Abstract
Protein degradation plays a critical role in cellular homeostasis, in regulating the cell cycle, and in the generation of peptides that are used in the immune response. The 20S proteasome core particle (CP), a barrel-like structure consisting of four heptameric protein rings stacked axially on top of each other, is central to this process. CP function is controlled by activator complexes that bind 75 Å away from sites catalyzing proteolysis, and biochemical data are consistent with an allosteric mechanism by which binding is communicated to distal active sites. However, little structural evidence has emerged from the high-resolution images of the CP. Using methyl TROSY NMR spectroscopy, we demonstrate that in solution, the CP interconverts between multiple conformations whose relative populations are shifted on binding of the 11S activator or mutation of residues that contact activators. These conformers differ in contiguous regions of structure that connect activator binding to the CP active sites, and changes in their populations lead to differences in substrate proteolysis patterns. Moreover, various active site modifications result in conformational changes to the activator binding site by modulating the relative populations of these same CP conformers. This distribution is also affected by the binding of a small-molecule allosteric inhibitor of proteolysis, chloroquine, suggesting an important avenue in the development of therapeutics for proteasome inhibition.
Keywords: NMR methyl TROSY, regulatory particle, allosteric network, proteasome inhibitors
The proteasome is a supramolecular machine that is central to protein degradation. The importance of the proteasome for cellular function is underscored by its role in the removal of misfolded or otherwise damaged proteins whose accumulation in the cell is cytotoxic, by its involvement in the immune response through the production of antigenic peptides, and by its regulation of the cell cycle through controlling the lifetime of key proteins that modulate gene expression (1). In addition, the proteasome serves as a critical drug target for the development of pharmaceuticals against a number of cancers (2).
The proteasome 20S core particle (CP) is a protein complex that catalyzes peptide bond hydrolysis of substrate proteins and is found in the cytoplasm and nucleus of eukaryotic cells as well as in Archaebacteria (1). The CP is constructed as a hollow cavity that sequesters catalytic sites involved in protein degradation, with access of substrates to these sites occurring through gated pores, 13 Å in diameter, that are located at opposing ends of the cavity. In the simplest type of CP, found in archaea, each heptameric ring is constructed from one of two structurally homologous proteins, α or β, arranged as α7β7β7α7, with gated pores and catalytic sites formed by α- and β-subunits, respectively (3). Eukaryotic CPs are more complex versions, whereby each ring is composed of seven distinct α- and β-subunits (4).
Important aspects of the mechanism of the peptide bond hydrolysis reaction are conserved in CPs from all species. Key chemical groups include the hydroxyl on the Thr1 side chain of the β-subunit (βThr1) that acts as a nucleophile in the peptide bond hydrolysis reaction, the βThr1 α-amino group that functions as a proton acceptor, and several ionizable groups located on the side chains of nearby residues that are important for maintaining the structural integrity and reactivity of the catalytic Thr. In addition, there are at least two identified substrate specificity pockets, S1 and S3, that bind amino acid side chains of the substrate at sites N terminal to the point of cleavage; the composition, and therefore the substrate preference, of the pockets varies among β-subunits in a eukaryotic CP, but their locations are conserved with respect to the other groups that directly participate in catalysis (2–4).
The similarities extend beyond the catalytic mechanism to include the processes by which substrates are granted access to catalytic sites. For example, the N-terminal tails of the α-subunits can adopt specific conformations that either expose or occlude the passageway through which substrates enter the barrel of the CP (5). Although the conformations of these tails vary among CPs from different species, the residues proximal to the gate that stabilize the open and closed conformations are highly conserved and gate opening can be induced on interaction of these residues with activator complexes (6–8), otherwise known as regulatory particles (RPs). Such activators include (i) the heptameric 11S RPs (otherwise known as PA28/PA26/REG) that stimulate hydrolysis of peptides and are implicated in the immune response, (ii) the 19S RP that recognizes substrates with ubiquitin tags and degrades them in an ATP-dependent manner, and (iii) Blm10/PA200, whose binding stimulates the degradation of peptides, although the function of this 250-kDa single polypeptide chain is unclear. Detailed structural studies of all three classes of RP rigorously establish that all known activators share a common binding mode, in which the C termini of subunits comprising the RP dock into a pocket that is located at the interface between α-subunits of the CP α-ring. Many of the interactions that form between the CP and RP are independent of the sequence composition of the activator C terminus, and include several hydrogen bonds between the main chains of the RP and α-subunits, as well as a hydrogen bond between the carboxylate group of the C-terminal amino acid of the RP and the side chain of a highly conserved residue in the α-subunit, αK66 (9). The conservation of the binding mechanism is further established by the fact that each 11S RP is able to activate CPs from many different species; for example, the 11S from Trypanosoma brucei activates rat and yeast proteasomes, as well as the 20S archaeal CP, even though archaea do not contain 11S RPs (6).
The degradation activities of the 20S CP are used for multiple biological functions for which the activity and sequence preference of the peptide bond hydrolysis reaction is tuned. As mentioned above, it has been clearly established that RPs both select substrates to be degraded and grant access of these substrates to catalytic sites by opening gates on binding to the CP (5, 9). However, it is also possible that RP binding could alter proteolysis by allosterically inducing a change to the catalytic sites (10). This possibility has not yet been supported by structural studies of the proteasome. No differences in the structures of the β-subunit are found in X-ray structures of complexes of the CP with the 11S (6, 11), Blm10 (12), or the C-termini of the PAN regulatory particle, in comparison to isolated CPs (13). However, biochemical studies show that proteolysis can be allosterically modulated (10, 14–16) and that modification of active sites leads to gate opening (17, 18), to changes in binding affinities for RPs, and to increases in the stability of CP–RP complexes (19, 20).
Here, we have used solution NMR spectroscopy to provide structural evidence of an allosteric pathway linking the binding sites of RP C termini with the active sites in the Thermoplasma acidophilum 20S CP through contiguous segments of structure. This pathway communicates RP binding to the active site, as we demonstrate in the case of binding of the 11S RP, or on mutation of αL81, a key residue that makes contact with the C termini of all known RPs (7). Furthermore, modifications to the CP active sites are communicated to the RP binding site and gate via this same pathway acting “in reverse.” These data demonstrate why high-resolution structural models that consist of a single conformation of the CP cannot rationalize biochemical data. Moreover, the discovery and characterization of an allosteric pathway in the proteasome offer unique opportunities for the design of novel proteasome drugs.
Results and Discussion
NMR Chemical Shift Perturbations of CP Methyl Groups upon 11S RP Binding or Mutation.
Because of the large size of the 20S CP (670 kDa) and of the CP–11S complex (1 MDa) we have used the T. acidophilum proteasome in all our studies. As described previously, this archaeal version presents a number of important advantages for NMR studies. First, because only single types of α- and β-subunits comprise the CP, rather than the seven different α,β-subunits that are found in eukaryotic CPs, there is both a significant simplification to NMR spectra and an important gain in sensitivity. Second, it derives from a thermophilic species and can withstand temperatures as high as 70 °C; this is conducive to NMR analysis because peaks in spectra, in general, become more intense with increasing temperature due to faster rates of macromolecular tumbling. We have exploited these properties in previous studies of the T. acidophilum CP, focusing almost exclusively on α-subunits of either the single ring (α7), double ring (α7α7), or full proteasome (α7β7β7α7) using Ile (δ1), Leu, and Val methyl groups as probes (13CH3-ILV) in an otherwise highly deuterated environment (16, 21, 22) and a methyl-TROSY effect that significantly improves both spectral resolution and sensitivity (23). Near-complete assignments were obtained for α-subunit methyl groups in the context of [2H,13CH3-ILV]-α–, [U-2H]-β–labeled CP samples (21), referred to in what follows as ILV-α, β CPs.
These assignments have been verified and extended here (89 of the 95 methyls assigned). These were supplemented by assignments for 72 of the 73 I,L,V methyls in the β-subunits, such that a large number of structural probes, dispersed throughout the complex, were available for analysis (Fig. 1A and SI Appendix, Fig. S1). Binding of the 11S activator (PA26) to the α, ILV-β CP ([U-2H]-α, [2H,13CH3-ILV]-β CP; KD = 10 μM) (21) produced significant chemical shift changes in 20 of the 38 peaks that were well resolved in the complex (Fig. 1 B–E). These changes do not result from direct contact with the activator because binding is to the α-ring; rather, they arise from conformational changes that are propagated from the α-rings to the β-rings.
Fig. 1.
Perturbations of the CP β-subunits on 11S binding or mutation at αL81. Selected regions of 13C-1H HMQC datasets of α, ILV-β CP without (A; 70 °C, 18.8 T) or with (B; 50 °C, 18.8 T) excess 11S RP. Assignments for some cross-peaks are indicated. (C) Chemical shift changes,  (Hz), where Δνi is the i = 1H, 13C chemical shift difference between corresponding methyls of WT CP and CP in complex with 11S (gray bars), αL81V CP (purple bars), and αL81A CP (blue bars). In addition to norm values quantifying amplitudes of peak shifts, the directions of peak movements were evaluated. Dot products of unit vectors extending from peak centers in 13C-1H HMQC spectra of WT CP to the corresponding correlations in spectra of either αL81V CP or αL81A CP have been calculated for peaks with shifts in excess of 5 Hz, as described in SI Appendix, Fig. S2 (normalized dot product). The ratios, as a function of methyl group, are plotted; values of 1 indicate that peaks shift in the same direction for αL81V CP and αL81A CP. Positions of methyl groups in spectra of the 11S-bound αL81 mutant and WT CPs are on a line, as exemplified by βV54γ1 (D) and βL87δ2 (E).
(Hz), where Δνi is the i = 1H, 13C chemical shift difference between corresponding methyls of WT CP and CP in complex with 11S (gray bars), αL81V CP (purple bars), and αL81A CP (blue bars). In addition to norm values quantifying amplitudes of peak shifts, the directions of peak movements were evaluated. Dot products of unit vectors extending from peak centers in 13C-1H HMQC spectra of WT CP to the corresponding correlations in spectra of either αL81V CP or αL81A CP have been calculated for peaks with shifts in excess of 5 Hz, as described in SI Appendix, Fig. S2 (normalized dot product). The ratios, as a function of methyl group, are plotted; values of 1 indicate that peaks shift in the same direction for αL81V CP and αL81A CP. Positions of methyl groups in spectra of the 11S-bound αL81 mutant and WT CPs are on a line, as exemplified by βV54γ1 (D) and βL87δ2 (E).
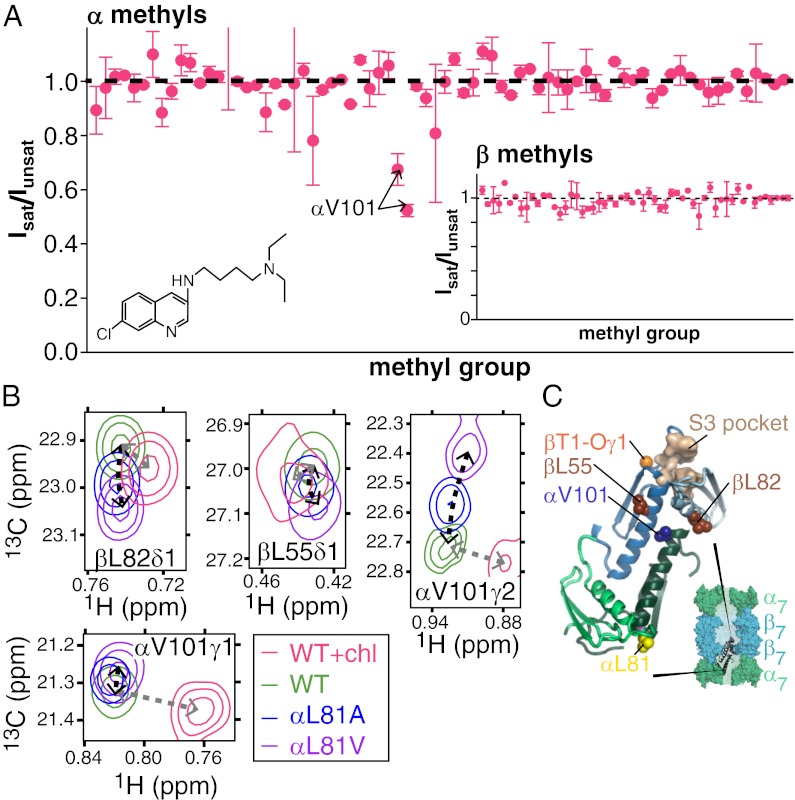
To determine whether or not the changes in the β-subunit are specific to 11S binding or occur more generally on perturbing the binding interface, CPs were made with mutations at αL81, a highly conserved residue whose side chain contributes to a hydrophobic surface patch that binds C termini of all known regulators. Studies by other groups have revealed the importance of αL81 in binding of the PAN RP that induces gate opening of the CP and promotes substrate hydrolysis (13). Remarkably, the same methyl groups shift in the same direction and with magnitudes that follow the relation 20S CP αL81A < 20S CP αL81V < 20S CP-11S to produce linear 13C chemical shift vs. 1H chemical shift profiles (Fig. 1 C–E and SI Appendix, Fig. S2). We have repeated these experiments, focusing on changes to the chemical shifts of methyl probes in the α-subunits of the CP, labeled as ILV-α, β CP. Because changes in chemical shifts may reflect the direct effect of the mutation rather than the long-range perturbations of interest, we have limited our analysis to resolved methyl groups further than 10 Å from the site of mutation (70 methyl groups; SI Appendix, Fig. S3). Again, linear chemical shift profiles are obtained, with the exception of a few residues localized to secondary structural elements containing the mutation. The simplest interpretation of these data is that the α,β-subunits of the CP interchange rapidly between two or more conformations, with exchange that is fast on the NMR chemical shift time scale so as to generate a single average NMR resonance for each probe, as has been observed in NMR studies of other protein systems (24–29). From the largest chemical shift difference (0.5 ppm for 13C, static magnetic field of 18.8 T), an exchange rate significantly in excess of 700 s−1 is estimated. To summarize, the effect of the αL81 mutations and 11S binding is to shift the position of an equilibrium involving a set of distinct conformers. This shift and the concomitant changes induced in the α,β-subunits are not specific to 11S binding but occur more generally on perturbing the RP binding pocket.
Our results show that perturbations introduced via binding of the 11S RP or mutations to αL81 extend from the α-ring to the β-subunits of the CP. To establish whether changes to the active sites could lead to global conformational changes, particularly extending to the α-rings, a number of modifications to catalytic residue βT1 have been made. These include (i) βT1A with a six-residue “GSGGGG” prosegment, βT1A-pro; (ii) βT1A with a two-residue “GM” prosegment, βT1A-GM; and (iii) βT1C (Fig. 2A). It has been shown that specific modifications of the active site can result in gate opening (17) or altered binding affinity of the CP for activators (19, 20), at least in the case of the Saccharomyces cerevisiae CP. We have measured the extent of gate opening for these three active site variants in the T. acidophilum CP by quantifying the relative intensities of correlations arising from the αM-1and αM1 residues that are part of the α-subunit N terminus that functions as a gate (Fig. 2B). As illustrated previously, separate peaks are observed in 13C,1H correlation spectra for Met gating residues corresponding to discrete conformations of N termini either inside or outside the lumen of the barrel (22) (Fig. 2C). For the WT CP, 25.0 ± 2.0% of the termini populate the “in” states, similar to βT1C, whereas for βT1A-pro and βT1A-GM, the “in” conformation decreases to 16.5 ± 1.0% and 15.0 ± 2.0%, respectively (Fig. 2D). These results suggest that gate opening does not result from the length or amino acid identity of the prosegment or from alteration of the amino acid side chain of βT1 but from modification of the α-amino group via attachment of the prosegment. Moreover, our results parallel observations from atomic force microscopy (AFM) studies of S. cerevisiae CPs, whereby binding of specific inhibitors or acetylation of the N terminus was shown to result in gate opening (17), further establishing the close similarities between CPs from different species.
Fig. 2.
Specific active site modifications lead to changes in the populations of the “in” and “out” states of the CP gate. (A) Table listing the constructs used to quantify the gate position shows both the identity of the amino acid residue at β1 that catalyzes the peptide bond hydrolysis reaction in WT CPs and the sequence of the prosegment. (B) Sequence of the gating residues that comprise the N termini of the α-subunits. The methionine residues that are probed by NMR are numbered. (C) Representative spectra show cross-peaks derived from methyl groups of Met residues that are part of the gate of the α-subunit; these data have been used to characterize the effect of active site modifications on the extent of gate opening. αM1 and αM-1 gate residues in B each give rise to multiple peaks. These include (i) “in” peaks that derive from the subset of α N termini occupying the space enclosed by the CP (i.e., the lumen of the CP), thereby occluding substrate entry, and (ii) “out” peaks that result from the gate conformation that places the α N-terminal residues near the outside surface of the CP, allowing substrates to enter (22). (D) Fraction of gating termini that are either outside (“out”) or inside (“in”) the lumen of the CP was quantitated for the CPs indicated in A from the relative intensities of 13CH3 peaks of Met residues at positions −1 and 1 of the N termini of α-subunits, using methods described previously (22).
For the 74 and 59 methyl correlations that could be quantified in spectra of the α- and β-subunits, respectively, of βT1A-pro and βT1C CPs, 33 and 42 shift in position compared with the WT CP by more than 5 Hz. Although it is not possible to determine the basis of differential gate opening of these active site perturbations (SI Appendix, Fig. S4), the effect on the CP conformation is related to that observed for the αL81 mutant and 11S-bound CPs. First, excluding residues located within 10 Å of each of the perturbations, the set of methyl group peaks that shift in position is effectively the same for all the CP samples. Moreover, the directions in which peaks in spectra of βT1A-pro and βT1C CPs are shifted with respect to the WT are identical to each other (SI Appendix, Fig. S4), and opposite to peak shifts observed for αL81 mutant and 11S-bound CPs relative to WT (Fig. 3 and SI Appendix, Fig. S5). Consequently, comparison of spectra across all CP samples, including those generated by binding the inhibitor chloroquine (see below), show that each methyl group peak position falls on a line (Fig. 3). Taken together, the αL81 mutations, 11S- or chloroquine binding, and the active site modifications shift the position of the 20S CP equilibrium between the same set of conformers. It is worth noting that for a relatively small fraction of the residues (∼20%), the 13C vs. 1H shift profiles trace curved lines as a function of perturbation (11S, chloroquine binding, or mutation). Such curvature likely reflects a process that is more complex than two-state. Alternatively, it can arise from an exchange involving a pair of states that interconvert outside the fast exchange regime, although this is unlikely because peaks do not show the characteristic broadening that would be expected if this scenario holds (30).
Fig. 3.
The CP is in rapid exchange between multiple conformers. Changes in chemical shifts of selected methyl group probes in α- and β-subunits as a function of binding 11S RP, a small molecule inhibitor (chloroquine), or mutations are color-coded as indicated. Dashed arrows are intended to guide the eye.
Contiguous Regions of Structure Give Rise to an Allosteric Network.
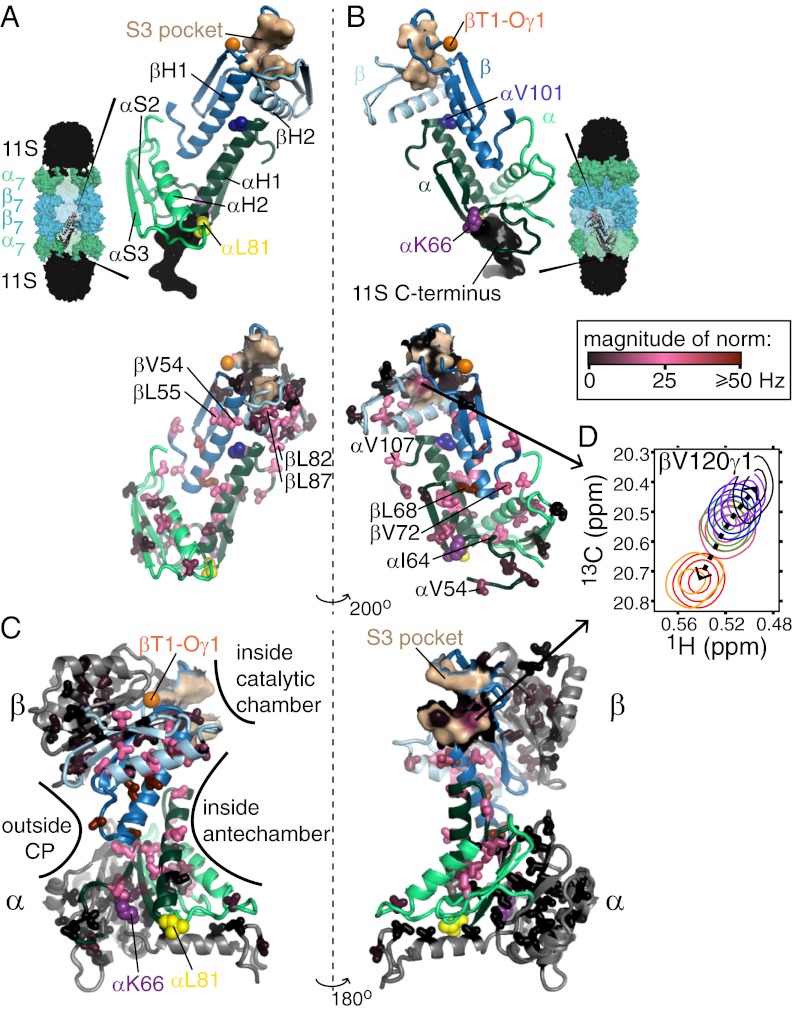
Insight into the nature of the conformational changes of the CP can be obtained by mapping the positions of the chemical shift changes onto the structure. For the β-subunit, changes in peak positions were observed for peaks derived from methyl groups localized to βH1, βH2, and the intervening loop, as well as both methyl groups that are located in the S3 substrate specificity pocket of the active site. For the α-subunit, changes in methyl shifts were localized to αH1, αH2, the intervening loop αH1-αH2, strand αS3, and some residues between αS2 and αS3. Intriguingly, all the I,L,V residues showing chemical shift changes localize to contiguous secondary structure elements, as illustrated in Fig. 4 A and B, showing portions of each of two pairs of adjacent α- and β-subunits. The fact that shift changes are observed on structural elements connecting RP binding, including key residues αK66 (6–8) and αL81 (13) that make important interactions with C termini of all regulators, extending to the S3 pockets of the active sites located ∼53 Å away, is made clear in Fig. 4C. Here full α- and β-subunits are illustrated, with all methyl group probes color-coded according to the magnitude of chemical shift changes (black, little change; red, large change) that are introduced by the αL81V (β-subunit shifts) or βT1A-pro (α-subunit shifts) perturbations. It is clear that the “red” methyl groups generate a path, whereas the “black” methyls line the outside of the CP or are localized to regions of structure that are not in direct contact with the path. Notably, βV120γ1, located in the S3 binding pocket of the active site, shows significant chemical shift changes as a function of perturbation (Fig. 4D), potentially leading to a modulation of the products of substrate cleavage. We show below that this is indeed the case.
Fig. 4.
Contiguous structural region connecting adjacent pairs of α- and β-subunits forms an allosteric network. (A and B) Significant chemical shift changes are localized only to structural elements connecting RP binding with CP active sites. Portions of the α- and β-subunits exhibiting the largest chemical shift changes (Upper), derived from pairs of adjacent α- (light and dark green) and β- (light and dark blue) subunits, are shown, with αH1 located at the interface between βH1 and βH2 from adjacent subunits and vice versa. Also shown are these same structural elements, along with the side chains of methyl-containing amino acids that were probed in the experiments (Lower), with side chains colored according to the magnitude of the chemical shift perturbations observed for αL81V CPs (β-subunit) and βT1A-pro CPs (α-subunit). Key residues/regions are highlighted, including conserved residues αK66 and αL81, which play critical roles in RP binding; the S3 substrate binding pocket that is modified on RP binding and αL81 mutations; and αV101 and βL82, which make up the chloroquine binding region. Views are from the inside (A) and outside (B) of the CP, as indicated schematically by the CP structures shown that are bound to the 11S RP (black). (C) Single β- and α-subunits (Upper and Lower, respectively) are shown in two orientations that differ by a 180° rotation, and the side chains of amino acids are colored according to the magnitude of the chemical shift perturbations, analogous to B. The structural elements colored in light/dark green and light/dark blue are the same as those depicted in A and B; however, in that case, they are shown in the context of two adjacent α-subunits and two adjacent β-subunits. (D) Chemical shift perturbations observed for βV120, which is part of the S3 substrate binding pocket, with coloring of peaks as in Fig. 3.
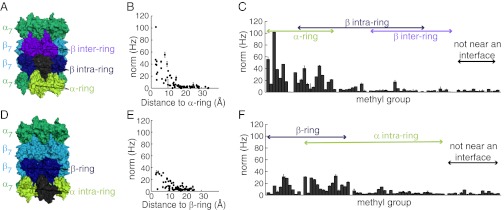
Notably, all residues that shift are located near interfaces between CP subunits. As an illustrative example, large shifts are noted for methyl groups of αH1, located between βH1 and βH2 of adjacent β-subunits, or βH1, located between αH1 and αH2 that reside on neighboring α-subunits (Fig. 4 A and B). These methyl groups do not form contacts with each other because, in general, only a few are present on each of the affected structural elements. Rather, as indicated above, they delineate the regions of contiguous structure that are affected by binding or mutations. Focusing on all methyl groups within the β-subunit, an approximate inverse correlation is observed between the shift difference of a given methyl in WT and αL81V CP samples and the distance to its nearest point of contact within the α-ring (Fig. 5B). All residues that shift are located within ∼12 Å of this interface. Each β-subunit contacts two α-subunits and two β-subunits in the same ring, as well as two β-subunits in the adjacent ring (Fig. 5A). Although most residues that undergo the strongest perturbations are located at the α/β-subunit interface, some localize to the β-intra-ring interface, as shown in Fig. 5C. This includes βV120 in the S3 substrate specificity pocket that is formed from adjacent β-subunits (Fig. 4D). Fig. 5 D–F shows the corresponding plots for residues in the α-ring, focusing on shift differences between WT and βT1A-pro. Again, the majority of the changes occur at the α/β interface, with methyl groups >12 Å from an interface showing no changes in chemical shifts. It is thus clear that the interfaces between subunits are fundamental to the conformational changes that occur in the CP. These changes are likely to be subtle. A recent X-ray structure of the T. acidophilum CP showed that binding of the C terminus of the PAN RP resulted in only a 4° rotation of the α-subunits about the CP symmetry axis (31).
Fig. 5.
Interchange between CP conformations involves subunit interfaces. (A) Each β-subunit (gray) is adjacent to a pair of α-subunits (lime) and pairs of β-intra-ring (dark blue) and β-inter-ring (purple) subunits. (B) β-Subunit methyl groups with the largest chemical shift differences (norm) between WT and αL81V CPs are within 12 Å of the α-ring. (C) β-Subunit chemical shift differences (WT vs. αL81V CPs) are largest for methyls at α/β interfaces, with some differences also at intra-β/β interfaces (methyl groups >12 Å from an interface are defined as “not near an interface”). (D–F) As above, but for the α-subunit, with norms computed as chemical shift differences of corresponding α-subunit methyl groups on WT and βT1A-pro CPs.
To explore the conformational changes in more detail, we have measured methyl 1H-13C residual dipolar couplings (RDCs) (32, 33) on samples of βT1A-pro and αL81V CPs. It is expected that the differences in structure will be largest for this pair of CPs because their corresponding methyl chemical shifts lie at opposite ends of 13C vs. 1H chemical shift profiles, as illustrated in Fig. 3. RDCs can be very sensitive reporters of structure, and in the case of an axially symmetrical complex, such as the CP, they provide a direct measure of the orientation of each methyl H3Cmethyl-C bond vector with respect to the symmetry axis. RDC data from βT1A-pro and αL81V CPs are in excellent agreement (R2 > 0.95), with a pairwise rmsd of ∼2.5 Hz, consistent with, at most, very small changes in orientations of methyl axis bond vectors between the different sets of conformers that are sampled by a CP (SI Appendix, Fig. S6). The small differences are in keeping with the observed changes in chemical shifts, which are also relatively small at 0.051 and 0.0063 ppm in 13C and 1H on average, respectively, with maximal changes of 0.54 and 0.04 ppm.
Modulation of CP Cleavage Products by Mutation or 11S Binding.
Because chemical shift changes extend into the S3 active site pocket (Fig. 4D), proteolysis assays were performed to determine whether this ultimately affects the cleavage products produced. WT, αL81A, αL81V, or 11S-bound 20S CPs were incubated in the presence of a large molar excess of α-synuclein, an intrinsically disordered substrate. Reaction times were kept sufficiently short, with less than 10% of the substrate consumed (SI Appendix, Fig. S7 A and B), so that the major degradation products reflect initial cleavage of intact α-synuclein rather than further proteolysis of products that reenter the CP after initially exiting (34, 35). Therefore, both the identities and the relative amounts of the different degradation products are characteristic of the reaction over a range of incubation times for which a small fraction of the total substrate is degraded. Reactions were quenched on addition of 0.1% TFA and 5.7 M guanidine-HCl, and peptide cleavage products were separated using reverse phase HPLC (Fig. 6 and SI Appendix, Fig. S7). Different CP variants produce HPLC profiles for which the dominant peaks exhibit the same retention times (Fig. 6A), indicating a common set of cleavage products for all CP types. However, the relative amount of each product is altered and unique to each CP variant (Fig. 6 A and B and SI Appendix, Fig. S7), suggesting that there are changes in propensities of the different substrate sites that are cleaved. To establish that the different cleavage patterns indeed reflect a change in CP preference for each cleavage site rather than differences in mean residence lifetimes for substrate in each of the CPs, we have measured the relative populations of the “in” vs. “out” states of the amino terminal gating residues, as described above and previously (22). Very similar relative populations are obtained for αL81A, αL81V, and WT CPs, consistent with very similar substrate lifetimes (SI Appendix, Fig. S7). Taken together, this indicates that the diverse patterns observed reflect differences in the proteasome hydrolysis reaction involving modifications to substrate specificity pockets, as seen by chemical shift changes for βV120γ1 (Fig. 4D), and consistent with shifts in the relative populations of exchanging conformers associated with each “type” of CP assayed. The complexity and multistep nature of substrate hydrolysis do lead, however, to a nontrivial relationship between amounts of cleavage products and relative populations of CP conformers that cannot be elucidated presently.
Fig. 6.
Relative populations of CP conformations modulate the distribution of cleavage products. (A) Reverse phase HPLC trace of cleavage products generated with different CP variants (0.3 μM WT, αL81A, αL81V, and 11S-bound) incubated with 300 μM α-synuclein for 30 min at 50 °C. (B) Comparison of reverse phrase HPLC profiles of α-synuclein degradation products shows that similar products are produced with the CP variants but that the relative amounts of each differ. Expansions of traces are shown for peaks 2, 3, and 5. mAu, milli Absorbance unit (UV-visible spectroscopy).
Intriguingly, the effect on substrate hydrolysis induced by binding of the T. brucei 11S RP to the archaeal CP parallels that which has been observed for related 11S–CP complexes, where the 11S and CP were derived from a number of eukaryotic organisms and characterized in both cognate and noncognate complexes (10, 36–39). In the case of eukaryotic CPs, the changes in substrate proteolysis on 11S binding play an important functional role because they result in enhanced production of peptides relevant to the immune response (37–39). Specifically, peptides for which the C termini are generated by proteasome cleavage events are displayed by the MHC class I complex (40). The C-terminal position has an enriched composition of hydrophobic and positively charged amino acids (41), and this coincides with the observed cleavage preferences of the β-subunits of eukaryotic CPs (40). The CP hydrolysis reaction is tuned to generate these antigenic peptides in a number of ways, one of which is through binding to the 11S RP (40, 41). Interestingly, it has been shown that 11S binding alters not only the proteolysis products produced by CPs from vertebrates that have an immune system but the products that are generated by noncognate complexes involving yeast or insect CPs for which an immune system is absent (38). It is of interest, therefore, to speculate about the possibility of a similar distribution of eukaryotic CP conformers to that observed for the archaeal T. acidophilum CP, with 11S binding shifting the relative populations of states here as well, leading to a modulation of activity and changes in cleavage products that are used in the immune response.
Chloroquine Binds Near the α,β Interface and Modulates Function Through the Allosteric Network.
The success of bortezomib, a potent proteasome inhibitor that has been used in the clinic for the treatment of multiple myeloma, has encouraged the design of further novel anticancer drugs that target the CP. Most of these pharmaceuticals, including bortezomib, are based on modification of function through binding to proteasome active sites (2, 42). We have previously described studies of the antimalaria drug chloroquine, which inhibits proteasome function in eukaryotic cell extracts and in preparations of purified 20S CP from T. acidophilum (16). Notably, biochemical data have established that chloroquine inhibits CP activity through binding to regions distal from proteolytic active sites (16), suggesting that similar compounds (albeit with higher affinity) could be used synergistically with bortezomib (42, 43).
Previous NMR characterization of chloroquine (Fig. 7A, Inset) binding to ILV-α, ILV-β CPs ([U-2H,13CH3-ILV]-α–, [U-2H,13CH3-ILV]-β–labeled CPs) showed large changes to the chemical shifts of many methyl groups, especially those located at the interface between α- and β-subunits (16). At that time, assignments for peaks in the β-subunit were not available and a binding site could not be identified. Here, we have used experiments to identify the binding site via magnetization transfer, whereby aromatic groups of chloroquine are selectively saturated, leading to a reduction in the intensity of CP peaks derived from methyl groups located near the site of binding (details are provided in SI Appendix). Both methyl groups from αV101 are affected (Fig. 7A), indicating proximity to chloroquine, although other methyls from α-subunits or from the β-ring (Fig. 7A, Inset) are not perturbed. As shown in Fig. 3, chloroquine binding shifts many peaks in the 13C,1H CP spectrum, analogous to what is observed on mutation at position αL81, modification to the active sites (βT1), or binding of 11S. Notably, however, several peaks from methyl groups that are located near αV101 shift in directions distinct from those observed previously, reflecting the formation of direct interactions with the drug or distortions in the structure of the CP near the binding site. For example, the two methyl groups of αV101 shift in a direction that is unequivocally different from what was observed in other CP samples (i.e., not on a line) and with a magnitude that is 4.8-fold larger than the maximum shift observed in all the other CP variants (Fig. 7B). The chemical shift perturbations for the βL55δ1 and βL82δ1 methyl groups, showing the next largest effects, are also distinct, but these are much smaller. βL55 and βL82 are two of the three methyl-containing residues in the β-subunit that are located closest to αV101 (methyl-methyl distances of 9.6 and 9.7 Å, respectively). The chemical shift perturbations are thus consistent with saturation transfer measurements, indicating that chloroquine binds proximally to αV101 (Fig. 7C), 23 Å removed from the nearest active site. Inhibition of CP function by chloroquine is thus allosteric, consistent with previous studies by our group showing that chloroquine inhibition is noncompetitive and that both chloroquine and MG132, a competitive inhibitor that binds to the CP active sites, can bind simultaneously (16).
Fig. 7.
Location of the chloroquine binding site. (A) Saturation transfer experiments indicate that chloroquine (structure in Inset) binds near αV101. Samples consisted of chloroquine mixed with α, ILV-β or ILV-α, β CPs with equimolar concentrations of chloroquine and CP α/β-subunits. Peak intensities of CP methyl groups were compared in pairs of spectra acquired, with the center of the saturation bandwidth (±1 ppm) applied either at 7.9 ppm (Isat), which selectively saturates chloroquine aromatic protons, or at −6.5 ppm (Iunsat), a site approximately the same distance from the methyl groups as the aromatics. To minimize artifacts resulting from a small amount of saturation in the control (unsat) experiment or magnetization transfer that is unrelated to binding (49, 50), Isat/Iunsat has been renormalized by the intensity ratio obtained by measurement in an analogously prepared sample that lacked chloroquine. Renormalized values are plotted along the y axes. (B) CP methyl group chemical shifts indicate that the chloroquine binding site is located near αV101. Notably, several peaks shift in a manner distinct from the perturbations that occur in methyl groups along the allosteric pathway (Fig. 3), reflecting chloroquine binding. (C) Locations of residues whose methyl chemical shift changes reflect binding are shown, where the CP is depicted in a manner analogous to that in Fig. 4A.
After omitting methyl groups proximal to the site of chloroquine binding, the largest shifts occur for residues located at the interface between the α- and β-subunits (16) (Fig. 8 and SI Appendix, Fig. S8), as has been observed for other CP variants as well (Fig. 5). In the majority of cases, those peaks that shift do so along the same curves defined by methyl groups from the αL81 mutant, the β-active site variant, and 11S-bound CP forms (Fig. 3 and SI Appendix, Fig. S8A). Moreover, the directions of the shifts are consistent with those observed for β-active site variants (SI Appendix, Fig. S8 B–G), which is expected because both chloroquine binding and βT1 modification result in the inactivation of the CP. Thus, chloroquine shifts the equilibrium between exchanging CP states in a direction that leads to inhibition.
Fig. 8.
Binding of the allosteric inhibitor chloroquine to the CP involves subunit interfaces. Histograms with bar heights corresponding to “norm” values but grouped according to the corresponding methyl group’s proximity to subunit interfaces [β-methyl groups (A) and α-methyl groups (B), respectively] show that the largest chemical shift changes occur for methyl groups located near the interfaces between CP subunits. A methyl group is considered to be proximal to an interface if the distance to its closest point of contact on an adjacent subunit is ≤12 Å. Each of the interfaces between subunits is defined with respect to the subunit colored in pink.
Concluding Remarks.
Although high-resolution structural analyses of the CP have indicated a single conformation (6), our results show that in solution, a distribution of structurally distinct states is present. These states link protein hydrolysis by and RP binding to the 20S CP via an allosteric pathway comprising contiguous elements of structure-connecting regions that are 80 Å apart. Manipulation of the pathway through mutation or binding of the 11S regulator shifts the relative populations of states that rapidly interconvert, leading to changes in substrate hydrolysis. The allosteric pathway described here for the T. acidophilum CP may well be operative in more complex CPs as well, because CPs from all species share many common features. For example, (i) both the mechanism of proteolysis (2) and the regulation of function through binding of RPs (9) are conserved across CPs; (ii) changes in relative amounts of substrate cleavage products that have been quantified here for T. acidophilum CP have also been observed for both cognate and noncognate 11S–CP complexes, particularly for CPs derived from eukaryotes (10, 36–39); (iii) similar changes to the in and out gate conformations have been observed in both archaeal and S. cerevisiae CPs via modifications to βT1; and (iv) both T. acidophilum and eukaryotic CPs have been shown to be inhibited by the allosteric inhibitor chloroquine. The identification of such a path offers a unique avenue for the design of new classes of inhibitors that may ultimately play an important role in the treatment of proteasome-related diseases (42, 44).
Materials and Methods
Constructs of the α- and β-subunits were expressed, purified, and assembled into CPs as described previously (45). Samples were produced with appropriate isotopic labeling for methyl TROSY NMR experiments by expressing the CP subunit of interest in M9 minimal media, 99% D2O (99 atom % D) with [12C, 2H] glucose as the sole carbon source. To introduce methyl labels, the appropriate precursors were added 1 h before induction [60 mg/L 2-keto-3,3-D2-4-13C-butyrate for Ile, 80 mg/L 2-keto-3-methyl-D3-3-D1-4-13C-butyrate for Leu/Val, and/or 100 mg/L methyl-13C L-Met for Met, as described previously (22, 46)].
All spectra were acquired on a Varian Inova spectrometer (18.8 T) equipped with a room temperature pulse field gradient triple resonance probe. Assignments of α-subunit ILV-methyl groups in the CP were published previously (21), and those for the β-subunit were determined by the process outlined in SI Appendix, Fig. S1. CPs were buffer-exchanged into 25 mM potassium phosphate (pH 6.7), 50 mM sodium chloride, 0.03% azide, 1 mM EDTA, and 99.9% D2O using a concentrator (30- or 50-kDa molecular mass cutoff). Chemical shifts are reported for methyl groups in the α-subunit from measurements on samples that had the following concentrations (expressed in terms of the α-subunit): 770 μM for αL81V CP, 790 μM for αL81A CP, 1,000 μM for WT CP, 500 μM for βT1A-pro CP, and 1,500 μM for βT1C CP. In the case of samples used to measure β-methyl group chemical shifts, the concentrations of the β-subunit were as follows: 280 μM for αL81V CP, 330 μM for αL81A CP, 550 μM for WT CP, 770 μM for βT1A-pro CP, and 750 μM for βT1C CP. For measurements of 11S binding, samples consisted of 300 μM CP and 1.2 mM 11S (conditions in which binding to the CP is saturated) in 25 mM Hepes (pH 8.5), 0.03% azide, 1 mM EDTA, and 99.9% D2O at 50 °C. The 13C,1H correlation spectra were recorded using a standard heteronuclear multiple quantum coherence (HMQC) pulse scheme that takes advantage of a methyl-TROSY effect (23). Typically, datasets for measuring Ile, Leu, and Val correlations were obtained with 24-ms and 64-ms acquisition times in t1 and t2, respectively, and with a relaxation delay of 1.5 s. For spectra in which gate populations were quantitated (Fig. 2C) by measuring volumes of α-subunit Met correlations, maximum t1 and t2 acquisition times were both set to 80 ms and a relaxation delay of 3 s was used. NMR spectra were processed using NMRPipe (47), and chemical shifts for peak centers were measured using the program SPARKY (48). In the case in which spectra of the CP-11S complex (50 °C) were compared with datasets from all other CPs (70 °C), control experiments were conducted to ensure that chemical shift perturbations were not significantly altered by differences in buffer composition or temperature. For each NMR sample, data were acquired as two to six separate successively measured HMQC spectra. Chemical shifts were extracted from each spectrum, and the values reported reflect the mean measured for each peak across all spectra, with error bars corresponding to the SD. All chemical shifts were referenced against 2,2-dimethyl-2-silapentane-5-sulphonic acid. Further experimental details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Prof. Julie Forman-Kay (Hospital for Sick Children, Toronto) for providing laboratory facilities for protein purification, and Dr. Ranjith Muhandiram (University of Toronto) for assistance in setting up some of the NMR experiments. A.M.R. was funded by a postdoctoral fellowship from the Canadian Institutes of Health Research. This work was supported by a grant from the Canadian Institutes of Health Research. L.E.K. holds a Canada Research Chair in Biochemistry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 20184 (volume 109, number 50).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213640109/-/DCSupplemental.
References
- 1.Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic mechanism and assembly of the proteasome. Chem Rev. 2009;109(4):1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 2.Borissenko L, Groll M. 20S proteasome and its inhibitors: Crystallographic knowledge for drug development. Chem Rev. 2007;107(3):687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- 3.Löwe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 4.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Y. Toward an atomic model of the 26S proteasome. Curr Opin Struct Biol. 2009;19(2):203–208. doi: 10.1016/j.sbi.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Förster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18(5):589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 7.Smith DM, et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27(5):731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadtmueller BM, et al. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J Biol Chem. 2010;285(1):13–17. doi: 10.1074/jbc.C109.070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41(1):8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, et al. Lysine 188 substitutions convert the pattern of proteasome activation by REGgamma to that of REGs alpha and beta. EMBO J. 2001;20(13):3359–3369. doi: 10.1093/emboj/20.13.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitby FG, et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408(6808):115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 12.Sadre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol Cell. 2010;37(5):728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabl J, et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30(3):360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidtke G, Emch S, Groettrup M, Holzhutter HG. Evidence for the existence of a non-catalytic modifier site of peptide hydrolysis by the 20 S proteasome. J Biol Chem. 2000;275(29):22056–22063. doi: 10.1074/jbc.M002513200. [DOI] [PubMed] [Google Scholar]
- 15.Myung J, Kim KB, Lindsten K, Dantuma NP, Crews CM. Lack of proteasome active site allostery as revealed by subunit-specific inhibitors. Mol Cell. 2001;7(2):411–420. doi: 10.1016/s1097-2765(01)00188-5. [DOI] [PubMed] [Google Scholar]
- 16.Sprangers R, et al. TROSY-based NMR evidence for a novel class of 20S proteasome inhibitors. Biochemistry. 2008;47(26):6727–6734. doi: 10.1021/bi8005913. [DOI] [PubMed] [Google Scholar]
- 17.Osmulski PA, Hochstrasser M, Gaczynska M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure. 2009;17(8):1137–1147. doi: 10.1016/j.str.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osmulski PA, Gaczynska M. Nanoenzymology of the 20S proteasome: Proteasomal actions are controlled by the allosteric transition. Biochemistry. 2002;41(22):7047–7053. doi: 10.1021/bi0159130. [DOI] [PubMed] [Google Scholar]
- 19.Kusmierczyk AR, Kunjappu MJ, Kim RY, Hochstrasser M. A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat Struct Mol Biol. 2011;18(5):622–629. doi: 10.1038/nsmb.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleijnen MF, et al. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat Struct Mol Biol. 2007;14(12):1180–1188. doi: 10.1038/nsmb1335. [DOI] [PubMed] [Google Scholar]
- 21.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445(7128):618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 22.Religa TL, Sprangers R, Kay LE. Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science. 2010;328(5974):98–102. doi: 10.1126/science.1184991. [DOI] [PubMed] [Google Scholar]
- 23.Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc. 2003;125(34):10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- 24.Li P, Martins IR, Amarasinghe GK, Rosen MK. Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nat Struct Mol Biol. 2008;15(6):613–618. doi: 10.1038/nsmb.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291(5512):2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 26.Masterson LR, et al. cAMP-dependent protein kinase A selects the excited state of the membrane substrate phospholamban. J Mol Biol. 2011;412(2):155–164. doi: 10.1016/j.jmb.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc Natl Acad Sci USA. 2011;108(15):6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzeng S-R, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488(7410):236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- 29.Zhuravleva A, Gierasch LM. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad Sci USA. 2011;108(17):6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavanagh J, Palmer AG, III, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. San Diego: Academic Press; 2007. [Google Scholar]
- 31.Yu Y, et al. Interactions of PAN’s C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 2010;29(3):692–702. doi: 10.1038/emboj.2009.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bax A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. 2003;12(1):1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprangers R, Kay LE. Probing supramolecular structure from measurement of methyl (1)H-(13)C residual dipolar couplings. J Am Chem Soc. 2007;129(42):12668–12669. doi: 10.1021/ja075846i. [DOI] [PubMed] [Google Scholar]
- 34.Masters EI, Pratt G, Förster A, Hill CP. Purification and analysis of recombinant 11S activators of the 20S proteasome: Trypanosoma brucei PA26 and human PA28 alpha, PA28 beta, and PA28 gamma. Methods Enzymol. 2005;398:306–321. doi: 10.1016/S0076-6879(05)98025-7. [DOI] [PubMed] [Google Scholar]
- 35.Cascio P, Goldberg AL. Preparation of hybrid (19S-20S-PA28) proteasome complexes and analysis of peptides generated during protein degradation. Methods Enzymol. 2005;398:336–352. doi: 10.1016/S0076-6879(05)98028-2. [DOI] [PubMed] [Google Scholar]
- 36.Harris JL, Alper PB, Li J, Rechsteiner M, Backes BJ. Substrate specificity of the human proteasome. Chem Biol. 2001;8(12):1131–1141. doi: 10.1016/s1074-5521(01)00080-1. [DOI] [PubMed] [Google Scholar]
- 37.Shimbara N, et al. Double-cleavage production of the CTL epitope by proteasomes and PA28: Role of the flanking region. Genes Cells. 1997;2(12):785–800. doi: 10.1046/j.1365-2443.1997.1610359.x. [DOI] [PubMed] [Google Scholar]
- 38.Niedermann G, et al. Potential immunocompetence of proteolytic fragments produced by proteasomes before evolution of the vertebrate immune system. J Exp Med. 1997;186(2):209–220. doi: 10.1084/jem.186.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dick TP, et al. Coordinated dual cleavages induced by the proteasome regulator PA28 lead to dominant MHC ligands. Cell. 1996;86(2):253–262. doi: 10.1016/s0092-8674(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 40.Rock KL, York IA, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 41.Rechsteiner M, Realini C, Ustrell V. The proteasome activator 11 S REG (PA28) and class I antigen presentation. Biochem J. 2000;345(Pt 1):1–15. [PMC free article] [PubMed] [Google Scholar]
- 42.Ruschak AM, Slassi M, Kay LE, Schimmer AD. Novel proteasome inhibitors to overcome bortezomib resistance. J Natl Cancer Inst. 2011;103(13):1007–1017. doi: 10.1093/jnci/djr160. [DOI] [PubMed] [Google Scholar]
- 43.Li X, et al. Effect of noncompetitive proteasome inhibition on bortezomib resistance. J Natl Cancer Inst. 2010;102(14):1069–1082. doi: 10.1093/jnci/djq198. [DOI] [PubMed] [Google Scholar]
- 44.Gräwert MA, Groll M. Exploiting nature’s rich source of proteasome inhibitors as starting points in drug development. Chem Commun (Camb) 2012;48(10):1364–1378. doi: 10.1039/c1cc15273d. [DOI] [PubMed] [Google Scholar]
- 45.Ruschak AM, Religa TL, Breuer S, Witt S, Kay LE. The proteasome antechamber maintains substrates in an unfolded state. Nature. 2010;467(7317):868–871. doi: 10.1038/nature09444. [DOI] [PubMed] [Google Scholar]
- 46.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1(2):749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 47.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 48. Goddard TD, Kneller DG (2008) SPARKY 3 (Univ of California, San Francisco)
- 49.Claasen B, Axmann M, Meinecke R, Meyer B. Direct observation of ligand binding to membrane proteins in living cells by a saturation transfer double difference (STDD) NMR spectroscopy method shows a significantly higher affinity of integrin alpha(IIb)beta3 in native platelets than in liposomes. J Am Chem Soc. 2005;127(3):916–919. doi: 10.1021/ja044434w. [DOI] [PubMed] [Google Scholar]
- 50.Bhunia A, Bhattacharjya S, Chatterjee S. Applications of saturation transfer difference NMR in biological systems. Drug Discov Today. 2012;17(9-10):505–513. doi: 10.1016/j.drudis.2011.12.016. [DOI] [PubMed] [Google Scholar]