Abstract
Circadian clocks provide a temporal structure to processes from gene expression to behavior in organisms from all phyla. Most clocks are synchronized to the environment by alternations of light and dark. However, many organisms experience only muted daily environmental cycles due to their lightless spatial niches (e.g., caves or soil). This has led to speculation that they may dispense with the daily clock. However, recent reports contradict this notion, showing various behavioral and molecular rhythms in Caenorhabditis elegans and in blind cave fish. Based on the ecology of nematodes, we applied low-amplitude temperature cycles to synchronize populations of animals through development. This entrainment regime reveals rhythms on multiple levels: in olfactory cued behavior, in RNA and protein abundance, and in the oxidation state of a broadly conserved peroxiredoxin protein. Our work links the nematode clock with that of other clock model systems; it also emphasizes the importance of daily rhythms in sensory functions that are likely to impact on organism fitness and population structure.
Keywords: chronobiology, circadian rhythm, lin-42, 1-octanol, G protein-coupled receptor kinase
Daily cycles of light and dark are at once highly predictable and extremely stressful to living organisms. Life has thus evolved numerous strategies to use the physical properties of light—for example, to maintain a distinct orientation in space (vision) and time (the circadian clock). Circadian biological clocks regulate processes from gene expression to behavior via a complex network—from molecule to cell to organ to organism to population—that requires exquisite coordination between all levels such that events occur at an optimal time (1–3). At the most basic, molecular level, many components that regulate daily timing are known (so-called clock genes), functioning as a transcriptional negative feedback loop. As more components are identified, forming additional and interlocked regulatory loops, the model of the molecular clock has become one of a complex molecular network (4, 5). In theoretical terms, the molecular network could be viewed as similar to the network that is formed by cells and organs with respect to their daily oscillations (3). We remain ill-equipped to probe the system as a network—whether on the molecular or cellular levels—although this is clearly essential for our understanding of circadian clock properties. To do so, we would benefit from using the simplest animal model genetic organism, Caenorhabditis elegans, whose developmental timing and cell connectivity are known in detail.
Previously, protocols used for clock research on the nematode, C. elegans (6–12), have shown circadian rhythms in behaviors (e.g., locomotor activity, defecation, and pharyngeal pumping rate), metabolism (e.g., resistance to osmotic stress), or the expression of hundreds of genes without a connection to rhythmic behaviors. Clearly, taken together, one would conclude that the nematode has a circadian system. How it relates to the system of other animals is not clear, however, and it is via a comparative approach that clocks research has made important advances. Our aim here is to investigate how or if the clock in the worm shares features with clocks in other animals.
As a starting point, we note that although the strongest zeitgeber for the circadian clock is usually light (13), likely due to its predictability from day to day and year to year, C. elegans is soil-dwelling and not generally exposed to light. Organisms, like the nematode, that evolved within spatial niches devoid of light often lack energetically costly and complex light-shielding or -detecting mechanisms such as pigmentation or eyes (14). How would a dark clock synchronize with the natural environment? Many clocks use nonphotic signals, such as temperature, that oscillate reliably each 24 h (15–17) as a consequence of the light cycle.
We have thus taken an ecological approach, invoking cyclic conditions similar to those found in soil (18). Specifically, we imposed low-amplitude temperature cycles in darkness on nematodes as they proceeded through development from egg to adult. With this protocol, we found rhythms in a previously reported clock-regulated RNA and in the oxidation state of peroxiredoxin (PRX), a widely conserved molecular marker of circadian rhythms, confirming our experimental design. Further, we show daily oscillations in a behavior, olfaction, under entrainment as well as in constant conditions. Olfaction is also clock-regulated in insects and mammals (19–21). Finally, we find rhythms in the amount of a key protein kinase that is involved in regulating olfaction in C. elegans, Drosophila, and mammals. This protein is rhythmic also in Drosophila, thus extending our observations of functional and molecular components that oscillate according to time of day in C. elegans as in other animals. The circadian program in the nematode operates despite the lack of clock gene orthologs that function as predicted.
Results
To discern daily oscillations in a population, it is critical that the individuals are synchronized relative to one another. If not, rhythmicity is not obvious due to an averaging effect. Concerning daily timing, synchronization is accomplished through a process called circadian entrainment (22, 23), whereby zeitgebers are used by the clock for information on time of day. Cultivation protocols were therefore designed to mimic regular, daily, zeitgeber cycles that would occur in soil. Temperature cycles were thus structured within a 24 h period, oscillating between 13 °C and 16 °C. Under these conditions, development from egg to adult takes 5 d (Fig. 1A and Fig. S1). As day 1 adults, the animals were either kept in the temperature cycle, as if in an entrainment situation, or they were released to constant conditions (as shown in Fig. 1A, starting at 120 h) to reveal the circadian property called free-running rhythm, whereby daily oscillations persist in the absence of, for example, light or temperature cycles (24).
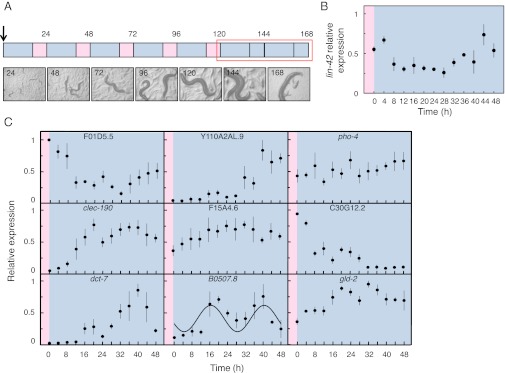
Fig. 1.
A protocol for supporting circadian rhythms in C. elegans. (A) Egg preps were inoculated onto a lawn of E. coli (arrow). The plates were placed in incubators that were programmed for temperature cycles of 16 h at 13 °C (shown in blue) and 8 h at 16 °C (shown in pink). On the sixth day, the plates were released to constant temperature (13 °C), unless otherwise indicated, and the experiments were started. Time point 120 h corresponds to the starting point for all of the experiments (time = 0 h). The pictures show the developmental stage of C. elegans at the end of each 24 h cycle. (B) The mRNA of the putative clock gene ortholog, lin-42, is not rhythmic in our cultivation protocol. lin-42 mRNA levels were measured by quantitative RT-PCR, and the results were normalized to the amount of act-4 mRNA and then to the sample with the highest expression level. The samples were collected for 48 h in constant conditions starting at 120 h after inoculation (red box in panel A). We express the timing of the harvesting protocol (x axis) as time from the end of the warm to cold transition at 120 h. The plot shows the average ± SEM of four biological replicates. (C) Quantitative RT-PCR was used to verify that the protocol supports circadian rhythms. The levels of mRNA corresponding to several transcripts, rhythmic under a different protocol (11), are shown. The plots show the mean ± SEM of three biological replicates. A sinusoidal curve was fitted to the data for B0507.8 using Circwave (P < 0.01). Additional statistical analysis with Circwave and JTK-Cycle is shown in Table S1. The results were normalized and x axes are labeled as for panel B.
We first tested if our protocol generated populations with synchronous, free-running circadian rhythms, by measuring RNA levels from nematodes harvested over 2 d after release into constant temperature (13 °C). There exists a putative clock gene ortholog in C. elegans, namely lin-42 (F47F6.1). To date, lin-42 mRNA has not been found to be expressed according to a circadian rhythm; rather, its expression correlates with developmental stages such as larval transitions (25, 26). In agreement with earlier work, we also found that this RNA species is expressed constitutively in adults, even after our synchronization protocol (Fig. 1B and Fig. S1). We then investigated expression of transcripts that had been reported as rhythmic in protocols that used higher amplitude temperature cycles (11). Although the expression of many of these genes was dynamic (decreasing or increasing on release to constant conditions), only one of nine (B0507.8) (11) was significantly rhythmic (Fig. 1C and Table S1), suggesting that this low-amplitude temperature cycle effectively entrains populations of C. elegans and that they have a free-running, circadian rhythm at the level of RNA transcription.
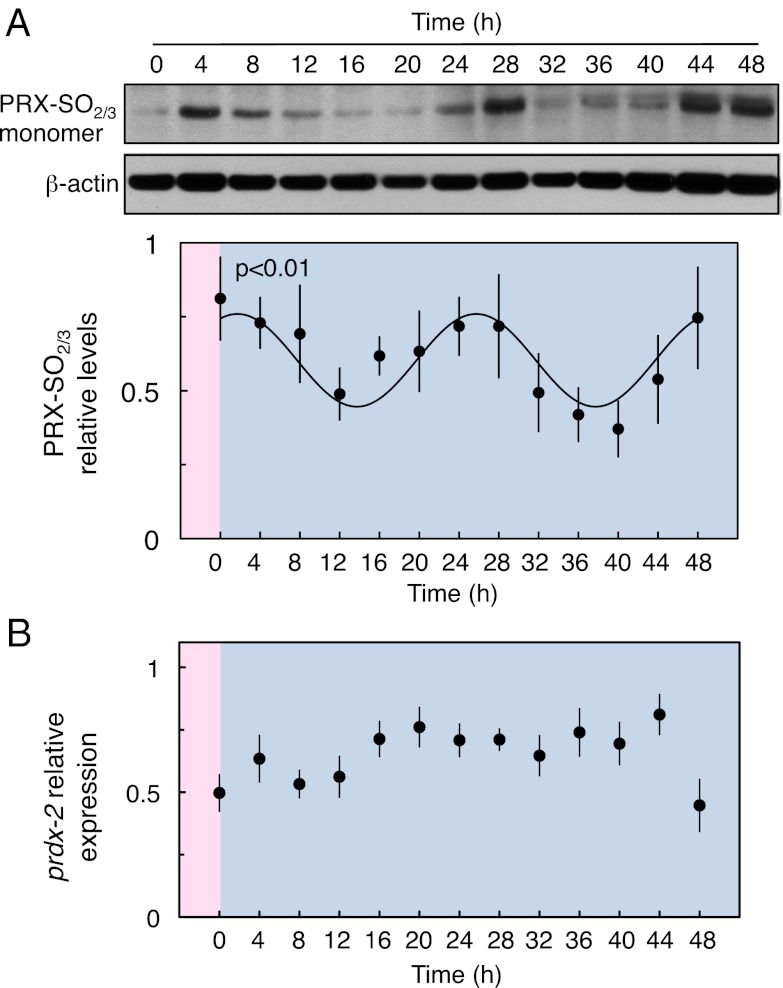
To extend our repertoire of rhythmic outputs, we asked if the redox state of PRX follows a circadian cycle. Recent reports show this protein, involved in hydrogen peroxide metabolism, oscillating between reduced and oxidized forms in organisms from all phyla, even in nonnucleated cells (27, 28). C. elegans expresses at least two typical 2-Cys PRX genes, prdx-2 (F09E5.15) and prdx-3 (R07E5.2). The PRX active site is highly conserved in these proteins (Fig. S2), and an antiserum raised against the oxidized peptide DFTFVC*PTEI detects predominantly PRDX-2 (Fig. S3). Protein extracts from animals released into constant conditions from temperature cycles showed statistically significant rhythms in this form of PRDX-2 (Fig. 2A and Table S1). This rhythm represents a phylogenetically conserved molecular marker of the circadian clock in C. elegans. Interestingly, the expression of prdx-2 mRNA is not clock regulated (Fig. 2B).
Fig. 2.
Oxidation state of PRX in constant conditions in C. elegans. (A) Representative immunoblot probed for over-/hyper-oxidized 2-Cys PRX (PRX-SO2/3). After entrainment to a temperature cycle (16 h at 13 °C and 8 h at 16 °C, darkness), the oxidation of PRDX-2 shows a circadian rhythm in free-running conditions (13 °C, darkness). Time point 0 h corresponds to the release from warm to cold. Loading control shows an image of the same blot probed for β-actin. For each time series, the data were quantified by densitometry and normalized to the highest value within each blot. The x axes are labeled as described in Fig. 1B. The mean ± SEM for three biological replicates is shown. A sinusoidal curve was fitted to the data using Circwave (P < 0.01). Additional statistical analysis with Circwave and JTK-Cycle is shown in Table S1. (B) prdx-2 mRNA levels are not rhythmic under the same protocol used for protein quantification. The expression of prdx-2 was measured by quantitative RT-PCR. The results were normalized to the amount of act-4 mRNA and then to the sample with the highest expression level. The x axes are labeled as described in Fig. 1B. The plot shows the mean ± SEM of three biological replicates.
Given that we find daily rhythms in RNA and metabolism, we wanted to know if the clock is regulating quantifiable behaviors. In seemingly all circadian systems, sensory functions are clock-regulated. Olfaction, for example, is regulated by the circadian clock in many animals (19–21, 29). Indeed, it is one of the most robust behaviors described in this nematode (30–33) and is a key component of fitness, as it guides between nutritious and pathogenic food choices (34). Olfaction is typically assessed in C. elegans with a chemotaxis assay, whereby the animals are harvested from their culture plates and washed several times to remove the interfering effects of Escherichia coli. We developed a unique, quantitative, in situ assay for olfaction so that we could measure olfaction without perturbing the animals, thus avoiding possible masking (35) that might occur due to the washing step. Eggs were inoculated on a small drop of food (Fig. S4A), and as described above, the developing animals were submitted to temperature cycles for 5 d (Fig. 1A). The olfaction assay was initiated by applying a drop of 1-octanol (a chemorepellant for C. elegans) to one side of the food source. This odorant was chosen because it was the only compound for which chemotaxis could be demonstrated in the presence of E. coli among many odorants that were tested (Table S2). At the end of the chemotaxis assay, a picture was taken showing the animals as they were distributed over the lawn of bacteria. Each picture was divided into a proximal and a distal half, and nematodes were counted to yield a Chemotaxis Index (CI; Fig. S4A). The in situ assay was optimized for quantitative studies by comparing a dilution series of the odorant over assay end points from 5 to 60 min (Fig. S4B). We determined that an end point of 15 min in combination with a 1/27 dilution of 1-octanol is optimal.
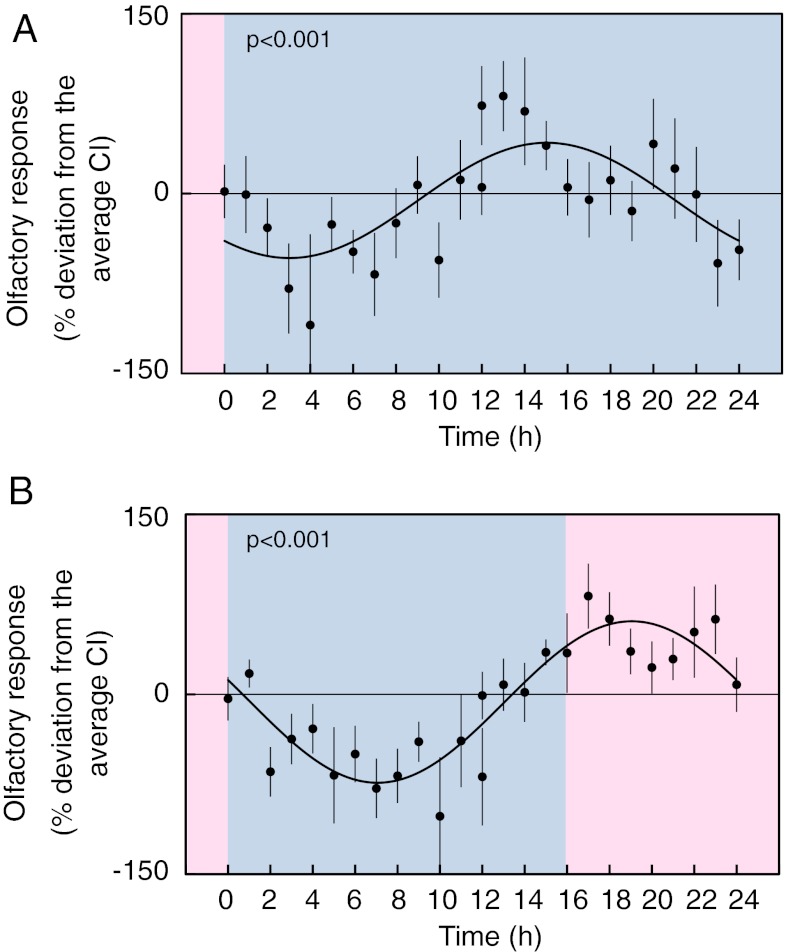
We applied this method to assay the aversive response to the chemical 1-octanol over the course of a day at constant temperature (Fig. 3A). The animals showed lower responses in the subjective night compared with the subjective day, yielding a fitted sine curve with high significance (P < 0.001). Interestingly, when the animals were assayed for chemotaxis under cycling temperature conditions, although they showed a similar pattern of increased responsiveness during the day, this increase in olfaction was extended longer and later than in constant conditions (Fig. 3B). This suggests that the increase in temperature itself induces a masking effect or possibly that the chemical is more volatile at the higher temperature. Importantly, the chemotaxis increased before the onset of warm temperature, showing anticipation of the zeitgeber transition, a feature that is expected under appropriately structured circadian entrainment conditions.
Fig. 3.
Circadian modulation of olfaction in response to 1-octanol. (A) Response to 1-octanol over 24 h in constant conditions. Blue panels represent cool temperature (13 °C); pink panels represent warm temperature (16 °C). After 5 d of growth in a temperature cycle (13 °C to 16 °C), the response to 1-octanol was measured for 24 h at constant temperature, 13 °C. The data are plotted as the percentage deviation from the average CI of the experiment. The x axes are labeled as described in Fig. 1B. The values for each time point are shown as the mean ± SEM calculated from 5 to 10 plates in three independent experiments. The average CIs for the three experiments are 0.13, 0.31, and 0.49. A sinusoidal curve was fitted to the data (P < 0.001) using Circwave. (B) Response to 1-octanol over 24 h in entraining conditions (a temperature cycle). Blue panels represent cool temperature (13 °C); pink panels represent warm temperature (16 °C). After 5 d of growth in a temperature cycle, chemotaxis was measured over 24 h at the same cycling conditions. The data are plotted as the percentage deviation from the average CI of the experiment. The x axes are labeled as described in Fig. 1B. The values for each time point are shown as the mean ± SEM calculated from 5–10 plates in three independent experiments. The average CIs for the three experiments are 0.19, 0.40, and 0.59. For a plot of the CI before normalization, see Fig. S5.
Another feature of circadian clocks is how they behave in non-24-h entraining cycles. For instance, in cycles that are slightly shorter than 24 h, a circadian program would be expected to entrain later, whereas a masked or driven rhythm should show similar phase reference points relative to zeitgeber transitions. The imposition of a 23 h temperature cycle on the nematodes perturbs the entrainment such that—on release to constant conditions—a later phase of 1-octanol olfaction is observed on the first day in constant conditions (Fig. S6). The data from entrainment (in two different cycle lengths) and release to a free run suggest the presence of an underlying circadian oscillator regulating olfaction.
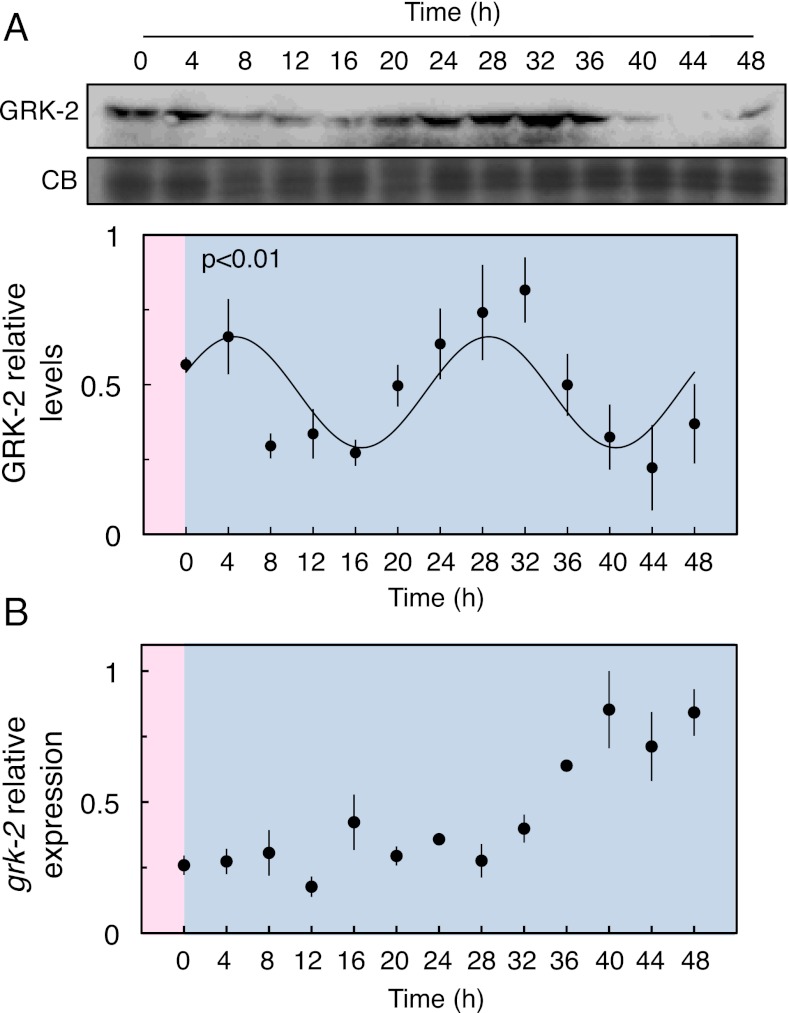
Much is known concerning the molecular aspects of olfaction. For instance, odorant receptors are a large family of G protein-coupled receptors (GPCRs) (36) [or highly homologous to these (37)]. GPCRs, in turn, are often regulated by G protein-coupled receptor kinases (GRKs) and arrestins that facilitate desensitization of these molecules in mammals (38). The genome of C. elegans contains two predicted GRKs, grk-1 (F19C6.1) and grk-2 (W02B3.2). GRK-2 regulates the response to 1-octanol in C. elegans, most likely through the phosphorylation of the correspondent GPCR (39), although the gene encoding the receptor for 1-octanol in C. elegans remains unknown. Furthermore, nematodes lacking GRK-2 function show disrupted chemosensation, including responses to 1-octanol (40) due to the activation of inhibitory pathways that dampen signaling in the absence of GRK-2 (41). 1-octanol is detected by the ASH neurons when animals are on food (42) and transgenic expression of grk-2 in these cells in mutant animals restores ASH-mediated aversive behavior (40). In Drosophila, the circadian rhythm in GPRK-2 abundance produces a rhythmic accumulation of odorant receptors that results in a corresponding circadian modulation of olfactory responses (21). We therefore focused on the circadian regulation of grk-2 in C. elegans. We measured GRK-2 protein levels from animals grown in temperature cycles and subsequently harvested over 2 d at constant temperature (13 °C). The abundance of GRK-2 shows a circadian pattern with a peak at the beginning of the subjective night (Fig. 4A and Table S1). Interestingly, GRK-2 protein abundance and the olfactory response to 1-octanol are in antiphase, supporting the idea that GRK-2 acts as a negative regulator of the response to 1-octanol. The mRNA for grk-2 does not show circadian regulation (Fig. 4B), demonstrating posttranscriptional regulation of protein abundance by the circadian clock in C. elegans. Oscillations in GRK-2 are consistent with a model in which rhythmic modulation of GRK-2 abundance results in circadian control of olfactory responses, as has been proposed for Drosophila (21).
Fig. 4.
GRK-2 protein and mRNA levels in constant conditions in C. elegans. (A) A representative immunoblot probed for GRK-2. After entrainment to a temperature cycle, the levels of GRK-2 protein show a circadian rhythm in free-running conditions (13 °C, darkness). The x axes are labeled as described for Fig. 1B. For each time series, the data were quantified by densitometry and normalized to the highest value. The mean ± SEM of four biological replicates is shown. To test for rhythmicity of the time series, a sinusoidal curve was fitted to the data using Circwave (P < 0.01). Statistic analysis with Circwave and JTK-Cycle is shown in Table S1. (B) grk-2 mRNA levels are not rhythmic under the same protocol used for protein quantification. The expression of grk-2 was measured by quantitative RT-PCR. The results were normalized to the amount of act-4 mRNA and then to the sample with the highest expression level. The x axes are labeled as described in Fig. 1B. The plot shows the mean ± SEM of three biological replicates.
Discussion
Circadian clocks confer a fitness advantage in the cycling light environment (43), presumably via the temporal structure that they create for processes ranging from gene expression to complex behavior such as sleep. The adaptive advantage of the clock in environments with neither light nor high-amplitude environmental cycles (such as in soil) is less obvious. Despite this, daily rhythms in blind cavefish were recently reported (44). The circadian system of the cavefish Phreatichthys andruzzii is not entrained by light/dark cycles but rather to feeding time (once per 24 h), showing a characteristic increase in activity in anticipation of food. In the context of such a feeding protocol, rhythms in gene expression were also observed. In constant conditions, gene expression of this light-blind oscillator shows a very long free-running rhythm (47 h).
Over the last decade, there have been numerous reports of daily rhythms in the soil-dwelling nematode, C. elegans (6–12), although the connection between molecules and behavior has been lacking. Reasons for this may include a lack of known clock gene homologs as starting points, a number of low-amplitude behavioral rhythms, and the use of conditions that are far from ecological. We noted that although C. elegans have no eyes, they retain some capability to sense light acutely (45–47). However, because they also have no pigmentation, we adopted the hypothesis that they do not regularly encounter light and hence would not use light to entrain their circadian clock. Here, we have developed a protocol using temperature as a zeitgeber that entrains the clock through development. Evidence of this synchronization is shown here as rhythms in molecules and behavior upon release to constant conditions. In contrast to the cavefish (44), the period of the nematode circadian rhythm is close to 24 h (Figs. 1C, 2A, 3A, and 4A and Table S1).
As has been reported for other animals, we found daily oscillations in olfaction (19, 29, 40, 48, 49), suggesting that this sensory function has an important status in the circadian system. Regulation of olfaction by the circadian clock in nematodes has far-reaching and ecologically important functions and is an indication of the adaptive nature of the clock in a dark-living animal, as they use olfaction to find appropriate food sources (34) and thus to orient themselves spatially in their environment. Like other animals, they use olfaction to find sexual partners (50). In contrast to others, however, they cannot compare olfactory cues with visual ones. We presume therefore that they rely almost exclusively on olfaction for these behaviors, and the daily rhythms therein are significant. For instance, C. elegans are usually hermaphrodites, with males occurring rarely; thus, the clock should impact successful mating in nature, as this is initiated based on olfactory cues. Obviously, genetic diversity in a population will derive mainly from sexual reproduction, and our experiments predict that it would be easier for males to find hermaphrodites at certain times of day.
Daily oscillations in the accumulation of RNA suggest that circadian regulation in C. elegans will show similarities to other clock model systems, insofar as the clock regulates countless cellular subsystems via transcription. The lack of rhythmicity of the clock gene ortholog, lin-42, shown here as well as by others (11, 26), is thus puzzling. However, even in metazoans that are more closely related, such as Drosophila and mice, orthologous “clock genes” can be regulated distinctly, with the proteins using alternative heterodimerization partners, for instance (51). Therefore, C. elegans may use clock gene orthologs posttranscriptionally (without relying on transcriptional regulation), or they may have a unique set of transcription factors that are involved in this level of the molecular mechanism. Importantly, at the level of metabolism (redox state of PDRX-2), behavior (olfaction), and posttranscriptional levels of a protein (GRK-2), we find circadian rhythms in components or functions that oscillate in other clock model systems. We conclude that the nematode shares clock mechanisms at numerous levels with other life forms.
This work demonstrates that life in the dark is clearly not a life without relevant and effective temporal cues (zeitgebers). The pervasive effect of the sun permeates into dark compartments—it may do so in soil, caves, or the ocean via temperature (higher during day, lower at night), food sources (strata of biota varying according to depth), or moisture (more at night, less in the day). These nonphotic zeitgebers will be somewhat less precise than the perfect predictability of sunlight, but they are nevertheless reliable markers of daily transitions. The primary use of nonphotic zeitgebers could support evolution of biological clocks that use molecular processes differently relative to the light-regulated clocks that are typical of most clock model organisms. Thus, in addition to serving as a powerful tool for analysis of a simple clock network, the nematode may provide tools to study posttranscriptionally regulated clock mechanisms and components.
Materials and Methods
Strains and Experiemental Conditions.
The N2 wild-type strain of C. elegans was used for all circadian experiments. The mutant strains VC289 and VC1151 (produced by the C. elegans Gene Knockout Consortium; www.celeganskoconsortium.omrf.org) were used to test the specificity of PRX-SO2/3 antibody. For stock animals, strains were cultured according to standard methods (52) and were routinely maintained at 18 °C on nematode growth medium (NGM) plates with a lawn of E. coli (OP50). For all experiments, the worms were grown to the stage of gravid adults, and eggs were prepared by the bleaching method. To start all of the experiments, eggs were placed in E. coli, and the plates were incubated in temperature cycles (16 h at 13 °C and 8 h at 16 °C). The same procedure was repeated after 12 h to create cultures in antiphase. This strategy allows collection of time points covering 24 h while harvesting over 12 h. After five cycles, the experiments were performed for 1 or 2 d in constant conditions (except when otherwise indicated). All of the manipulations with live animals in the “dark” were performed under a red safe light.
Quantitative Real-Time PCR (qPCR)
Nematodes were collected from the plates every 4 h for 2 d in constant conditions (13 °C, darkness) using cold M9 buffer, centrifuged, and washed one time to remove bacteria. Total RNA was isolated using TRIzol (Invitrogen), and cDNA synthesis was performed (30 min at 48 °C) using Random Hexamers and MultiScribe Reverse Transcriptase (Applied Biosystems). Quantitative PCR analyses (SYBR Green PCR Master Mix, Applied Biosystems) were performed using a 7500 Real Time PCR System thermal cycler (Applied Biosystems). The reaction included denaturation (10 min at 95 °C) and 40 PCR cycles (15 s at 95 °C and 1 min at 60 °C). The results for each gene were normalized to the corresponding results obtained with act-4 by the ΔCt method and then to the sample with the highest RNA level. Primers were designed using Primer Express Software (Applied Biosystems) (Table S3). Circadian rhythmicity was evaluated for each gene, and a sinusoidal curve was fitted to the data for B0507.8 using Circwave (by R. Hut, available at www.euclock.org).
In Situ Chemotaxis Assay.
For the 1-octanol assay, NGM plates were inoculated with 15 μL of a concentrated E. coli culture (100 g wet weight/L) and allowed to dry overnight at room temperature. To start the experiment, about 100 eggs were placed in the E. coli drop, and the worms were grown for 5 d in temperature cycles (13 °C to 16 °C; as in Fig. 1A) in constant dim light. The chemotaxis assay was performed either during the sixth day in a temperature cycle or on the sixth day following release to constant temperature. Every hour, chemotaxis assays were initiated on naïve plates by placing 1 μL of 1-octanol [3.7% (vol/vol) in ethanol] at a distance of 0.8 cm from the center of the E. coli drop. The plates were incubated for 15 min, at which point a picture was taken using a light microscope (10×, Stereo Discovery V8, Zeiss). The CI was calculated for each time point and each plate (Fig. S4). A sinusoidal curve was fitted to the data using Circwave (by R. Hut, available at www.euclock.org).
Protein Preparation.
Nematodes were collected using cold M9 buffer, centrifuged, and washed one time to remove bacteria. Proteins were extracted in Nonidet P-40 protein lysis buffer (150 mM NaCl, 1% Nonidet P-40, 50 mM Tris pH 8.0) containing protease inhibitors (Complete Mini, Roche) and a mixture of phosphatase inhibitors (PhosSTOP, Roche).
PRX Gel Electrophoresis and Western Blotting.
We used NuPAGE Novex 4–12% Bis-Tris gradient gels (Life Technologies) and ran them using the manufacturer’s protocol with a nonreducing Mes SDS buffer system. Protein transfer to nitrocellulose for blotting was performed using the iBlot system (Life Technologies), with a standard (P3, 7 min) protocol. Nitrocellulose was then washed briefly and then blocked for 30 min in 0.5% wt/wt BSA/nonfat dried milk (Marvel) in Tris buffered saline/0.05% Tween-20 (TBST). After three brief washes in TBST, membranes were incubated in antibody diluted in blocking buffer (0.5% milk/BSA) overnight at 4 °C. The following day, membranes were washed for 5 min three times (in TBST) and then incubated with 1:10,000 HRP-conjugated secondary antibody (Sigma-Aldrich) for 30 min. Four additional 10 min washes were performed before chemiluminescence detection using Immobilon reagent (Millipore) or ECL Plus reagent (GE Healthcare). To check for even protein loading, blots were probed for β-actin (Santa Cruz Antibodies, sc-47778), used at 1:5,000 in 0.5% milk/BSA. Antiserum against PRX-SO2/3 (Abcam, ab16830) was used at 1:10,000 in blocking buffer.
GRK-2 Gel Electrophoresis and Western Blotting.
Total protein (50 μg per lane) was subjected to SDS/PAGE on 7.5% gels and transferred to nitrocellulose using a Mini Trans-Blot Cell (Bio-Rad). Nitrocellulose was washed briefly, blocked for 1 h in 5% nonfat dried milk in TBST, and then incubated in 1:1,000 anti-GRK2/3 antibody (Upstate Biotechnology, clone C5/1.1) diluted in blocking buffer (5% milk in TBST) overnight at 4 °C. The following day, membranes were washed three times in TBST for 15 min for each wash and then incubated with 1:5,000 HRP-conjugated anti-mouse antibody (Bio-Rad) for 1 h. After three 15 min washes with TBST, chemiluminescence detection was performed using SuperSignal West Pico Reagent (Pierce). To check for even protein loading, gels were stained with Coomassie Blue.
Supplementary Material
Acknowledgments
We thank M. van der Pol and D. Allardyce for critical technical assistance; R. Schnabel, E. Nollen and J.-C. Billeter for comments on an earlier version of this manuscript; P. Hardin for helpful suggestions; T. Roenneberg (Ludwig-Maximilians-Universität-München), J. Hogenesch (University of Pennsylvania), and R. Hut (University of Groningen) for input on data analysis; and the Caenorhabditis Genetics Center supported by the National Institutes of Health for supplying animals. Our work is supported by the European Commission (Euclock, 6th Framework Programme Integrated Project), the Netherlands Organization for Scientific Research (Dutch Science Foundation, Vici, and Open Programme), and the Rosalind Franklin Fellowships of the University of Groningen. A.B.R. is supported by the Wellcome Trust (083643/Z/07/Z), the European Research Council (ERC) (Starting Grant 281348, MetaCLOCK), and European Molecular Biology Organization Young Investigators Programme, as well as the Medical Research Council Centre for Obesity and Related Metabolic Disorders (MRC CORD), and the National Institute of Medical Research Cambridge Biomedical Research Centre. J.S.O. is supported by the Wellcome Trust (093734/Z/10/Z).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211705109/-/DCSupplemental.
References
- 1.McWatters HG, Devlin PF. Timing in plants—A rhythmic arrangement. FEBS Lett. 2011;585(10):1474–1484. doi: 10.1016/j.febslet.2011.03.051. [DOI] [PubMed] [Google Scholar]
- 2.Ripperger JA, Jud C, Albrecht U. The daily rhythm of mice. FEBS Lett. 2011;585(10):1384–1392. doi: 10.1016/j.febslet.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 3.Merrow M, Spoelstra K, Roenneberg T. The circadian cycle: Daily rhythms from behaviour to genes. EMBO Rep. 2005;6(10):930–935. doi: 10.1038/sj.embor.7400541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baggs JE, et al. Network features of the mammalian circadian clock. PLoS Biol. 2009;7(3):e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roenneberg T, Merrow M. The network of time: Understanding the molecular circadian system. Curr Biol. 2003;13(5):R198–R207. doi: 10.1016/s0960-9822(03)00124-6. [DOI] [PubMed] [Google Scholar]
- 6.Migliori ML, Simonetta SH, Romanowski A, Golombek DA. Circadian rhythms in metabolic variables in Caenorhabditis elegans. Physiol Behav. 2011;103(3–4):315–320. doi: 10.1016/j.physbeh.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 7.Simonetta SH, Migliori ML, Romanowski A, Golombek DA. Timing of locomotor activity circadian rhythms in Caenorhabditis elegans. PLoS ONE. 2009;4(10):e7571. doi: 10.1371/journal.pone.0007571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonetta SH, Romanowski A, Minniti AN, Inestrosa NC, Golombek DA. Circadian stress tolerance in adult Caenorhabditis elegans. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2008;194(9):821–828. doi: 10.1007/s00359-008-0353-z. [DOI] [PubMed] [Google Scholar]
- 9.Saigusa T, et al. Circadian behavioural rhythm in Caenorhabditis elegans. Curr Biol. 2002;12(2):R46–R47. doi: 10.1016/s0960-9822(01)00669-8. [DOI] [PubMed] [Google Scholar]
- 10.Kippert F, Saunders DS, Blaxter ML. Caenorhabditis elegans has a circadian clock. Curr Biol. 2002;12(2):R47–R49. doi: 10.1016/s0960-9822(01)00670-4. [DOI] [PubMed] [Google Scholar]
- 11.van der Linden AM, et al. Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 2010;8(10):e1000503. doi: 10.1371/journal.pbio.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Migliori ML, et al. Daily variation in melatonin synthesis and arylalkylamine N-acetyltransferase activity in the nematode Caenorhabditis elegans. J Pineal Res. 2012;53(1):38–46. doi: 10.1111/j.1600-079X.2011.00969.x. [DOI] [PubMed] [Google Scholar]
- 13.Roenneberg T, Foster RG. Twilight times: Light and the circadian system. Photochem Photobiol. 1997;66(5):549–561. doi: 10.1111/j.1751-1097.1997.tb03188.x. [DOI] [PubMed] [Google Scholar]
- 14.Jeffery WR. Regressive evolution in Astyanax cavefish. Annu Rev Genet. 2009;43:25–47. doi: 10.1146/annurev-genet-102108-134216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12(18):1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 16.Merrow M, Brunner M, Roenneberg T. Assignment of circadian function for the Neurospora clock gene frequency. Nature. 1999;399(6736):584–586. doi: 10.1038/21190. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann K. Zur Beziehung zwischen phasenlage und spontanfrequenz bei der endogenen tagesperiodik [On the relationship between phase angle and free running period of the endogenous circadian rhythm] Z Naturforschg. 1963;18(b):154–157. German. [Google Scholar]
- 18.Robinson AF. Movement of five nematode species through sand subjected to natural temperature gradient fluctuations. J Nematol. 1994;26(1):46–58. [PMC free article] [PubMed] [Google Scholar]
- 19.Granados-Fuentes D, Tseng A, Herzog ED. A circadian clock in the olfactory bulb controls olfactory responsivity. J Neurosci. 2006;26(47):12219–12225. doi: 10.1523/JNEUROSCI.3445-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granados-Fuentes D, et al. Daily rhythms in olfactory discrimination depend on clock genes but not the suprachiasmatic nucleus. J Biol Rhythms. 2011;26(6):552–560. doi: 10.1177/0748730411420247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanoue S, Krishnan P, Chatterjee A, Hardin PE. G protein-coupled receptor kinase 2 is required for rhythmic olfactory responses in Drosophila. Curr Biol. 2008;18(11):787–794. doi: 10.1016/j.cub.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roenneberg T, Merrow M. Entrainment of the human circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:293–299. doi: 10.1101/sqb.2007.72.043. [DOI] [PubMed] [Google Scholar]
- 23.Johnson CH, Elliott JA, Foster R. Entrainment of circadian programs. Chronobiol Int. 2003;20(5):741–774. doi: 10.1081/cbi-120024211. [DOI] [PubMed] [Google Scholar]
- 24.Pittendrigh CS. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–184. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Temmerman L, et al. C. elegans homologs of insect clock proteins: A tale of many stories. Ann N Y Acad Sci. 2011;1220:137–148. doi: 10.1111/j.1749-6632.2010.05927.x. [DOI] [PubMed] [Google Scholar]
- 26.Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286(5442):1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469(7331):498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400(6742):375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- 30.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74(3):515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 31.Firestein S. Olfaction: Scents and sensibility. Curr Biol. 1996;6(6):666–667. doi: 10.1016/s0960-9822(09)00444-8. [DOI] [PubMed] [Google Scholar]
- 32.Nuttley WM, Atkinson-Leadbeater KP, Van Der Kooy D. Serotonin mediates food-odor associative learning in the nematode Caenorhabditiselegans. Proc Natl Acad Sci USA. 2002;99(19):12449–12454. doi: 10.1073/pnas.192101699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hart AC, Chao MY. In: From odors to behaviors in Caenorhabditis elegans. The Neurobiology of Olfaction (Frontiers in Neuroscience) Menini A, editor. Boca Raton, FL: CRC Press; 2010. pp. 1–33. [PubMed] [Google Scholar]
- 34.Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438(7065):179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- 35.Mrosovsky N, Thompson S. Negative and positive masking responses to light in retinal degenerate slow (rds/rds) mice during aging. Vision Res. 2008;48(10):1270–1273. doi: 10.1016/j.visres.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 36.Buck L, Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65(1):175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 37.Sato K, et al. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452(7190):1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 38.Reiter E, Lefkowitz RJ. GRKs and beta-arrestins: Roles in receptor silencing, trafficking and signaling. Trends Endocrinol Metab. 2006;17(4):159–165. doi: 10.1016/j.tem.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Wood JF, Wang J, Benovic JL, Ferkey DM. Structural domains required for Caenorhabditis elegans G protein-coupled receptor kinase 2 (GRK-2) function in vivo. J Biol Chem. 2012;287(16):12634–12644. doi: 10.1074/jbc.M111.336818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuto HS, et al. G protein-coupled receptor kinase function is essential for chemosensation in C. elegans. Neuron. 2004;42(4):581–593. doi: 10.1016/s0896-6273(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 41.Ezak MJ, Hong E, Chaparro-Garcia A, Ferkey DM. Caenorhabditis elegans TRPV channels function in a modality-specific pathway to regulate response to aberrant sensory signaling. Genetics. 2010;185(1):233–244. doi: 10.1534/genetics.110.115188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chao MY, Komatsu H, Fukuto HS, Dionne HM, Hart AC. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc Natl Acad Sci USA. 2004;101(43):15512–15517. doi: 10.1073/pnas.0403369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95(15):8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavallari N, et al. A blind circadian clock in cavefish reveals that opsins mediate peripheral clock photoreception. PLoS Biol. 2011;9(9):e1001142. doi: 10.1371/journal.pbio.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, et al. C. elegans phototransduction requires a G protein-dependent cGMP pathway and a taste receptor homolog. Nat Neurosci. 2010;13(6):715–722. doi: 10.1038/nn.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward A, Liu J, Feng Z, Xu XZ. Light-sensitive neurons and channels mediate phototaxis in C. elegans. Nat Neurosci. 2008;11(8):916–922. doi: 10.1038/nn.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards SL, et al. A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol. 2008;6(8):e198. doi: 10.1371/journal.pbio.0060198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Decker S, McConnaughey S, Page TL. Circadian regulation of insect olfactory learning. Proc Natl Acad Sci USA. 2007;104(40):15905–15910. doi: 10.1073/pnas.0702082104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rymer J, Bauernfeind AL, Brown S, Page TL. Circadian rhythms in the mating behavior of the cockroach, Leucophaea maderae. J Biol Rhythms. 2007;22(1):43–57. doi: 10.1177/0748730406295462. [DOI] [PubMed] [Google Scholar]
- 50.Edison AS. Caenorhabditis elegans pheromones regulate multiple complex behaviors. Curr Opin Neurobiol. 2009;19(4):378–388. doi: 10.1016/j.conb.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2(9):702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 52.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.