Abstract
Riboswitches are mRNA regulatory elements that control gene expression by altering their structure in response to specific metabolite binding. In bacteria, riboswitches consist of an aptamer that performs ligand recognition and an expression platform that regulates either transcription termination or translation initiation. Here, we describe a dual-acting riboswitch from Escherichia coli that, in addition to modulating translation initiation, also is directly involved in the control of initial mRNA decay. Upon lysine binding, the lysC riboswitch adopts a conformation that not only inhibits translation initiation but also exposes RNase E cleavage sites located in the riboswitch expression platform. However, in the absence of lysine, the riboswitch folds into an alternative conformation that simultaneously allows translation initiation and sequesters RNase E cleavage sites. Both regulatory activities can be individually inhibited, indicating that translation initiation and mRNA decay can be modulated independently using the same conformational switch. Because RNase E cleavage sites are located in the riboswitch sequence, this riboswitch provides a unique means for the riboswitch to modulate RNase E cleavage activity directly as a function of lysine. This dual inhibition is in contrast to other riboswitches, such as the thiamin pyrophosphate-sensing thiM riboswitch, which triggers mRNA decay only as a consequence of translation inhibition. The riboswitch control of RNase E cleavage activity is an example of a mechanism by which metabolite sensing is used to regulate gene expression of single genes or even large polycistronic mRNAs as a function of environmental changes.
Keywords: gene regulation, RNA degradosome, translational control
Since the first demonstration that translation attenuation regulates the expression of the tryptophan operon (1), accumulating evidence has revealed the importance of posttranscriptional regulation in prokaryotes and eukaryotes alike. Posttranscriptional regulators include RNA molecules that operate through several mechanisms to control a wide range of physiological responses (2). Among newly identified RNA regulators are riboswitches, which are located in untranslated regions of several mRNAs and that modulate gene expression at the level of transcription, translation, or splicing (3). Riboswitches are highly structured regulatory domains that directly sense cellular metabolites such as amino acids, carbohydrates, coenzymes, and nucleobases. These genetic switches are composed of two modular domains consisting of an aptamer and an expression platform. The aptamer is involved in the specific recognition of the metabolite, and the expression platform is used to control gene expression by altering the structure of the mRNA. Recent bioinformatic analyses have reported the existence of several new RNA motifs exhibiting unconventional expression platforms (4, 5), suggesting that riboswitches using different regulation mechanisms are still likely to be discovered (6).
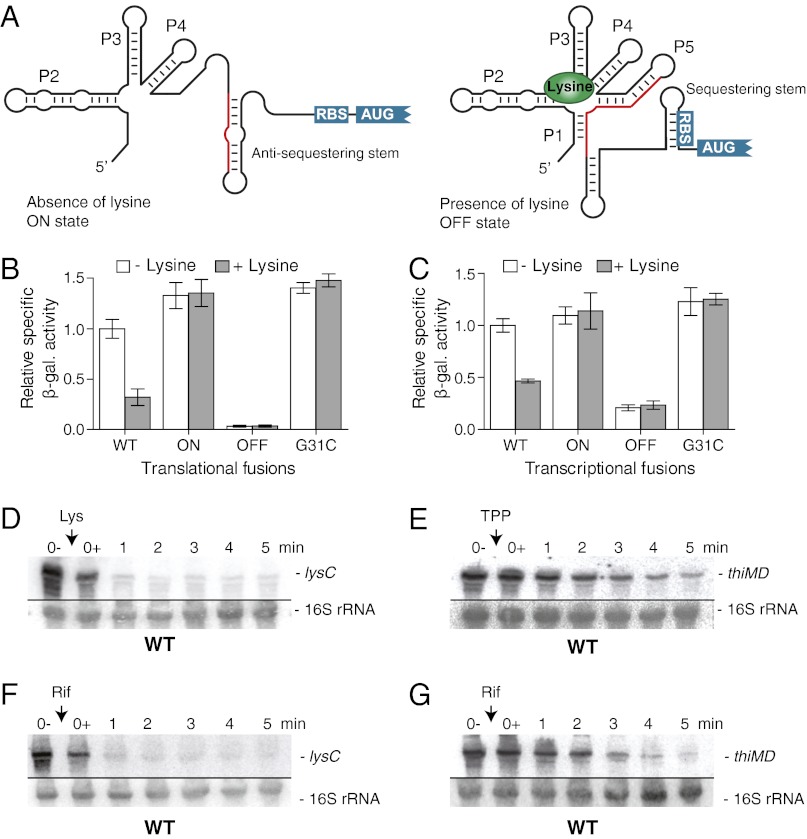
The lysine riboswitch was first characterized in Bacillus subtilis, where it is located upstream of a lysine transporter (yvsH) and a lysine-sensitive aspartokinase (lysC) (7–9). Sequence alignments predicted that the secondary structure of the aptamer domain is arranged around a five-way junction that is important for aptamer architecture (7, 9). Lysine binding to the riboswitch results in the stabilization of the anti-antiterminator, the P1 stem, which induces premature transcription termination (7, 9, 10). Helices P2–P5 are involved in the formation of the global riboswitch structure that is required in the formation of the binding pocket specifically recognizing lysine (11, 12). Sequence alignments also predicted that, in contrast to B. subtilis, the Escherichia coli lysC riboswitch could control gene expression at the translational level (8). According to this model (Fig. 1A), the E. coli lysC riboswitch adopts an ON state in the absence of lysine to allow ribosome access to the ribosome-binding site (RBS). However, it is predicted that, when bound to lysine, the riboswitch folds into an OFF state that sequesters the RBS into a stem–loop structure, thereby inhibiting translation initiation.
Fig. 1.
The E. coli lysC riboswitch modulates the mRNA level upon lysine binding. (A) Schematic representation of the predicted lysine riboswitch translational control. In the absence of lysine, the ON state exhibits an antisequestering stem, which exposes the RBS and allows translation initiation. However, when bound to lysine, the riboswitch adopts the OFF state in which the presence of a sequestering stem masks the RBS and inhibits gene expression. The region shared by the antisequestering stem (ON state) and the aptamer (OFF state) is represented in red. (B) β-Galactosidase assays of translational LysC-LacZ fusions for wild type and ON, OFF, and G31C mutants (Tables S1–S3). Enzymatic activities were measured in the absence and presence of 10 µg/mL lysine. Values were normalized to the enzyme activity obtained for the wild type in the absence of lysine. (C) β-Galactosidase assays of transcriptional lysC-lacZ fusions for the wild type and for the ON, OFF, and G31C mutants. Enzymatic activities were measured in the absence and presence of 10 µg/mL lysine. Values were normalized to the activity obtained for the wild type in the absence of lysine. (D) Northern blot analysis of lysC mRNA level. Wild-type E. coli strain MG1655 was grown to midlog phase in M63 minimal medium with 0.2% glucose at 37 °C, and total RNA was extracted at the indicated times immediately before (0−) and after (0+) the addition of lysine (10 µg/mL). The probe was designed to detect the riboswitch region (positions 42–201) of the lysC mRNA (SI Appendix, Fig. S4A). 16S rRNA was used as a loading control. (E) Northern blot analysis of the thiMD mRNA level. Bacterial growth culture and RNA extractions were performed as described in D. TPP was added at a final concentration of 500 µg/mL. The probe was designed to detect the riboswitch region (positions 4–141) of thiMD mRNA. 16S rRNA was used as a loading control. (F) Determination of the lysC mRNA stability. Bacterial growth culture and RNA extractions were performed as described in D, but without the addition of lysine. Rifampicin was added at a final concentration of 250 µg/mL. 16S rRNA was used as a loading control. (G) Determination of thiMD mRNA stability. Bacterial growth culture and RNA extractions were performed as described in D. Rifampicin was added at a final concentration of 250 µg/mL. 16S rRNA was used as a loading control.
Despite recent efforts dedicated to characterizing in vivo riboswitch regulation, very little is known about the actual cellular processes involved in riboswitch control mechanisms. In this work, we have studied the in vivo regulation mechanism of the E. coli lysC riboswitch. As expected from earlier predictions, our results are consistent with the riboswitch modulating translation initiation as a function of lysine. However, we observed that lysine binding to lysC also targets RNase E cleavage in the riboswitch, significantly reducing the lysC mRNA level. RNase E cleavage sites were mapped within the riboswitch expression platform, which become accessible to RNase E uniquely when the riboswitch is bound to lysine. The conformation of the lysine-free riboswitch prevents mRNA cleavage, thereby allowing lysC mRNA accumulation and translation. We also found that both regulatory mechanisms can be individually inhibited, directing the riboswitch to regulate expression at the level of either translation initiation or mRNA decay. These results are in contrast to other riboswitches, such as thiM and btuB, for which mRNA decay occurs only as a consequence of the inhibition of translation initiation. Our study provides a clear molecular mechanism in which riboswitches directly control mRNA decay by selectively modulating the access to RNase E cleavage sites as a function of metabolite binding.
Results
lysC Riboswitch Controls Gene Expression at the mRNA Level.
We first investigated the regulatory mechanism of the lysC riboswitch using transcriptional and translational lacZ chromosomal fusions containing the riboswitch domain with the first 57 nucleotides (19 codons) of lysC ORF (see constructs in SI Appendix, Fig. S1A). In these experiments, although the LysC-LacZ translational fusion reports on the LysC protein level, the transcriptional lysC-lacZ fusion has two independent RBS for lysC and lacZ, which monitor the lysC mRNA level. Because the lysC promoter is regulated, to a small extent, by lysine (SI Appendix, Fig. S1 B and C) (13), we used an arabinose-inducible promoter to prevent lysine-dependent promoter regulation.
In the context of a translational construct, we observed that the β-galactosidase activity of the wild-type riboswitch was reduced by 70% in the presence of 10 µg/mL lysine (Fig. 1B, WT). The extent of regulation is significantly larger than that obtained at the promoter level (SI Appendix, Fig. S1C), suggesting that genetic control is performed mostly via the lysC riboswitch. To verify that the regulation was dependent on riboswitch conformational changes, we used reporter constructs in which the riboswitch secondary structure was stabilized in either the ON or OFF state (see constructs in SI Appendix, Fig. S2). As expected, β-galactosidase activities were consistent with predicted structural states, in which ON and OFF constructs showed elevated and reduced activities, respectively (Fig. 1B). The introduction of a single-point mutation (G31C) in the core domain of the riboswitch completely abolished the lysine-induced regulation (Fig. 1B), in agreement with ligand binding being important for riboswitch activity (9). We next used transcriptional reporter constructs to establish whether the lysC riboswitch controls gene expression by modulating the mRNA level. Unexpectedly, we found a strong decrease in β-galactosidase activity when cells were grown in the presence of lysine (Fig. 1C, WT). Furthermore, when using constructs stabilizing the riboswitch in either the ON or OFF state or having the G31C mutation, we observed a clear correlation between reporter gene expression and riboswitch conformation, clearly indicating that the lysC riboswitch modulates the lysC mRNA level upon lysine binding. To determine whether the lysC ORF region is required to modulate the mRNA level, we designed a transcriptional fusion containing only the riboswitch domain up to the AUG start codon (SI Appendix, Fig. S1D). A very similar lysine-dependent decrease in β-galactosidase activity was obtained (SI Appendix, Fig. S1E), indicating that the lysC riboswitch is sufficient to modulate the mRNA level as a function of lysine binding.
Because mRNA stability may be decreased when translation initiation is inhibited (14, 15), we next verified whether a decrease in the mRNA level (Fig. 1C) is observed generally among riboswitches that control translation when inhibiting translation initiation. To address this question, we used the E. coli thiM thiamin pyrophosphate (TPP) riboswitch that previously was shown to act at the translational level (16, 17). Although a strong TPP-dependent decrease in β-galactosidase activity was observed for the translational fusion in the presence of 500 µg/mL TPP, no such decrease was detected when the transcriptional construct was used (SI Appendix, Fig. S3). However, when a transcriptional fusion containing a significant portion of the thiM ORF (100 codons) was used, a large reduction of β-galactosidase activity was obtained in the presence of TPP (SI Appendix, Fig. S3B). These results show that the thiM riboswitch requires a section of the ORF to decrease the mRNA level upon ligand binding, most probably because of putative regulatory elements, such as ribonuclease sites or transcription terminators, that are located in the coding region (14, 15). Thus, these results support our data indicating that the lysC riboswitch domain contains a regulatory signal that is modulated in a lysine-dependent manner to control lysC mRNA decay.
Level of lysC mRNA Decreases Rapidly in the Presence of Lysine.
To analyze further the influence of lysine on the lysC mRNA, we performed Northern blot experiments using a probe directed against the riboswitch domain of lysC (see probe details in SI Appendix, Fig. S4A). In these experiments, cells were grown in minimal medium, and total RNA was extracted before and after the addition of lysine at several time points. A very rapid decrease in the lysC mRNA level was observed upon the addition of ligand (Fig. 1D), in agreement with lysine modulating the expression of a transcriptional lacZ fusion (Fig. 1C). A similar lysine-induced rapid decrease in the mRNA level also was obtained when using a probe directed against the lysC coding region (SI Appendix, Fig. S4B), supporting the idea that lysine binding to the riboswitch modulates the lysC mRNA level. However, in contrast to lysC, Northern blot experiments showed that the addition of TPP results in a slower decrease of the thiMD mRNA level (Fig. 1E). These results show that ligand binding to the lysC and thiM riboswitches does not modulate the mRNA level to the same extent, as is consistent with the two riboswitches exhibiting mechanistic differences in regulating gene expression.
Recently, it was reported that two riboswitches control gene expression via a Rho-dependent transcription termination mechanism (18). Because no obvious Rho-independent terminator could be identified within the lysC riboswitch, we wondered if the riboswitch could rely on Rho to terminate transcription prematurely upon lysine binding. We thus performed Northern blot assays in the presence of bicyclomycin, which is known to inhibit specifically the Rho ATP-dependent RNA-binding activity (19), and assessed whether the lysC mRNA level still would be decreased as a function of lysine. No effect of bicyclomycin was observed on the lysine-dependent decrease in the lysC mRNA level (SI Appendix, Fig. S5), suggesting that Rho is not involved in the lysC mRNA regulation. In contrast, we observed that bicyclomycin had a strong effect on the level of rho mRNA (SI Appendix, Fig. S5), which is known to be autoregulated via a Rho-dependent mechanism (20). Therefore, our results show that the level of the lysC mRNA is not controlled via a Rho-dependent transcription regulation mechanism.
The rapid decrease of the lysC mRNA level in the presence of lysine suggests an active mechanism of mRNA destabilization. To determine whether lysine binding to lysC mRNA alters transcript stability, we used rifampicin to inhibit transcription initiation (21). Upon the addition of rifampicin, the level of lysC mRNA decreased drastically, with only trace amounts being detected after 1 min (Fig. 1F). This result indicates that the lysC mRNA half-life is too short to be determined experimentally, unlike most E. coli mRNAs, which exhibit half-lives between 3 and 9 min (22). However, a half-life of ∼2 min was observed for thiMD mRNA (Fig. 1G), indicating that it is more stable than lysC mRNA. Therefore, because the lysC mRNA is a highly unstable transcript, our data suggest that lysC could be a target for a ribonuclease in which lysine binding to the riboswitch could modulate the rate of mRNA cleavage and degradation.
RNase E and RNA Degradosome Are Involved in lysC mRNA Decay.
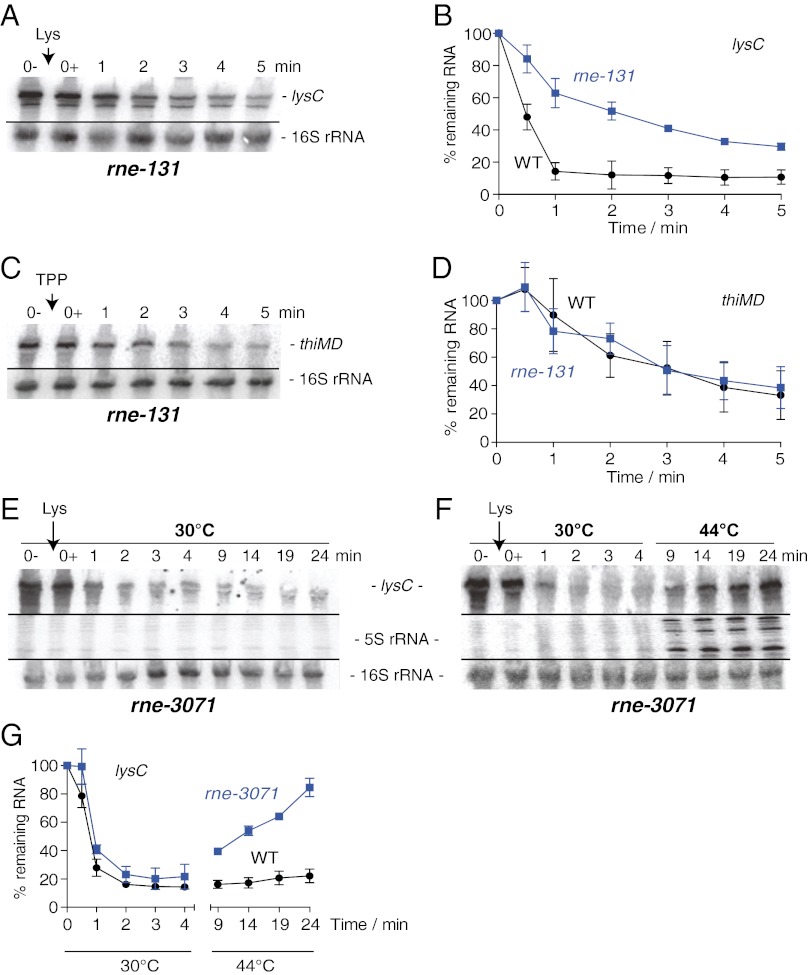
In E. coli, the initiation of mRNA decay is caused primarily by rate-limiting endonucleolytic cleavage, generally mediated by the essential enzyme RNase E (23). RNase E is part of a multiprotein complex, the RNA degradosome, which also contains an exoribonuclease (PNPase), an RNA helicase (RhlB), and an enolase (24). We speculated that RNase E and/or the RNA degradosome could be involved in the rapid degradation of lysC mRNA. To verify the implication of the degradosome, we used the bacterial strain rne-131, which retains the ribonucleolytic activity of RNase E while preventing the formation of the RNA degradosome (25). We observed that, as compared with the wild-type strain (Fig. 1D), the level of lysC mRNA in the rne-131 strain was increased significantly in the presence of lysine (Fig. 2 A and B), suggesting that the RNA degradosome is involved in the lysine-dependent degradation of lysC mRNA. In agreement with this notion, we found that the half-life of lysC mRNA was increased to ∼3 min in rne-131 cells, significantly longer than in the wild-type strain (SI Appendix, Fig. S6). In contrast, the thiMD mRNA level remained very similar in both wild-type and rne-131 strains (Fig. 2 C and D), indicating that the RNA degradosome is not implicated in the regulation of thiMD.
Fig. 2.
RNase E and the RNA degradosome are involved in the rapid decrease of lysC mRNA. (A) Northern blot analysis of the lysC mRNA level in the context of the rne-131 strain. The E. coli strain rne-131 was grown to midlog phase in M63 minimal medium with 0.2% glucose at 37 °C, and total RNA was isolated at the indicated times immediately before (0−) and after (0+) the addition of lysine (10 µg/mL). 16S rRNA was used as a loading control. (B) Quantification analysis of Northern blots shown in A and Fig. 1D. The average values of three independent experiments with SDs are shown. (C) Northern blot analysis of the thiMD mRNA level in the context of the rne-131 strain. Bacterial growth culture and RNA extractions were performed as described in A. Total RNA was isolated at the indicated times immediately before (0−) and after (0+) the addition of TPP (500 µg/mL). 16S rRNA was used as a loading control. (D) Quantification analysis of Northern blots shown in C and Fig. 1E. The average values of three independent experiments with SDs are shown. (E) Northern blot analysis of lysC mRNA in the context of the rne-3071 [RNase E(TS)] strain grown at 30 °C (permissive temperature). The E. coli strain rne-3071 was grown to midlog phase in M63 minimal medium with 0.2% glucose at 30 °C, and total RNA was isolated at the indicated times immediately before (0−) and after (0+) the addition of lysine (10 µg/mL). 16S rRNA was used as a loading control. (F) Northern blot analysis of lysC mRNA in the context of the rne-3071 strain grown at 30 °C followed by a temperature shift at 44 °C (restrictive temperature). Bacterial growth culture and RNA extractions were performed as described in E. Total RNA was isolated at the indicated times immediately before (0−) and after (0+) the addition of lysine (10 µg/mL). Cells were incubated at 30 °C from 0–4 min and at 44 °C from 4–24 min. 16S rRNA was used as a loading control. Note that unprocessed 5S rRNA intermediates are detected at 44 °C, as is consistent with the inactivation of RNase E (26). (G) Quantification analysis of Northern blots shown in F and SI Appendix, Fig. S7B. The quantification represents the extent of recovery of lysC mRNA in the context of the rne-3071 strain when grown at 44 °C. Such a recovery is not observed in wild-type cells. The average values of three independent experiments with SDs are shown.
To determine whether the endoribonucleolytic activity of RNase E is involved in the lysC mRNA decay, we used the mutant strain rne-3071, which carries a thermosensitive RNase E [RNase E(TS)] that is inactivated at 44 °C (26). As expected, when cells were grown at 30 °C, very efficient lysC mRNA degradation was observed in the presence of lysine (Fig. 2E). However, when RNase E(TS) cells were incubated with lysine and shifted from 30 °C to 44 °C, the level of lysC mRNA recovered to ∼85% after 24 min (Fig. 2F). These results show that heat inactivation of RNase E(TS) resulted in the accumulation of lysC mRNA, even in the presence of lysine. For comparison, we performed similar experiments using the wild-type strain (SI Appendix, Fig. S7) and observed a slight accumulation of the lysC mRNA when cells were grown at 44 °C (∼22% at 24 min; SI Appendix, Fig. S7B). The significantly higher recovery yield of lysC mRNA at 44 °C in the RNase E(TS) strain (∼85%) as compared with wild-type (∼22%) (Fig. 2G) strongly suggests that RNase E is involved in lysine-dependent degradation of lysC mRNA.
lysC Riboswitch Exposes RNase E Cleavage Sites When Bound to Lysine.
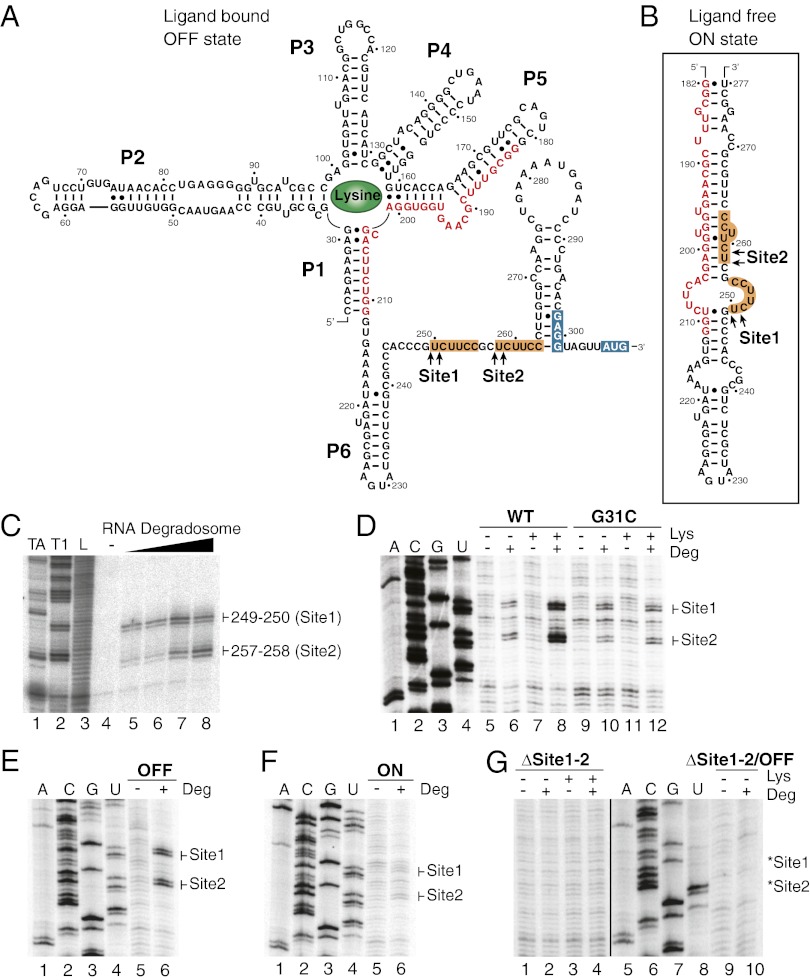
Because the lysC mRNA level depends on both the riboswitch conformational changes (Fig. 1C) and RNase E activity (Fig. 2G), we reasoned that the riboswitch could modulate the lysC mRNA level by selectively controlling the access to critical RNase E cleavage sites located in the riboswitch expression platform. We therefore set out to map cleavage sites precisely by using an in vitro cleavage assay using purified RNA degradosomes (27). As an effort to use minimal substrate sequences for RNase E activity, we first used an RNA molecule corresponding to the expression platform domain, comprising positions 212–309 (Fig. 3 A and B). When incubating the 3′-end–radiolabeled expression platform in the presence of increasing amounts of purified RNA degradosomes, two cleavage sites were observed corresponding to nucleotides 249–250 and 257–258 (Site1 and Site2; Fig. 3C). Close examination of surrounding sequences showed that both cleaved regions exhibit an identical sequence, UCUUCC, in which the two cleavage sites flank the first uracil (Fig. 3A). The cleaved sequence is similar to previously reported RNase E cleavage sites, which are often, but not always, restricted to AU-rich sequences (14).
Fig. 3.
The RNase E cleavage sites are located in the lysC riboswitch expression platform. (A) Predicted secondary structure of the lysC riboswitch in the presence of lysine (OFF state). Nucleotides shared by the antisequestering stem (ON state) and the aptamer (OFF state) are shown in red. RNase E cleavage Site1 (positions 249–250) and Site2 (positions 257–258) determined in vitro are indicated by arrows. Identical sequences surrounding both cleavage sites are in orange boxes. The RBS and AUG start codon are in blue boxes. (B) Predicted secondary structure of the antisequestering stem of the lysC riboswitch (positions 182–277). RNase E cleavage sites 1 and 2 are shown by arrows. (C) In vitro mapping of the RNA degradosome cleavage sites using 3′-end–radiolabeled riboswitch expression platform (positions 212–309). Increasing amounts of purified RNA degradosomes (0, 0.5, 0.76, 1.0, and 1.5 ng/µL) were used in the cleavage buffer (lanes 4–8). RNase TA (lane 1), RNase T1 (lane 2), and alkaline hydrolysis (L; lane 3) were used to generate molecular markers for gel migration. (D) In vitro mapping of the RNA degradosome cleavage sites using wild-type (lanes 5–8) or G31C riboswitch mutant (lanes 9–12). Riboswitch molecules were incubated in the absence and in the presence of lysine (68 µM) and/or RNA degradosomes (1 ng/µL). Reaction products were analyzed by reverse transcription using the EM1444 oligonucleotide (SI Appendix, Table S2). Lanes 1–4 represent the sequencing ladder. (E) In vitro mapping of the RNA degradosome cleavage sites using the OFF state riboswitch mutant in the absence (lane 5) and presence (lane 6) of RNA degradosomes (1 ng/µL). Cleavage sites were detected using reverse transcription as indicated in D. See SI Appendix, Fig. S2 for details about the OFF state riboswitch mutant. (F) In vitro mapping of the RNA degradosome cleavage sites using the ON state riboswitch mutant in the absence (lane 5) and presence (lane 6) of RNA degradosomes (1 ng/µL). Cleavage sites were detected using reverse transcription as indicated in D. See SI Appendix, Fig. S2 for details about the ON state riboswitch mutant. (G) In vitro mapping of the RNA degradosome cleavage sites using the ∆Site1-2 (lanes 1–4) and the ∆Site1-2/OFF state (lanes 9 and 10) riboswitch mutants. Experiments were performed in the absence and presence of lysine (68 µM) and/or RNA degradosomes (1 ng/µL). Cleavage sites were detected using reverse transcription as indicated in D. The expected positions of cleaved products are indicated on the right of the gel by asterisks. See SI Appendix, Fig. S2 for details about the ∆Site1-2 riboswitch mutant. The ∆Site1-2/OFF construct contains mutations used to obtain the ∆Site1-2 and OFF riboswitch mutants. Note that the gels shown for ∆Site1-2 and ∆Site1-2/OFF cleavage reactions were taken from different experiments.
To determine whether the complete riboswitch sequence could be a substrate for the RNA degradosome, we next used the lysine riboswitch in our degradation assays and used primer extension to detect cleavage sites. In the absence of lysine, a low cleavage activity was observed for regions Site1 and Site2 (Fig. 3D, lane 6), which correspond to determined positions using the expression platform sequence (Fig. 3C). However, a significant increase in RNase E cleavage was observed for both regions in the presence of lysine (Fig. 3D, lane 8), suggesting that lysine binding promotes RNase E cleavage. No such lysine-dependent increase was observed when using the G31C mutant (Fig. 3D, compare lanes 10 and 12), confirming that lysine binding to the riboswitch is important to facilitate RNase E cleavage. Moreover, when using constructs corresponding to OFF and ON riboswitch mutants, we observed high and low cleavage levels for OFF (Fig. 3E) and ON (Fig. 3F) state mutants, respectively, indicating that RNase E cleavage is modulated by lysC riboswitch conformational changes.
Because no clear consensus sequence has been determined for RNase E cleavage sites (24), we performed additional mutations to determine the specificity of the cleavage reaction. To avoid unforeseen production of cleavage sites caused by site-directed mutagenesis, we engineered a riboswitch mutant in which both sites were partially deleted (∆Site1-2; SI Appendix, Fig. S2). As expected, no cleavage was detected for both sites when this construct was used in our in vitro degradation assays, suggesting that the identity of Sites 1 and 2 is important for RNase E cleavage (Fig. 3G, lanes 1–4). In principle, because sequences surrounding both cleavage sites were removed, it is possible that the mutant inherently folds into the ON state, thus precluding the access to cleavage sites. To verify this possibility, we introduced the ∆Site1-2 mutation in the context of an OFF state mutant that should prevent the riboswitch from folding into the ON structure. As expected, no cleavage activity was observed when using this mutant (Fig. 3G, ∆Site1-2/OFF, lanes 9 and 10), indicating that the inability of RNase E to cleave is caused by the removal of the cleavage site. Thus, our data clearly suggest that lysine binding induces the riboswitch to adopt the OFF state secondary structure in vitro, in turn allowing RNase E to cleave the riboswitch expression platform. Interestingly, when performing 3′ RACE experiments, we detected lysC mRNA species that were truncated at Site1, consistent with the lysC riboswitch being targeted by RNase E in vivo (The cleavage site represented by an arrow, CCCGU↓CUUCC, was detected at least three times in our conditions).
Coupling Between Translation and mRNA Decay Is Not Required for lysC Riboswitch Regulation.
To evaluate the in vivo importance of RNase E cleavage sites in the riboswitch expression platform, we engineered lacZ constructs in which both RNase E cleavage sites were removed (∆Site1-2). Although the β-galactosidase activity of the translational fusion (trL) construct was reduced strongly in the presence of lysine, no such effect was observed with the transcriptional fusion (trX) construct (Fig. 4A), indicating that the mRNA level no longer is modulated as a function of lysine. Furthermore, to establish the relative importance of Site1 and Site2, we made reporter constructs in which both sites were mutated (SI Appendix, Fig. S2). When the Site1 sequence was altered, the effects observed were very similar to those observed with the ∆Site1-2 mutant (Fig. 4A), as is consistent with Site1 being essential for the lysine-induced mRNA decay. In contrast, the removal of Site2 still allowed the mRNA level to be reduced (Fig. 4A, ∆Site2, trX construct), suggesting that Site2 is less important than Site1 in modulating the mRNA level. Therefore, although our results clearly suggest that the lysine riboswitch directly controls the mRNA decay by selectively allowing RNase E cleavage at Site1, they also indicate that the lysC riboswitch does not require RNase E cleavage for regulation at the translational level.
Fig. 4.
The lysC riboswitch can be directed to control either translation initiation or mRNA decay. (A) β-Galactosidase assays using translational (trL) and transcriptional (trX) fusions of the lysC riboswitch for the wild-type, ∆Site1, ∆Site2, and ∆Site1-2 constructs. β-Galactosidase enzymatic activities were measured in the absence and presence of 10 µg/mL lysine. Values obtained for translational and transcriptional fusions were normalized to enzymatic activities obtained for WT translational and transcriptional fusions, respectively, in the absence of lysine. See SI Appendix, Fig. S2 for details about riboswitch mutant constructs. (B) β-Galactosidase assays using translational and transcriptional fusions of the lysC riboswitch for the wild-type, AAG, and GACG constructs. Values obtained for translational and transcriptional fusions were normalized to enzymatic activities obtained for WT translational and transcriptional fusions, respectively, in the absence of lysine. See SI Appendix, Fig. S2 for details about riboswitch mutant constructs.
We next evaluated whether the lysC riboswitch could control gene expression exclusively by modulating mRNA decay. To do so, we first altered the AUG initiation codon sequence by introducing a U308A mutation (AAG mutant; SI Appendix, Fig. S2). When this mutation was introduced in a translational fusion, virtually no β-galactosidase activity was detected (Fig. 4B, AAG mutant, trL construct), in agreement with the inability of ribosomes to initiate translation. However, a significant decrease in gene expression was observed in the presence of lysine in the context of a transcriptional fusion (Fig. 4B, AAG mutant, trX construct), suggesting that the riboswitch is able to modulate the lysC mRNA level even in the absence of efficient translation regulation. Similar results were obtained when a G300C mutation was introduced into the RBS to disrupt ribosome binding (Fig. 4B, GACG mutant). These results concur with the lysC riboswitch being able to modulate mRNA decay even in the absence of translational control, in stark contrast to results for the thiM riboswitch (SI Appendix, Fig. S3B).
RNase E Cleavage of the lysC Riboswitch Is Limiting for lysC mRNA Decay.
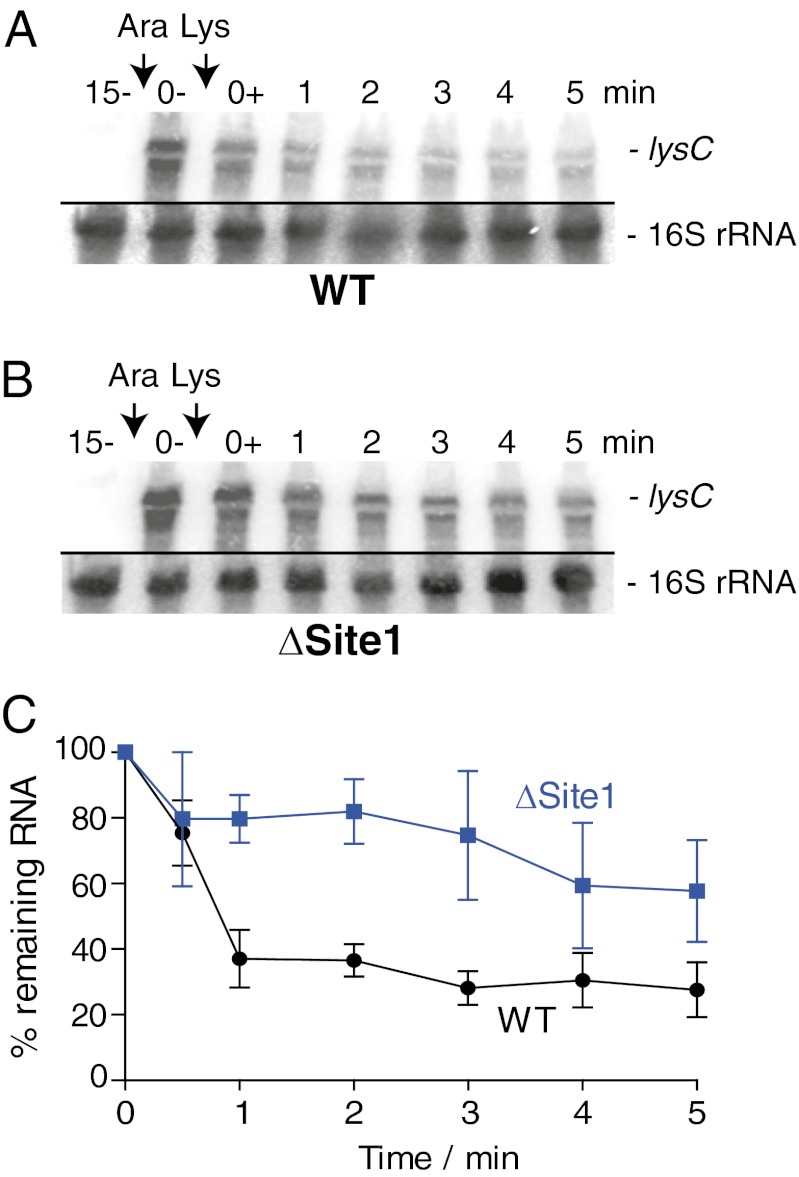
Our results suggest that RNase E cleavage at Site1 is essential to modulate the decay of the lysC-lacZ mRNA construct (Fig. 4A). To verify whether cleavage at Site1 also is important to control the decay in the context of the complete lysC mRNA, we engineered plasmids carrying an arabinose-inducible wild-type lysC or a lysC ΔSite1 gene. Both wild-type and ΔSite1 plasmids were introduced into a ΔlysC strain (see SI Appendix, SI Materials and Methods for details). According to our results, the lysC ΔSite1 mRNA exhibited a significantly (30–50%) stronger steady-state signal than the wild-type lysC mRNA after 15 min of arabinose induction (Fig. 5 A and B, compare lanes 0−). In addition, although both mRNAs responded to lysine (added at time 0+), the lysC ΔSite1 mRNA was significantly less sensitive than wild-type lysC mRNA to lysine addition. Quantification of our results indicated that lysC ΔSite1 mRNA was consistently more abundant (20–40%) than wild-type mRNA in the presence of lysine (Fig. 5C). Thus, the increased steady state and reduced turnover of the lysC ΔSite1 mRNA suggest that RNase E cleavage of the lysC riboswitch is rate limiting for lysC mRNA degradation, as is consistent with the cleavage at Site1 being important to initiate the decay of the lysC mRNA.
Fig. 5.
Deletion of Site1 affects the lysine-dependent turnover of the lysC mRNA. (A) Northern blot analysis of the lysC mRNA expressed from a pBAD-lysC plasmid in the context of a ΔlysC strain. The plasmid was transformed into a strain in which the endogenous lysC gene was deleted (EM1055∆lysC::cat; SI Appendix, SI Materials and Methods). The resulting MPC70 strain was grown at 37 °C to midlog phase in M63 medium with 0.2% glycerol. Total RNA extractions were performed 15 min (lane 15−) before lysC mRNA induction with 0.1% arabinose (Ara) and before (lane 0−) and after (0+) the addition of lysine (10 μg/mL). The probe was designed to detect the riboswitch region (positions 42–201) of the lysC mRNA. 16S rRNA was used as a loading control. (B) Northern blot analysis of the lysC ∆Site1 mRNA expressed from a pBAD-lysC-∆Site1 plasmid. Bacterial growth cultures and RNA extractions were performed as described in A. 16S rRNA was used as a loading control. (C) Quantification analysis of Northern blots shown in A and B. The average values of three independent experiments are shown with SD.
Discussion
Our work has revealed a regulation mechanism involving a lysine-sensing riboswitch that controls mRNA decay directly through the selective folding of its expression platform. The large substrate recognition spectrum of RNase E suggests that riboswitch control of mRNA decay may be a widespread mechanism for regulating gene expression. Moreover, the lysC riboswitch modulates gene expression at two different genetic levels, namely translation initiation and mRNA decay.
Riboswitch Modulation of Translation Initiation and mRNA Decay in E. coli.
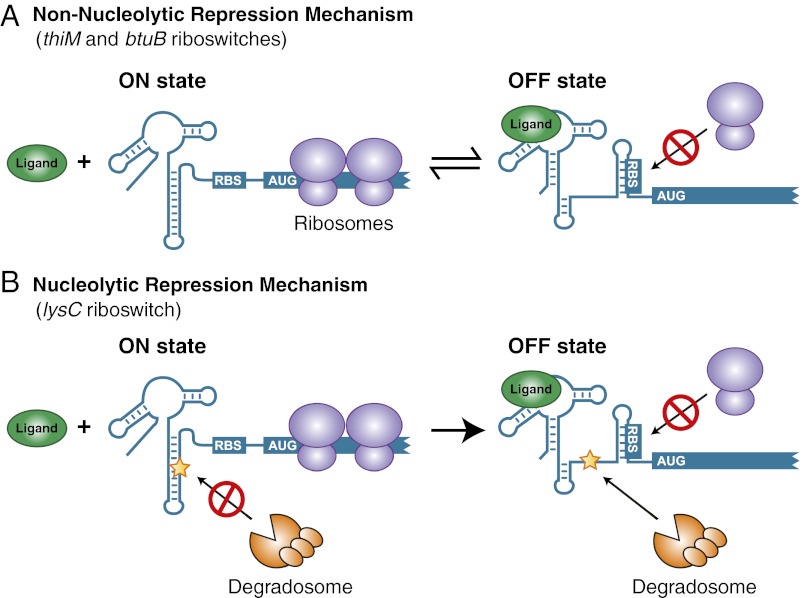
Various studies have shown that riboswitches repress the initiation of translation upon ligand binding (17, 28–31). As expected for most translationally repressed mRNAs, the absence of translating ribosomes can reveal critical cleavage site(s) located in the ORF domain that are prone to nuclease attack, ultimately leading to mRNA degradation (32). A clear example has been described for the translationally acting btuB riboswitch from E. coli, for which transcriptional fusions containing only the riboswitch domain showed no modulation as a function of ligand (33). However, the β-galactosidase activity was decreased in the presence of ligand only when using fusions containing significant portions of the coding region (34), suggesting the presence of regulatory element(s) within the btuB ORF that negatively modulate the mRNA level in the absence of translating ribosomes (34). Together with recent studies (16, 17), our data suggest that the thiM riboswitch uses a similar genetic regulating mechanism (SI Appendix, Fig. S3). There is considerable precedence for such a interplay between translation and degradation in which mRNA lifetime is influenced mostly by the time during which it can support protein synthesis (32). In such cases, mRNA decay occurs only as a consequence of translation inhibition and is involved only as a scavenging process, which is referred to as a “nonnucleolytic repression mechanism” (15). Therefore, for both btuB and thiM riboswitches, available data are consistent with the translational process being central to mRNA regulation in which ligand binding results in the nonnucleolytic repression of the regulated gene (Fig. 6A).
Fig. 6.
Regulation mechanisms of riboswitches controlling at the levels of translation initiation and mRNA decay. (A) Regulation mechanism of btuB and thiM riboswitches controlling translation initiation (nonnucleolytic repression). In the absence of ligand, the riboswitch adopts the ON state in which translation initiation is allowed and gene expression ensues. However, ligand binding to the riboswitch leads to the adoption of the OFF state in which the RBS is sequestered, resulting in the inhibition of translation initiation and ultimately to mRNA degradation. (B) Regulation mechanisms of the lysC riboswitch controlling translation initiation and mRNA decay (nucleolytic repression). In the absence of ligand, the riboswitch folds into the ON state that sequesters the RNase E cleavage site (indicated by a star) and that allows translation initiation for gene expression. Upon ligand binding, the riboswitch adopts the OFF state that exposes an RNase E cleavage site and sequesters the RBS, both ensuring definitive gene repression.
In contrast to btuB and thiM, our results show that the lysC riboswitch employs a different regulation mechanism, by directly modulating the cleavage of RNase E (Fig. 6B). Our data indicate that, in the absence of ligand, the riboswitch folds into the ON state that not only allows ribosome binding but also sequesters RNase E cleavage sites, thus ensuring efficient mRNA translation. However, in its ligand-bound form, the lysC riboswitch adopts the OFF state that concomitantly sequesters the RBS and exposes RNase E cleavage sites, thus effectively inhibiting translation and initiating mRNA decay. The location of RNase E cleavage sites in the riboswitch expression platform, and not in the ORF, strongly suggests that the lysC riboswitch directly controls the cleavage of the mRNA as a function of ligand binding. In such cases, the transcript stability and concomitant translation are reduced directly through endoribonucleolytic action, a situation referred to as “nucleolytic repression” (Fig. 6B) (15). In addition to the various characterized systems using such a regulation mechanism, it has been observed recently that small RNAs (sRNAs) also can use nucleolytic repression to control gene expression (27). However, although these observations suggest that riboswitches and sRNAs may exhibit similarities in their regulatory mechanisms, it also is likely that they possess fundamental differences such as the requirement for Hfq, which often is found in complex with RNase E. Indeed, when the lysine-induced lysC mRNA decay in the context of an hfq mutant strain was assayed, no apparent change in the decay was observed compared with a wild-type strain, suggesting that Hfq is not involved in lysC riboswitch-dependent mRNA decay (SI Appendix, Fig. S8A).
For the lysC riboswitch, it is possible that the presence of a bound 30S ribosome subunit at the RBS would prevent RNase E cleavage at Site1 by steric hindrance, thereby increasing the lysC mRNA level. Accordingly, a transcriptional lysC-lacZ fusion blocking translation elongation (AUG mutant) is expressed significantly more than a construct inhibiting translation initiation (RBS mutant) (Fig. 4B). Similarly, we found that the steady-state level of lysC mRNA is increased significantly in the presence of the antibiotic kasugamycin (SI Appendix, Fig. S8B), which specifically blocks initiating ribosomes at the RBS (35). However, even in the complete absence of ribosome binding (RBS mutant), a similar lysine-dependent reduction of the mRNA level is still observed (approximately twofold as compared with the wild-type) (Fig. 4B). These results suggest that the riboswitch does not rely mainly on ribosome binding to alter the lysC mRNA level but rather relies on conformational changes to modulate RNase E cleavage activity selectively. In addition, because the initiation of mRNA decay occurs within seconds after the addition of lysine to the medium (Fig. 1D, compare lanes 0− and 0+), it is not immediately clear why the lysC riboswitch also performs translational control. Most likely, the repression of initiating ribosomes may unmask ribonuclease cleavage sites located in the ORF, thereby allowing efficient mRNA degradation as observed in nonnucleolytic inactivation. Moreover, it is interesting to consider why the lysC riboswitch regulates mRNA decay in addition to translation initiation (Fig. 6B). One obvious explanation may be that it is important to shut down lysC expression irreversibly in the presence of lysine. For instance, we have shown previously that translation regulation is performed under thermodynamic control for the add adenine riboswitch (36, 37), allowing structural reversibility between both ligand-free and ligand-bound riboswitch conformations. However, in contrast to add, ligand binding to the riboswitch ensures rapid mRNA degradation and consequently definitive repression of gene expression, a function we consider to be dominant over translation inhibition. Moreover, our study strongly suggests that RNase E cleavage at Site1 is crucial for the rapid decay of the lysC mRNA (Figs. 4A and 5B), in agreement with the RNase E cleavage in the riboswitch expression platform being rate limiting and probably among the initial steps involved in the lysC mRNA degradation process, as previously observed for other RNase E targets (14). Additional cleavage sites are likely to be present in lysC mRNA, because the deletion of Site1 does not completely abolish the mRNA decay.
Although the action of RNase E has been shown previously to be favored by an accessible 5′-end terminus carrying a monophosphate residue (38), it also was suggested that RNase E cleavage might occur through internal entry (39). The sequence upstream of the lysC riboswitch contains 22 nucleotides and does not exhibit any strong secondary structure, suggesting that RNase E may recognize this sequence as a recruitment site. However, in addition to region containing Site1, the lysC riboswitch contains a single-stranded region (positions 276–288) in the expression platform that could constitute an RNase E entry site in the context of the OFF state (Fig. 3A). However, deletion of a major part of this region (∆279–283 mutant) did not significantly alter the lysine-dependent transcriptional repression (SI Appendix, Fig. S8C), suggesting that the 5′ region of the riboswitch may be important for RNase E recruitment.
Riboswitches and Control of mRNA Decay.
No other translationally acting riboswitch has been described that selectively modulates the access of ribonuclease cleavage sites to regulate gene expression. Still, several studies have reported that ligand binding to riboswitches may be linked to the modification of mRNA stability. For example, tRNA binding to the B. subtilis thrS leader has been shown to result in transcription antitermination (40). However, tRNA binding subsequently was found also to produce a cleavage upstream of the leader terminator, ultimately promoting mRNA stabilization and gene expression (41). Moreover, a recent study in B. subtilis has shown that the transcriptionally acting yitJ SAM riboswitch is targeted rapidly by RNase Y (42). In contrast to lysC, the antiterminated full-length mRNA is not a substrate for RNase Y in vitro or in vivo, suggesting that RNase Y is not involved in the regulation of the expressed gene but rather in riboswitch turnover (42). Another study performed in Salmonella enterica has suggested that the mgtA transcript is targeted by RNase E in the presence of high magnesium ion concentrations (43). Although additional experiments are required to establish how the transcriptionally acting mgtA riboswitch controls ribonuclease cleavage and whether putative cleavage site(s) occur in the riboswitch domain, it is possible that the mgtA riboswitch may use a regulation mechanism similar to that of lysC to control gene expression. In addition, a very recent study has found that ribB and mgtA riboswitch regulation mechanisms are more complex than previously thought, because both were found to control transcription termination using the transcription cofactor Rho (18). Interestingly, the modulation of Rho-dependent termination exhibits some mechanistic similarity to the regulation performed by the lysC riboswitch. Indeed, for both mechanisms, alternate riboswitch conformations selectively sequester or exhibit sequences that are recognized by external protein cofactors. Last, it also is important to consider the glmS ribozyme/riboswitch performing autocatalytic mRNA cleavage using glucosamine-6-phosphate (44). Even if the glmS ribozyme does not rely on a protein complex such as RNase E to perform mRNA cleavage, it uses a mechanism similar to the lysC riboswitch, because it also relies on ligand-induced endonucleolytic activity to achieve mRNA cleavage, thus providing access to the 5′-3′ exoribonuclease J1 (45).
Together with existing literature, our findings describe a probably widespread mechanism whereby riboswitches directly control the mRNA level as a function of ligand binding. From an evolutionary perspective, it is plausible that RNase E cleavage sites appeared in already existing riboswitches controlling only translation initiation, giving rise to more modern dual-acting riboswitches modulating at the levels of translation initiation and mRNA decay. In this sense, dual-acting riboswitches may represent more evolved representatives than transcriptionally or translationally regulating riboswitches and could consist of intermediate riboregulators toward a protein-based world.
Materials and Methods
DNA Oligonucleotides, Bacterial Strains, and Plasmids.
DNA oligonucleotides were purchased from Integrated DNA Technologies. All bacterial strains were derived from E. coli MG1655. Mutations performed in lysC were made essentially as described previously (46). Transcriptional and translational fusions were performed using a PCR method (47, 48) described in SI Appendix, SI Materials and Methods.
β-Galactosidase Assays.
Kinetic assays for β-galactosidase experiments were performed using a SpectraMax 250 microtiter plate reader (Molecular Devices) as described previously (49). Briefly, an overnight bacterial culture grown in M63 0.2% glycerol minimal medium was diluted 50× into 50 mL of fresh medium. The culture was incubated at 37 °C until an OD600 of 0.1 was achieved, and then arabinose (0.1% final concentration) was added to induce expression of the lacZ constructs. Lysine (10 µg/mL final concentration) was added when indicated. For the lysC-promoter fusions, cells were grown in M63 0.2% glucose minimal medium at 37 °C until an OD600 of 0.1 was achieved, and the lysine (10 µg/mL) was added when indicated. For all experiments, specific β-galactosidase activities were calculated as described previously (27). Each specific β-galactosidase activity has been relativized to the specific activity of the WT construct obtained in the absence of lysine. Reported results represent data of at least three experimental trials.
In Vitro Degradation Assays.
Reactions were performed with 5 nM of unlabeled or [3′-32P]-labeled RNA corresponding, respectively, to the complete riboswitch sequence or to the riboswitch expression platform (see SI Appendix, Table S4 for oligonucleotides used to generate DNA templates). RNA molecules were mixed with tRNA (6.6 ng/µL), denatured at 70 °C, and slowly cooled to 30 °C before the addition of the degradation buffer (13 mM Tris⋅HCl, 0.33 mM DTT, 73 mM NH4Cl, 3.33 mM MgOAc, 0.1 mM EDTA, 0.7% glycerol). Purified RNase E-FLAG degradosomes (27) or degradation buffer [5 mM Tris⋅HCl, 50% (vol/vol) glycerol, 75 mM NaCl] was added to the mixture, which was incubated at 37 °C for 30 min. Reactions using [3′-32P]-labeled RNA corresponding to the riboswitch expression platform were stopped with an equal volume of phenol. Reactions using unlabeled transcripts corresponding to the complete riboswitch sequence were extracted using phenol-chloroform-isoamyl, precipitated and annealed with 0.4 pmol of 5′-end–radiolabeled oligonucleotide (EM1444). The mixture then was heated at 90 °C for 1 min and chilled on ice. Reverse transcription was performed using 50 U of SuperScript II (Invitrogen) according to the manufacturer’s protocol. The reaction was stopped by adding the stop buffer [41 mM Tris⋅HCl (pH 8.0), 0.083% SDS, 8 mM EDTA], and the cDNA was extracted once with phenol-chloroform, pH 8.0. The aqueous phase was treated with 125 mM KOH, heated at 90 °C for 5 min for RNA degradation, and precipitated with ethanol. Reaction products were separated on 8% polyacrylamide 8 M urea gel. The gel was scanned with a Typhoon Trio (GE Healthcare) and analyzed by ImageQuant software (Molecular Dynamics). The sequencing ladder was performed with the DNA T7 template used for in vitro RNA synthesis and 5′-end–radiolabeled oligonucleotide EM1444. The generation of the RNA ladder has been described previously (27).
Northern Blot Analysis and mRNA Half-Life Determination.
Bacterial cultures were grown at 30 °C or 37 °C in M63 0.2% glucose minimal medium to midlog phase. Cells then were centrifuged and resuspended with sterile water, and total RNA was extracted immediately by the hot phenol procedure (50). Lysine (10 µg/mL), TPP (500 µg/mL), bicyclomycin (20 µg/mL), or kasugamycin (500 µg/mL) was added at the indicated time. For half-life determination, RNA transcription was blocked by the addition of 250 µg/mL rifampicin. Northern blot experiments were performed as described previously (27).
Supplementary Material
Acknowledgments
This work was funded by Canadian Institute for Health Research (CIHR) Operating Grants MOP69005 (to E.M.) and MOP82877 (to D.A.L.). E.M. is a Fonds de Recherche Santé Québec Senior Scholar, and D.A.L. is a CIHR New Investigator Scholar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 20182 (volume 109, number 50).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214024109/-/DCSupplemental.
References
- 1.Morse DE, Baker RF, Yanofsky C. Translation of the tryptophan messenger RNA of Escherichia coli. Proc Natl Acad Sci USA. 1968;60(4):1428–1435. doi: 10.1073/pnas.60.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4(2):a003566. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberg Z, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35(14):4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irnov I, Sharma CM, Vogel J, Winkler WC. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010;38(19):6637–6651. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastet L, Dubé A, Massé E, Lafontaine DA. New insights into riboswitch regulation mechanisms. Mol Microbiol. 2011;80(5):1148–1154. doi: 10.1111/j.1365-2958.2011.07654.x. [DOI] [PubMed] [Google Scholar]
- 7.Grundy FJ, Lehman SC, Henkin TM. The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci USA. 2003;100(21):12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Regulation of lysine biosynthesis and transport genes in bacteria: Yet another RNA riboswitch? Nucleic Acids Res. 2003;31(23):6748–6757. doi: 10.1093/nar/gkg900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17(21):2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blouin S, Lafontaine DA. A loop loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA. 2007;13(8):1256–1267. doi: 10.1261/rna.560307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garst AD, Héroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283(33):22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455(7217):1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao HH, Hseu TH. Analysis of the regulatory region of the lysC gene of Escherichia coli. FEMS Microbiol Lett. 1998;168(1):31–36. doi: 10.1111/j.1574-6968.1998.tb13251.x. [DOI] [PubMed] [Google Scholar]
- 14.Carpousis AJ, Luisi BF, McDowall KJ. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- 15.Dreyfus M. Killer and protective ribosomes. Prog Mol Biol Transl Sci. 2009;85:423–466. doi: 10.1016/S0079-6603(08)00811-8. [DOI] [PubMed] [Google Scholar]
- 16.Ontiveros-Palacios N, et al. Molecular basis of gene regulation by the THI-box riboswitch. Mol Microbiol. 2008;67(4):793–803. doi: 10.1111/j.1365-2958.2007.06088.x. [DOI] [PubMed] [Google Scholar]
- 17.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419(6910):952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 18.Hollands K, et al. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci USA. 2012;109(14):5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohn H, Widger W. The molecular basis for the mode of action of bicyclomycin. Curr Drug Targets Infect Disord. 2005;5(3):273–295. doi: 10.2174/1568005054880136. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto Y, Shigesada K, Hirano M, Imai M. Autogenous regulation of the gene for transcription termination factor rho in Escherichia coli: Localization and function of its attenuators. J Bacteriol. 1986;166(3):945–958. doi: 10.1128/jb.166.3.945-958.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartmann G, Honikel KO, Knüsel F, Nüesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta. 1967;145(3):843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 22.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA. 2002;99(15):9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kushner SR. mRNA decay in Escherichia coli comes of age. J Bacteriol. 2002;184(17):4658–4665, discussion 4657. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpousis AJ. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 25.Kido M, et al. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J Bacteriol. 1996;178(13):3917–3925. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apirion D, Lassar AB. A conditional lethal mutant of Escherichia coli which affects the processing of ribosomal RNA. J Biol Chem. 1978;253(5):1738–1742. [PubMed] [Google Scholar]
- 27.Prévost K, Desnoyers G, Jacques JF, Lavoie F, Massé E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25(4):385–396. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99(25):15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol. 2006;13(3):226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 30.Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci USA. 2000;97(13):7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs RT, Grundy FJ, Henkin TM. S-adenosylmethionine directly inhibits binding of 30S ribosomal subunits to the SMK box translational riboswitch RNA. Proc Natl Acad Sci USA. 2007;104(12):4876–4880. doi: 10.1073/pnas.0609956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deana A, Belasco JG. Lost in translation: The influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19(21):2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- 33.Franklund CV, Kadner RJ. Multiple transcribed elements control expression of the Escherichia coli btuB gene. J Bacteriol. 1997;179(12):4039–4042. doi: 10.1128/jb.179.12.4039-4042.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nou X, Kadner RJ. Coupled changes in translation and transcription during cobalamin-dependent regulation of btuB expression in Escherichia coli. J Bacteriol. 1998;180(24):6719–6728. doi: 10.1128/jb.180.24.6719-6728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuwirth BS, et al. Structural analysis of kasugamycin inhibition of translation. Nat Struct Mol Biol. 2006;13(10):879–886. doi: 10.1038/nsmb1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemay JF, et al. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7(1):e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem. 2007;8(8):896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 38.Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395(6703):720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 39.Baker KE, Mackie GA. Ectopic RNase E sites promote bypass of 5′-end-dependent mRNA decay in Escherichia coli. Mol Microbiol. 2003;47(1):75–88. doi: 10.1046/j.1365-2958.2003.03292.x. [DOI] [PubMed] [Google Scholar]
- 40.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74(3):475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 41.Condon C, Putzer H, Grunberg-Manago M. Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93(14):6992–6997. doi: 10.1073/pnas.93.14.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009;28(22):3523–3533. doi: 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spinelli SV, Pontel LB, García Véscovi E, Soncini FC. Regulation of magnesium homeostasis in Salmonella: Mg(2+) targets the mgtA transcript for degradation by RNase E. FEMS Microbiol Lett. 2008;280(2):226–234. doi: 10.1111/j.1574-6968.2008.01065.x. [DOI] [PubMed] [Google Scholar]
- 44.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428(6980):281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 45.Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21(24):3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97(11):5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandin P, Gottesman S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol. 2009;72(3):551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99(7):4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prévost K, et al. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol. 2007;64(5):1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 50.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256(22):11905–11910. [PubMed] [Google Scholar]