Abstract
Identifying targets is critical for understanding the biological effects of microRNA (miRNA) expression. The challenge lies in characterizing the cohort of targets for a specific miRNA, especially when targets are being actively down-regulated in miRNA– RNA-induced silencing complex (RISC)–messengerRNA (mRNA) complexes. We have developed a robust and versatile strategy called RISCtrap to stabilize and purify targets from this transient interaction. Its utility was demonstrated by determining specific high-confidence target datasets for miR-124, miR-132, and miR-181 that contained known and previously unknown transcripts. Two previously unknown miR-132 targets identified with RISCtrap, adaptor protein CT10 regulator of kinase 1 (CRK1) and tight junction-associated protein 1 (TJAP1), were shown to be endogenously regulated by miR-132 in adult mouse forebrain. The datasets, moreover, differed in the number of targets and in the types and frequency of microRNA recognition element (MRE) motifs, thus revealing a previously underappreciated level of specificity in the target sets regulated by individual miRNAs.
Keywords: GW182, argonaute
Understanding how microRNAs (miRNAs) regulate cellular pathways is necessary for appreciating how signaling networks contribute to biology and disease. Typically, mature microRNAs target specific transcripts for down-regulation through translational silencing and messenger RNA (mRNA) destabilization. MicroRNAs are incorporated into the multimeric protein–RNA complex, RNA-induced silencing complex (RISC), for target recognition, and cellular effectors are recruited to silence and degrade the targeted transcripts (1). Each miRNA can potentially target hundreds of mRNA transcripts, thus one of the most important challenges is to identify the cohort of direct mRNA targets for a particular microRNA in a cell.
Global analyses have demonstrated that individual miRNAs can have substantial impact on regulated targets at the transcriptome level (2–7). Thus, many highly regulated miRNA target transcripts—which may be present at low abundance due to mRNA destabilization—are likely missed or underrepresented with current methods to detect miRNA–mRNA interactions, e.g., Ago2 immunoprecipitations or crosslinking immunoprecipitation (CLIP)-Seq approaches (8–10). To overcome this challenge, we have developed an approach called RISCtrap that couples stabilization of actively degraded targets with the purification of RISC–miRNA–mRNA intermediates. Multiple studies have cumulated to identify a conserved mechanism for RISC-dependent mRNA destabilization through the actions of GW182/trinucleotide repeat-containing (TNRC) 6 family members. GW182 is recruited to targeted transcripts as a core component of RISC through a direct interaction between its N-terminal domain and Argonaute (11–16). GW182 then binds to polyadenylate-binding protein 1 (PABP) and recruits cytoplasmic deadenylase complexes that destabilize the transcript through deadenylation (16–24). Moreover, several studies have provided evidence that translational repression is often coupled to and precedes mRNA destabilization (5, 20, 25, 26).
The RISCtrap approach uses a C-terminal truncation of GW182 (dnGW182) that acts as a dominant negative by retaining its ability to bind Argonaute but can no longer recruit effectors to silence and degrade the targeted mRNA (11, 13, 15, 24, 27). Transcripts are thus “trapped” in this intermediary protein–RNA complex, easily copurified with a tagged dnGW182, and targets are identified with gene-specific primers, cloning, microarray, or deep sequencing. Stabilization of actively down-regulated targets provides significant robustness to target identification; the high signal-to-noise ratio following purification of a direct target eliminated the need to account for varying input levels of transcripts or enrichment of canonical seed sites. Moreover, a single RISCtrap experiment can capture miRNA targets mediated by multiple Argonaute family members and is not limited specifically to associations with Ago2. To identify transcripts corresponding to a particular microRNA, we expressed the microRNA of interest so that the total cellular pool of microRNA–RISC is preferentially programmed toward desired complexes. We then compared the enriched transcripts to those obtained using a different microRNA to identify differentially enriched candidate targets.
To demonstrate the utility of RISCtrap, we screened for targets of three well-studied microRNAs. MicroRNA-124 expression is limited to neuronal cells where it contributes to the differentiation of neural progenitors by targeting nonneuronal transcripts (28). MicroRNA-132 was first discovered by our laboratory as an activity-dependent miRNA in excitatory neurons (29), but its roles have now expanded to activity-dependent processes in a variety of nonneuronal cell types (30–36). We also examined targets of miR-181 whose functions have been characterized primarily in nonneural cells (2, 37–40).
Combining the RISCtrap approach with RNA-seq permitted identification of unique and characteristic target datasets for all three miRNAs that differed from each other with regard to number of targets and microRNA recognition element (MRE) motifs. Moreover, we identified previously unrecognized miR-132 targets—adaptor protein CT10 regulator of kinase (CRK) and tight junction associated protein 1 (TJAP1)—and established that they were functionally regulated in adult mouse forebrain.
Results
Validation of the RISCtrap Assay.
We initially generated and tested dominant-negative versions of all three human GW182 paralogs—hTNRC6A1-1213, hTNRC6B1-1223, and hTNRC6C1-1215—and found that each acted similarly in a dose-dependent and dominant manner with no additive effects. Ultimately, hTNRC6A1-1213 (dnGW182) was chosen for subsequent use. Without its C-terminal silencing domain, hTNRC6A1-1213 was surmised to act as a dominant negative by binding Argonaute but not being able to recruit the necessary effectors for transcript silencing and destabiliziation (11, 13, 15, 24, 27).
To confirm that dnGW182 properly incorporated into RISC, we examined its ability to associate with the other RISC subunits, particularly the integral Argonaute proteins. Flag–dnGW182 was immunoprecipitated from HEK293T cells and associated proteins were assayed by Western blot (Fig. 1A). A specific interaction was detected with both endogenous Argonaute proteins 1 and 2 (Ago1 and Ago2, Fig. 1A), suggesting that the RISCtrap approach can capture targets from these different versions of RISC (41–43). Moreover, dnGW182 associated with Ago2 with similar efficiencies in the presence of different microRNAs (Fig. 1B).
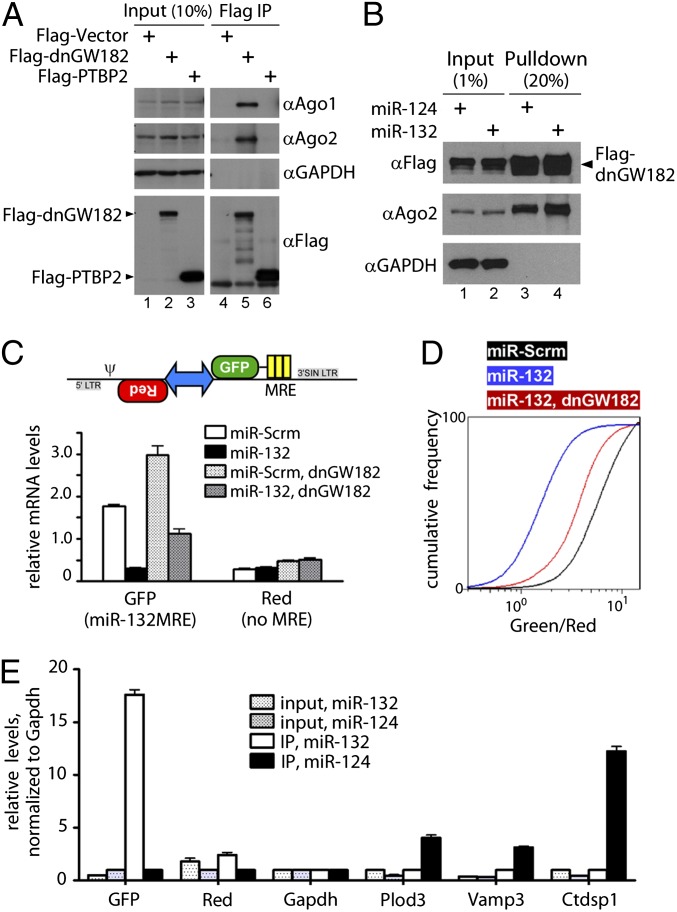
Fig. 1.
microRNA targets were stabilized by dnGW182, facilitating their enrichment by immunoprecipitation. (A and B) Flag–dnGW182 associated with endogenous Argonaute family members, and this association was similar with different microRNAs. Flag–PTBP2 is an unrelated control. (C, Upper) Schematic of the cassette encoding synthetic targets to monitor miR-132–RISC. (Lower) Relative abundance of transcripts in the presence of miR-132 and dnGW182, assessed by qPCR (2-∆Ct, normalized to Gapdh). (D) Ratiometic analysis using flow cytometry revealed that dnGW182 stabilized GFP target transcripts and increased expression in individual cells. (E) Endogenous miR-124 targets (Plod3, Vamp3, and Ctdsp1) were specifically copurified with Flag–dnGW182 and miR-124. Similarly, the mRNA for GFP–132MRE was specifically enriched with Flag–dnGW182 in the miR-132 IP condition. In contrast, transcripts for negative control DsRed (Red) were not enriched. qPCR was used to analyze relative enrichment and abundance of transcripts in both IP and input samples, normalized to Gapdh.
We next tested the ability of dnGW182 to stabilize synthetic transcripts that represented ideal positive and negative control targets for miR-132. A stable HEK293T cell line was created that constitutively expressed two synthetic transcripts cotranscribed from a bidirectional promoter (Fig. 1C). As the positive control target for miR-132, one transcript encoded green fluorescent protein (GFP) with three reiterated bulged MREs in its 3′ untranslated region (3′UTR). The other transcript encoded DsRedExpress1 but lacked any MREs in its 3′UTR. Coexpression of these two transcripts allowed the use of quantitative measurements, such as flow cytometry analysis, to obtain ratiometric values in individual cells that reflected miR-132 activity (44). Expression of miR-132 decreased levels of the GFP transcript compared with a scrambled microRNA (miR-Scrm), without changing the abundance of the red transcript (Fig. 1C). Introduction of dnGW182 stabilized the GFP transcript in the presence of miR-132. We also observed an increase in basal GFP transcript levels upon addition of dnGW182, likely due to a block of endogenous miR-132 activity in these cells (Fig. S1). To confirm that the stabilized GFP transcript correlated with increased GFP expression in individual cells, we analyzed 10,000 cells from each condition with flow cytometry and plotted the cumulative frequency of the green/red ratio (Fig. 1D). Ectopic expression of miR-132 caused a leftward shift of the plot compared with miR-Scrm, representing a decrease of green fluorescence compared with red. Introduction of dnGW182 partially rescued the ratio to control levels. Together, the data demonstrated that dnGW182 could stabilize targeted transcripts.
To determine whether dnGW182 could facilitate the enrichment of targets, we expressed Flag–dnGW182 with either miR-132 or miR-124 in the cell line that constitutively expressed GFP–132MRE and red transcripts. Following a Flag immunoprecipitation (IP), co-enriched mRNAs were examined with qPCR. We observed a robust enrichment of GFP transcript specifically in the miR-132 IP sample and not in the miR-124 IP sample (Fig. 1E). Moreover, we saw a specific enrichment of previously reported endogenous miR-124 targets in the miR-124 IP sample. The red and Gapdh transcripts, neither of which were expected to be targets of either microRNA, were not enriched in either miR-132 or miR-124 IP conditions. Most notably, the fold enrichment we observed using RISCtrap was easily discernible over background by simply comparing the IP samples without having to normalize to input levels or identify canonical MREs.
Unbiased Screen for miR-124, miR-132, and miR-181 Targets.
To develop the RISCtrap assay into an unbiased screen for targets, we coupled it with deep sequencing (Fig. 2). The screen was performed as biological triplicates in HEK293T cells. We hypothesized that this cell line expressed many neuronal microRNA targets (45, 46). Illumina TruSeq libraries were prepared from enriched mRNAs and sequenced using a HiSeq v3 platform. We obtained ∼40–50 million reads per sample; on average, 75% uniquely mapped to the RefSeq annotation and >90% of those to exonic regions (Fig. S2). Our RISCtrap datasets were composed of unique and variably sized datasets, so we developed a normalization strategy that would ensure retention of these distinct properties that likely reflected the specific targeting of each miRNA, while still allowing application of statistical methods for cross-comparison of current and future datasets (Figs. S3–S5 and SI Materials and Methods). This would allow us to compare datasets from different experiments (i.e., replicates and different miRNAs) and even incorporate future RISCtrap datasets with previously obtained datasets. Significantly enriched transcripts for each microRNA were determined with pairwise comparisons among the triplicates using ANOVA (FDR < 0.15) and combined with an experimentally determined twofold enrichment cutoff. Our high confidence lists of targets for each miRNA (Dataset S1) were finalized by requiring more than one pairwise comparison indicating enrichment for the target (e.g., for miR-124: miR-124 vs. miR-181 and miR-124 vs. miR-132). Ultimately, some of the filtered candidates may be actual targets; however, candidates that demonstrated enrichment in only a single pairwise comparison validated at ∼50%, which was not sufficiently rigorous for us to include in our high-confidence list but they are included in Dataset S2.
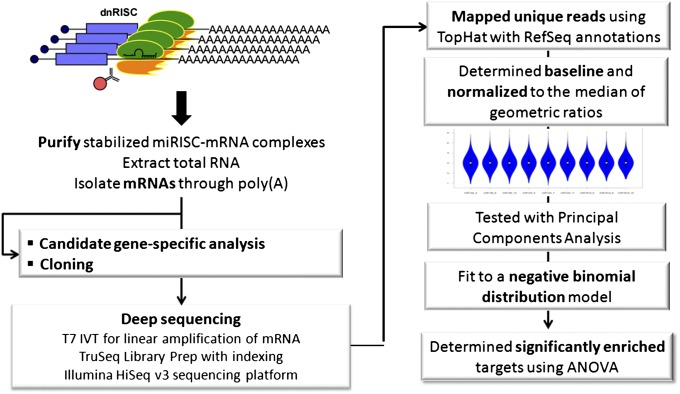
Fig. 2.
Use of RISCtrap as an unbiased screen for microRNA targets. This workflow depicts steps associated with the RISCtrap screen, including preparation of RISCtrap samples (Left) and our bioinformatic methodology to analyze differentially enriched targets (Right).
Among our high-confidence miRNA targets, we found many published and also unknown hits (Fig. 3). Overall, we obtained 281 miR-124 targets, 262 miR-181 targets, and 94 miR-132 targets. Analysis of miR-124 targets revealed substantial overlap between our dataset and previously published miR-124 datasets, despite the assays being performed in different cell types using different strategies. Comparison with related Ago2-immunoprecipitation approaches (9, 10) revealed 86 and 99 overlapping targets, respectively, and comparison with high-throughput sequencing (HITS)-CLIP biological complexity (BC) 4 (8) revealed 53. The latter is particularly notable considering that the HITS-CLIP dataset was obtained from murine brain. Moreover, many targets also overlapped with microarray (6) and proteomic datasets (2) from HeLa cells (45 and 64 targets, respectively). This further supported the ability of RISCtrap to enrich targets that are actively down-regulated. Of particular interest was our miR-181 dataset in which we observed that almost half of the targets (125/262) were C2H2 class zinc-finger proteins. Moreover, 95 of these 125 targets shared the specific domain architecture of having an N-terminal KRAB domain followed by multiple tandem C2H2 motifs. Data from several recent papers using microarray and luciferase assays to study miR-181 similarly reported the regulation of this unusual cohort of targets (2, 38, 40).
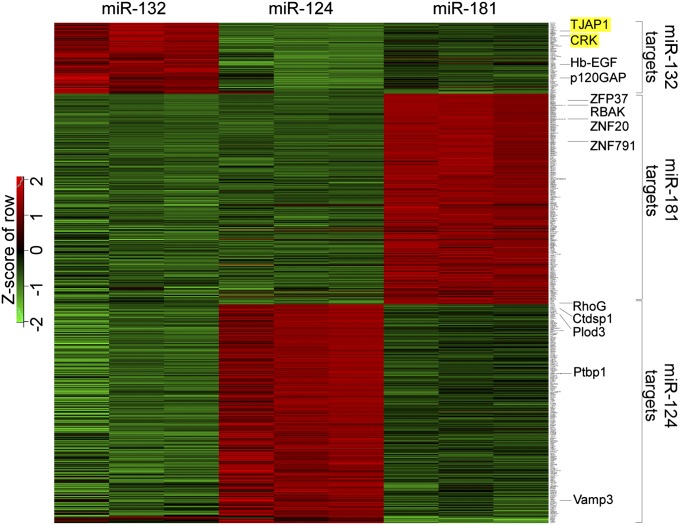
Fig. 3.
RISCtrap screens in HEK293T cells for miR-124, mir-132, and miR-181 identified known and unique targets. All targets identified from the RISCtrap screens with miR-124, miR-132, and miR-181 are organized in a heatmap by biological replicates. (FDR < 0.15, fold enrichment ≥2.) Selected known miRNA targets are labeled and examples of previously unknown miR-132 targets are highlighted in yellow.
Examination of the target datasets revealed several interesting features (Fig. 3). First, grouping of targets by biological replicates produced an enrichment profile for each transcript that was extremely reproducible (Fig. S4) and the cohorts of candidate targets clearly segregated depending on the particular microRNA. Second, the number of targets for each microRNA depended on the identity of the microRNA, ranging from ∼100–300. Third, we found surprisingly minimal overlap among the target sets, suggesting that each miRNA may have its own characteristic set of targets (Fig. 3).
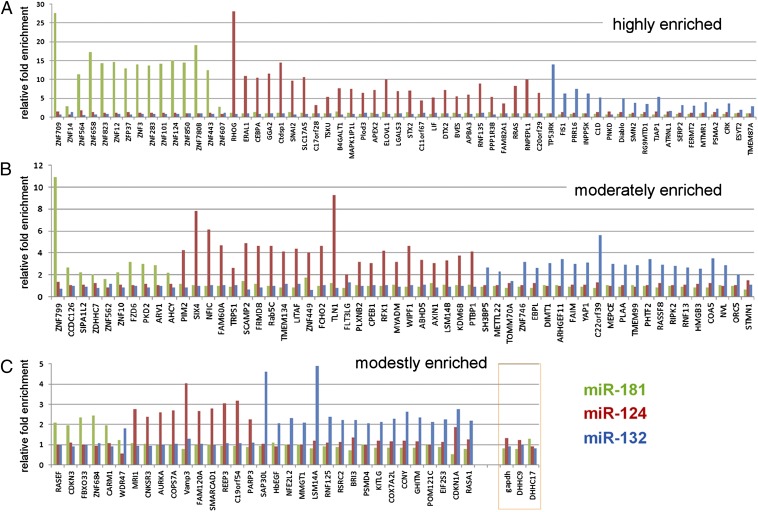
We selected candidate targets from the top, middle, and bottom of each microRNA target list, sorted by fold enrichment—representing highly, moderately, and modestly enriched candidates—for validation of binding using qPCR from an independent RISCtrap experiment (Fig. 4). Overall, we examined 149 targets and the validation rate was 96%, i.e., 143 targets ≥twofold enrichment. In particular, the observed fold enrichment of a target correlated with that detected by the RISCtrap screens (note different scales of the y axes). Analysis of RISCtrap miR-124 candidate targets with available microarray data from HeLa cells (6) suggested a trend where fold enrichment generally correlated with fold repression; however, it was not statistically significant. Importantly, we additionally examined three transcripts that did not enrich in any of our RISCtrap screens to test as negative controls, Gapdh, Asp-His-His-Cys (DHHC)9, and DHHC17; none of these transcripts showed any enrichment in the miR-181, mir-124, or miR-132 RISCtrap screens (Fig. 4).
Fig. 4.
Validation of identified candidate targets confirmed microRNA-specific enrichment. Approximately 150 candidate targets from the three microRNA RISCtrap screens—representing candidates that were classified as highly enriched (A), moderately enriched (B), and modestly enriched (C)—were tested in an independent experiment for their copurification with RISCtrap using qPCR. Overall validation rate was 96%. Also included were three negative controls GAPDH, DHHC9, and DHHC17 (orange box). Y axes represent relative fold enrichment.
Characterization of MREs.
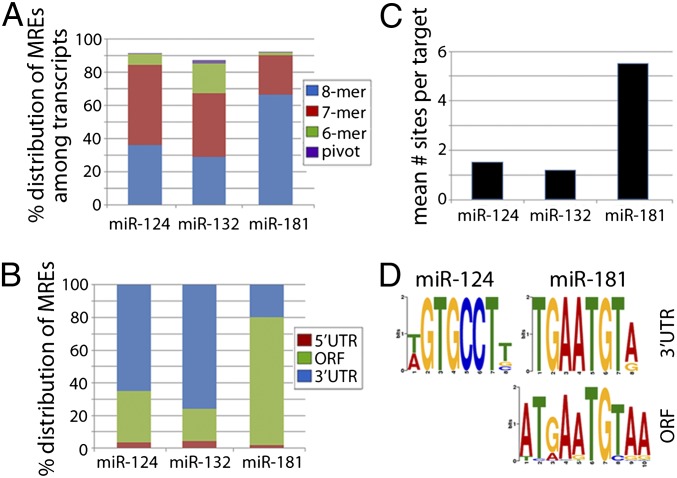
Using directed searches, we examined whether the target datasets obtained by RISCtrap contained expected microRNA binding motifs. Both canonical MREs and the recently described pivot or hinged MREs were examined (47). Approximately 90% of all targets contained an MRE corresponding to the targeting microRNA: 91.5% of miR-124 targets, 87.2% of miR-132 targets, and 92.4% miR-181 targets (Fig. 5A). The majority of targets contained at least an 8-mer, 7-mer-m8, or 7mer-a1 type motifs; fewer had only a 6-mer or pivot MRE (7% miR-124, 20% miR-132, and 2% miR-181). Moreover, the frequency of 7mer-m8 motifs among nontargeted transcript pools was low, suggesting that the appropriate MRE motifs were specifically enriched among targeted transcripts. Appropriately, we also found that 82% of targets predicted to be coregulated by at least two miRNAs examined here contained MRE motifs for both microRNAs (Fig. S6A). We next examined the distribution of canonical 7mer-m8 sites among the miR-124 and miR-132 targets and discovered that the majority of MREs (60–80%) were located in the 3′UTR, with ∼20–30% in ORFs (Fig. 5B). In these cases, the relative position of MRE motifs along 3′UTRs appeared evenly dispersed (Fig. S6B). Conversely, the majority of miR-181 targets contained MREs in the ORFs and, in agreement with previously published reports, were specifically encoded within the C2H2 motif repeats (38, 40). Targets of miR-124 and miR-132 averaged ∼1 MRE per target; in contrast, miR-181 targets averaged 5.5 MREs per target (Fig. 5C). Lastly, we performed de novo multiple expectation maximization for motif elicitation (MEME) analyses to identify overly represented sequences. This identified motifs that corresponded to canonical MREs for both miR-124 and miR-181 in the 3′UTRs of their respective targets, as well as many more in the ORFs of miR-181 targets (Fig. 5D). We did not identify the miR-132 motif with our de novo analysis, despite its high representation when performing a directed search. Most likely, this is due to a relatively higher reliance on 6-mer motifs compared with miR-124 and miR-181 (Fig. 5A) and this shorter motif is not easily distinguished by de novo analysis.
Fig. 5.
Identification of microRNA recognition element (MRE) motifs enriched among RISCtrap targets. (A) Percentages of transcripts in each dataset were classified by inclusion of at least 1 MRE motif and distribution of motif types. Each transcript is counted only once and classified according to inclusion of the following motifs in this order: 8-mer > 7mer-m8 > 7mer-a1 > 6-mer > pivot. (B) Distribution of candidate MREs is shown for each microRNA target dataset, based on its position in the target’s 5′UTR, ORF, or 3′UTR. (C) The mean number of 7mer-m8 MRE motifs per target is depicted for each microRNA dataset. (D) De novo MEME analysis using all targets from the miR-124 and miR-181 target datasets revealed canonical MRE motifs in the 3′UTR of miR-124 targets (296 motifs, P = 2.6 × 10−108), in the 3′UTR of miR-181 targets (151 motifs, P = 1.3 × 10−54), and in the ORF of miR-181 targets (1,000 motifs, P = 9.4 × 10−1488)
Identification of Previously Unrecognized miR-132 Targets, CRK and TJAP1.
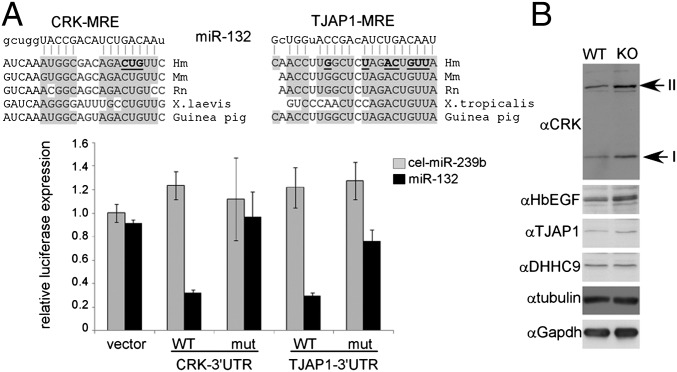
RISCtrap identified many previously known targets. However, our datasets also identified previously unrecognized targets, many of which exhibited fold enrichments exceeding those of known targets. We selected two putative miR-132 targets, CRK and TJAP1, for further investigation. CRK is an adaptor protein for receptor tyrosine kinases and TJAP1 associates with tight junctions. Both candidates validated for specific enrichment in the miR-132 RISCtrap screen (Fig. 4) and available microarray data indicated that both were expressed at high levels in brain (48, 49). Moreover, each has a well-conserved MRE site in their 3′UTR (Fig. 6A). Incorporation of the 3′UTR sequence for either CRK or TJAP1 downstream of renilla luciferase in a dual luciferase assay conferred miR-132 regulation (WT) and mutation of the putative MRE (mut) caused it to be refractory to this regulation (Fig. 6A). We next asked whether these targets were stabilized in a miR-132 knockout mouse model. Examination of endogenous protein from whole cell lysates from the forebrain of litter-matched male siblings revealed an accumulation of CRK and TJAP1 in the miR-132 knockout animal (Fig. 6B). The abundance of negative controls DHHC9 (Fig. 4), α-tubulin, and Gapdh remained equivalent between wild-type and knockout samples. These data demonstrated that RISCtrap analysis of a nonneuronal cell type can identify previously unrecognized targets that are functionally regulated in the brain.
Fig. 6.
Two previously unknown miR-132 targets are regulated by miR-132 in mouse brain. (A) Conserved miR-132 MRE sequences were identified in the 3′UTR of both unique candidate targets CRK and TJAP1 (Top). Luciferase assays in HEK293T cells demonstrated that each of their 3′UTR sequences conferred regulation by miR-132. Mutation of the predicted MRE (mutated sequences are bold and underlined) blocked miR-132 regulation. (B) Whole cell lysates from forebrains of litter-matched siblings of miR-132(+/+) and miR-132(−/−) mice were probed for endogenous protein levels of previously unknown targets, CRK (isoforms I and II) and TJAP1, as well as known target Hb-EGF, and negative controls DHHC9, α-tubulin, and GAPDH.
Discussion
RISCtrap provides a robust strategy for the unbiased identification of specific microRNA–mRNA target interactions and may be adaptable for in vivo studies through viral induction or generation of inducible animal models. For this proof-of-principle screen, we chose HEK293T cells to identify targets of miR-132, miR-124, and miR-181 that turned out to be relevant in a variety of cell types. We were able to identify published miR-132 targets, e.g., p120RasGAP (30) and Hb-EGF (33), as well as previously unknown targets CRK and TJAP1 that were regulated both in HEK293T cells and murine forebrain. Of note, these two transcripts were not identified as miR-132 targets by HITS-CLIP in murine brain (8). MicroRNA-124 targets Ctdsp1 and PTBP1—both identified in this screen—have known roles in the embryonic development of the central nervous system in chick and mouse, as well as the nervous system development in Ciona intestinalis (50–52). The top candidate target in our miR-124 screen was RhoG, which showed an ∼20-fold enrichment. It was recently reported that miR-124–dependent regulation of RhoG significantly contributed to dendritic and axonal complexity in hippocampal neurons (53).
The bioinformatic strategy used here for RISCtrap provides a platform for comparison of current and future datasets under identical experimental conditions. It also limited false positives, preventing an overestimation of the number of identified targets. Our high confidence lists of targets overall validated at 96% for binding and appropriate MRE motifs were overrepresented among the identified targets. Additional analyses may be able to identify novel factors contributing to target recognition, possibly accounting for the ∼10% of targets that did not contain canonical MRE motifs.
Any screen is subject to the possible omission of a few actual microRNA targets. A technical reason why a target may have been missed with this particular screen is if the transcript is not expressed in HEK293T cells, e.g., miR-132 targets p250GAP (29) and acetylcholinesterase (34). Another possibility is if the specific regulation of targets depended on cellular context, such as organism, tissue, activity, timing, or age, suggesting an additional level of regulation. One example of this is methyl CpG binding protein MeCP2, which is a miR-132 target in neural cells (54). The neural-specific isoform of MeCP2 encodes a long 3′UTR that contains the miR-132 MRE. The isoform found in HEK293T cells, however, has a shorter 3′UTR that excludes this MRE (55). Thus, MeCP2 escapes miR-132 regulation in nonneural cells and did not register as a hit in our current screen. Another reason for missing targets is if there was too much variability among biological replicates, e.g., miR-124 target, Baf53A (56), and miR-132 target, p300 (31), or if the enrichment was just under twofold, e.g., miR-181 target, KLF6 (57). Despite these specific occurrences, the substantial overlap with the miR-124 HITS-CLIP dataset and our studies of the forebrain of the miR-132 knockout mouse (Fig. 6B) lead us to conclude that RISCtrap can yield substantial information about target recognition that is applicable across cell types and species.
Another potential concern might be false positives resulting from having to ectopically express the microRNA to preprogram the dnRISCs. In actuality, our datasets contain fewer targets than several others. Importantly, a previous comparison of mouse brain and HeLa cells using HITS-CLIP demonstrated no spurious binding interactions after miR-124 ectopic expression (8). To test whether addition of exogenous miRNA caused spurious interactions, we selected 13 miR-124 candidate targets identified in HITS-CLIP (BC = 5) that were also expressed in HEK293T cells but absent from the miR-124 RISCtrap dataset, to assay whether we could detect their binding to dnGW182–RISC with qPCR (Fig. S7). None of these candidate targets demonstrated enrichment despite ectopic miR-124 expression, suggesting that we were not forcing illegitimate interactions. Although cross-linking approaches have the theoretical advantage of not requiring addition of exogenous miRNAs, this did not appear to be a limitation of our approach.
Materials and Methods
RISCtrap screens were performed by cotransfecting 6 × 106 HEK293T cells in a 10-cm dish with 20 μg of expression plasmid for Flag–dnGW182 and 50 nM miRNA mimics, using either Lipofectamine 2000 (Invitrogen) or a standard calcium phosphate method. Twenty-four hours posttransfection, cells were rinsed with cold PBS and harvested in cold lysis buffer (20 mM Tris pH 7.5, 200 mM NaCl, 1 mM DTT, 0.05% Nonidet P-40, 2.5 mM MgCl2, 60 units/mL RNase inhibitor, and EDTA-free protease inhibitor). Cleared lysates were incubated for 2 h at 4 °C with 10 μL of preblocked Flag–M2 agarose (2 h with 1 mg/mL yeast tRNA and 1 mg/mL BSA). After washing the beads with lysis buffer, bound RNA was eluted by Trizol extraction following the manufacturer’s protocol (Invitrogen). Messenger RNA was enriched from 200 ng of total RNA by generation of double-stranded cDNA using an oligo-dT-T7 primer (GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGGT24) during first-strand synthesis and a standard second strand synthesis. This was followed by a 16-h T7 IVT reaction to linearly enrich for mRNAs. The TruSeq v2 library protocol was used on 200 ng of mRNA-enriched material from each sample to generate Illumina-compatible indexed libraries. Samples were pooled into three lanes, with each biological replicate sequenced in a separate lane on a HiSeq v3 platform using a single-read 100-bp protocol. Reads were then uniquely mapped using TopHat v1.4.0 to a human GRCh37/hg19 reference genome and RefSeq gene annotation guidance (as of Oct. 9, 2011). We determined a baseline for the dataset (minimum of 200 counts per target), bioinformatically removed nonpolyadenylated transcripts, and normalized to the median of geometric ratios broadly following the DESeq approach (58). At each step, we analyzed the distribution and evaluated the effects of our normalization using violin plots, as well as principal components analyses (PCA) to evaluate the retention of clustering among conditions (Fig. S3). Transcript-specific variance estimates were obtained by fitting the negative binomial model implemented in DESEq (58) (Fig. S5), and significantly enriched targets were determined with ANOVA among the biological replicates.
Supplementary Material
Acknowledgments
We thank Y. Wang for help with the luciferase assays; E. Makeyev for providing the Flag–PTBP1 expression construct; B. Leighton for technical help with cloning dnGW182; Y. Cao for help with principal components analysis; B. Wilmot, C. Zheng, and B. Searles for discussions about RNA sequencing; G. Mandel and G. Hannon for critical reading of the manuscript; R.H.G. labortatory members for discussions; S. Chai for editorial help; and the Oregon Health and Science University Massively Parallel Sequencing Shared Resource core for preparation of TruSeq libraries and HiSeq sequencing. This work was supported by funding from MH094416 and HL111397 (to R.H.G.) and NS076094 (to X.A.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218887109/-/DCSupplemental.
References
- 1.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 2.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eulalio A, et al. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15(1):21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7(11):e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 7.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 8.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS ONE. 2008;3(5):e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karginov FV, et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA. 2007;104(49):19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009;15(6):1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15(4):346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 13.Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15(6):1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao B, et al. Divergent GW182 functional domains in the regulation of translational silencing. Nucleic Acids Res. 2011;39(7):2534–2547. doi: 10.1093/nar/gkq1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15(5):781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20(14):1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44(1):120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Chekulaeva M, et al. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18(11):1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabian MR, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18(11):1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 20.Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35(6):868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huntzinger E, Braun JE, Heimstädt S, Zekri L, Izaurralde E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 2010;29(24):4146–4160. doi: 10.1038/emboj.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol. 2010;17(2):238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuzuoglu-Öztürk D, Huntzinger E, Schmidt S, Izaurralde E. The Caenorhabditis elegans GW182 protein AIN-1 interacts with PAB-1 and subunits of the PAN2-PAN3 and CCR4-NOT deadenylase complexes. Nucleic Acids Res. 2012;40(12):5651–5665. doi: 10.1093/nar/gks218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zekri L, Huntzinger E, Heimstädt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol. 2009;29(23):6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol. 2012;19(6):603–608. doi: 10.1038/nsmb.2309. [DOI] [PubMed] [Google Scholar]
- 27.Baillat D, Shiekhattar R. Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol Cell Biol. 2009;29(15):4144–4155. doi: 10.1128/MCB.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103(7):2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vo N, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102(45):16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anand S, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16(8):909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagos D, et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12(5):513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 32.Mellios N, et al. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci. 2011;14(10):1240–1242. doi: 10.1038/nn.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molnár V, et al. MicroRNA-132 targets HB-EGF upon IgE-mediated activation in murine and human mast cells. Cell Mol Life Sci. 2012;69(5):793–808. doi: 10.1007/s00018-011-0786-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaked I, et al. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31(6):965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tognini P, Putignano E, Coatti A, Pizzorusso T. Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nat Neurosci. 2011;14(10):1237–1239. doi: 10.1038/nn.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 38.Huang S, et al. MicroRNA-181a modulates gene expression of zinc finger family members by directly targeting their coding regions. Nucleic Acids Res. 2010;38(20):7211–7218. doi: 10.1093/nar/gkq564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39(4):493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnall-Levin M, et al. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome Res. 2011;21(9):1395–1403. doi: 10.1101/gr.121210.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landthaler M, et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14(12):2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012;40(19):9850–9862. doi: 10.1093/nar/gks705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juvvuna PK, Khandelia P, Lee LM, Makeyev EV. Argonaute identity defines the length of mature mammalian microRNAs. Nucleic Acids Res. 2012;40(14):6808–6820. doi: 10.1093/nar/gks293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magill ST, et al. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci USA. 2010;107(47):20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16(8):869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 46.Zhu G, Zhang Y, Xu H, Jiang C. Identification of endogenous outward currents in the human embryonic kidney (HEK 293) cell line. J Neurosci Methods. 1998;81(1-2):73–83. doi: 10.1016/s0165-0270(98)00019-3. [DOI] [PubMed] [Google Scholar]
- 47.Chi SW, Hannon GJ, Darnell RB. An alternative mode of microRNA target recognition. Nat Struct Mol Biol. 2012;19(3):321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu C, et al. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen JS, Pedro MS, Zeller RW. miR-124 function during Ciona intestinalis neuronal development includes extensive interaction with the Notch signaling pathway. Development. 2011;138(22):4943–4953. doi: 10.1242/dev.068049. [DOI] [PubMed] [Google Scholar]
- 51.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27(3):435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21(7):744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franke K, et al. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 2012;31(13):2908–2921. doi: 10.1038/emboj.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein ME, et al. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10(12):1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 55.Reichwald K, et al. Comparative sequence analysis of the MECP2-locus in human and mouse reveals new transcribed regions. Mamm Genome. 2000;11(3):182–190. doi: 10.1007/s003350010035. [DOI] [PubMed] [Google Scholar]
- 56.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460(7255):642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X, et al. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumour Biol. 2012;33(5):1589–1597. doi: 10.1007/s13277-012-0414-3. [DOI] [PubMed] [Google Scholar]
- 58.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.