–dockerin complex. The first nonlinear rise in force was caused by stretching of the PEG and protein linker regions. Afterward, a series of up to three sudden drops in force with decreasing height was observed, corresponding to xylanase (Xyn) unfolding. Finally, the cohesin–dockerin interface ruptured in a single step (Single), or in a two-step process characterized by an 8-nm contour length increment (Double). In ∼7% of the cases, an additional high-force peak was observed consistent with the unfolded length of the cellulose-binding module (CBM). The last peak in all traces was attributed to rupture of the cohesin–dockerin binding interface. A total of 880 force–distance traces were analyzed to arrive at the ratio values shown on the right.
The cohesin under investigation comprised the C. thermocellum CipA cohesin2 module (Coh2). This protein was expressed in E. coli as an artificial C-terminal fusion to the CBM from the CipA scaffoldin of C. thermocellum to facilitate purification via a cellulose affinity column and to improve expression levels, as described previously (5). The CBM was modified with an A2C site-specific mutation for surface attachment before force spectroscopy. The CBM crystal structure is shown in Fig. S1B.
We used the SWISS-MODEL workspace (7) in conjunction with the crystallized cohesin and dockerin structures (PDB ID codes 2CCL and 1OHZ) (2, 8) to model the structure of our cohesin–dockerin pair based on structural homology. The results from this model for dockerin binding mode 1 are shown in Fig. 1C. Equilibrated structural models of the cohesin–dockerin complex in each binding mode side-by-side for comparison are found in Fig. S2.
Unfolding Fingerprints of Fusion Proteins.
Force spectroscopy investigations were undertaken in which one of the binding partners was covalently immobilized onto an aminosilanized cantilever and the other onto a glass surface via NHS-PEG-maleimide spacers at the engineered cysteine residues. The stability of single-molecule binding interfaces between the protein constructs was probed by contacting the surface repeatedly with the cantilever. After each approach–retract cycle, the x–y piezo stage was actuated, exposing new surface molecules to the same molecule on the cantilever.
The recorded force–distance traces exhibited sawtooth-like peaks if successful binding of cohesin and dockerin was established. Each peak in the force–distance trace corresponded to the unfolding of a single protein domain or folded subdomain, whereas the last peak always corresponded to rupture of the cohesin–dockerin binding interface. Positions along the amino acid chain that resisted the applied load represented energy barriers to unfolding. The specific positions of these energy barriers could be used as a fingerprint to identify the various protein domains of interest. This analysis method based on contour lengths has been previously used to identify globular protein domains, interrogate key residues involved in the folding of membrane proteins (9–17), and probe the sequence-dependent stability of nucleic acid hairpins (18, 19).
We probed our binding partners in two different experimental configurations to obtain a complete picture of the reversibility of domain unfolding and binding interface generation. Fig. 2A depicts the pulling configuration, which we refer to as pulling configuration (i), where the more stable CBM–cohesin was attached to the cantilever and probed repeatedly. With each force–distance trace, a new dockerin module was probed by the same cohesin, giving rise to unfolding fingerprints shown in Fig. 2B. In configuration (ii), which will be discussed below, the positions of the two fusion proteins were exchanged.
An initial two- to three-peaked sawtooth pattern containing peaks of decreasing height between distances of 50 and 100 nm was observed in configuration (i). The sequentially decreasing peak heights indicated that the first barrier shielded the subsequent ones, suggesting assignment of these rupture events to a single protein domain. The sawtooth feature was followed by a high-force “single” or “double” peak. Thirty-three percent of the data traces exhibited a single peak upon cohesin–dockerin rupture (Fig. 2B, single), whereas 60% exhibited a double peak (Fig. 2B, double). A small fraction (∼7%) exhibited an additional barrier representing CBM unfolding (Fig. 2B, CBM). The fractional occurrences of single, double, and CBM-type unfolding events were calculated from 880 force–distance traces.
To measure the contour lengths of the various protein domains, the force–distance data were transformed into contour length space using a worm-like chain (WLC) model (20). We assumed a fixed persistence length of 0.4 nm, which was previously found to be appropriate for modeling protein unfolding in a high force (>50 pN) regime (21). After WLC transformation, cross-correlation (22) of the data traces was performed to align the traces and correct for the polydispersity of the PEG spacers. The transformed data traces were then combined to produce a barrier position histogram that exhibited characteristic contour length increments. This data transformation and analysis process is depicted in Fig. S3.
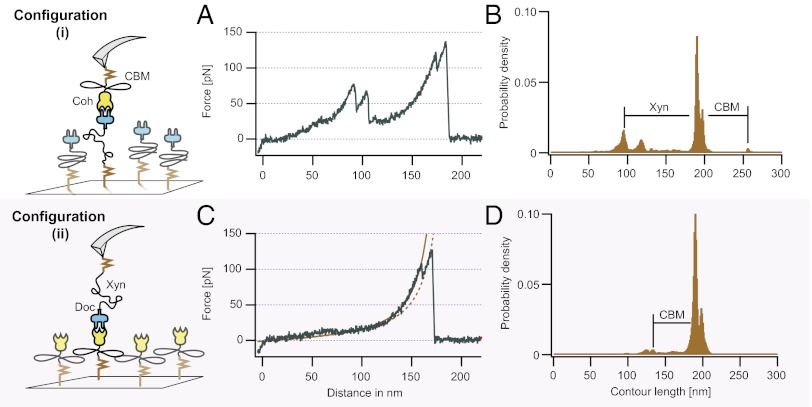
Shown in Fig. 3 is a comparison of unfolding traces obtained in the two different pulling configurations (i) and (ii). A typical single-molecule unfolding trace for configuration (i) is found in Fig. 3A. The corresponding barrier position histogram shown in Fig. 3B was assembled from 351 WLC-transformed and cross-correlated force–distance traces. The distances between the histogram peaks correspond to the end-to-end contour lengths of the various unfolded segments of the fusion proteins. By comparing the measured contour length increments in the barrier position histogram with the known lengths of the protein domains, assuming a length per amino acid of 0.365 nm (23), we could make module assignments as shown in Table 1. We were able to confirm that the initial sawtooth peaks corresponded to unfolding the xylanase module, with a resulting contour length increment of 89 nm, as shown in Fig. 3B (Xyn) (Table 1). In some of the traces, a third substep with a poorly defined location was detected at low forces (< 30 pN) during the xylanase unfolding. The comparatively rare unfolding of the CBM produced a small peak with a contour length increment of 57 nm, as shown in Fig. 3B (CBM). Due to the rarity of the CBM unfolding events, manual transformation and alignment were performed with the resulting histogram shown in Fig. S4.
Fig. 3.
Unfolding traces and contour length histograms in configurations (i) (Upper) and (ii) (Lower). (A) Xylanase unfolding was followed by cohesin–dockerin rupture in configuration (i). (B) Worm-like chain (WLC) transformation produced a contour length histogram with peak increments that matched the expected xylanase (Xyn) and CBM contour length increments. (C) The xylanase module was irreversibly unfolded in the first few approach–retract cycles in configuration (ii). Only single- and double-type cohesin–dockerin rupture events were observed. Shown is a double event with WLC fits. (D) The contour length histogram exhibited the double peak and CBM increments.
Table 1.
Domain assignment of contour length increments
| Increment (peaks) | 0–1 | 1–2 | 2–3 | 3–4 | Not observed |
| Observed contour length increment, nm | 19 ± 1 | 70 ± 1 | 8 ± 1 | 57 ± 1 | — |
| Combined increments, nm | 89 ± 1 | 8 ± 1 | 57 ± 1 | — | |
| Assigned protein module | Xylanase | Dockerin | CBM | Cohesin | |
| Amino acids (total) Nt | 260 (378) | 76 | 159 | 146 | |
| Folded length, nm, Lf | 6 | <2 | 2 | <4 | |
| Expected increment, nm, ΔLc | 89 | <28 | 56 | <54 | |
Contour length increments are labeled according to the numbered peaks shown in Fig. 2B (Bottom). Protein module assignments were made by comparing the expected increment lengths (ΔLc) with the observed increments determined from barrier position histograms. Expected contour length increments for the single modules in the protein constructs were calculated as follows: ∆Lc = Nt * 0.365 nm − Lf . For the 378-aa xylanase module, only 260 aa located C-terminally from the mutated cysteine are included for calculation of ∆Lc.
A typical force–distance trace and barrier position histogram obtained in pulling configuration (ii) are shown in Fig. 3 C and D. In configuration (ii), the same xylanase–dockerin attached to the cantilever was probed with each approach–retract cycle. Within the first few approach–retract cycles, we observed the three-peaked sawtooth pattern associated with xylanase unfolding. After the xylanase module was unfolded, however, the three-peaked sawtooth pattern was not again observed, indicating that the xylanase module was not able to refold during an experimental run. This resulted in the remainder of the force–distance traces lacking the xylanase unfolding fingerprint in configuration (ii), and a barrier position histogram that only showed the high-force double peak and rare CBM increments. CBM unfolding events in configuration (ii) contributed in the barrier position histogram to two small peaks before the double peak as a result of the applied cross-correlation analysis, because the highest correlation value can be reached by aligning to either of the double-event peaks depending on the particular curve. Several examples of force–distance traces exhibiting CBM increments are shown in Fig. S5.
The contour length increment between the double peaks that unfolded at forces >110 pN was found to be ∼8 nm, or ∼22–24 aa. The fact that it was observed throughout data acquisition in both configurations (i) and (ii) indicated that this was a reversible event. This increment is consistent with the dual mode of dockerin binding, as discussed in further detail below.
Probing the Binding Energy Landscape.
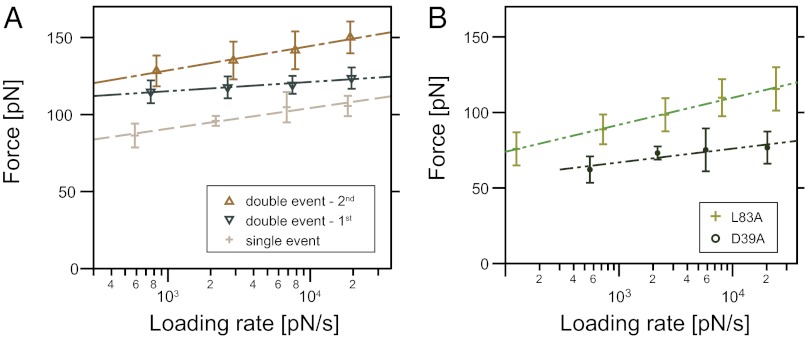
In configuration (i), we characterized the loading rate dependency of cohesin–dockerin rupture events at four pulling speeds (0.2, 0.7, 2, and 5 µm/s). We observed two types of rupture events: (i) single rupture events at 90–100 pN; and (ii) double events at 120–150 pN consisting of the “double-event-first” and “double-event-second” substeps, as shown in Fig. 4. We used the conventional linear two-state Bell–Evans model (24–26) to estimate the koff and ∆x for the three barriers of interest in Fig. 4A. The koff and ∆x represent the dissociation rate in the absence of force and the distance to the transition state along the reaction coordinate, respectively. These parameters were calculated using the y-axis intercept and slope of the dynamic force spectrum shown in Fig. 4. The single events showed a ∆x = 0.7 nm, and koff = 3e−5 s−1. Analysis of the double-event-first ruptures produced a ∆x = 1.6 nm, and koff = 2e−17 s−1, whereas the double-event-second exhibited a ∆x = 0.6 nm, and koff = 1e−6 s−1. The results suggest that the double-event-first is a longer range interaction with a very low off-rate, compared with either the single event or double-event-second peak, which have similar slopes and thus comparable ∆x values. It should be noted that the close proximity of the double-event peaks could give rise to deviations from the Bell–Evans model.
Fig. 4.
Dependency of rupture forces on loading rate. Shown is the loading rate dependency of (A) WT cohesin–dockerin rupture events and (B) mutant cohesin–dockerin events. Error bars represent SEM.
The remarkably high rupture forces, particularly for the double-event-second peaks (>125 pN), are consistent with the known high affinity of the cohesin–dockerin pair. Prior surface plasmon resonance studies reported Kd values of <10−11 M (27). This reported value represented an instrumental limit of the surface plasmon resonance method, so the true Kd of the type I cohesin–dockerin interaction from C. thermocellum could be significantly lower. Our measurements on the cohesin–dockerin interaction are among the highest rupture forces for a receptor–ligand pair reported to date, on par with those measured for the streptavidin–biotin interaction, which at a loading rate of 104 pN/s ruptures at ∼100–125 pN (24, 28–30).
The rupture forces and loading rate dependencies for the two mutant CBM–cohesin fusion proteins (D39A and L83A) were measured, as shown in Fig. 4B. For the D39A mutant, we found ∆x = 1.0 nm and koff = 1e−5 s−1. For the L83A mutant, we found ∆x = 0.5 nm and koff of 1e−4 s−1. Prior work from our group demonstrated that a cohesin point mutation of Asp-39 to asparagine yielded a dramatic decrease in affinity toward the corresponding dockerin (31). The D39A mutation used here performs similarly and is more destabilizing than the L83A mutation. When the dockerin is bound in binding mode 1, the D39A mutation eliminates hydrogen bonding contacts between the cohesin and the serine residue located at the end of calcium binding loop 2. Furthermore, other indirect hydrogen bonds directed toward residues in helix 1 and 2 might be affected (31). When the dockerin is bound in binding mode 2, the D39A mutation results in a weakened cohesin interaction with the serine located toward the end of calcium binding loop 1. This loss of hydrogen bonding resulted in a sharp drop in rupture forces for the D39A mutation compared with the WT. The L83A mutation resulted in the loss of hydrophobic contacts between the cohesin and the hydrophobic residues (L, I, A, V) located in the dockerin α-helices 1 or 3, depending on the binding mode. High-force double events were extremely rare for both cohesin mutants, likely due to the fact that the destabilized cohesin–dockerin interface dissociated before reaching forces sufficiently high to induce structural changes in the dockerin module, which are suspected to be the cause of the double events (see below).
Force-Induced Dissociation of Calcium.
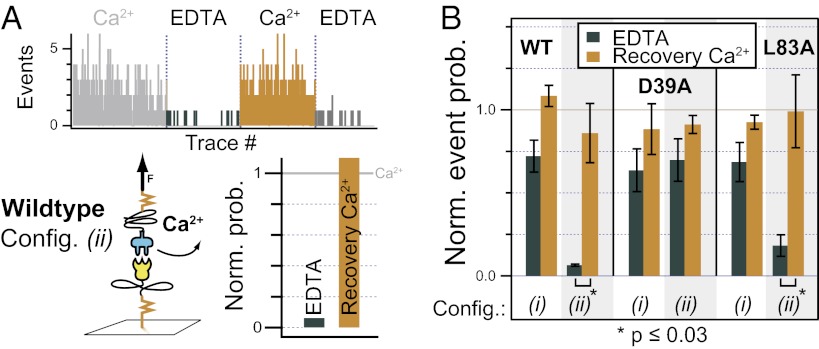
The dockerin module is known to bind two calcium ions via a repeated motif that resembles the EF-hand motif found in several calcium-dependent regulatory proteins, such as calmodulin (32) and recoverin (33). The dockerin module differs, however, from the typical EF-hand motif in that the E helix that precedes the calcium binding loop is absent in the dockerin. Two conserved pairs of ST residues located toward the C-terminal end of each calcium binding loop are known to be key residues involved in molecular recognition by the cohesin (2). We used a cantilever with a larger tip curvature radius (MLCT-AUHW-B; Bruker) for calcium dependency experiments, which allowed us to obtain force–distance traces that exhibited multiple interactions in each trace. The larger number of bound cohesin–dockerin pairs obtained with each approach–retract cycle allowed us to use the number of rupture peaks per force curve as a measure of dockerin binding activity. We then compared the events per trace obtained in calcium buffer with those obtained in EDTA buffer in different experimental pulling configurations, and with different cohesin mutants (WT, L83A, and D39A).
Fig. 5A depicts a typical experimental run for the calcium dependency experiments. A basal level of activity was first determined by obtaining ∼200 force traces in TBS containing 1 mM Ca2+, and measuring the average number of rupture events per trace. Next, the 1 mM Ca2+ in the sample buffer was replaced with 1 mM EDTA, and ∼200 more force traces were obtained. The sample buffer was then changed again to 1 mM Ca2+, and an additional data set was obtained to characterize recovery of activity after EDTA exposure. The average events per trace in EDTA and during the second calcium exposure were both normalized to the basal value obtained during the initial Ca2+ phase to correct for differential functionalization densities on the cantilevers between experiments. Normalization to the basal level produced the calculated “normalized event probability” shown in Fig. 5 A and B. All experimental runs were performed numerous times (n ≥ 4) with freshly prepared cantilevers and surface samples. Error bars in Fig. 5 represent SEM. Examples of several force–distance traces resulting from a multiply loaded cantilever tip are provided in Fig. S6.
Fig. 5.
Force-induced dissociation of Ca2+ from the dockerin module. Cantilevers with larger tips that showed multiple interactions were used. Rupture events with F > 35 pN and x > 40 nm were detected with an automated software tool. (A) In configuration (ii), the number of events per trace dropped drastically when Ca2+ was replaced with EDTA. The events per trace were recoverable after switching back to Ca2+ buffer. (B) For comparability, the events per trace in EDTA and during the second Ca2+ exposure (“Recovery Ca2+”) were normalized by the first Ca2+ cycle to provide the normalized event probability. A statistically significant decrease in the presence of EDTA was observed for the WT and the L83A mutant in configuration (ii). The minor decrease of activity for all other configurations showed that the Ca2+ mainly stays bound to the dockerin in EDTA buffer but could be dissociated under applied force.
As shown in Fig. 5B for the WT cohesin in configuration (i), where a new surface-bound dockerin module was probed with each force trace, exposure to EDTA resulted in only a ∼25% drop in activity. Upon returning the binding partners to the calcium ambient, activity was completely recovered. These differences in binding activity between EDTA and Ca2+ exposure for the WT cohesin–dockerin in pulling configuration (i) were not statistically significant. This suggested that the surface-bound dockerin modules mostly retain calcium ions in the presence of EDTA, remain correctly folded, and exhibit activity that does not significantly differ from the native basal activity in Ca2+ buffer. In configuration (ii), however, the results shown in Fig. 5B for the WT cohesin indicated that repeated application of force to the xylanase–dockerin construct in EDTA buffer resulted in a dramatic loss of activity. Application of force to the dockerin module pried open the calcium binding loops and allowed sequestration of the calcium ions by EDTA. After loss of calcium, the same dockerin modules on the lever were probed repeatedly, and because no calcium was present in the buffer, refolding did not occur, resulting in a dramatic loss of activity within the first few data traces. Upon returning to Ca2+ buffer, dockerin-binding activity was recovered nearly completely, indicating that, in the Ca2+ buffer, unfolding and refolding of the dockerin’s calcium binding loops were reversible.
The behavior of the L83A cohesin mutant was similar to that of the WT. In configuration (i) with the dockerin modules bound to the surface, no statistically significant loss of activity was observed in EDTA, and recovery in Ca2+ was high. In configuration (ii), however, when the dockerin was on the cantilever in EDTA buffer, a dramatic loss of activity was measured within the first few force traces. This was attributable to unfolding of the calcium binding loops and sequestration of the calcium ions from the dockerin modules bound to the cantilever. The remaining data traces in EDTA showed only nonspecific surface interaction without any cohesin–dockerin binding. Recovery during the second Ca2+ exposure was meanwhile high, similar to WT.
The D39A cohesin mutant exhibited a dramatically different calcium dependency profile [Fig. 5B, D39A (i) and (ii)]. No statistically significant loss in binding activity was observed in an EDTA ambient, regardless of the experimental pulling configuration. This observation is fully understandable given that the D39 residue on the cohesin is known to make hydrogen-bonding contacts with the calcium binding loops of the dockerin in both binding modes (31). The destabilizing D-to-A mutation decreased the amount of force borne by the calcium loop on dockerin during pulling, leading to lower overall rupture forces (Fig. 4), and maintenance of binding activity in EDTA.
Discussion
In this work, we measured the extremely high mechanical strength of the cellulosomal components of C. thermocellum. Furthermore, we observed a force hierarchy that is well adapted to the conditions in which the cellulosome-expressing bacteria operate. The weakest component was the enzymatic subunit xylanase T6. It unfolded in multiple steps at comparatively low forces around 50–80 pN. For enzymes, however, this is already a comparatively high stability, and indeed this enzyme is known to be thermostable. Enzymes in nature have to be flexible during operation to accommodate and process their substrates (34). Prior work by our group had found considerable reorganization in other mechanically loaded enzymes even at lower forces (35, 36). It should be noted that the pulling geometry applied to the xylanase in this work is not physiologically relevant. The attachment point at T129C was chosen by searching for a Thr or Ser on the protein surface distant from the C-terminal dockerin module.
The binding interface of the cohesin–dockerin complex was found to be remarkably strong. The measured force levels exceeded conventional receptor–ligand interactions like those between antibodies and their target peptides (37), and are comparable to the strongest known biomolecular interactions [e.g., streptavidin–biotin (24, 28–30)]. Although giant multiprotein complexes with large interfacial areas such as the titin–telethonin complex are known to withstand higher forces (38), the comparatively small dockerin module exerts a huge amount of load resistance per amino acid.
The scaffoldin components were found to possess the highest stability of all of the cellulosomal components examined here. Again, this matches the requirements of the cellulosome in nature because mainly the scaffoldin is strained if there are shear forces acting between the bacterial cell and the cellulosic substrate. In our pulling configuration, the CBM only rarely unfolded before rupture of the cohesin–dockerin interface, and the cohesin modules themselves never unfolded. Prior work had shown that cohesins unfolded between 200 and 500 pN when pulled from the N and C termini, and this high strength was dependent on their position within the scaffoldin (11).
Regarding the double event with a contour length increment of 8 nm that appeared in 60% of the traces, our results suggest that this unfolding event was located in the dockerin module, possibly due to the unfolding of calcium binding loop 1 when the dockerin was in binding mode 1. In principle, it is possible the 8-nm increment is located elsewhere in the structure. However, the data preclude association of the 8-nm increment with the CBM or xylanase modules, because the observed contour length increments accounted for these modules’ lengths in their entirety. The 8-nm increment could be caused by a partial unfolding of the cohesin module, but Valbuena et al. (11) did not observe any folding intermediates when unfolding the same cohesin2 module from C. thermocellum CipA. Although we cannot completely exclude the possibility that the different pulling geometry in our case exhibits intermediates, we treat this scenario as extremely unlikely.
The observed data support the notion that the cohesin–dockerin interface itself ruptures under force in two steps, with the dockerin undergoing substantial conformational changes that are reversible if calcium is present in the ambient. We know from the calcium dependency experiments that there is a high chance of force-induced Ca2+-dissociation during the unfolding process, and a linkage between this event and the observed additional double event giving rise to the 8-nm increment seems likely. The loading rate dependencies suggest that the single and the “double-second” events were caused by the same unfolding pathway. The question remains whether the nonbinding calcium loop or the cohesin-bound calcium loop caused the event. Dockerin mutations that are known to have a preferential binding mode or exhibit only one binding geometry seem to be promising candidates to provide answers to these questions in the future.
Our data have shown that cellulosomes are protected against loss of their enzymatic components not only by a single energy barrier, but by two extremely high barriers that makes the probability of thermally induced dissociation under ambient conditions vanishingly small. This safety belt mechanism of enzyme assembly makes it unlikely that the bacteria are able to repair defective enzyme modules in situ. More likely, entirely new cellulosomes during growth and propagation of the parent bacteria are produced as old ones are shed in response to changes in the cellulosic substrate. In general, single-molecule force spectroscopy is uniquely suited for revealing the mechanical underpinnings of the extremely high-affinity cohesin–dockerin interaction that dictates cellulosome architecture, and application of this technique in future work seems promising.
Methods
Protocols describing material preparation, including site-directed mutagenesis and protein expression, are included in SI Methods. AFM cantilever and cover glass substrates were prepared according to previously published procedures (39). Cantilever calibration was performed using the equipartition theorem (40, 41). Pulling speeds ranged from 50 nm/s to 5 µm/s for loading rate analysis, and were set to 700 nm/s for calcium dependency experiments. Contour length transformations were performed as previously described (20). For calcium dependency experiments shown in Fig. 5, error bars were determined by performing independent experiments numerous times (n = 4 [WT (i)], n = 5 [WT (ii), D39A (ii), L83A (i), L83A (ii)], n = 6 [D39A (i)]). Error bars represent SEM, weighted according to absolute interaction numbers in the first calcium buffer. In Fig. 5, differences were considered to be statistically significant for P ≤ 0.03 using Welch’s t tests.
Supplementary Material
Acknowledgments
This research was supported by an advanced grant from the European Research Council and by the Nanosystems Initiative Munich. M.A.N. acknowledges funding from an Alexander von Humboldt Foundation Postdoctoral Research Fellowship and from the Society in Science–Branco Weiss Fellowship program. This research was also supported by grants from the United States–Israel Binational Science Foundation (Jerusalem, Israel), by The Helmsley Foundation, by Israel Science Foundation Grant 966/09, and by establishment of an Israeli Center of Research Excellence (I-CORE Center 152/11) managed by the Israel Science Foundation. E.A.B. is the incumbent of The Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.O.Y. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211929109/-/DCSupplemental.
References
- 1.Himmel ME, et al. Microbial enzyme systems for biomass conversion: Emerging paradigms. Biofuels. 2010;1(2):323–341. [Google Scholar]
- 2.Carvalho AL, et al. Evidence for a dual binding mode of dockerin modules to cohesins. Proc Natl Acad Sci USA. 2007;104(9):3089–3094. doi: 10.1073/pnas.0611173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer EA, Belaich JP, Shoham Y, Lamed R. The cellulosomes: Multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 4.Karpol A, Barak Y, Lamed R, Shoham Y, Bayer EA. Functional asymmetry in cohesin binding belies inherent symmetry of the dockerin module: Insight into cellulosome assembly revealed by systematic mutagenesis. Biochem J. 2008;410(2):331–338. doi: 10.1042/BJ20071193. [DOI] [PubMed] [Google Scholar]
- 5.Barak Y, et al. Matching fusion protein systems for affinity analysis of two interacting families of proteins: The cohesin-dockerin interaction. J Mol Recognit. 2005;18(6):491–501. doi: 10.1002/jmr.749. [DOI] [PubMed] [Google Scholar]
- 6.Demishtein A, Karpol A, Barak Y, Lamed R, Bayer EA. Characterization of a dockerin-based affinity tag: Application for purification of a broad variety of target proteins. J Mol Recognit. 2010;23(6):525–535. doi: 10.1002/jmr.1029. [DOI] [PubMed] [Google Scholar]
- 7.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho AL, et al. Cellulosome assembly revealed by the crystal structure of the cohesin–dockerin complex. Proc Natl Acad Sci USA. 2003;100(24):13809–13814. doi: 10.1073/pnas.1936124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Q, Fang J, Wang M, Li H. Kinetic partitioning mechanism governs the folding of the third FnIII domain of tenascin-C: Evidence at the single-molecule level. J Mol Biol. 2011;412(4):698–709. doi: 10.1016/j.jmb.2011.07.049. [DOI] [PubMed] [Google Scholar]
- 10.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276(5315):1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 11.Valbuena A, et al. On the remarkable mechanostability of scaffoldins and the mechanical clamp motif. Proc Natl Acad Sci USA. 2009;106(33):13791–13796. doi: 10.1073/pnas.0813093106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puchner EM, Gaub HE. Force and function: Probing proteins with AFM-based force spectroscopy. Curr Opin Struct Biol. 2009;19(5):605–614. doi: 10.1016/j.sbi.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Shank EA, Cecconi C, Dill JW, Marqusee S, Bustamante C. The folding cooperativity of a protein is controlled by its chain topology. Nature. 2010;465(7298):637–640. doi: 10.1038/nature09021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oesterhelt F, et al. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288(5463):143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 15.Ainavarapu SRK, et al. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys J. 2007;92(1):225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunes JM, et al. A “force buffer” protecting immunoglobulin titin. Angew Chem Int Ed Engl. 2010;49(20):3528–3531. doi: 10.1002/anie.200906388. [DOI] [PubMed] [Google Scholar]
- 17.Puchner EM, Gaub HE. Exploring the conformation-regulated function of titin kinase by mechanical pump and probe experiments with single molecules. Angew Chem Int Ed Engl. 2010;49(6):1147–1150. doi: 10.1002/anie.200905956. [DOI] [PubMed] [Google Scholar]
- 18.Woodside MT, et al. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314(5801):1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodside MT, et al. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci USA. 2006;103(16):6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puchner EM, Franzen G, Gautel M, Gaub HE. Comparing proteins by their unfolding pattern. Biophys J. 2008;95(1):426–434. doi: 10.1529/biophysj.108.129999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rief M, Gautel M, Schemmel A, Gaub HE. The mechanical stability of immunoglobulin and fibronectin III domains in the muscle protein titin measured by atomic force microscopy. Biophys J. 1998;75(6):3008–3014. doi: 10.1016/S0006-3495(98)77741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietz H, Rief M. Detecting molecular fingerprints in single molecule force spectroscopy using pattern recognition. Jpn J Appl Phys. 2007;46(8B):5540–5542. [Google Scholar]
- 23.Dietz H, Rief M. Protein structure by mechanical triangulation. Proc Natl Acad Sci USA. 2006;103(5):1244–1247. doi: 10.1073/pnas.0509217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397(6714):50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 25.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72(4):1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 27.Mechaly A, et al. Cohesin-dockerin recognition in cellulosome assembly: Experiment versus hypothesis. Proteins. 2000;39(2):170–177. doi: 10.1002/(sici)1097-0134(20000501)39:2<170::aid-prot7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 28.Yuan C, Chen A, Kolb P, Moy VT. Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy. Biochemistry. 2000;39(33):10219–10223. doi: 10.1021/bi992715o. [DOI] [PubMed] [Google Scholar]
- 29.Moy VT, Florin EL, Gaub HE. Intermolecular forces and energies between ligands and receptors. Science. 1994;266(5183):257–259. doi: 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- 30.Florin EL, Moy VT, Gaub HE. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264(5157):415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 31.Handelsman T, et al. Cohesin-dockerin interaction in cellulosome assembly: A single Asp-to-Asn mutation disrupts high-affinity cohesin-dockerin binding. FEBS Lett. 2004;572(1–3):195–200. doi: 10.1016/j.febslet.2004.07.040. [DOI] [PubMed] [Google Scholar]
- 32.Junker JP, Ziegler F, Rief M. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science. 2009;323(5914):633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 33.Desmeules P, Grandbois M, Bondarenko VA, Yamazaki A, Salesse C. Measurement of membrane binding between recoverin, a calcium-myristoyl switch protein, and lipid bilayers by AFM-based force spectroscopy. Biophys J. 2002;82(6):3343–3350. doi: 10.1016/S0006-3495(02)75674-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ringe D, Petsko GA. Biochemistry. How enzymes work. Science. 2008;320(5882):1428–1429. doi: 10.1126/science.1159747. [DOI] [PubMed] [Google Scholar]
- 35.Puchner EM, et al. Mechanoenzymatics of titin kinase. Proc Natl Acad Sci USA. 2008;105(36):13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gumpp H, et al. Triggering enzymatic activity with force. Nano Lett. 2009;9(9):3290–3295. doi: 10.1021/nl9015705. [DOI] [PubMed] [Google Scholar]
- 37.Morfill J, et al. Affinity-matured recombinant antibody fragments analyzed by single-molecule force spectroscopy. Biophys J. 2007;93(10):3583–3590. doi: 10.1529/biophysj.107.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertz M, Wilmanns M, Rief M. The titin-telethonin complex is a directed, superstable molecular bond in the muscle Z-disk. Proc Natl Acad Sci USA. 2009;106(32):13307–13310. doi: 10.1073/pnas.0902312106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmermann JL, Nicolaus T, Neuert G, Blank K. Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nat Protoc. 2010;5(6):975–985. doi: 10.1038/nprot.2010.49. [DOI] [PubMed] [Google Scholar]
- 40.Cook S, et al. Practical implementation of dynamic methods for measuring atomic force microscope cantilever spring constants. Nanotechnology. 2006;17(9):2135–2145. [Google Scholar]
- 41.Butt HJ, Jaschke M. Calculation of thermal noise in atomic-force microscopy. Nanotechnology. 1995;6(1):1–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.