Abstract
7,8-Dihydro-8-oxoguanine (8-oxo-G) is a highly abundant and mutagenic lesion. Replicative DNA polymerases (pols) are slowed down at 8-oxo-G and insert both correct cytosine (C) and incorrect adenine (A) opposite 8-oxo-G, but they preferentially extend A:8-oxo-G mispairs. Nevertheless, 8-oxo-G bypass is fairly accurate in vivo. Thus, the question how correct bypass of 8-oxo-G lesions is accomplished despite the poor extension of C:8-oxo-G base pairs by replicative pols remains unanswered. Here we show that replicative pol δ pauses in front of 8-oxo-G and displays difficulties extending from correct C:8-oxo-G in contrast to extension from incorrect A:8-oxo-G. This leads to stalling of pol δ at 8-oxo-G after incorporation of correct C. This stalling at C:8-oxo-G can be overcome by a switch from pol δ to pols λ, β, or η, all of which are able to assist pol δ in 8-oxo-G bypass by translesion synthesis (TLS). Importantly, however, only pol λ selectively catalyzes the correct TLS past 8-oxo-G, whereas pols β and η show no selectivity and even preferentially enhance incorrect TLS. The selectivity of pol λ to promote the correct bypass depends on its N-terminal domain. Furthermore, pol λ−/− mouse embryonic fibroblast extracts display reduced 8-oxo-G TLS. Finally, the correct bypass of 8-oxo-G in gapped plasmids in mouse embryonic fibroblasts and HeLa cells is promoted in the presence of pol λ. Our findings suggest that even though 8-oxo-G is not a blocking lesion per se, correct replication over 8-oxo-G is promoted by a pol switch between pols δ and λ.
Keywords: DNA polymerase switch, DNA repair, DNA replication
Cells are perpetually exposed to reactive oxygen species (ROS), which originate from either exogenous (e.g., smoking, pollution, and UV) or endogenous sources (e.g., by-products of cellular respiration and components of the inflammatory responses). ROS can give rise to DNA lesions such as 7,8-dihydro-8-oxoguanine (8-oxo-G), a very abundant DNA lesion (reviewed in ref. 1). 8-oxo-G is particularly deleterious because of its ability to functionally mimic thymine (T) in the syn conformation, giving rise to the formation of very stable A(anti):8-oxo-G(syn) Hoogsteen base pairs (2). Thus, the majority of DNA polymerases (pols), including the three replicative pols α, δ, and ε, bypasses 8-oxo-G often in an inaccurate way by incorporating the incorrect A instead of the correct C, thus giving rise to A:8-oxo-G mismatches (3). This potentially results in CG→AT transversion mutations, which can transform cells and eventually lead to different cancers (4). The majority of 8-oxo-G is removed by the 8-oxo-G DNA glycosylase 1 that initiates short-patch base-excision repair (5). Removal of A from A:8-oxo-G mismatches is processed by the MutYH DNA glycosylase and AP endonuclease 1, after which the repair pol λ efficiently catalyzes the incorporation of a correct C opposite 8-oxo-G in the presence of replication protein A (RP-A) and proliferating cell nuclear antigen (PCNA) (3, 6). Pol λ has also been found to preferentially extend C:8-oxo-G and to strongly discriminate against extension of A:8-oxo-G base pairs (7). Despite the presence of this repair system, the replication fork encounters 8-oxo-G bases that have not been removed before replication, especially under conditions of oxidative stress. 8-oxo-G is not considered as a blocking lesion per se (8–10). Nevertheless, transient inhibition of chain extension occurring 3′ to 8-oxo-G has been observed, and the Klenow fragment of Escherichia coli pol I, calf thymus pol α (8), as well as pol δ purified from calf thymus (11) were shown to extend A:8-oxo-G mispairs much more efficiently than the correct C:8-oxo-G base pairs. Similarly, human pol δ has been found to stall at 8-oxo-G sites (12). The overall in vivo mutation frequency of 8-oxo-G without postreplicative repair mechanisms has been estimated to be around 19% (9). Taken together, an important question that remains unanswered is: How can correct bypass of 8-oxo-G by the replication fork be accomplished in view of the fact that the extension of correct C:8-oxo-G base pairs is so difficult to achieve for the replicative pols?
In the work presented here, we set out to investigate the contribution, dynamics, and mode of bypass of 8-oxo-G by replicative pol δ. We show with primer extension assays that, after pausing in front of the lesion, pol δ readily incorporates both C and A opposite 8-oxo-G. Importantly, however, pol δ displays difficulties in extending from correct C:8-oxo-G base pairs, leading to stalling of pol δ at 8-oxo-G after incorporation of the correct C. In stark contrast, A:8-oxo-G mispairs are readily extended by pol δ without substantial stalling. We found that this stalling of pol δ at 8-oxo-G base pairs could be overcome by pol λ as well as pols β and η by performing translesion synthesis (TLS), thereby assisting pol δ to overcome the lesion. Most importantly, however, only pol λ assisted pol δ in performing correct TLS over 8-oxo-G in vitro by selectively enhancing exclusively correct bypass with C. This interplay of pols λ and δ in the correct bypass of 8-oxo-G was confirmed in mouse embryonic fibroblasts (MEFs) pol λ+/+ and pol λ−/− crude cell extracts. Also, pol λ−/− MEFs and pol λ siRNA-treated HeLa cells show more error-prone replication over 8-oxo-G in vivo. Taken together, our data suggest the existence of a pathway during DNA replication that involves a switch between the replicative pol δ and the repair pol λ to promote the error-free bypass of an 8-oxo-G in an efficient, accurate, and low-cost manner to decrease the mutational burden of 8-oxo-G.
Results
Principle of the Assay System.
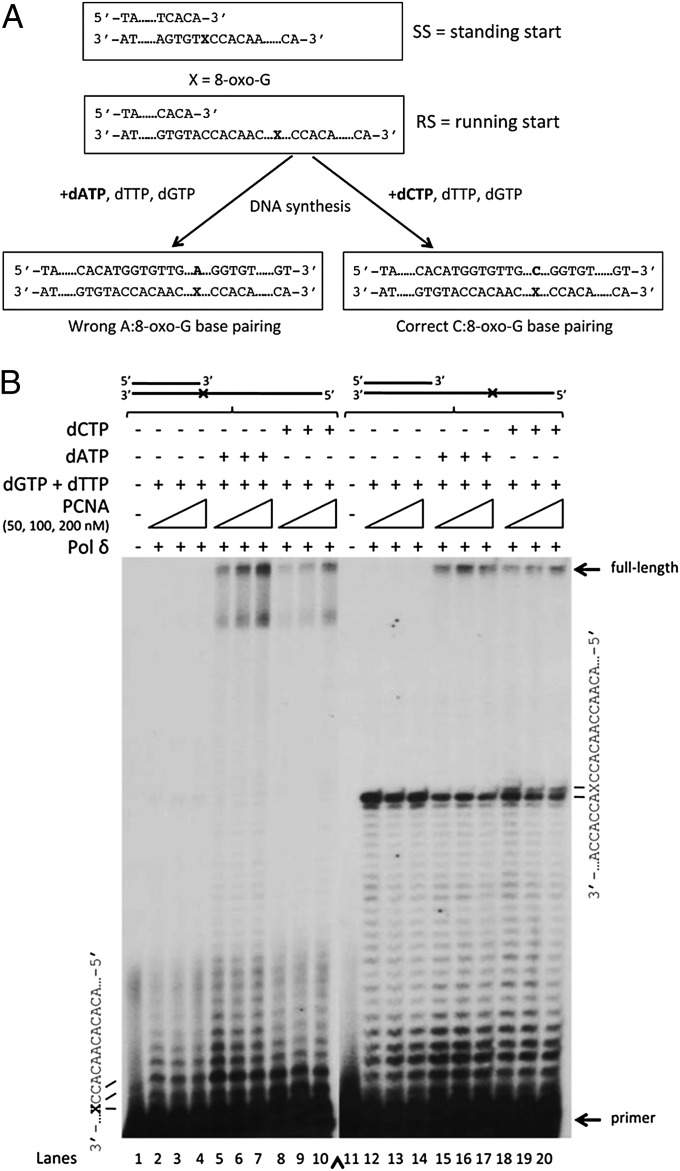
We aimed to assess the dynamics, the fidelity and the extent of 8-oxo-G bypass by pol δ in an in vitro assay. For this, we used a primer/template combination previously developed in our laboratory (6) that allows quantitative monitoring of the incorporation of incorrect A versus correct C opposite 8-oxo-G. This approach is schematically depicted in Fig. 1A. In addition to the single 8-oxo-G, the template strand consists only of C and A bases. Therefore, the only position where C or A can be incorporated is opposite 8-oxo-G. By performing two parallel reactions with the three nucleotides A + T + G or C + T + G, it can be assessed how much of C or A is incorporated opposite 8-oxo-G. If the pol incorporates more A opposite 8-oxo-G, more product will be generated by the addition of A + T + G, whereas the reverse is true for the addition of C + T + G. This allows a direct correlation between C or A incorporation opposite 8-oxo-G with the signal intensity of the polymerization products after extension. For all of the studies presented here, the templates that were used contained an 8-oxo-G placed either immediately after the 3′-OH of the primer, defined as standing start reaction (SS), or upstream of the primer (at position +26 from the 3′-OH), defined as running start reaction (RS) (Fig. 1A and Table S1). The RS conditions possibly mimic better a physiological situation of DNA replication.
Fig. 1.
DNA polymerase δ incorporates both A and C opposite 8-oxo-G, but displays difficulties when extending from C:8-oxo-G base pairs. (A) Schematic depiction of the primer-extension reactions. The sequence context of the template allows incorporation of C or A only opposite 8-oxo-G, as the rest of the sequence consists exclusively of C and A. (B) Selectivity of pol δ to incorporate A or C opposite 8-oxo-G and the influence of PCNA on this reaction. PCNA in indicated amounts was titrated to 1 nM pol δ on 8-oxo-G (SS) template (lanes 1–10) or on 8-oxo-G (RS) template (lanes 11–20) in the presence of either dATP + dGTP + dTTP (lanes 15–17) or dCTP + dGTP + dTTP (lanes 18–20).
DNA Polymerase δ Pauses at 8-oxo-G Before Bypassing It.
Analysis of 8-oxo-G bypass by human recombinant four-subunit pol δ in the presence of all four dNTPs showed that pol δ efficiently synthesized full-length (FL) products in a PCNA-dependent manner on the control (Fig. S1A) as well as on the 8-oxo-G template. However, FL formation was reduced by 40% on the 8-oxo-G template (Fig. S1A: compare lanes 2–5 to lanes 7–10, quantified in Fig. S1B). Under the conditions tested here and due to the sequence context of the template, the stimulation of pol δ by PCNA was relatively weak. Thus, we confirmed the PCNA dependency of pol δ on a different primer/template containing all four nucleotides (Fig. S1C: compare lanes 2–8 to lanes 9–15). A distinct pausing of pol δ at the nucleotide preceeding 8-oxo-G was observed, as indicated by an accumulation of a prominent band (Fig. S1A, lanes 7–10). Nevertheless, FL product was detected, indicating an intrinsic capacity of pol δ to bypass 8-oxo-G. A 20-min incubation time was determined to be optimal and used thereafter (Fig. S1D).
DNA Polymerase δ Incorporates both A and C Opposite 8-oxo-G, but Displays Difficulties When Extending from Correct C:8-oxo-G Base Pairs.
Next, we addressed the selectivity of pol δ for incorporation of A or C opposite 8-oxo-G by incubating pol δ with labeled primer/template and either A + T + G (which yields products only when wrong A is incorporated opposite 8-oxo-G) or C + T + G (which yields products only when correct C is incorporated opposite 8-oxo-G) (Fig. 1). In an SS reaction, pol δ bypassed the lesion in the presence of either A (incorrect bypass; Fig. 1B, lanes 5–7) or C (correct bypass; lanes 8–10). Clearly, 8-oxo-G bypass by pol δ was much more efficient in the presence of A than C, as visualized by the amount of FL product formed (Fig. 1B: compare lanes 5 and 8). As expected, the bypass of 8-oxo-G by pol δ could be stimulated by higher concentrations of PCNA (Fig. 1B: compare lanes 5–7 and 8–10). Under RS conditions, pol δ promoted formation of FL product by either A+T+G or C+T+G, even though pol δ stalled at the nucleotide preceeding 8-oxo-G (Fig. 1B, lanes 15–17 to 18–20), as previously shown by Fazlieva et al. (12). Interestingly, the incorporation of C opposite 8-oxo-G led to a pausing of pol δ, as visualized by the presence of a band opposite 8-oxo-G (Fig. 1B, lanes 18–20), which could not be seen with A (lanes 15–17, quantified in Fig. S1E). This pausing upon incorporation of C indicated that pol δ experienced difficulties in the extension of C:8-oxo-G base pairs, whereas it did easily extend from A:8-oxo-G base pairs, which is also consistent with the higher amount of FL product formed with A compared with C (Fig. 1B, lanes 15–17 and 18–20, quantified in Fig. S1F). These data suggested that pol δ incorporates both A and C opposite 8-oxo-G, but can preferentially extend A:8-oxo-G base pairs.
DNA Polymerase λ Assists DNA Polymerase δ in Correct TLS over 8-oxo-G by a Polymerase Switch Reaction.
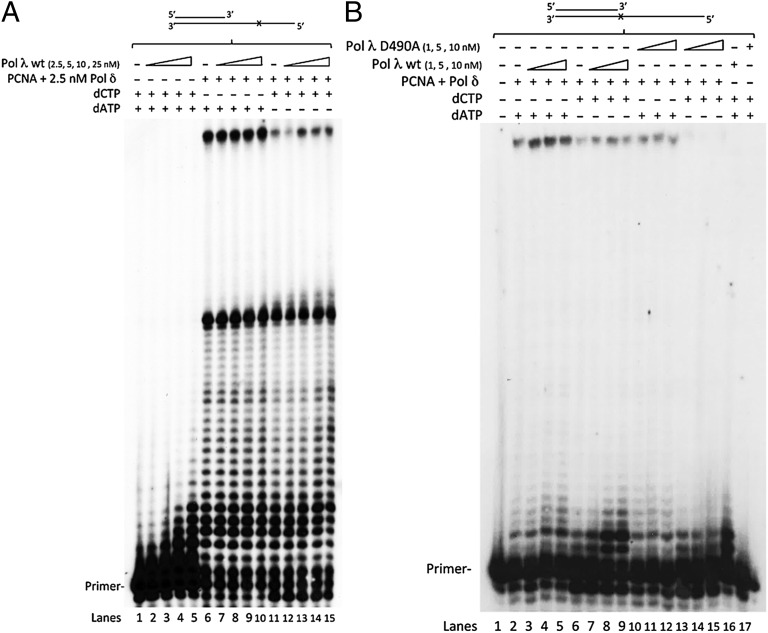
As pol δ seemed to pause at 8-oxo-G, we investigated whether the efficiency of pol δ in the bypass of 8-oxo-G could be modulated by the presence of other pols. Pol λ has been shown to play a role in the correct bypass of 8-oxo-G (3, 6) and to preferentially extend C:8-oxo-G base pairs (7). Addition of pol λ to the reaction with pol δ led to the formation of extension products of a few nucleotides from the pause site opposite 8-oxo-G. Importantly, however, this occurred only with C and not with A and depended on the amounts of pol λ in the reactions (Fig. 2A: compare lanes 6–10 to lanes 11–15). Also, the FL product generated by pol δ increased as a function of pol λ, but only in the presence of C (Fig. 2A: compare lanes 6–10 to lanes 11–15, quantified in Fig. S2A), suggesting that pol λ can help pol δ in performing the correct TLS over 8-oxo-G by participating in a pol switch. The use of a catalytically inactive mutant of pol λ, pol λ D490A, confirmed that the observed effect derived from a dynamic exchange between pols δ and λ and was not due to a simple protein-crowding effect (Fig. S2B, lanes 4, 7, and 9). The pol λ D490A mutant could still bind to DNA, indicating that this mutant pol λ was correctly folded (Fig. S2C). The assistance of pol λ to specifically promote the error-free, but not the incorrect, TLS of 8-oxo-G by pol δ was further verified under SS conditions (Fig. 2B: compare lanes 2–5 with lanes 6–9). As pol λ alone did not incorporate more than a few nucleotides under these conditions (Fig. 2B, lane 16), the FL products could only be the product of pol δ activity, suggesting a dynamic pol switch from pol λ to pol δ after pol λ had extended the primer end by several nucleotides past 8-oxo-G. Again, this stimulatory effect of pol λ was dependent on its catalytic activity, as the D490A mutant failed to show a similar effect (Fig. 2B: compare lanes 3–5 and 6–9 with lanes 10–12 and 13–15). In fact, pol λ D490A greatly reduced the FL product formation in the reactions containing C (Fig. 2B: compare lane 6 with lanes 13–15), suggesting that it bound to the primer terminus at 8-oxo-G, but could not dissociate anymore as efficiently as pol λ wild type, thus blocking pol δ from accessing the primer terminus. Again, this effect was much more pronounced in the presence of C and was hardly visible with A. Analysis of the influence of PCNA on both pols on the SS or the RS template suggested that PCNA did not affect the bypass catalyzed by pol λ but enhanced pol δ’s utilization of the 8-oxo-G bypass products generated by pol λ (Fig. S2D). In summary, these data suggest that pol λ assists pol δ in the correct TLS 8-oxo-G by a dynamic pol switch occurring at the lesion itself by incorporating the correct C opposite 8-oxo-G and/or extending the correct C:8-oxo-G base pair, thus assisting pol δ in performing TLS in a correct manner.
Fig. 2.
DNA polymerase λ assists DNA polymerase δ in correct TLS over 8-oxo-G by a polymerase switch reaction. (A) Influence of pol λ on the C- or A-mediated bypass of 8-oxo-G by pol δ. Pol λ was titrated in the presence of 100 nM PCNA and all four dNTPs alone (lanes 2–5) or in the presence of 100 nM PCNA, 2.5 nM pol δ, and dATP + dGTP + dTTP (lanes 6–10) or dCTP + dGTP + dTTP (lanes 11–15). (B) Titration of pol λ wild type (wt) and pol λ D490A in a SS reaction with 1 nM pol δ, 100 nM PCNA, and dATP + dGTP + dTTP (lanes 3–5 and 10–12) or dCTP + dGTP + dTTP (lanes 7–9 and 13–15). All lanes contain G and T.
Selectivity to Assist DNA Polymerase δ in Error-Free Translesion Synthesis Is Specific for Full-Length DNA Polymerase λ.
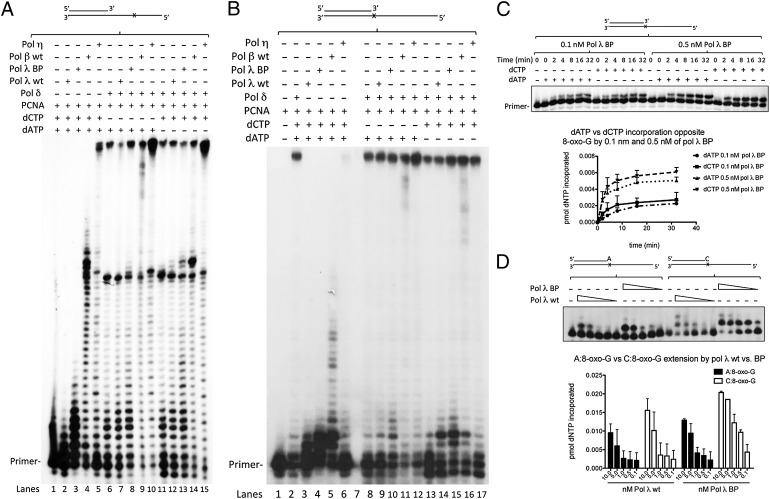
Pol λ contains three major domains: an N-terminal breast cancer susceptibility gene 1 C terminus (BRCT) domain, a proline-rich domain (PRD), and the C-terminal pol domain (13). We next investigated the importance of the BRCT and PRD of pol λ in TLS of 8-oxo-G, comparing wild-type pol λ and a truncated pol λ (called pol λ BP) lacking these two domains and thus closely resembling pol β (see Fig. S3A for a schematic representation of the domains of pol λ, pol λBP, and pol β). Like pol λ wild type, pol λ BP also assisted pol δ in the bypass of 8-oxo-G, as shown by the increase in lesion bypass and FL products (Fig. S3B: compare lanes 12–14 and 15–17, quantified in Fig. S3C). Generally, pol λ BP was more active than pol λ wild type at equimolar amounts, as expected (14). However, in strong contrast to pol λ, pol λ BP not only stimulated TLS by correct C incorporation, but even preferentially enhanced the bypass by wrong A, as demonstrated by the accumulation of short extension products after the lesion as well as FL products (Fig. 3A: compare lanes 6–8 and 11–13). The same effect could be observed in a SS reaction setup (Fig. 3B: compare 8–10 and 13–15). It has been previously shown that the N-terminal domain of pol λ plays a pivotal role in modulating the fidelity of the enzyme on undamaged DNA (15). The failure of pol λ BP to discriminate between the correct C incorporation opposite 8-oxo-G and the incorrect A incorporation prompted us to investigate its incorporation fidelity opposite 8-oxo-G. Indeed, single-nucleotide incorporation studies revealed that pol λ BP catalyzed incorporation of both incorrect A and correct C opposite 8-oxo-G to a similar extent (Fig. 3C and Fig. S3F). Assays with primer/template combinations that harbored either A:8-oxo-G or C:8-oxo-G base pairs at the 3′ primer terminus showed no strong differences between the pol λ wild type and pol λ BP, suggesting that the N-terminal mutant of pol λ retains its ability to discriminate against incorrect extension (Fig. 3D). These results suggested that, although the catalytic domain of pol λ was sufficient for assisting pol δ in TLS over 8-oxo-G, it seems to display a lower selectivity in discriminating against insertion of the incorrect A opposite 8-oxo-G, thus denoting a possible role of the N-terminal part in maintaining the fidelity in 8-oxo-G bypass. Also, these results indicate that pol λ not only might help pol δ to bypass 8-oxo-G by extending from C:8-oxo-G base pairs, but also possibly performs the incorporation step of C opposite 8-oxo-G and thus supports pol δ in overcoming the lesion correctly. Because RP-A has been shown to modulate the fidelity of 8-oxo-G bypass by pol λ (3, 16), we tested the influence of RP-A on the pol switch between pols δ and λ in a SS setup (Fig. S3G). We found that addition of RP-A does not selectively promote insertion of C or A by pol λ under these conditions. Similarly, monoubiquitination of PCNA, known to promote TLS of blocking lesions, did not influence the TLS by pol λ (Fig. S4 A and B), suggesting that monoubiquitinated PCNA was dispensable for a pol switch between pols λ and δ at 8-oxo-G.
Fig. 3.
Selectivity to assist DNA polymerase δ in error-free translesion synthesis is specific for DNA polymerase λ. (A) Influence of the N-terminal deletion of pol λ, pol β, or pol η on the selectivity of the TLS in a RS reaction. A total of 10 nM pol λ, pol λBP, or pol β or 4 nM pol η was assayed on the 8-oxo-G (RS) template alone with 100 nM PCNA and all four dNTPs (lanes 2–5) or in the presence of 100 nM PCNA, 1 nM pol δ, and dATP + dGTP + dTTP (lanes 6–10) or dCTP + dGTP + dTTP (lanes 11–15). Lane 6: pol δ alone with 100 nM PCNA and dATP + dGTP + dTTP; lane 11: pol δ alone with 100 nM PCNA and dCTP + dGTP + dTTP. (B) Influence of the N-terminal deletion of pol λ, pol β, or pol η on the selectivity of the TLS in a SS reaction. A total of 10 nM pol λ, 10 nM pol λBP, 10 nM pol β, and 4 nM pol η was assayed on the 8-oxo-G (SS) template in the presence of dATP + dGTP + dTTP (lanes 8–12) or dCTP + dGTP + dTTP (lanes 13–17) in the presence of 100 nM PCNA (lanes 3–6) or in the presence of 1 nM pol δ and 100 nM PCNA (lanes 8–17). (C) Time course of single-nucleotide incorporation specificity by 0.1 or 0.5 nM pol λ BP (0–32 min) in a SS reaction with either 0.5 μM C or A, respectively, in the presence of 1 mM MgCl2. (Lower) Quantification of two independent experiments showing mean ± SD. (D) Extension of A:8-oxo-G or C:8-oxo-G primer termini by 10, 5, 1, 0.5, and 0.1 nM pol λ wild type (wt) or pol λ BP in the presence of all four dNTPs. (Lower) Quantification of two independent experiments showing mean ± SD.
The X-family member pol β shares 30% homology with pol λ (17) (Fig. S3A). At 10 nM, pol β alone was able to synthesize past 8-oxo-G whereas pol λ was not (Fig. S3B: compare lanes 3–5 to 9–11). In the presence of pol δ and PCNA, pol β accumulated more products immediately after 8-oxo-G than pol λ (Fig. S3B: compare lane 12–14 to lanes 18–20, quantified in Fig. S3C). However, in contrast to pol λ, pol β enhanced bypass of 8-oxo-G by incorporating more of the wrong A and less of the correct C compared with pol λ (Fig. 3A: compare lane 7 with lane 9 and lane 12 with lane 14; Fig. S3D: compare lanes 12–15 and 16–19). This effect was also visible under SS conditions (Fig. 3B: compare lane 9 with lane 11 and lane 14 with lane 16; note the primer use). Therefore, pol β was capable of assisting pol δ in bypassing 8-oxo-G, but, in contrast to pol λ, without discriminating between correct C or incorrect A.
Pol η, a Y-family pol, has been implicated in 8-oxo-G TLS (18–20). At the highest concentrations used, pol η was able to bypass 8-oxo-G much more efficiently than pol λ (Fig. S3E: compare lanes 7–8 to lane 17). However, pol η seemed to not only assist pol δ in bypassing 8-oxo-G by a pol switch in this context, but also to perform FL primer extension past 8-oxo-G on its own, as the polymerization signature of pol η was very reminiscent of an overlap of the individual activities of pols δ and η, unlike for pol λ (Fig. S3E: compare lanes 8 and 15 and lanes 9–15 to lane 18). Moreover, similarly as seen for pol λ BP and pol β, pol η also enhanced bypass of 8-oxo-G without discriminating between correct C and incorrect A (Fig. 3A: compare lane 7 with lane 10 and lane 12 with lane 15; Fig. S3D: compare lanes 12–15 and 16–19). This was further verified in the SS setup (Fig. 3B: compare lane 9 with lane 12 and lane 14 with lane 17). This evidence suggests that pol η does not participate in an error-free switching mechanism with pol δ under our assay conditions. In conclusion, these results substantiated the idea of a unique role for pol λ in the correct bypass of 8-oxo-G by participating in a pol switch with pol δ at the lesion to promote correct TLS.
Switch Between the Replicative DNA Polymerase δ and the TLS DNA Polymerase λ Influences the Correct Bypass of 8-oxo-G Lesions both in Cellular Extracts and in Vivo.
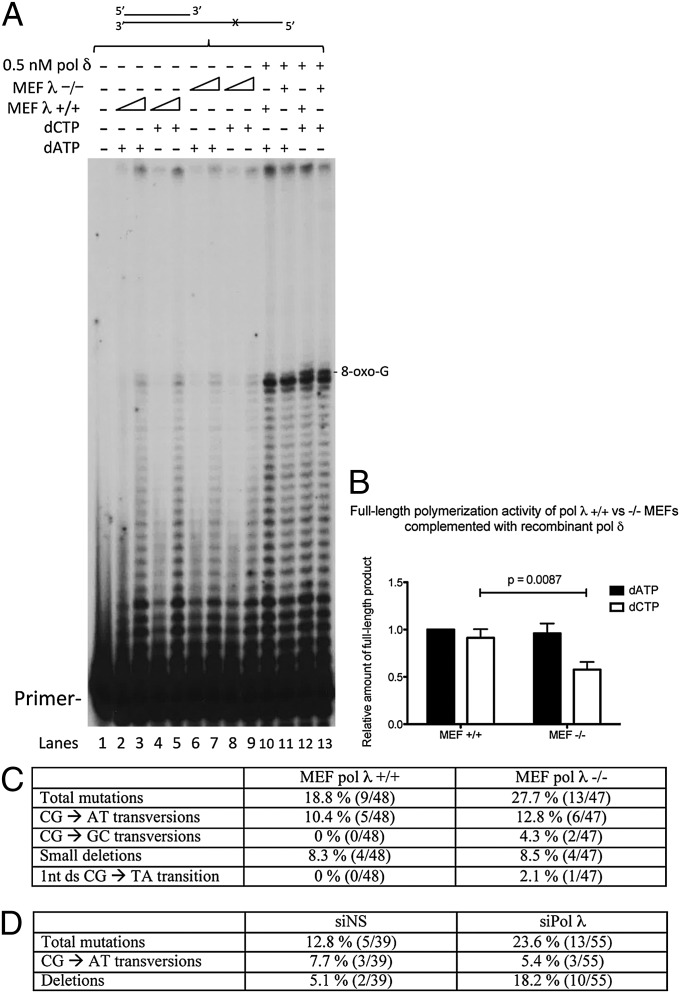
To see whether this presumable in vitro switch between pols δ and λ played a role in TLS of 8-oxo-G in vivo, we performed experiments on the RS template by using extracts from pol λ+/+ and pol λ−/− MEFs. Indeed, we found a more pronounced FL product formation by pol λ+/+ extracts compared with pol λ−/− extracts (Fig. 4A: compare lanes 3 and 5 to lanes 7 and 9). Complementation of these extracts with recombinant pol δ further emphasized that, although the FL product formation with the incorrect A was comparable between pol λ+/+ and pol λ−/− extracts, pol λ−/− extracts exhibited a significant decrease in FL product formation with correct C compared with the pol λ+/+ extracts (Fig. 4A: compare lanes 10–11 and 12–13 and quantification in Fig. 4B). To further substantiate the role of this switch in vivo, we used a gapped plasmid containing an 8-oxo-G at a defined site, as developed by the Livneh laboratory (9). This plasmid was transfected into MEFs, where the cellular pols performed TLS to close the gap. Transformation of extracted plasmids into E. coli mutY−/− yielded single colonies, of which the plasmid sequence was analyzed to reveal how accurately the cellular machinery had bypassed 8-oxo-G. Importantly, the short incubation time (5 h) of the plasmid in MEFs allowed only the closure of the gap, but no recognition of eventual A:8-oxo-G mismatches by MutYH (9, 21). The absence of pol λ in the MEF pol λ−/− was verified by Western blot analysis (Fig. S5A). Analysis of the plasmids revealed a total mutation frequency of 18.8% for pol λ+/+ MEFs (Fig. 4C), a value very close to previous findings (9). Importantly, however, the mutation frequency in pol λ−/− MEFs increased to 27.7% (Fig. 4C). To further validate these results in a human cell line, we knocked down pol λ by siRNA in HeLa cells (Fig. 4D and Fig. S5B). Similarly, HeLa cells with siRNA knockdown of pol λ also showed an increase of the total mutations to 23.6% compared with 12.8% in the control cells. In summary, these data indicate that pol λ indeed plays an important role in the correct bypass of 8-oxo-G lesions in vivo.
Fig. 4.
Switch between the replicative DNA polymerase δ and the TLS DNA polymerase λ influences the correct bypass of 8-oxo-G lesions both in cellular extracts and in vivo. (A) Activity of MEF pol λ+/+ or pol λ−/− extracts on RS primer extension assays in the absence or the presence of recombinant pol δ. All reactions contain dGTP + dTTP. (B) Quantification of full-length product in A in the presence of recombinant pol δ. The full-length signal was normalized to the total intensity of the polymerization products and the amount of full-length product formed by MEF pol λ+/+ extracts in presence of A. Values are the results from three independent experiments showing mean ± SD. The P value was calculated using the Student’s t test. (C) Sequence analysis of gapped plasmids extracted from MEF pol λ+/+ or pol λ−/− cells. The absolute numbers of the respective mutations followed by the total amount of analyzed sequences are indicated in parentheses. (D) Sequence analysis of gapped plasmids extracted from HeLa cells treated with siRNA against pol λ or an unspecific siRNA. The absolute numbers of the respective mutations followed by the total amount of analyzed sequences are indicated in parentheses.
Discussion
During DNA replication the fork can be stalled or paused at sites of DNA damage. One of the mechanisms to overcome such a replication block is the recruitment of specific pols to overcome the lesion in the process called TLS (reviewed in ref. 22). In eukaryotes, this encompasses the replacement of the replicative pol δ or ε by a pol that is capable of bypassing the damaged nucleotide, a process called pol switch.
8-oxo-G lesions do not greatly block replicative pols (8, 9). However, several studies have found that replicative pols extend incorrect A:8-oxo-G base pairs much more efficiently than C:8-oxo-G (8, 11). Nevertheless, 8-oxo-G has been shown to be bypassed correctly in up to 81% of the cases in vivo (9). The key question of this work was how the correct bypass of 8-oxo-G could be accomplished when the extension of C:8-oxo-G base pairs poses such a problem for the replicative pols. Consistent with previous results, pol δ elongation was initially stalled on the nucleotide located 3′ to 8-oxo-G (8, 11), but it could ultimately bypass the lesion. Pol δ incorporated, as expected, both the correct C and the incorrect A opposite 8-oxo-G in the presence of PCNA (3). The incorporation of the correct C opposite 8-oxo-G induced a stalling of pol δ opposite the lesion, which was less evident upon incorporation of the incorrect A (Figs. 1 and 2). This difficulty of pol δ to extend a correct C:8-oxo-G base pair compared with an incorrect A:8-oxo-G Hoogsteen base pair might be due to the fact that C:8-oxo-G base pairs induce template and pol distortions as they are seen when the active site of a pol encounters a mismatch (23; discussed in ref. 2). Thus, upon formation of C:8-oxo-G base pairs, pol δ might enter a futile cycle of removal of C by its 3′→5′ exonuclease activity, followed by renewed incorporation of C (in our setup, as no A is provided), thereby hindering it from further elongation. Pol λ can discriminate between mismatched A:8-oxo-G and correct C:8-oxo-G pairs at the extension step (7). Furthermore, pol λ is by far the most faithful pol in bypassing 8-oxo-G, incorporating the correct C 1,200-fold more efficiently than the incorrect A in the presence of RP-A and PCNA (3). In addition to pol λ, pol η has been suggested to be involved in the bypass and correct repair of 8-oxo-G in Saccharomyces cerevisiae and in mammalian cells (18–20). In view of our findings that pol δ paused at 8-oxo-G (Figs. 1–3), we tested whether an exchange between the replicative pol δ and the pols λ, β, and η at 8-oxo-G could be observed. Our experiments suggest the existence of a pathway that promotes error-free TLS by a specific interplay of pol δ with pol λ, but not pols β or η, at 8-oxo-G (Figs. 2 and 3). This pathway consists of a specific switch from pol δ at 8-oxo-G to pol λ, which incorporates correct C opposite 8-oxo-G and subsequently extends the resulting C:8-oxo-G base pair by two or more nucleotides, after which pol λ is again replaced by pol δ. Therefore, even though 8-oxo-G is not considered a blocking lesion per se, it can be bypassed by a pol switch reaction. The effect to only promote the correct 8-oxo-G bypass is specific to pol λ wild type, as pol λ BP (lacking the N-terminal proline-rich and BRCT domains; Fig. S3A), pol β, and pol η could all assist pol δ in the bypass of 8-oxo-G, but were unable to discriminate between correct and incorrect bypass (Fig. 3 and Fig. S3). These findings are in agreement with data showing that the deletion of the pol λ N terminus greatly decreases its fidelity (15) as well as with a recent structural report showing that pol β does indeed extend both A:8-oxo-G and C:8-oxo-G base pairs with a similar efficiency (24). We have found that pol λ can catalyze the excision of wrong A base-paired to 8-oxo-G by means of pyrophosphorolysis (25). This mechanism could further contribute to the correct bypass of 8-oxo-G by promoting removal of wrong A that was inserted by pol δ, followed by insertion of correct C by pol λ. The amount of FL product formation in the reactions with correct C could be stimulated by the addition of pol λ (Fig. 2A and Fig. S2A), but it never surpassed the amount that was generated by wrong A incorporation by pol δ alone. This can be explained by the fact that it presumably takes more time to switch between pols δ and λ at the lesion compared with replicating past 8-oxo-G without switching. Therefore, even though the switch clearly promotes the error-free bypass of 8-oxo-G, this reaction always generated less FL product than the wrong A-containing reaction, which was simply due to the necessity to switch between pols. Also, the efficiency of this in vitro reaction is possibly much less efficient than a similar switch occurring in vivo due to the lack of one or more yet-unknown switch factors that coordinate and control such events in the cellular context. This is also consistent with the fact that the impact of the pol λ presence on FL product formation is much more profound in MEF extracts than in the reactions with purified proteins (Fig. 4 A and B). Correct TLS over 8-oxo-G was significantly more efficient in extracts from pol λ+/+ compared with pol λ−/− MEFs complemented with pol δ, supporting the concept that our findings are also relevant in cells (Fig. 4 A and B). Additionally, analysis of cellular repair of gapped plasmids harboring an 8-oxo-G lesion in pol λ+/+ MEFs revealed a total mutation frequency of 18.8% occurring at 8-oxo-G (Fig. 4C). This mutation frequency reflects only bypass of 8-oxo-G, as the conditions that were assayed prevent the contribution of MutYH-initiated repair (9, 21). This result is consistent with a previous report of 8-oxo-G plasmids in H1299 cells (9). Importantly, however, the mutation frequency in pol λ−/− MEFs was increased by 50% (Fig. 4C), consistent with a role of pol λ in the bypass of 8-oxo-G in vivo. Interestingly, although the CG→AT transversion rate seemed unaffected by the pol λ background of the cells, only pol λ−/− MEFs showed the occurrence of CG→GC transversions, in line with previous results by the Livneh laboratory (9). The rather frequent occurrence of small deletions (1–6 nt) found in our study was surprising and might be a cell and/or a sequence-specific effect. These results were further validated by analysis of gapped plasmids in HeLa cells depleted of pol λ by siRNA (Fig. 4D), where the results were very similar. Further work aimed at investigating whether this switch between pols δ and λ at 8-oxo-G lesions happens in vivo will be necessary to further confirm our data obtained thus far.
Concluding, the data of this work suggest the existence of a pol switch from replicative pol δ to the X-family pol λ at 8-oxo-G during replication to enhance the correct bypass of 8-oxo-G. This model is supported by previous evidence from our laboratory showing that addition of pol λ to the pol λ−/− MEFs extract restored the preferential incorporation of the C versus A in the presence of PCNA and RP-A (3). Moreover, pol λ is recruited to the sites of oxidative DNA damage (9), and it is recruited to chromatin upon induction of oxidative stress in S-phase cells (26, 27) as we showed very recently. The accumulating evidence supports the notion that pol λ acts as a key player in the prevention and processing of A:8-oxo-G mismatches by a dual role: first, pol λ is involved in the correct MutYH-initiated repair of A:8-oxo-G mispairs that occurs post replication (6, 16, 26, 27), and, second, it is also important during DNA replication itself, as demonstrated in this study. The pol switch between pol δ and pol λ during TLS of 8-oxo-G that we propose might contribute to prevention of replication fork stalling and to enhanced accuracy of bypass of this very frequent lesion [estimations are about 1,000–2,000 8-oxo-G per cell per day under physiological conditions (28)]. This switch could physiologically function as an accurate, efficient, and low-cost system to counteract the mutagenic effect of the many 8-oxo-G lesions that are constantly present in cells.
Materials and Methods
Antibodies.
Antitubulin was purchased from Sigma, anti-pol λ for human cells was obtained from Bethyl laboratories, and the antibody for detection of murine pol λ was a kind gift of L. Blanco, Centro de Biologica Molecular Severo Ochoa, Madrid.
Chemicals.
Deoxyribonucleoside triphosphates were purchased from Sigma. Labeled γ[32P] ATP was purchased from Hartmann Analytic. All other reagents were of analytic grade and purchased from Fluka, Sigma, or Merck.
Oligonucleotide DNA Substrates.
The control oligonucleotides were ordered PAGE-purified from Microsynth, the ones with 8-oxo-G were HPLC-purified from Purimex. Annealing of 100-mer containing an 8-oxo-G with the 5′-labeled 39-mer was carried out as described (6). A complete description of materials and methods used can be found in SI Materials and Methods.
Figures.
For some main and SI Text figures, parts of the gels that were not important for this work were removed to save space. The sites of removal are indicated by arrowheads at the bottom of the respective gels. The DNA substrates used are indicated on the top of the figure, and for some figures the sequence of the template around the lesion (X) is shown next to the panel.
Supplementary Material
Acknowledgments
We thank Z. Livneh and S. Shachar for material, advice, and help with the construction of the gapped plasmids and L. Blanco for the antibody against murine pol λ. This work was supported by Oncosuisse Grant KLS-02339-02-2009 (to B.C. and U.H.); by Swiss National Science Foundation Grant 31003A_133100/1 (to B.C., E.M., and U.H.); by an MD-PhD grant from the Swiss National Science Foundation and “Forschungskredit” of the University of Zurich (to E.M.); and by the University of Zurich.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211532109/-/DCSupplemental.
References
- 1.van Loon B, Markkanen E, Hübscher U. Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst) 2010;9(6):604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 2.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447(7147):941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maga G, et al. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447(7144):606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 4.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nash HM, et al. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6(8):968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 6.van Loon B, Hübscher U. An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase lambda. Proc Natl Acad Sci USA. 2009;106(43):18201–18206. doi: 10.1073/pnas.0907280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picher AJ, Blanco L. Human DNA polymerase lambda is a proficient extender of primer ends paired to 7,8-dihydro-8-oxoguanine. DNA Repair (Amst) 2007;6(12):1749–1756. doi: 10.1016/j.dnarep.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349(6308):431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 9.Avkin S, Livneh Z. Efficiency, specificity and DNA polymerase-dependence of translesion replication across the oxidative DNA lesion 8-oxoguanine in human cells. Mutat Res. 2002;510(1–2):81–90. doi: 10.1016/s0027-5107(02)00254-3. [DOI] [PubMed] [Google Scholar]
- 10.Mozzherin DJ, Shibutani S, Tan CK, Downey KM, Fisher PA. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase delta. Proc Natl Acad Sci USA. 1997;94(12):6126–6131. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase delta. Steady-state and pre-steady-state kinetic analysis. J Biol Chem. 2001;276(6):3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 12.Fazlieva R, et al. Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009;37(9):2854–2866. doi: 10.1093/nar/gkp155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Díaz M, et al. DNA polymerase lambda, a novel DNA repair enzyme in human cells. J Biol Chem. 2002;277(15):13184–13191. doi: 10.1074/jbc.M111601200. [DOI] [PubMed] [Google Scholar]
- 14.Shimazaki N, et al. Over-expression of human DNA polymerase lambda in E. coli and characterization of the recombinant enzyme. Genes Cells. 2002;7(7):639–651. doi: 10.1046/j.1365-2443.2002.00547.x. [DOI] [PubMed] [Google Scholar]
- 15.Fiala KA, Duym WW, Zhang J, Suo Z. Up-regulation of the fidelity of human DNA polymerase lambda by its non-enzymatic proline-rich domain. J Biol Chem. 2006;281(28):19038–19044. doi: 10.1074/jbc.M601178200. [DOI] [PubMed] [Google Scholar]
- 16.Maga G, et al. Replication protein A and proliferating cell nuclear antigen coordinate DNA polymerase selection in 8-oxo-guanine repair. Proc Natl Acad Sci USA. 2008;105(52):20689–20694. doi: 10.1073/pnas.0811241106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiala KA, Abdel-Gawad W, Suo Z. Pre-steady-state kinetic studies of the fidelity and mechanism of polymerization catalyzed by truncated human DNA polymerase lambda. Biochemistry. 2004;43(21):6751–6762. doi: 10.1021/bi049975c. [DOI] [PubMed] [Google Scholar]
- 18.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet. 2000;25(4):458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 19.Lee DH, Pfeifer GP. Translesion synthesis of 7,8-dihydro-8-oxo-2′-deoxyguanosine by DNA polymerase eta in vivo. Mutat Res. 2008;641(1–2):19–26. doi: 10.1016/j.mrfmmm.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCulloch SD, Kokoska RJ, Garg P, Burgers PM, Kunkel TA. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases delta and eta. Nucleic Acids Res. 2009;37(9):2830–2840. doi: 10.1093/nar/gkp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Page F, Guy A, Cadet J, Sarasin A, Gentil A. Repair and mutagenic potency of 8-oxoG:A and 8-oxoG:C base pairs in mammalian cells. Nucleic Acids Res. 1998;26(5):1276–1281. doi: 10.1093/nar/26.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9(4):729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 23.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431(7005):217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 24.Batra VK, Shock DD, Beard WA, McKenna CE, Wilson SH. Binary complex crystal structure of DNA polymerase β reveals multiple conformations of the templating 8-oxoguanine lesion. Proc Natl Acad Sci USA. 2012;109(1):113–118. doi: 10.1073/pnas.1112235108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crespan E, Maga G, Hübscher U. A new proofreading mechanism for lesion bypass by DNA polymerase-λ. EMBO Rep. 2012;13(1):68–74. doi: 10.1038/embor.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markkanen E, Hübscher U, van Loon B. Regulation of oxidative DNA damage repair: The adenine:8-oxo-guanine problem. Cell Cycle. 2012;11(6):1070–1075. doi: 10.4161/cc.11.6.19448. [DOI] [PubMed] [Google Scholar]
- 27.Markkanen E, et al. Regulation of oxidative DNA damage repair by DNA polymerase λ and MutYH by cross-talk of phosphorylation and ubiquitination. Proc Natl Acad Sci USA. 2012;109(2):437–442. doi: 10.1073/pnas.1110449109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. 2nd ed. pp xxix, 1118 pp. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.