Abstract
Covalently conjugating multiple copies of the drug zanamivir (ZA; the active ingredient in Relenza) via a flexible linker to poly-l-glutamine (PGN) enhances the anti-influenza virus activity by orders of magnitude. In this study, we investigated the mechanisms of this phenomenon. Like ZA itself, the PGN-attached drug (PGN-ZA) binds specifically to viral neuraminidase and inhibits both its enzymatic activity and the release of newly synthesized virions from infected cells. Unlike monomeric ZA, however, PGN-ZA also synergistically inhibits early stages of influenza virus infection, thus contributing to the markedly increased antiviral potency. This inhibition is not caused by a direct virucidal effect, aggregation of viruses, or inhibition of viral attachment to target cells and the subsequent endocytosis; rather, it is a result of interference with intracellular trafficking of the endocytosed viruses and the subsequent virus-endosome fusion. These findings both rationalize the great anti-influenza potency of PGN-ZA and reveal that attaching ZA to a polymeric chain confers a unique mechanism of antiviral action potentially useful for minimizing drug resistance.
Keywords: polymeric inhibitor, viral trafficking, inhibition constant, mode of drug action, IC50
Influenza A viruses cause epidemics and pandemics in human populations, inflicting enormous suffering and economic losses (1). Currently, two distinct strategies—vaccines and small-molecule drugs—are used to control the spread of influenza (1). Vaccination offers limited protection and is hampered by logistical challenges, such as accurate prediction of future circulating strains and production of sufficient quantities of vaccine for large populations within a short time (2, 3). Two of the four antiviral drugs approved in the United States for the treatment and prophylaxis of influenza, amantadine and rimantadine, inhibit the viral M2 ion-channel protein (4); the other two, zanamivir (ZA) and oseltamivir, inhibit the viral neuraminidase (NA) enzymatic activity (5, 6). These drugs have limited therapeutic windows, side effects, and high costs (7–9), and most circulating viruses are already resistant to the M2 inhibitors (10, 11). Furthermore, resistance to the NA inhibitors is spreading rapidly (12, 13). Thus, the need to develop novel influenza therapeutics that can prevent viral resistance or significantly reduce its incidence is urgent (14, 15).
An alternative approach to conventional antivirals is the use of multivalent polymeric inhibitors (16). In particular, small-molecule inhibitors covalently conjugated to a biocompatible polymer have been reported to inhibit human influenza strains (17) and prevent influenza binding to red blood cells (18, 19). We have previously shown that the antiviral efficacy of ZA is dramatically enhanced when multiple copies thereof are attached via a flexible linker to the benign and biodegradable polymer poly-l-glutamine (PGN) (20): the resultant PGN-attached drug (PGN-ZA) is 1,000- to 10,000-fold more potent than monomeric ZA in plaque reduction assays and, importantly, is effective even against ZA- and oseltamivir-resistant influenza viruses.
Herein we have investigated mechanisms underlying the dramatically higher antiviral potency of this multivalent drug conjugate. We show that, like ZA itself, PGN-ZA binds to NA and inhibits its activity and the release of newly synthesized virions from the infected cells. In addition, PGN-ZA interferes with intracellular trafficking of endocytosed viruses and the subsequent virus-endosome fusion. Thus, attaching ZA to PGN gives rise to a previously undescribed mode of drug action. The synergistic inhibition of both the early and late stages of influenza virus infection accounts for the markedly enhanced antiviral potency of PGN-ZA compared with the monomeric ZA precursor.
Results
PGN-ZA Binds to, and Inhibits, Viral NA.
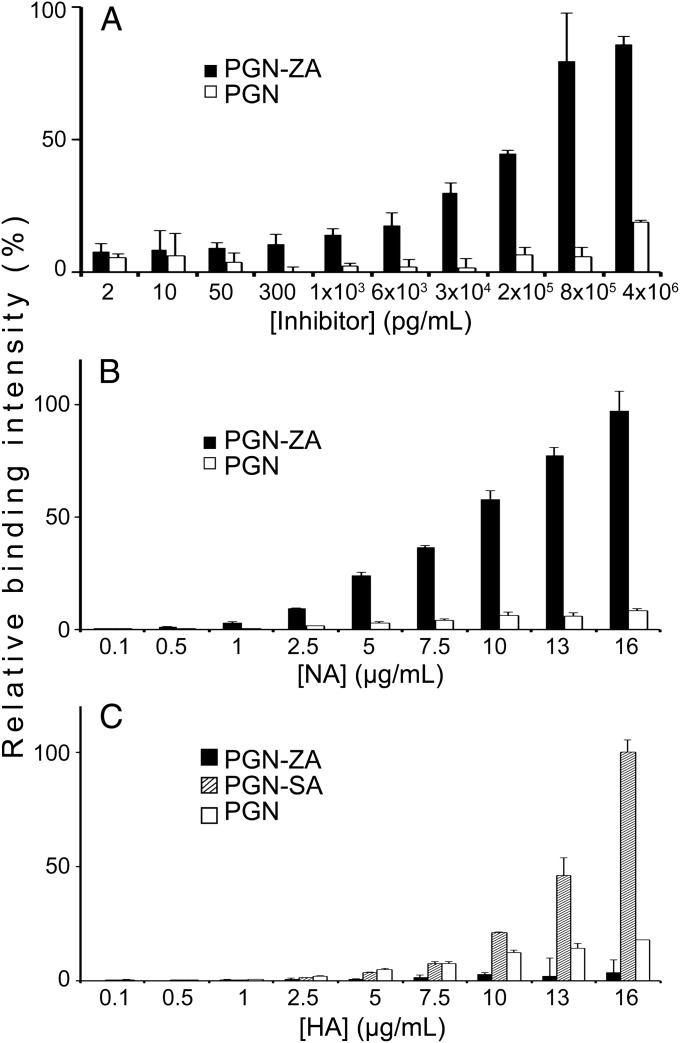
Influenza virus has two main surface glycoproteins, hemagglutinin (HA) and NA (21). Both of these glycoproteins bind to the terminal sialic acid (SA) of cell-surface glycans (22–24). Because ZA is a SA derivative and inhibits the enzymatic activity of NA, we sought to (i) determine how its conjugation to PGN via a flexible linker raised its binding and inhibitory activities and (ii) exclude nonspecific effects by the PGN chain itself. To characterize binding of PGN-ZA to whole virions, we performed whole-virus ELISA binding assays where PGN-ZA or PGN were immobilized to 96-well plates by UV cross-linking, incubated with influenza A/WSN/33 (H1N1) (WSN), and then quantified using HRP-conjugated anti-H1 antibodies. As seen in Fig. 1A, PGN-ZA exhibited a concentration-dependent binding with saturation to the viruses in the therapeutic range, whereas PGN itself showed no significant virus binding under the same conditions.
Fig. 1.
PGN-ZA binding to influenza virus. PGN-ZA and PGN were first covalently attached to 96-well plates via UV cross-linking. The relative binding of whole influenza A/WSN/33 virions (A), and the two major influenza surface proteins, NA (B) and HA (C), to the inhibitors were determined using ELISA. Bare PGN was included as a control of nonspecific binding and PGN-SA as a positive control for HA binding. Error bars represent the SEM from two independent experiments.
Next, we examined PGN-ZA’s specific site of action by measuring its binding to purified HA and NA proteins by means of ELISA. The polymer-attached drug displayed a dose-dependent binding to NA, but not to HA (Fig. 1 B and C, and Fig. S1). In contrast, multivalent polymeric SA conjugates (PGN-SA) exhibited specific binding to HA, as SA is the cognate ligand of HA (Fig. 1C). Importantly, PGN by itself bound to neither HA nor NA. PGN-ZA was 3- and 10-fold more potent than ZA modified with the linker (ZA-linker) [the antiviral activity of which is similar to that of ZA itself (20)] in inhibiting the NA activity of WSN and A/PR/8/34 (PR8) influenza viruses, respectively (Table 1). Hence, bare PGN has no appreciable interactions with HA, NA, or whole virions, and PGN-ZA specifically binds to NA and inhibits its enzymatic activity.
Table 1.
Inhibition constants (Ki) of viral neuraminidase by ZA-linker and PGN-ZA against WSN and PR8 influenza strains
|
Ki (nM, based on ZA) |
||
| Strain | ZA-linker | PGN-ZA |
| A/WSN/33 | 0.92 ± 0.19 | 0.27 ± 0.06 |
| A/PR/8/34 | 6.8 ± 1.3 | 0.72 ± 0.04 |
The Ki values, expressed in concentrations of ZA whether free or conjugated to PGN, were obtained from experiments run in triplicate.
PGN-ZA Synergistically Inhibits both Early and Late Stages of Influenza Virus Infection.
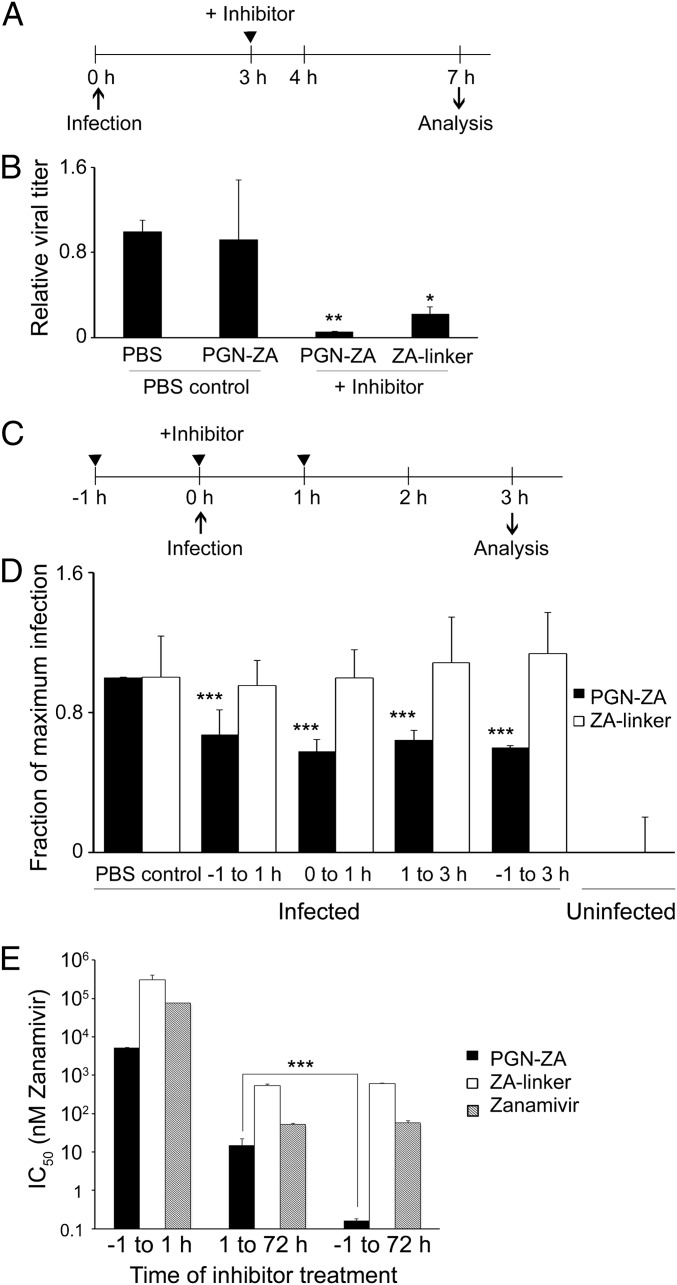
Because PGN-ZA inhibits NA, as does the monomeric ZA, we expected PGN-ZA to inhibit the release of newly synthesized virions. Madin-Darby canine kidney (MDCK) cells were infected at a multiplicity of infection (MOI) of 2. Because newly synthesized viruses were released after about 4 h, PGN-ZA and ZA-linker were added 3 h postinfection (hpi) to restrict inhibitory activity to the late phase of virus replication (Fig. 2A). At 7 hpi, the culture supernatant was harvested and the viral titer was measured by the plaque assay. Compared with the PBS control, addition of PGN-ZA and ZA-linker reduced the virus titer by some 90% and 80%, respectively (Fig. 2B). To control for the presence of leftover inhibitors in the collected supernatants (albeit at concentrations below IC50 upon serial dilution), some PBS control samples were spiked with the same concentration of PGN-ZA just before the plaque assay. No significant reduction of virus titer was detected in those cases compared with the PBS control, confirming no interference from low concentrations of inhibitors remaining in the supernatants. These results show that PGN-ZA specifically inhibits the release of newly synthesized viruses from infected cells.
Fig. 2.
PGN-ZA synergistically inhibits early and late steps of influenza virus infection. (A) Experimental design to detect the release of newly synthesized viruses from infected cells. (B) MDCK cells were inoculated with WSN virus in a synchronized infection, and unbound virus was then removed. The inhibitors were added 3 hpi. The supernatant was collected at 7 hpi., serially diluted, and titrated using the plaque assay. As a control for any remaining inhibitor in the plaque assay, some PBS control samples were spiked with the same concentration of PGN-ZA and titrated in parallel with the original PBS controls. (C) Scheme of time-of-addition experiment to assay inhibition in the early phase of virus infection in a single replication cycle assay. (D) MDCK cells were infected with WSN, and the inhibitors were added in at −1, 0, or 1 hpi. The cells were trypsinized, fixed at 3 hpi., stained for intracellular viral NP and M1, and analyzed by flow cytometry. The gating for virus-infected cells was drawn based on the expression level of a mock-infected control, and the fraction of the cells infected was normalized to the untreated, infected sample. (E) Synergistic inhibition of early and late steps of influenza infection by PGN-ZA. MDCK cells were infected with A/Turkey/MN/80, and the inhibitors were added either −1 to 1 hpi, 1 to 72 hpi, or −1 to 72 hpi. The bars represent IC50 values (i.e., the concentration of inhibitor reducing the plaque number in untreated controls by half). For the samples in the −1 to 1 h condition, the IC50s are higher than the values indicated, as we did not observe any significant plaque number reduction at this concentration. Error bars in B, D, and E represent SEM from three to five independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
To test whether PGN-ZA inhibits early events of influenza virus infection, we performed time-of-addition experiments in a single-cycle infection (Fig. 2C). To this end, MDCK cells were infected with WSN virus at a MOI of 20, and the inhibitors were added at different time points: −1, 0, or 1 h. The cell culture supernatants were harvested at 3 hpi before the completion of a single infection cycle. The cells were fixed, and expression of the viral proteins NP and M1 was quantified by flow cytometry. The fraction of infected cells decreased by 30–50% upon the addition of PGN-ZA (Fig. 2D). In contrast, for all of the conditions tested, ZA-linker did not affect the fraction of cells infected. Thus, surprisingly, PGN-ZA also specifically inhibits an early stage of influenza virus infection.
To explore the relationship between PGN-ZA’s inhibitory effects in the early and late stages of infection, we performed a time-of-addition plaque assay with the avian strain A/Turkey/MN/80 (TKY) of the virus. The inhibitors were added at different time points of the assay: (i) early (−1 to 1 hpi), (ii) late (1 to 72 hpi), or (iii) both early and late (−1 to 72 hpi). When added during the late phase of plaque assay, PGN-ZA significantly reduced the number of plaques with an IC50 of 14.8 nM (Fig. 2E). Remarkably, when the virus was exposed to PGN-ZA throughout the assay in both the early and late stages, the potency of PGN-ZA improved almost 100-fold to an IC50 of 0.16 nM. The IC50 values for the monomeric ZA and ZA-linker remained the same under both conditions, thereby revealing no additional benefit from introducing the monomeric inhibitors in the early phase of the infection. As expected, the drop in the IC50 value was also associated with a reduction in the size of the plaques.
Taken together, the foregoing results indicate that: (i) the multivalent PGN-ZA potently inhibits at least two distinct stages in influenza infection, an event early during the infection process and the release of newly synthesized virions; (ii) monomeric ZA inhibits only virus release; and (iii) PGN-ZA’s dual mechanism of action produces a synergistic inhibition of virus replication.
PGN-ZA Inhibits Influenza Infection Through Neither Direct Virucidal Effect Nor Virus Aggregation.
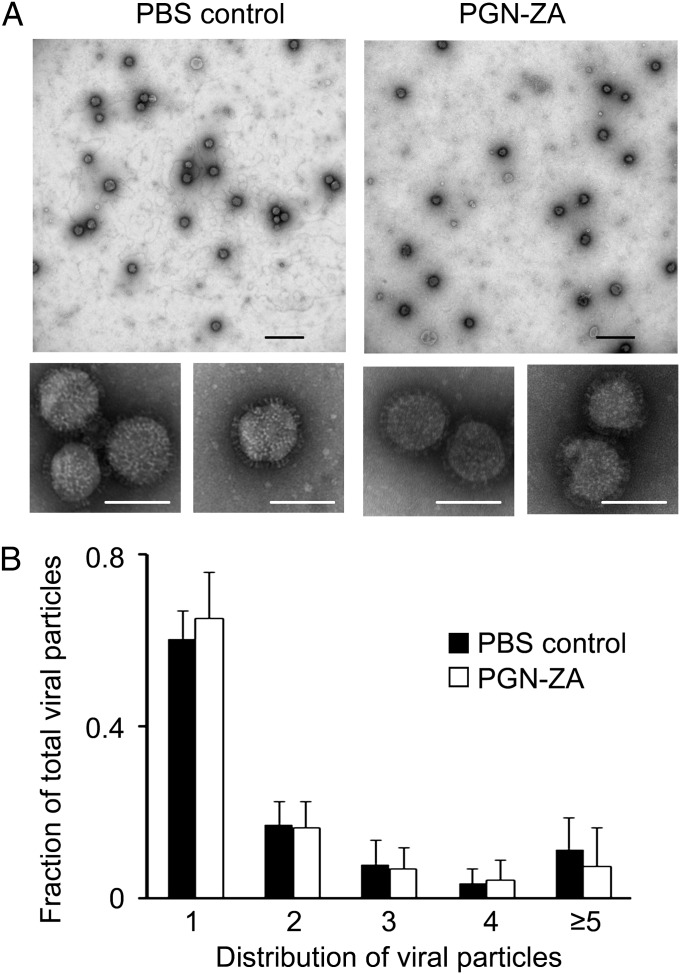
PGN-ZA may inhibit an early step of influenza virus infection through a direct virucidal effect or by aggregating viruses and thus preventing them from infecting target cells. To test these mechanisms, we used transmission electron microscopy (TEM) imaging to look for changes in viral envelope integrity and morphology upon PGN-ZA treatment. Purified WSN virus was filtered through a 0.2-μm filter and treated with either PGN-ZA or PBS for 1 h before staining with uranyl formate, followed by TEM imaging. As seen in high-magnification micrographs depicted in the lower panel of Fig. 3A, PGN-ZA did not affect the morphology or envelope integrity of viral particles. In addition, low-magnification micrographs (Fig. 3A, Upper) were taken to determine the distribution of viral particles in clusters. With over 5,000 viral particles analyzed, no significant increase was observed in virus aggregation (clustering of two or more viruses together) upon PGN-ZA treatment (Fig. 3B), consistent with the corresponding dynamic light scattering results (Fig. S2). To rule out staining artifacts, phosphotungstic acid was also used to visualize the samples, and the data obtained corroborated those of the uranyl formate-stained samples (Fig. S3). Thus, somewhat surprisingly, inhibition of the early stage of influenza infection by PGN-ZA is not through a direct virucidal effect or aggregation of viral particles.
Fig. 3.
PGN-ZA causes neither viral aggregation nor any direct virucidal activity. (A) WSN viruses were visualized by TEM in the presence or absence of 1.8 μM PGN-ZA (10-fold IC50). Representative TEM images were taken at 4,800× (Upper) and 49,000× (Lower) magnifications, respectively. (B) The number of viral particles (1, 2, 3, etc.) in each viral cluster in images of 4,800× magnification were enumerated and normalized to the total number of virions in each image. The number of viruses counted for each group is as follows: 1: n = 3,303; 2: n = 909; 3: n = 393; 4: n = 208; ≥5: n = 516. (Scale bars: black, 500 nm; white, 100 nm.)
PGN-ZA Does Not Affect Virus Attachment and Endocytosis.
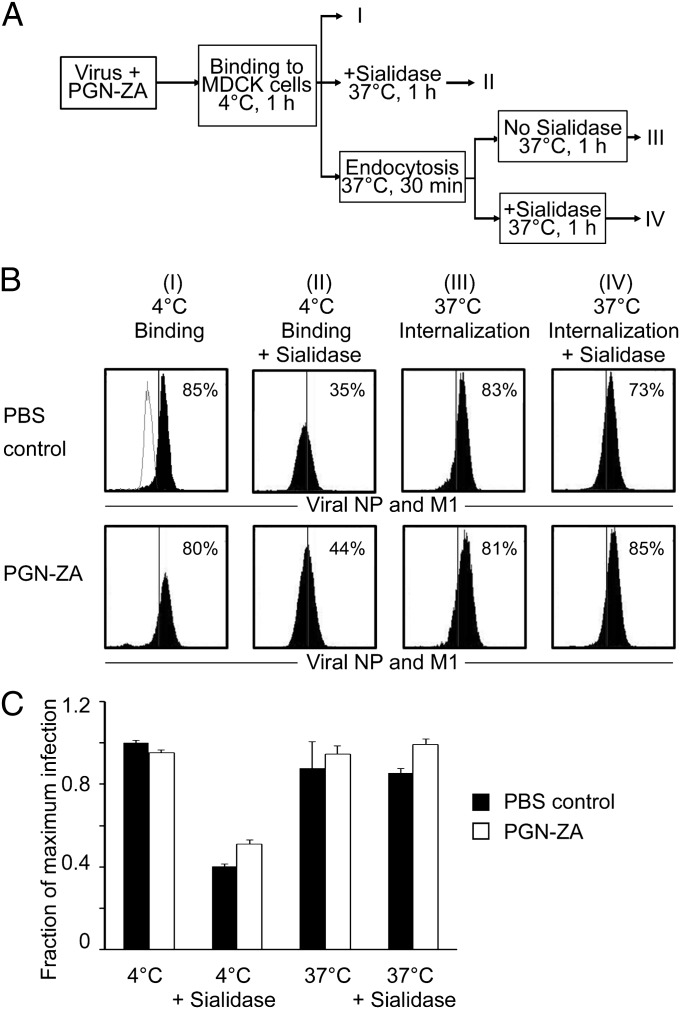
To examine whether PGN-ZA affects virus binding and endocytosis, we performed a flow-cytometry assay using labeled antibodies against viral NP and M1 (Fig. 4A). Virus attachment was measured by incubating WSN virus at a MOI of 20 with MDCK cells at 4 °C, at which temperature no endocytosis occurs (Fig. 4A, group I). To assay for endocytosis, the same cells were incubated at 37 °C for 30 min to allow the surface-bound virions to be endocytosed. Bacterial sialidase was later introduced into the system to remove surface-bound virions (Fig. 4A, groups II and IV). Because internalized viruses are protected from sialidase activity, any cell-associated virus remaining after the sialidase treatment would presumably be that which has been internalized (Fig. 4A, group IV). As shown in the left panel of Fig. 4B, PGN-ZA did not inhibit virus binding to MDCK cells. Expectedly, there was a significant drop in cell-associated viruses following sialidase treatment (Fig. 4B, group II). PGN-ZA also did not affect virus endocytosis, as evidenced by the similar levels of cell-associated viruses with or without sialidase treatment of 37 °C-incubated cells (Fig. 4A, groups III and IV). Statistical analysis of all four sets of conditions confirmed that the presence of PGN-ZA does not affect virus attachment and internalization (Fig. 4C). Consistently, hemagglutination inhibition assays also revealed that PGN-ZA did not affect virus binding to red blood cells (Fig. S4). These results indicate that PGN-ZA does not inhibit binding of influenza viruses to the target cells or endocytosis of influenza viruses into the target cells.
Fig. 4.
PGN-ZA does not inhibit endocytosis of influenza viruses. (A) An experimental scheme to study the effect of PGN-ZA on viral binding to target cells and the subsequent endocytosis. (B) MDCK cells were inoculated with virus in the absence (Upper) or presence of PGN-ZA (Lower) at 4 °C for 1 h to allow for virus binding to cells. To study the effect of PGN-ZA on binding, the samples were fixed directly after the 4 °C incubation and stained for viral proteins NP and M1 (group I). For assaying endocytosis, the cells were then incubated at 37 °C for 30 min to allow for the bound virus to be internalized. Some samples were treated with sialidase to remove surface-bound virions (groups II and IV). All samples were fixed and stained for viral NP and M1. Flow-cytometry gating was determined based on the uninfected control (shown as gray overlay in group I PBS control panel), and the percentage of cells exceeding the gate for each sample was normalized to the untreated control to determine virus binding and endocytosis. (C) The results represent the mean ± SEM of the fraction of cells infected from two to four independent experiments normalized to the mean of untreated Group I samples.
PGN-ZA Interferes with Intracellular Trafficking of the Endocytosed Viruses.
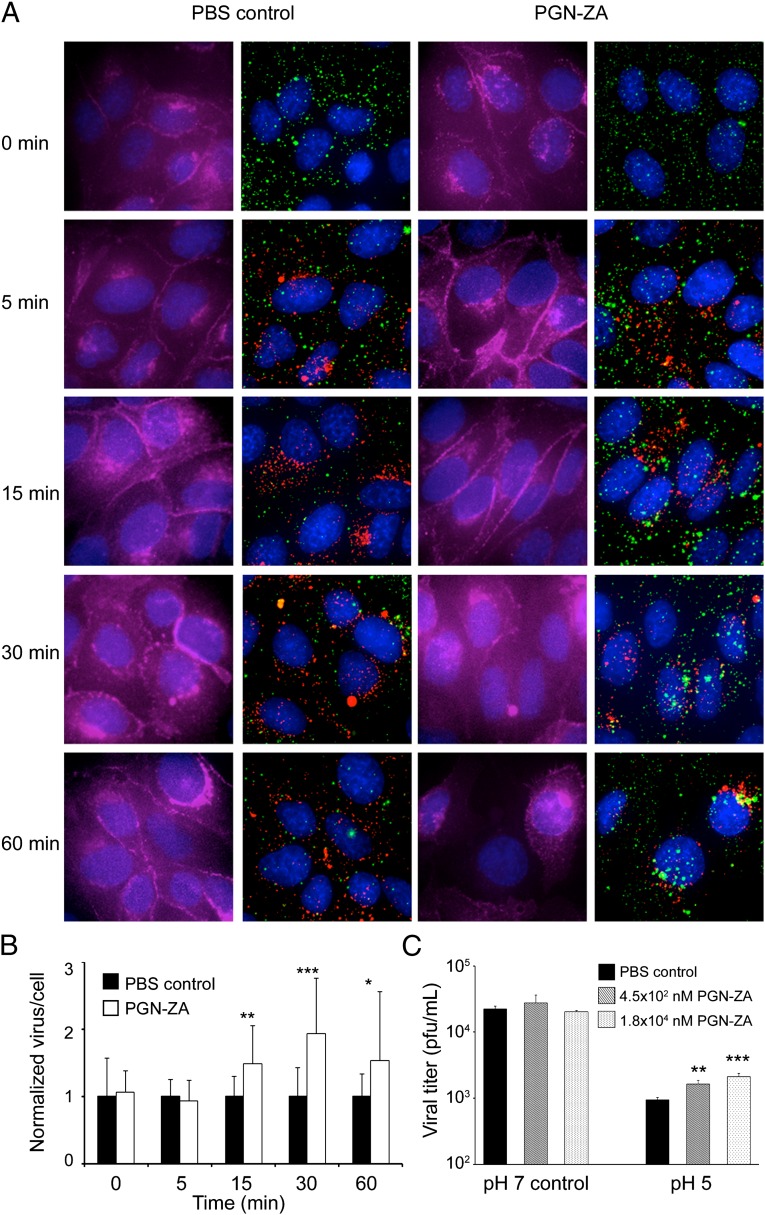
To investigate PGN-ZA’s effect on early steps of influenza virus infection, we imaged by fluorescence microscopy individual viral particles in MDCK cells fixed at different time points postinfection. The WSN viruses were labeled with amine-reactive Alexa Fluor 647 dye; the virus retained infectivity and binding to red blood cells (25, 26). To synchronize infection, the viruses were first incubated with MDCK cells on ice for 60 min in the absence or presence of PGN-ZA. The mixture was then rapidly warmed to 37 °C to initiate infection. The MDCK cells were then fixed at 0, 5, 15, 30, or 60 min postinfection and stained with E-cadherin, Lysotracker, and DAPI to visualize the cell boundary, the acidic compartments and nuclei, respectively (Fig. 5A). No apparent difference in the abundance of labeled viral particles inside the cells was observed between the samples with or without PGN-ZA at t = 0 and 5 min, concordant with the results of the flow cytometry-based binding experiments (Fig. 4B). However, from t = 15 min onwards, a significant accumulation of viral particles was observed inside the cells with the PGN-ZA-treated samples, compared with the PBS control (Fig. 5 A and B). Notably, although most of the viral particles did not colocalize with acidic compartments at t = 15 and 30 min, by t = 60 min the accumulation of viral particles in the perinuclear region was clearly evident. Similarly, we observed an accumulation of viral particles inside the cells at t = 15 min in the presence of amantadine, a known inhibitor of influenza virus acidification and fusion (Fig. S5).
Fig. 5.
PGN-ZA inhibits intracellular trafficking of endocytosed viruses. (A) Influenza virus was labeled with Alexa 647 succinimidyl ester (green) and mixed with PGN-ZA. The mixture was then added to MDCK cells at 4 °C for 1 h and washed away with PBS. Medium containing Lysotracker (red) and either PBS or PGN-ZA were added to the samples and were immediately moved to a 37 °C. Samples were taken at time points 0, 5, 15, 30, and 60 min, and cells were fixed and stained with E-cadherin (magenta) and DAPI, and imaged by fluorescent microscopy at 60× magnification. For each sample, the left panel shows E-cadherin (cell boundary) and DAPI (nuclei) staining and the right panel shows viruses, Lysotracker (acidic compartments) and DAPI staining. Representative images are shown. (B) The mean ± SEM of the number of viral particles per cell was quantified from the microscopy images and normalized to the PBS control. (C) Influenza virus was incubated at pH 5 or pH 7 (as a control) in the presence or absence of PGN-ZA for 15 min at 37 °C. The level of infectious viruses remaining after the treatment was quantified by serial dilution and the plaque assay. Two concentrations of PGN-ZA were used. The mean ± SD of infectious viruses shown here are representative of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
When an influenza virus is exposed to an acidic environment, its HA undergoes a conformational change. In the presence of a membrane, fusion occurs; in the absence of a membrane, the HA is irreversibly inactivated abolishing the viral infectivity (27). To investigate the ability of PGN-ZA to inhibit this process, the TKY virus was incubated at pH 5 in the presence or absence of PGN-ZA at 37 °C for 15 min. The level of infectious virus remaining after this acidic treatment was determined by serial titrations using the plaque assay. PGN-ZA blocked the pH 5-induced inactivation of virions two- to threefold compared with the PBS control (Fig. 5C). In contrast, the viral titer did not change following a pH 7 incubation. Taken together, these observations suggest that PGN-ZA inhibits the early steps of influenza virus infection by interfering with the intracellular trafficking of the endocytosed viruses and virus-endosome fusion.
Discussion
In this report, we have investigated the mechanism underlying the greatly enhanced antiviral potency of the polymer-attached drug zanamivir. Compared with its small-molecule parent, PGN-ZA is three-to-four orders-of-magnitude more potent in inhibiting influenza virus infection, as determined by plaque reduction assays (20). We have found that, like ZA, PGN-ZA specifically binds to NA and inhibits its enzymatic activity and the release of the newly synthesized viruses from infected cells. PGN-ZA is more potent in inhibiting virus release than ZA itself, likely because of an increased avidity to NA from polymeric binding and hence an increased inhibition of NA’s activity. Although inhibition of virus release by PGN-ZA was expected, that PGN-ZA also inhibits an early step of influenza infection is surprising. Compared with the inhibition of virus release, which reduces virus titer by over 90% (Fig. 2B), inhibition of the early step of influenza infection by PGN-ZA lowers infection by 30–50% (Fig. 2D), indicating that the former process is still the dominant mechanism of inhibition. More importantly, the two antiviral mechanisms act synergistically (Fig. 2E), accounting for the greatly enhanced (∼1,000-fold) antiviral potency of PGN-ZA over monomeric ZA.
Our observations afford further mechanistic insights. A PGN-ZA–induced viral aggregation may lead to a direct virucidal effect or interfere with infection. However, we detected no obvious violation of virus integrity or significant aggregation of viruses caused by PGN-ZA. Nor did we see any significant effect of PGN-ZA on attachment of viruses to the cell surface and their subsequent endocytosis into target cells. What we did observe was the prolonged accumulation of viruses inside the cells, including in the perinuclear region. Between the initial endocytosis and virus-endosome fusion to release the viral genomic content into the cytosol, viral particles were transported inside the cell in three separate stages (25). Stage I lasted for an average of 6 min and was characterized by movement in the cell periphery near the initial site of viral binding. In stage II, the virus-bearing endocytic compartment was transported to the perinuclear region in a few seconds. In stage III, the virus-bearing endocytic compartment moved around the perinuclear region and underwent maturation. The maturing endosomes underwent an initial acidification to pH 6, followed by a second one to pH 5. After exposure to the low pH in the endosomes, viral HA is subject to a conformation change, leading to fusion of the viral envelope with the endosomal membrane and subsequent release of viral genome into the cytosol (28).
Our finding that viral particles accumulate inside the cells in the presence of PGN-ZA suggests that PGN-ZA interferes with intracellular trafficking of the endocytosed viruses. Furthermore, the accumulation of viral particles in the perinuclear region from t = 15 min onwards suggests a block in virus-endosome fusion. How does PGN-ZA inhibit virus-endosome fusion? We showed that at t = 15 and 30 min, most accumulated viral particles did not colocalize with Lysotracker, the marker for acidic cellular compartments, suggesting a possible block of acidification of virus-bearing endosomes to pH 5. PGN-ZA also protects influenza virus from low pH-induced inactivation (i.e., HA does not undergo a conformational change in response to lowering pH in the presence of PGN-ZA). The combined effect of PGN-ZA on endosome acidification and HA conformational change underscores the inhibition of virus-endosome fusion by PGN-ZA. Intriguingly, we still observed some inhibitory effects on viral protein production when PGN-ZA was added at time 1 hpi (Fig. 2D), when most early infection processes ought to have been completed, raising the possibility that the multivalent PGN-ZA may interfere with additional intracellular processes of infection beyond the initial viral trafficking and virus-endosome fusion. Although the nature of these additional mechanisms remains to be elucidated, to our knowledge our study is unique in showing that attaching monomeric inhibitors to a polymeric backbone confers new mechanisms of action.
All existing influenza antivirals have only one mode of action, and a rapid emergence of drug-resistant variants is a major challenge in the control of influenza (13–15). The data presented here show that PGN-ZA can synergistically inhibit both viral fusion and release at subnM concentrations of ZA. This dual mechanism of inhibition is unique among known influenza antivirals and consistent with our previous observation that PGN-ZA remains effective against ZA- or oseltamivir-resistant influenza virus isolates (20). Multivalent antivirals thus offer an alternative to conventional combination therapy by not only protecting against influenza virus infection but also potentially minimizing the emergence of drug resistance.
Materials and Methods
Inhibitors.
Poly-l-glutamic acid (molecular weight of 50,000–100,000 Da) and all other chemicals, biochemicals, and solvents were from Sigma-Aldrich. 4-Guanidino-Neu5Ac2en (4-guanidino-2,4-dideoxy-2,3-dehydro-N-acetylneuraminic acid) was obtained from Bioduro. The ZA-linker derivative was synthesized as described previously (29). PGN-ZA and the bare PGN were prepared from poly-L-glutamic acid and characterized as described previously (20). Concentrations of PGN-ZA and ZA-linker used in the mechanistic studies were 100 × IC50 (18 μM and 50 μM of ZA, respectively), unless indicated otherwise.
Viruses and Cells.
Influenza virus A/WSN/33 (WSN), subtype H1N1, was kindly provided by Peter Palese (Mount Sinai School of Medicine, New York, NY). Influenza A/Turkey/MN/80 (TKY), subtype H4N2, was obtained from the Centers for Disease Control and Prevention (Atlanta, GA). Influenza A/PR/8/34 virus was purchased from Charles River Laboratories. The WSN virus was cultured in MDCK cells from the ATCC. The cells were routinely passaged in Eagle’s MEM containing 10% (vol/vol) FBS. The TKY virus was propagated in 11-d-old embryonated chicken eggs. The grown viruses were clarified by low-speed centrifugation and concentrated before sucrose gradient purification using a SW41 Ti rotor at 24,000 rpm (Beckman Coulter). Viruses were resuspended in PBS and stored at −80 °C.
The other experimental methods used in this study are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Eliza Vasile of the Massachusetts Institute of Technology Koch Institute Microscopy and Imaging Core Facility for her generous assistance; Maria Ericsson and Elizabeth Benecchi of the Harvard Medical School Electron Microscopy Facility for their help with transmission electron microscopy; Deborah Pheasant of the Massachusetts Institute of Technology Biophysical Instrumentation Facility for technical support; members of Prof. Xiaowei Zhuang’s laboratory (Harvard University) for advice on virus labeling; and members of the J.C. and A.M.K. laboratories for helpful discussions and assistance. This study was supported by the National Institutes of Health Grant U01-AI074443.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219155109/-/DCSupplemental.
References
- 1.Fiore AE, et al. Centers for Disease Control and Prevention Prevention and control of influenza with vaccines: Recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Recomm Rep. 2010;59(RR-8):1–62. [PubMed] [Google Scholar]
- 2.Webby RJ, Webster RG. Emergence of influenza A viruses. Philos Trans R Soc Lond B Biol Sci. 2001;356(1416):1817–1828. doi: 10.1098/rstb.2001.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(1):36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 4.Pinto LH, Lamb RA. Controlling influenza virus replication by inhibiting its proton channel. Mol Biosyst. 2007;3(1):18–23. doi: 10.1039/b611613m. [DOI] [PubMed] [Google Scholar]
- 5.Kim CU, et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: Design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J Am Chem Soc. 1997;119(4):681–690. doi: 10.1021/ja963036t. [DOI] [PubMed] [Google Scholar]
- 6.von Itzstein M, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363(6428):418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 7.De Clercq E. Antiviral agents active against influenza A viruses. Nat Rev Drug Discov. 2006;5(12):1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Itzstein M. The war against influenza: Discovery and development of sialidase inhibitors. Nat Rev Drug Discov. 2007;6(12):967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- 9.Lipatov AS, et al. Influenza: Emergence and control. J Virol. 2004;78(17):8951–8959. doi: 10.1128/JVI.78.17.8951-8959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bright RA, et al. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: A cause for concern. Lancet. 2005;366(9492):1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 11.Fiore AE, et al. Centers for Disease Control and Prevention Antiviral agents for the treatment and chemoprophylaxis of influenza: Recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2011;60(1):1–24. [PubMed] [Google Scholar]
- 12.Moscona A. Global transmission of oseltamivir-resistant influenza. N Engl J Med. 2009;360(10):953–956. doi: 10.1056/NEJMp0900648. [DOI] [PubMed] [Google Scholar]
- 13.WHO Global monitoring of antiviral resistance in currently circulating human influenza viruses, November 2011. Wkly Epidemiol Rec. 2011;86(45):497–508. [PubMed] [Google Scholar]
- 14.Das K, Aramini JM, Ma LC, Krug RM, Arnold E. Structures of influenza A proteins and insights into antiviral drug targets. Nat Struct Mol Biol. 2010;17(5):530–538. doi: 10.1038/nsmb.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayden F. Developing new antiviral agents for influenza treatment: What does the future hold? Clin Infect Dis. 2009;48(Suppl 1):S3–S13. doi: 10.1086/591851. [DOI] [PubMed] [Google Scholar]
- 16.Mammen M, Choi S-K, Whitesides GM. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed. 1998;37(20):2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Honda T, Yoshida S, Arai M, Masuda T, Yamashita M. Synthesis and anti-influenza evaluation of polyvalent sialidase inhibitors bearing 4-guanidino-Neu5Ac2en derivatives. Bioorg Med Chem Lett. 2002;12(15):1929–1932. doi: 10.1016/s0960-894x(02)00330-x. [DOI] [PubMed] [Google Scholar]
- 18.Mammen M, Dahmann G, Whitesides GM. Effective inhibitors of hemagglutination by influenza virus synthesized from polymers having active ester groups. Insight into mechanism of inhibition. J Med Chem. 1995;38(21):4179–4190. doi: 10.1021/jm00021a007. [DOI] [PubMed] [Google Scholar]
- 19.Lees WJ, Spaltenstein A, Kingery-Wood JE, Whitesides GM. Polyacrylamides bearing pendant alpha-sialoside groups strongly inhibit agglutination of erythrocytes by influenza A virus: Multivalency and steric stabilization of particulate biological systems. J Med Chem. 1994;37(20):3419–3433. doi: 10.1021/jm00046a027. [DOI] [PubMed] [Google Scholar]
- 20.Weight AK, et al. Attaching zanamivir to a polymer markedly enhances its activity against drug-resistant strains of influenza A virus. J Pharm Sci. 2011;100(3):831–835. doi: 10.1002/jps.22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. 2010;285(37):28403–28409. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klenk E, Faillard H, Lempfrid H. [Enzymatic effect of the influenza virus] Hoppe Seylers Z Physiol Chem. 1955;301(4–6):235–246. German. [PubMed] [Google Scholar]
- 23.Gottschalk A. Neuraminidase: The specific enzyme of influenza virus and Vibrio cholerae. Biochim Biophys Acta. 1957;23(3):645–646. doi: 10.1016/0006-3002(57)90389-x. [DOI] [PubMed] [Google Scholar]
- 24.Gottschalk A. On the mechanism underlying initiation of influenza virus infection. Ergeb Mikrobiol Immunitatsforsch Exp Ther. 1959;32:1–22. doi: 10.1007/978-3-662-42618-0_1. [DOI] [PubMed] [Google Scholar]
- 25.Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci USA. 2003;100(16):9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumari K, et al. Receptor binding specificity of recent human H3N2 influenza viruses. Virol J. 2007;4:42. doi: 10.1186/1743-422X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scholtissek C. Stability of infectious influenza A viruses at low pH and at elevated temperature. Vaccine. 1985;3(3, Suppl):215–218. doi: 10.1016/0264-410x(85)90109-4. [DOI] [PubMed] [Google Scholar]
- 28.Lamb RA, Krug RM. Orthomyxoviridae: The Viruses and Their Replication. Philadelphia: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 29.Haldar J, et al. Bifunctional polymeric inhibitors of human influenza A viruses. Pharm Res. 2010;27(2):259–263. doi: 10.1007/s11095-009-0013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.