Abstract
Copy number variations are genomic structural variants that are frequently associated with human diseases. Among these copy number variations, duplications of DNA segments are often assumed to lead to dosage effects by increasing the copy number of either genes or their regulatory elements. We produced a series of large targeted duplications within a conserved gene desert upstream of the murine HoxD locus. This DNA region, syntenic to human 2q31-32, contains a range of regulatory elements required for Hoxd gene transcription, and it is often disrupted and/or reorganized in human genetic conditions collectively known as the 2q31 syndrome. Unexpectedly, one such duplication led to a transcriptional down-regulation in developing digits by impairing physical interactions between the target genes and their upstream regulatory elements, thus phenocopying the effect obtained when these enhancer sequences are deleted. These results illustrate the detrimental consequences of interrupting highly conserved regulatory landscapes and reveal a mechanism where genomic duplications lead to partial loss of function of nearby located genes.
Keywords: chromatin architecture, enhancer-promoter interaction
Genetic variation is both a source of phenotypic diversity and a cause of many human diseases. Such variations range from single-nucleotide exchanges to large rearrangements, including inversions, translocations, or copy number variations (CNVs), collectively referred to as structural variants (1). CNVs are either a gain (duplication) or a loss (deletion) of DNA segments that can span from kilobase- to megabase-sized intervals. CNVs may include genes and/or noncoding sequences, and they account for a large part of the genetic diversity in animal populations. Over the last years, increasing evidence has associated them with a number of diseases in humans (2, 3).
However, although the consequences of point mutations are relatively well-understood for many genetic conditions, even when the causative variant is located in noncoding intervals (4, 5), the mechanisms where CNVs can cause diseases or malformations are more elusive. In particular, duplications are generally expected to cause dosage effects because of gene duplications within the DNA segment, leading to a global increase in protein products. However, the comparison of tissue transcriptomes from different mouse inbred strains has suggested that CNVs can also affect the expression of genes located nearby the rearrangements at distances of up to several hundred kilobases (6). Such large-scale effects are in agreement with current models of gene regulation, involving complex sets of control elements that can span large genomic distances, particularly for genes with special roles during embryonic development (7, 8). These genes often display complex expression patterns and accordingly, duplications of either known or putative associated enhancers were reported to cause various malformations in humans (for example, at the SHH, IHH, or BMP2 loci) (9–11). However, except for some rare cases (12), the relevant human material could not be assessed, thus calling for the development of animal models of CNVs-induced pathologies.
We have used the murine HoxD gene cluster as a model locus to investigate the impact of structural variation on gene regulation. In mammals, 39 Hox genes are grouped at four genomic loci, referred to as the HoxA to HoxD gene clusters (13). These genes encode transcription factors, which are critical for proper patterning of the embryonic anterior to posterior axis, as shown by genetic evidence in vivo. In addition to this ancestral function, specific Hox clusters have evolved novel functions along with the emergence of diverse embryonic structures (14). The evolution of these new global specificities were often associated to cluster-wide regulations (i.e., to the presence of strong enhancer sequences controlling several Hox genes at one time). For example, the expression of Hoxd genes was co-opted to organize the development of both the proximal (forearm and lower leg) and distal (hands and feet) limb segments (15), as shown by genetic and biochemical studies in mice.
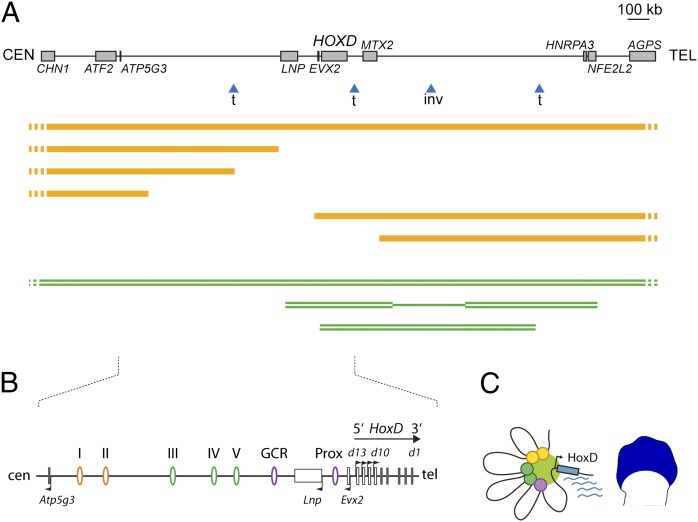
In humans, the HOXD cluster is in a several megabase-sized syntenic region, which expectedly contains all sequences identified as important for the regulation of Hoxd genes during murine limb development. Interestingly, many human genetic syndromes displaying limb malformations involve structural variants overlapping with either the HOXD cluster itself or the conserved gene deserts flanking this gene cluster (Fig. 1A). For example, the 2q31 microdeletion syndrome is caused by different deletions of various sizes, overlapping with the LNP-ATP5G3 gene desert on the centromeric side of the HOXD cluster. This syndrome is associated with hand malformations resembling mutations into the HOXD13 gene, even in patients where the HOXD gene cluster itself is not deleted (16), suggesting that such deletions affect regulatory elements controlling HOXD gene expression in developing limbs rather than the genes themselves.
Fig. 1.
Long-range transcriptional control of Hoxd genes during limb development. (A) The mammalian HOXD cluster is flanked by two conserved gene deserts on its centromeric (CEN) and telomeric (TEL) sides (Upper). In humans, multiple structural variants are found within this interval and are associated with various limb malformations. Some of them are shown with blue arrowheads indicating breakpoints for either translocations (t) or an inversion (inv), which modified this interval. (Lower) Examples of haplo-insufficient deletions (orange lines) and duplications (double green lines) are depicted. (B) Enlargement of the mouse syntenic region, including the HoxD cluster and the centromeric gene desert. In developing digits (schematized limb bud; blue territory), the coordinated expression of the Hoxd13 to Hoxd10 genes as well as of Lnp and Evx2 is under the control of a regulatory archipelago, which consists of multiple regulatory islands located either within the gene desert (ovals I–V and GCR) or between Lnp and Hoxd13 (Prox). (C) Chromatin looping brings these various elements to the vicinity of their target gene promoters, thus forming a transcriptionally active conformation (19).
Within the syntenic mouse genomic interval, multiple enhancer sequences were recently described, including a global control region (GCR), several regulatory islands (I to V) dispersed within the Lnp-Atp5g3 gene desert, and the Prox element, which is located between Lnp and Evx2 (17–19) (Fig. 1B). These enhancers collectively form a regulatory archipelago spanning over 800 kb on the centromeric side of the gene cluster and controlling the coordinated transcription of Hoxd13 to Hoxd10, Lnp, and Evx2 in developing digits. In these cells, these various elements are brought to the vicinity of the HoxD cluster by the formation of chromatin microarchitecture, such as looping (Fig. 1C). Furthermore, the genetic dissection of this interval has revealed that each of these regulatory islands participate, in a partially redundant fashion, in the transcriptional activation of the target genes (19).
In addition to deletions, other rearrangements, such as inversions, translocations, or duplications involving sequences flanking the gene cluster, are observed in human patients, which are often linked to various limb anomalies (20–23) (Fig. 1A). In these latter cases, however, the variability of the clinical outcomes and the extent of the genomic modifications, which can be sometimes very large, make the determination of a molecular mechanism linking genotype to phenotype problematic.
To understand better the molecular origins of the phenotypes displayed by these human syndromes, we engineered a series of inversions and duplications within the regulatory archipelago controlling mouse Hoxd gene transcription in digits. These rearrangements led to diverse morphological outcomes depending on the genomic topology of the modified locus. Surprisingly, a duplication of a subset of the enhancer sequences displayed a phenotype similar to the phenotype associated with a nonoverlapping and partial deletion within the gene desert. We show that this duplication induces a reorganization of the spatial conformation of the regulatory interval in developing digits. This reorganization leads to a loss in the functional contribution of some distal enhancer sequences, resulting in a concurrent down-regulation in the expression of Hoxd genes similar to the down-regulation observed upon deletion of these same sequences. We discuss the relevance of such mechanisms to our understanding of the molecular etiologies of CNVs-associated diseases.
Results
Large Inversions Interrupt a Regulatory Landscape.
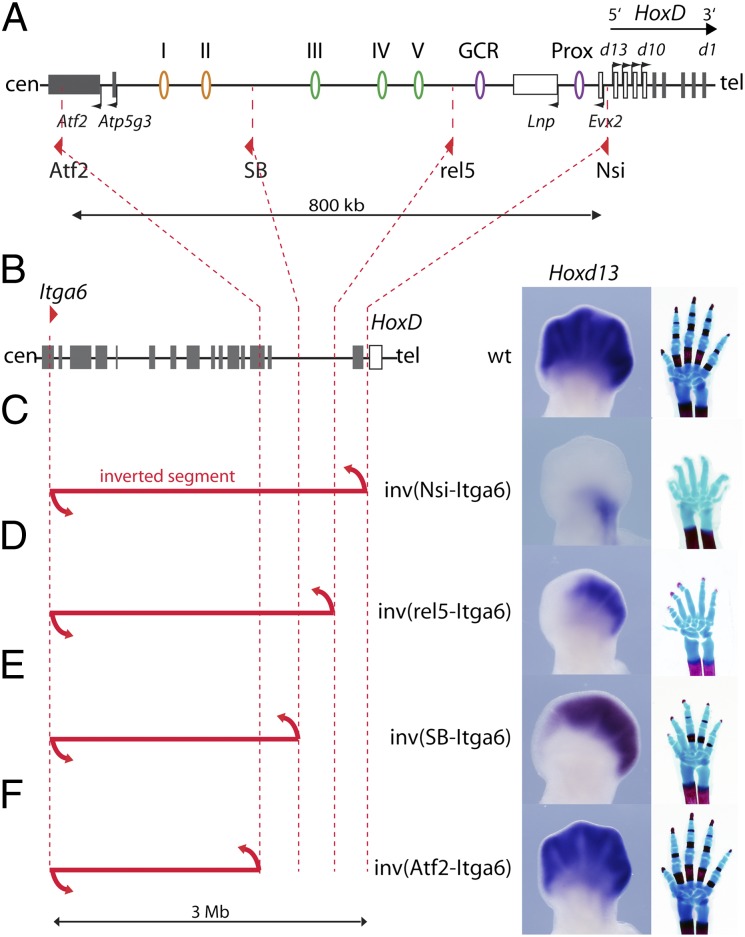
To characterize the extent of the DNA interval necessary for Hoxd gene transcription in developing digits with more precision, we used a set of inversions with one and the same breakpoint within the Itga6 gene (i.e., 3 Mb centromeric to the gene cluster) and a variety of second breakpoints at distinct positions within the regulatory archipelago (Fig. 2 A and B). In the inverted configurations, DNA sequences normally located centromeric to the proximal breakpoints were repositioned at a large distance from the HoxD cluster. We had reported previously that a 3-Mb large inversion with a breakpoint immediately upstream of Hoxd13 led to a complete loss of Hoxd13 expression in developing digits at embryonic day 12.5 (E12.5). Accordingly, animals of this genotype displayed severe shortening of digits at birth, a phenotype similar to the phenotype obtained upon deletion of the gene cluster itself (24) (Fig. 2C). A shorter inversion, with a proximal breakpoint near the telomeric extremity of the gene desert, separated the cluster from regulatory islands I–V, but it maintained the linkage between the GCR, Prox, and Hoxd genes. In this configuration, Hoxd13 expression was limited to a faint domain at the posterior margin of the hand plate (19) (Fig. 2D). This configuration revealed the functional outcome of both the GCR and Prox sequences alone when left in their endogenous context, and thus, it emphasized the importance of the islands I–V in the transcriptional readout of the system.
Fig. 2.
A set of nested inversions disrupts the regulatory archipelago controlling Hoxd gene transcription in digits. (A) Map of the centromeric gene desert along with the positions of the various LoxP sites located within the HoxD regulatory archipelago (red triangles) used for the inversions, deletions, and duplications shown in this study. (B) The WT genomic context of the HoxD cluster is shown in Left, with the location of a remote loxP site within the Itga6 gene used for the set of nested inversions described in C–F. Gray rectangles represent genes, and the HoxD cluster is in white. (Right) The expression of Hoxd13 in an E12.5 limb bud is depicted as well as a WT hand skeleton at birth. (C) A 3-Mb large inversion separates the HoxD cluster from its regulatory elements and thus, abrogates all Hoxd13 expression in developing digits. The resulting phenotype is identical to a full deletion of the HoxD cluster (24). (D) A smaller inversion separating the gene desert from the HoxD cluster maintains a limited Hoxd13 expression to a faint posterior domain. Only the GCR and Prox elements keep their vicinity to Hoxd13. (E) An inversion separating the distal half of the gene desert leads to a decreased anterior expression of Hoxd13 and a shortening of digit II at birth. In this inversion, islands I and II have been removed from the archipelago. (F) An inversion leaving the gene desert uninterrupted had no detectable impact on either Hoxd13 expression or limb morphology. All specimens are homozygous for the various indicated inversions.
We generated inversions using the STRING approach (25), with breakpoints either between islands II and III in the middle of the gene desert [Inv(SB-Itga6)] or on the other side of the desert within the Atf2 gene [Inv(Atf2-Itga6)]. The (SB-Itga6) inversion led to a loss of Hoxd13 expression in the anterior hand plate, including both presumptive digit I and a part of digit II. At birth, affected animals displayed a shorter digit II with a missing phalange (Fig. 2E). In contrast, the Inv(Atf2-Itga6) configuration did not elicit any detectable change in Hoxd13 expression or alterations in the limb phenotype compared with the WT situation (Fig. 2F). From this series, we concluded that (i) sequences farther centromeric from Atf2 are not critical for Hoxd gene expression in developing digits and (ii) the interruption of the regulatory archipelago affects Hoxd gene regulation with a severity that depends on the extent of the DNA segment, which was disconnected from the gene cluster (19).
Duplications Within the Regulatory Interval.
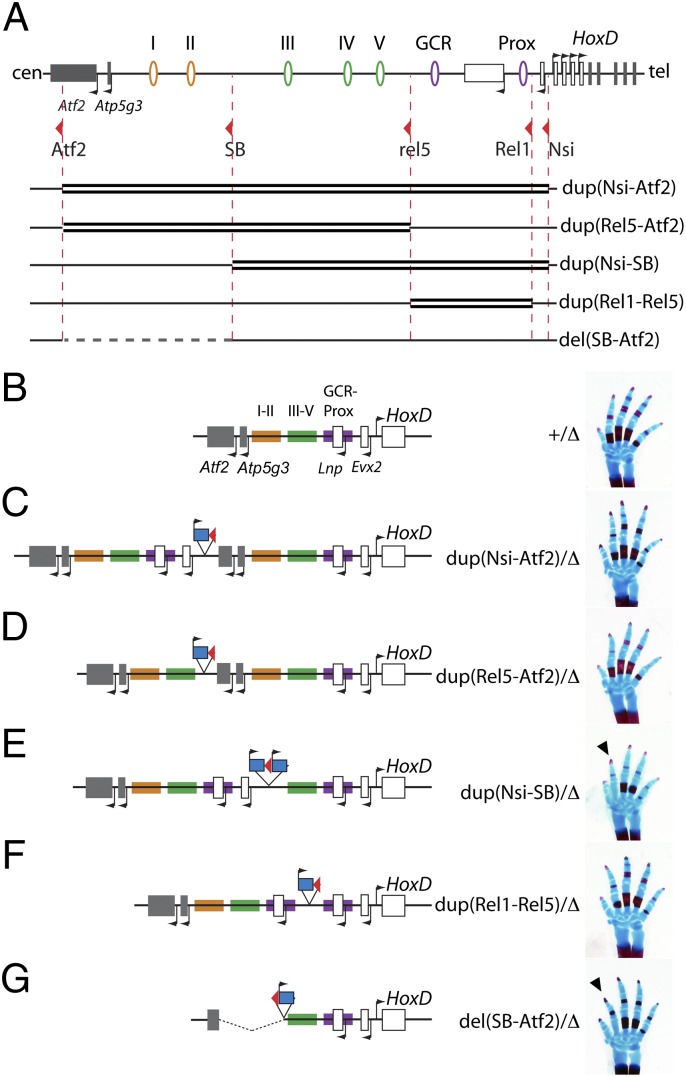
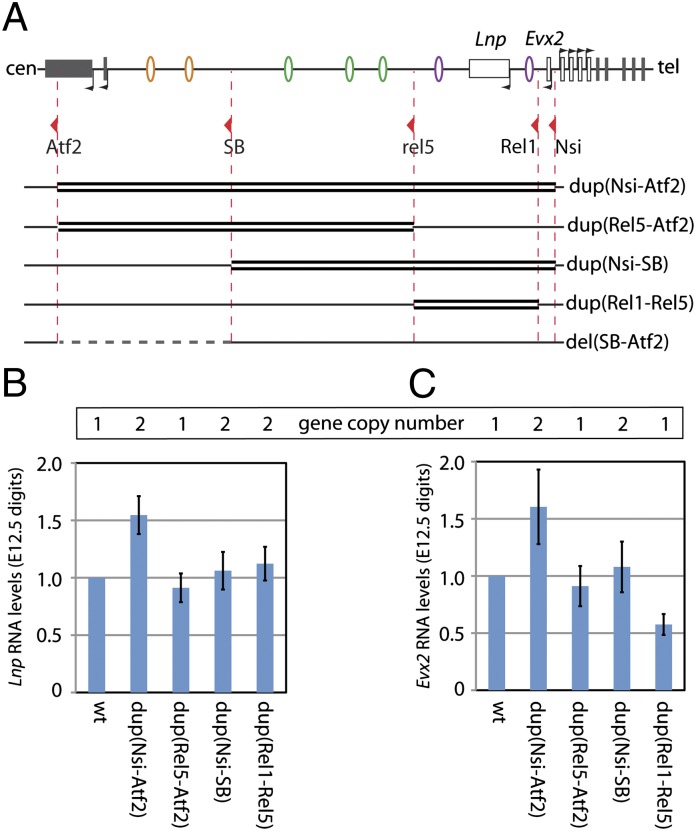
To investigate the specificity of these regulatory islands (i.e., to assess whether they could replace one another) as well as to evaluate the impact of their copy number on the transcription of Hoxd genes in digits, we applied TAMERE (26) to generate a series of large duplications. We used the same breakpoints as described above as well as a loxP site located immediately centromeric of Evx2 (27). These duplications included either parts or all of the gene desert, the Evx2, Lnp, and Atp5g3 genes and the first exons of Atf2 (Fig. 3A). Consequently, they modified both the number of copies of these regulatory elements and their organization relative to the gene cluster. Each duplication was balanced with a chromosome carrying a targeted deletion from Hoxd13 to Hoxd8 [Del(8–13)] to compare animals with a single copy of Hoxd genes in cis with the various modified configurations.
Fig. 3.
Morphological effects of inducing duplications within the regulatory archipelago. (A) The various duplications were produced by using the same LoxP sites as for Fig. 2 (red arrowheads), and they are depicted by double thick black lines. Below the set of duplications, the position of the del(SB-Atf2) is indicated by a dashed gray line. (B–G) Schematics of the locus after the various duplications (or deletions) were produced are shown in Left, with a hand skeleton at birth shown in Right. In vivo, each configuration was balanced by a chromosome carrying a deletion of Hoxd13 to Hoxd8 [the Del(8–13) allele, indicated as ∆]. For the sake of simplicity, the three segments of the regulatory archipelago, as defined by the positions of LoxP sites, are highlighted using different colors (control in B). In B–G, these colors are used to identify the parts of the archipelago that are duplicated (C–F) or deleted (G). In all cases, LacZ reporter genes were associated with the various configurations. They are indicated on the schemes by a blue rectangle along with the presence of the associated LoxP site (red arrowheads). In E, two such LacZ reporters are present (in the text). (B) WT configuration. (C) Duplication of the full archipelago from Evx2 to Atf2. (D) Duplication of islands I–V. (E) A duplication extending from Evx2 until the proximal half of the gene desert is associated with a shortening of digit II at birth (arrowhead). (F) Duplication of the Prox-GCR segment. (G) Deletion of the distal half of the gene desert complementary to the duplication in E. Note the similar shortening of digit II (arrowhead).
Animals carrying a duplication of the full regulatory interval from Evx2 to Atf2 did not display any detectable defects at birth compared with controls (Fig. 3 B and C). Likewise, animals with a duplication of the whole-gene desert from Rel5 to Atf2 were phenotypically normal (Fig. 3D). In contrast, a duplication extending from upstream Hoxd13 to the SB position (i.e., in the middle of the gene desert) induced a shortening of digit II, with a missing phalange [Dup(Nsi-SB)] (Fig. 3E). Digit I was also affected but with an incomplete penetrance. Such a phenotype was not observed, however, when a shorter duplication was analyzed, including Prox, Lnp, and the GCR but without additional copies of regulatory islands III–V (Fig. 3F). Surprisingly, animals carrying the Dup(Nsi-SB) duplication displayed a phenotype strikingly similar to the phenotype associated with a deletion of the SB to Atf2 DNA segment [Del(SB-Atf2)], a deletion of the distal half of the gene desert that removes regulatory islands I and II with no overlap with the duplicated Nsi-SB fragment (Fig. 3G). In addition, the same shortening of digit II was also observed in Inv(SB-Itga6) animals (i.e., in a configuration where the same DNA segment containing islands I and II was disconnected from the gene cluster) (Fig. 2E). Altogether, these genetic approaches indicated that the duplication of the Nsi to SB DNA interval had an impact on the function of distal regulatory elements, including islands I and II.
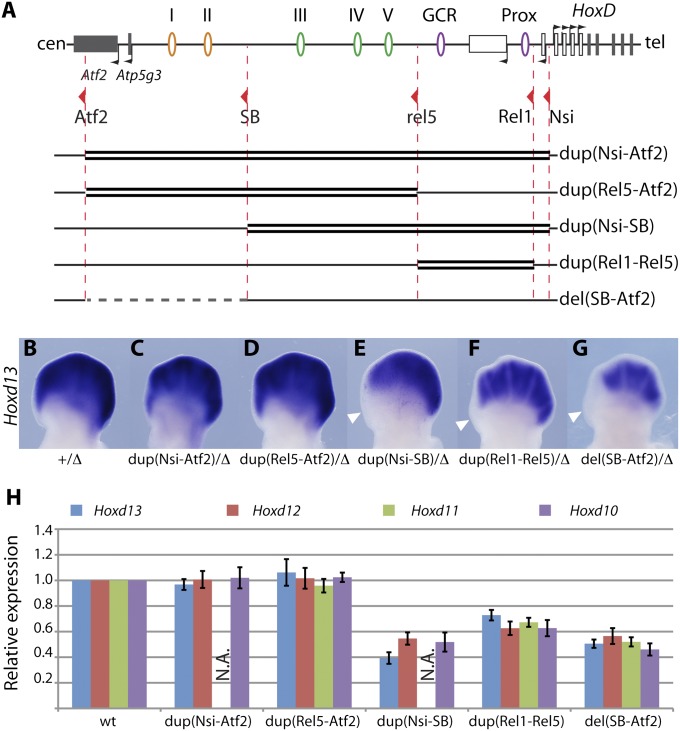
Proximal Duplications Affect the Expression of Remote Target Genes.
To address the origin of these phenotypes, we looked at the effect of the duplications on the transcription of Hoxd genes in developing digits. Although the expression profile of Hoxd13 was not affected in either Dup(Nsi-Atf2) or Dup(Rel5-Atf2) animals (Fig. 4 A–D), Hoxd13 transcripts were lost from the anterior digits of Dup(Nsi-SB) embryos in a territory precisely corresponding to presumptive digit I and part of digit II (Fig. 4E). A milder reduction was observed in the distal limbs of Dup(Rel1-Rel5) embryos (Fig. 4F). Interestingly, a similar loss of expression of Hoxd13 in the anterior aspect of the developing limb bud was observed in the Del(SB-Atf2) specimen (Fig. 4G), pointing again to a convergent effect of both the deletion of two upstream regulatory islands and the duplication of a nonoverlapping piece of DNA located between these two islands and the target promoters.
Fig. 4.
Duplications induce a partial loss of Hoxd gene expression. (A) Schemes of the genetic configurations are as for Fig. 3. (B–G) Hoxd13 expression at E12.5 in the various mutant configurations. Each allele is balanced with a Del(8–13) chromosome (∆). Dup(Nsi-Atf2) (C) and Dup(Rel5-Atf2) (D) do not affect the Hoxd13 expression domain. In contrast, Hoxd13 expression is lost in the anterior part of Dup(Nsi-SB) distal limbs (E) and significantly decreased in Dup(Rel1-Rel5) (F). Similar reductions are observed in embryos carrying a deletion of the distal gene desert (G). (H) RT-qPCR analysis of Hoxd gene expression levels in E12.5 developing digits of embryos homozygous for the various genetic rearrangements. Dup(Nsi-SB) and Del(SB-Atf2) elicit a similar down-regulation of Hoxd13 to Hoxd10. Milder decreases are observed in Dup(Rel1-Rel5). Dup(Nsi-Atf2) and Dup(Rel5-Atf2) are not associated with significant changes in Hoxd gene expression levels. A supernumerary copy of Hoxd11 is associated with Dup(Nsi-Atf2) and Dup(Nsi-SB) as an Hoxd11LacZ reporter gene, and Hoxd11 levels were, thus, not assessed in these configurations (N.A.). The WT levels are set to one for each gene. Error bars indicate SD (n = 4).
We used quantitative RT-PCR (RT-qPCR) to quantify the steady-state levels of mRNAs in these various configurations and found a 60% reduction in the amount of Hoxd13 mRNA in Dup(Nsi-SB) homozygous digits, whereas Hoxd12 and Hoxd10 were reduced to ∼50% (Fig. 4H). These values are close to the values that we observed in Del(SB-Atf2) digits, where Hoxd genes are expressed at approximately half their WT levels (19). In Dup(Rel1-Rel5) animals, a milder down-regulation was scored, with posterior Hoxd genes expressed at ca. 60–70% of WT levels. In contrast, neither the Dup(Nsi-Atf2) nor the Dup(Rel5-Atf2) allele caused any significant change in these steady-state levels. In addition, none of these latter duplications did elicit an elevation of mRNAs copies, suggesting that supernumerary regulatory elements did not work more efficiently to activate gene transcription. Altogether, these results indicated that duplications of the proximal part of the regulatory archipelago led to a partial loss of Hoxd gene expression, likely by interfering with the activity of more distally located elements (islands I and II), and their normal activities were not compensated for by other regulatory islands when duplicated.
Expression of the Duplicated Genes.
This down-regulation of Hoxd gene transcription in the two duplicated configurations could be caused by the presence of additional copies of target promoters within the duplicated intervals. The duplicated copies of Lnp and Evx2, as well as their promoters, might, indeed, titrate the activity of the various enhancers at the expense of Hoxd gene expression (28). We investigated this possibility by quantifying RNA levels of these duplicated genes in the various configurations and observed increased steady-state levels of Lnp and Evx2 mRNAs in Dup(Nsi-Atf2) digits (Fig. 5). Surprisingly, however, their expression levels were comparable with the WT situation in the Dup(Nsi-SB) configuration, which is also associated with supernumerary copies of these genes. Likewise, the Dup(Rel1-Rel5) allele did not lead to an increased expression of the duplicated Lnp gene. Therefore, gene copy number was not predictive of expression levels in digits, suggesting that the global organization of the interval was critical in defining the final transcriptional activity.
Fig. 5.
Expression levels of the two duplicated genes Evx2 and Lnp. (A) Schemes of the various genetic configurations. (B and C) RT-qPCR analysis of Lnp and Evx2 expression levels in E12.5 developing digits dissected from embryos homozygous for the various rearrangements. For each allele, the number of copies of these two genes is indicated above the graphs. Both genes are clearly expressed at higher levels in the Dup(Nsi-Atf2) allele but not other configurations, where they are present in two copies, indicating an influence of genomic topology rather than a mere consequence of copy number. Lnp and Evx2 mRNAs levels do not correlate with the impact of these modified configurations on Hoxd gene expression. The WT levels are set to one for each gene. Error bars indicate SD (n = 4).
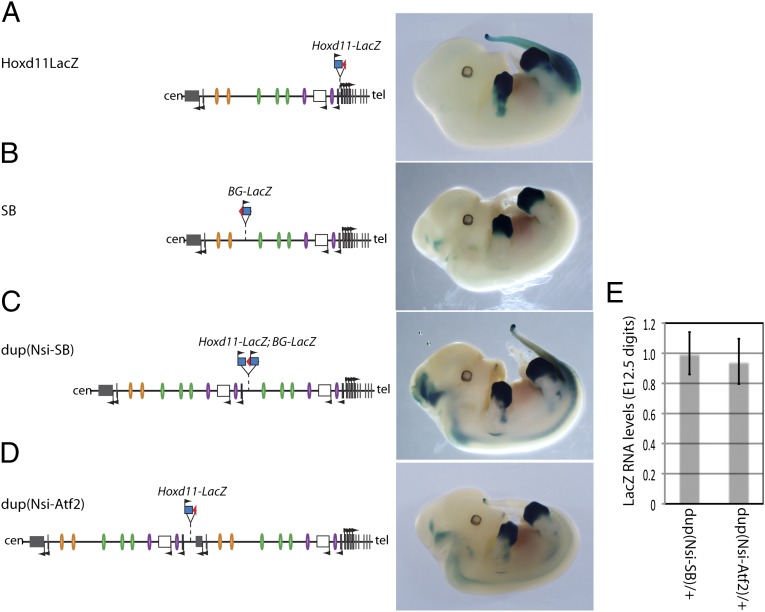
In the same context, several of the parental loxP sites, which were used to produce the duplicated alleles, are associated with a LacZ reporter gene. As a consequence, LacZ reporter transgenes are present within each duplication allele. In particular, the Dup(Nsi-SB) allele, which lead to the strongest transcriptional interference, displayed two neighboring LacZ copies in between the duplicated fragments (Fig. 3E). The potential impact of these transgene insertions was evaluated by comparing their expression profiles in the various genetic configurations (Fig. 6).
Fig. 6.
Reporter gene expression in the modified configurations. (A–D) Scheme of the genetic configurations (Left) along with the expression profiles of the associated LacZ reporter genes (Right). The parental alleles used to produce the duplications are associated with different LacZ insertions within the regulatory landscape (A and B). Although the expression of these reporter transgenes slightly varies with insertion sites, with specific domains in the posterior mesoderm, the proximal limb (presumptive forearm), and the CNS, they all display the same expression pattern in developing digits. (C and D) The Dup(Nsi-SB) and Dup(Nsi-Atf2) rearrangements are associated with a distinct pattern in the CNS, but they do not display an altered digit expression compared with parental configurations. (E) RT-qPCR analysis of LacZ expression levels in E12.5 developing digits of embryos heterozygous for the duplicated configurations indicated similar RNA levels in both duplications, although only the Dup(Nsi-SB) elicited a phenotype. Error bars indicate SD (n = 4).
In the parental strains, the reporter genes displayed expression profiles specific for their insertion sites within the regulatory landscape. For example, when an Hoxd11LacZ gene was located immediately upstream of Hoxd13, its transcription was detected in both the posterior trunk and proximal limb, corresponding to the future forearm (Fig. 6A), in addition to the strong staining observed in distal limb buds. In contrast, although a LacZ transgene integrated in the middle of the gene desert was expectedly expressed in distal limb buds as well, staining also appeared in limited cell populations within the neural tube (Fig. 6B). Likewise, the Dup(Nsi-SB) and Dup(Nsi-Atf2) alleles were associated with slightly different patterns of transcriptional activity in the neural tube, but LacZ staining was always detected in developing digits in the same domain (Fig. 6 C and D) and at comparable levels (Fig. 6E).
Altogether, the expression of additional transcription units within our regulatory interval could not be correlated with the decrease in Hoxd gene transcription, which was observed in our two proximal duplications. Consequently, we concluded that this regulatory interference was unlikely caused by promoter competition for shared enhancers but rather by an altered spatial organization of the regulatory landscape.
Impaired Long-Range Interactions.
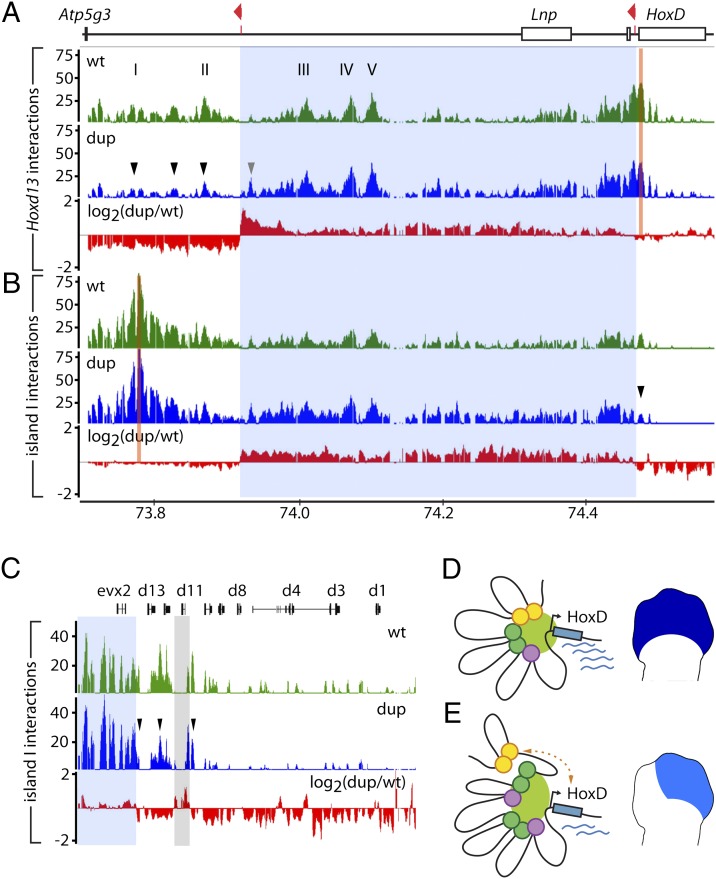
To challenge this hypothesis, we applied chromosome conformation capture (4C) (29, 30) to establish the long-range interaction profile of Hoxd13 in both WT and Dup(Nsi-SB) developing digits. In WT digit cells, Hoxd13 is brought to the vicinity of the various distal regulatory elements, likely through chromatin looping (19) (Fig. 7A), suggesting that a tight physical proximity is established between the promoter of this gene and the various regulatory islands. In the duplicated configuration, this profile of interactions was substantially altered. The detected interactions between Hoxd13 and sequences located within the duplicated segment were globally stronger, particularly with DNA segments displaying relatively weak interactions in the WT situation, such as the vicinity of the centromeric breakpoint (Fig. 7A, gray arrowhead). In contrast, sequences located farther centromeric of the duplicated segment (i.e., whose genomic distance to Hoxd13 had been increased) displayed decreased interaction frequencies, including regulatory islands I and II (Fig. 7A, black arrowheads).
Fig. 7.
The duplication interferes with regulatory interactions. (A) 4C analysis with a viewpoint in Hoxd13 (orange bar) showing long-range interactions over the centromeric gene desert in developing digits of E12.5 WT (green) or Dup(Nsi-SB) (blue) embryos. The red profile displays the ratio (log2 scale) of Dup/WT intensities. The duplicated interval is highlighted in light blue over the profiles. Hoxd13 interactions are increased with sequences within the duplicated segment (gray arrowhead) and decreased with sequences located farther centromeric (black arrowheads). The x axis shows chromosomal coordinates (mm8, 2006 University of California, Santa Cruz assembly) in megabases and the y axes are the ratio of 4C-amplified/genomic DNA intensities. (B) Similar analysis with a viewpoint taken within island I. (C) Enlargement of the HoxD cluster from B. The interactions of island I with the 5′ HoxD cluster are reduced (arrowheads). The light gray bar highlights the sequence corresponding to the Hoxd11LacZ reporter gene present at two positions on this genetic configuration (Fig. 3). (D and E) Distinct conformations in WT or Dup(Nsi-SB) digits, with schematic representation of the transcriptional output in developing digits. (D) In the WT situation, the locus adopts an active conformation, bringing the various islands in the vicinity of the HoxD cluster. (E) The duplication impairs the association of the distal elements (orange circles) with the cluster. The scheme represents one of the possible conformations of the locus, because the interactions experienced by each of the duplicated copies of the islands (purple and green circles) cannot be discriminated with our approach.
This result suggests that the phenotypes observed in the duplications derive from reduced interactions between Hoxd target promoters and islands I and II because of either an increase in the absolute distance in between, or an increase in the total number of islands. We controlled this experiment by using island I instead of Hoxd13 as a viewpoint in a 4C approach. In WT developing digits, island I interacted with both other regulatory elements and the centromeric part of the HoxD cluster. In Dup(Nsi-SB) digits, these contacts were stronger with sequences located within the duplicated segment (Fig. 7B). As for the Hoxd13 interaction profile, the interactions observed for island I seemed generally less specific for the regulatory elements than in the WT situation. Instead, interactions were reinforced within most of the duplicated interval. In agreement with what we observed when using Hoxd13 as a viewpoint, the interactions of island I with the centromeric part of the cluster, including sequences located in the Hoxd13 to Hoxd10 interval, were reduced (Fig. 7C). We concluded that the duplication led to an altered conformation of the regulatory landscape, impairing the association of Hoxd genes with distal regulatory elements and thus, leading to a localized loss of Hoxd gene expression in the digit-forming area with concurrent morphological defects (Fig. 7 D and E).
Discussion
Copy Numbers and Genomic Topology.
The transcription of Hoxd genes in developing digits relies on the activity of multiple regulatory elements dispersed over an interval of ca. 800 kb, overlapping with a conserved gene desert that extends from Lnp to Atp5g3 (19). A duplication covering the proximal part of this regulatory archipelago led to an unexpected and localized loss of Hoxd gene transcription, with the associated morphological defects in digits. This effect was strikingly similar to the effect elicited by a deletion of the most distal third of the landscape, suggesting that both types of rearrangements had a common impact on Hox genes transcription.
In contrast, a duplication of the entire regulatory interval had no detectable effect on Hox genes regulation, indicating that it is the genomic organization of the regulatory elements relative to their target genes rather than their absolute number of copies that is of importance for the transcriptional output of the system. Accordingly, we did not observe any correlation between the impact of duplications on Hoxd gene regulation, on the one hand, and the expression levels of either duplicated target genes (Lnp, Evx2) or exogenous transcription units within the landscape, on the other hand, arguing against an interference caused by enhancer/promoter competition, which was described in other contexts (31). This effect is also distinct from known cases of copy number-induced silencing, in which multicopy transgenes or repeat elements are themselves repressed rather than their neighbor genes (32). Only duplications increasing the distance between the HoxD cluster and distal enhancers induced a decreased transcription in distal limbs; these regulatory interferences were a function of the size of the duplicated segment, because a short proximal duplication, including both Prox and the GCR but not regulatory islands III–V, caused a down-regulation of Hoxd genes milder than the down-regulation observed with a larger duplication, insufficient to elicit a detectable morphological alteration.
This observed decrease in the transcriptional readout whenever the distal islands I and II are moved away from their target further highlights the specific requirement for all of the various regulatory elements to establish the genuine expression profile of Hoxd genes in WT condition. Additional copies of a subset of these enhancers, indeed, cannot compensate for the relocation of others at a distance. This observation indicates that multiple regulatory elements are not merely required to provide a sufficient number of binding sites for a similar set of trans-acting factors but instead, that various islands may recruit (at least partially) distinct molecular complexes leading to subtle qualitative and quantitative differences.
Modified Architecture of the Regulatory Landscape.
The down-regulation in Hoxd gene transcription scored with the large proximal duplication was associated with a modified spatial organization of the genomic regulatory interval. The contacts established both by Hoxd13 and island I, which are located telomeric and centromeric of the duplicated segment, respectively, were strengthened with those sequences lying within the duplicated segment, whereas they were clearly weakened with more distally located sequences (Fig. 7 D and E). Because our 4C approach cannot discriminate between contacts experienced by each of the two copies of any duplicated DNA sequences, we do not know whether the increased levels of interactions observed with these sequences reflect an association of the viewpoint with both copies of the same island or alternatively, a reinforced interaction with one of them only.
Likewise, decreased interactions between the viewpoints and sequences distal to the duplicated segment (such as islands I and II when assessed from Hoxd13) could be because of either a larger genomic distance between the target genes and the enhancer sequences or alternatively, a competition between the various elements for the formation of long-range interactions, such that additional copies of islands within the duplicated sequences would compete out the contacts between Hoxd genes and distal sites. In the former case, the intercalation of any similar-sized piece of DNA would lead to the same effect. However, the concomitant increase in the quantity of contacts with duplicated enhancers suggests that these sequences participate in the spatial organization of the landscape in the duplicated mutant, as if the reiterated segment would now actively take part to this regulatory architecture. As a consequence, distal islands I and II would be somehow left out of the structure (Fig. 7 D and E).
Genome Evolution and Human Disease.
Interestingly, Hoxd gene regulation seemed unaffected in all of the mutant configurations where at least one complete regulatory archipelago was maintained upstream of the gene cluster. In contrast, any condition interrupting this interval, by either an inversion or a duplication intercalating some DNA sequences within the regulatory landscape, resulted in a partial loss of expression. Therefore, it is critical for the proper transcriptional control of these genes that an integral regulatory block be preserved, including the gene desert. Such a regulatory constraint likely provided a selective pressure to maintain this highly syntenic region uninterrupted, because the Lnp-Atp5g3 gene desert is present upstream of the HoxD cluster in all vertebrate genomes that have been sequenced so far. Similar constraints might participate in the stability of other gene deserts, since such DNA intervals have been associated with long-range regulation in several instances (33–40).
These observations also suggest a mechanism where duplications overlapping regulatory regions may lead to a decreased expression of critical target gene(s) by disturbing the intricate organization of complex regulatory landscapes. Such a mechanism could underlie the molecular etiology of some CNV-associated diseases in humans. This possibility is rarely discussed, because an increase in copy number of putative regulatory elements is usually expected to result in an increased expression of their target genes. More precise and exhaustive analyses may, thus, reveal a higher complexity in the organization of regulatory landscapes, and hence, the effects of CNVs affecting such regions may have to be integrated into a global topographic context rather than using mere quantitative parameters.
The complexity of CNV-associated diseases is highlighted by three overlapping duplications, including the human HOXD cluster and flanking sequences. Two such duplications are associated with mesomelic dysplasia (a shortening of the forearm and lower leg), whereas a third and larger duplication causes syndactyly (fused digits) (21–23). Such distinct clinical outcomes as well as the present report illustrate the difficulty in elucidating those molecular mechanisms underlying the pathological consequences of complex genetic conditions in humans in the absence of a proper and adapted animal model.
Materials and Methods
Mouse Strains.
The Del(8–13), Inv(Nsi-Itga6), Del(SB-Atf2), and Inv(Rel5-Itga6) alleles were described previously (19, 24, 41). The inversions (SB-Itga6) and (Atf2-Itga6) where generated by STRING (25) using an loxP site inserted into the Itga6 gene (42) and a second loxP site located either at the SB position within the gene desert (43) or in the Atf2 gene (44). Recombinant offspring with both loxP sites in cis were crossed with Hprt-Cre mice (45). Duplications were produced by TAMERE (26) using the loxP sites in Nsi (46), Rel1 (27), Rel5 (19), SB, and Atf2. Genotyping of mice and embryos was performed by PCR analysis (SI Materials and Methods).
LacZ Staining, in Situ Hybridization, and Skeletal Preparation.
Detection of LacZ reporter activity and in situ hybridization were performed according to standard protocols. The Hoxd13 probe was previously described (47). For skeletal preparation, newborns were stained with standard Alcian blue/Alizarin red protocols.
RT-qPCR Analyses.
Presumptive digits were dissected from E12.5 embryos and stored in RNAlater reagent (Qiagen) before genotyping. RNA was isolated from individual embryos using the RNeasy microkit (Qiagen); 500 ng RNA were reverse-transcribed using random primers and SuperScript III RT (Invitrogen). cDNA was PCR-amplified using SYBR green containing qPCR master mix (Invitrogen) with a CFX96 Real-Time System (Bio-Rad). A mean quantity was calculated from triplicate reactions for each sample. Expression changes were normalized to Rps9. Primers used were as described in ref. 48.
4C Analysis.
Presumptive digits were dissected from E12.5 embryos, dissociated by collagenase, and fixed in 2% formaldehyde for 10 min at room temperature. Nuclei were stored at −80 °C until genotyped. 4C libraries were produced as described (49) using NlaIII and DpnII (New England Biolabs) as primary and secondary restriction enzymes, respectively. Digits samples from 10 embryos were pooled for each library. Religated sequences were amplified by inverse PCR with AmpliTaq DNA polymerase (Applied Biosystems) using 200 ng 4C library per reaction and the following primers: Hoxd13-F 5′-AAAATCCTAGACCTGGTCATG-3′; Hoxd13-R 5′-GGCCGATGGTGCTGTATAGG-3′; island I-F 5′-AAGTAGCAAAGCAACCACAGTAAAG-3′; and island I-R 5′-GGCAGAAATGTGGAAAGGTCA-3′. For each condition, 16 reactions were pooled and purified using the Qiagen PCR Clean-Up Kit, fragmented and labeled using the GeneChip WT Double-Stranded DNA Terminal Labeling Kit (Affymetrix), and hybridized to custom-made tiling arrays (50). Arrays were processed according to the manufacturer’s instructions. For each genotype and fragment of interest, two independent samples were analyzed.
Tiling Array Data Analyses.
Array data were quantile-normalized within 4C-amplified/input replicate groups and scaled to medial feature intensity of 100 using TAS software (Affymetrix). For each genomic position, a dataset was generated consisting of all (PM-MM) pairs mapping within a sliding window of 2 kb (broad view) or 500 bp (close view). Average ratios were plotted along the genomic DNA sequence using Integrated Genome Browser software (Affymetrix) (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank B. Mascrez for help with mice, E. Joye for technical assistance, and G. Andrey, N. Soshnikova, and members of the D.D. laboratories for discussions and reagents. This work was supported by a fellowship from the Human Frontier Science Program Organization (HFSPO) (to L.T.) and funds from the Ecole Polytechnique Fédérale de Lausanne (EPFL), the University of Geneva, the Swiss National Research Fund (SNF), the National Research Center (NCCR) Frontiers in Genetics, the European Research Council (ERC), and the FP7 European Union Program Integrated research on Developmental determinants of Ageing and Longevity (IDEAL).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217659109/-/DCSupplemental.
References
- 1.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 2.Klopocki E, Mundlos S. Copy-number variations, noncoding sequences, and human phenotypes. Annu Rev Genomics Hum Genet. 2011;12:53–72. doi: 10.1146/annurev-genom-082410-101404. [DOI] [PubMed] [Google Scholar]
- 3.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12(14):1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 5.Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132(4):797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 6.Henrichsen CN, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41(4):424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 7.Kleinjan DA, van Heyningen V. Long-range control of gene expression: Emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76(1):8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montavon T, Duboule D. Landscapes and archipelagos: Spatial organization of gene regulation in vertebrates. Trends Cell Biol. 2012;22(7):347–354. doi: 10.1016/j.tcb.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Klopocki E, et al. A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J Med Genet. 2008;45(6):370–375. doi: 10.1136/jmg.2007.055699. [DOI] [PubMed] [Google Scholar]
- 10.Dathe K, et al. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am J Hum Genet. 2009;84(4):483–492. doi: 10.1016/j.ajhg.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klopocki E, et al. Copy-number variations involving the IHH locus are associated with syndactyly and craniosynostosis. Am J Hum Genet. 2011;88(1):70–75. doi: 10.1016/j.ajhg.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaeger E, et al. Hereditary mixed polyposis syndrome is caused by a 40-kb upstream duplication that leads to increased and ectopic expression of the BMP antagonist GREM1. Nat Genet. 2012;44(6):699–703. doi: 10.1038/ng.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78(2):191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 14.Tschopp P, Duboule D. A genetic approach to the transcriptional regulation of Hox gene clusters. Annu Rev Genet. 2011;45:145–166. doi: 10.1146/annurev-genet-102209-163429. [DOI] [PubMed] [Google Scholar]
- 15.Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17(4):359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Mitter D, et al. Genotype-phenotype correlation in eight new patients with a deletion encompassing 2q31.1. Am J Med Genet A. 2010;152A(5):1213–1224. doi: 10.1002/ajmg.a.33344. [DOI] [PubMed] [Google Scholar]
- 17.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113(3):405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez F, Duboule D, Spitz F. Transgenic analysis of Hoxd gene regulation during digit development. Dev Biol. 2007;306(2):847–859. doi: 10.1016/j.ydbio.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 19.Montavon T, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147(5):1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Dlugaszewska B, et al. Breakpoints around the HOXD cluster result in various limb malformations. J Med Genet. 2006;43(2):111–118. doi: 10.1136/jmg.2005.033555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho TJ, et al. A dominant mesomelic dysplasia associated with a 1.0-Mb microduplication of HOXD gene cluster at 2q31.1. J Med Genet. 2010;47(9):638–639. doi: 10.1136/jmg.2009.074690. [DOI] [PubMed] [Google Scholar]
- 22.Ghoumid J, et al. Duplication at chromosome 2q31.1-q31.2 in a family presenting syndactyly and nystagmus. Eur J Hum Genet. 2011;19(11):1198–1201. doi: 10.1038/ejhg.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kantaputra PN, et al. Mesomelic dysplasia Kantaputra type is associated with duplications of the HOXD locus on chromosome 2q. Eur J Hum Genet. 2010;18(12):1310–1314. doi: 10.1038/ejhg.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tschopp P, Duboule D. A regulatory ‘landscape effect’ over the HoxD cluster. Dev Biol. 2011;351(2):288–296. doi: 10.1016/j.ydbio.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 25.Spitz F, Herkenne C, Morris MA, Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat Genet. 2005;37(8):889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- 26.Hérault Y, Rassoulzadegan M, Cuzin F, Duboule D. Engineering chromosomes in mice through targeted meiotic recombination (TAMERE) Nat Genet. 1998;20(4):381–384. doi: 10.1038/3861. [DOI] [PubMed] [Google Scholar]
- 27.Kondo T, Duboule D. Breaking colinearity in the mouse HoxD complex. Cell. 1999;97(3):407–417. doi: 10.1016/s0092-8674(00)80749-7. [DOI] [PubMed] [Google Scholar]
- 28.Monge I, Kondo T, Duboule D. An enhancer-titration effect induces digit-specific regulatory alleles of the HoxD cluster. Dev Biol. 2003;256(2):212–220. doi: 10.1016/s0012-1606(02)00136-7. [DOI] [PubMed] [Google Scholar]
- 29.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38(11):1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38(11):1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 31.De Gobbi M, et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312(5777):1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 32.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18(1):56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 33.Benko S, et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41(3):359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 34.Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133(4):761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 35.Kokubu C, et al. A transposon-based chromosomal engineering method to survey a large cis-regulatory landscape in mice. Nat Genet. 2009;41(8):946–952. doi: 10.1038/ng.397. [DOI] [PubMed] [Google Scholar]
- 36.Navratilova P, et al. Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev Biol. 2009;327(2):526–540. doi: 10.1016/j.ydbio.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 37.Tena JJ, et al. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nat Commun. 2011;2:310. doi: 10.1038/ncomms1301. [DOI] [PubMed] [Google Scholar]
- 38.Sagai T, et al. A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development. 2009;136(10):1665–1674. doi: 10.1242/dev.032714. [DOI] [PubMed] [Google Scholar]
- 39.Ovcharenko I, et al. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15(1):137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302(5644):413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 41.Tarchini B, Huynh TH, Cox GA, Duboule D. HoxD cluster scanning deletions identify multiple defects leading to paralysis in the mouse mutant Ironside. Genes Dev. 2005;19(23):2862–2876. doi: 10.1101/gad.351105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gimond C, et al. Cre-loxP-mediated inactivation of the alpha6A integrin splice variant in vivo: Evidence for a specific functional role of alpha6A in lymphocyte migration but not in heart development. J Cell Biol. 1998;143(1):253–266. doi: 10.1083/jcb.143.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruf S, et al. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet. 2011;43(4):379–386. doi: 10.1038/ng.790. [DOI] [PubMed] [Google Scholar]
- 44.Shah M, et al. A role for ATF2 in regulating MITF and melanoma development. PLoS Genet. 2010;6(12):e1001258. doi: 10.1371/journal.pgen.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32(3):199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- 46.van der Hoeven F, Zákány J, Duboule D. Gene transpositions in the HoxD complex reveal a hierarchy of regulatory controls. Cell. 1996;85(7):1025–1035. doi: 10.1016/s0092-8674(00)81303-3. [DOI] [PubMed] [Google Scholar]
- 47.Dollé P, Izpisúa-Belmonte JC, Boncinelli E, Duboule D. The Hox-4.8 gene is localized at the 5′ extremity of the Hox-4 complex and is expressed in the most posterior parts of the body during development. Mech Dev. 1991;36(1–2):3–13. doi: 10.1016/0925-4773(91)90067-g. [DOI] [PubMed] [Google Scholar]
- 48.Montavon T, Le Garrec JF, Kerszberg M, Duboule D. Modeling Hox gene regulation in digits: Reverse collinearity and the molecular origin of thumbness. Genes Dev. 2008;22(3):346–359. doi: 10.1101/gad.1631708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simonis M, Kooren J, de Laat W. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods. 2007;4(11):895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 50.Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324(5932):1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.