Abstract
Distinct subsets of thymic epithelial cells (TECs) support T-cell development and selection. Isolated TECs contain multicellular complexes that enclose many viable thymocytes. However, the functions of those TECs, termed thymic nurse cells (TNCs), are unclear and the idea that TNCs are present in vivo is questioned. Here, we show that TNCs represent a fraction of cortical (c)TECs that are defined by the expression of thymoproteasomes. Intravital imaging revealed TNCs in the thymic cortex in situ, whereas TNCs were detected neither during embryogenesis nor in the postnatal thymuses of various “positive-selector” T-cell receptor (TCR)-transgenic mice, indicating that TNCs are not essential for T-cell differentiation, including positive selection. Rather, cells within TNCs were enriched for long-lived CD4+CD8+ thymocytes that underwent secondary TCR-Vα rearrangement. Thus, TNC complexes are formed in vivo by persistent cTEC–thymocyte interactions that then provide a microenvironment that optimizes T-cell selection through secondary TCR rearrangement.
Keywords: repertoire selection, thymic microenvironment, beta5t
Nurse cells were originally defined as cells that neighbor and nurture oocytes in invertebrates (1, 2). Sertoli cells in the mammalian testis are also called nurse cells because the cells provide nutrients to and protection for, as well as regulate, developing sperm cells (3, 4). Skeletal muscle cells can become nurse cells for Trichinella spiralis when the parasite induces surrounding muscle cells to provide a long-lasting niche that would supply nutrients and protect the parasite from the host’s immune system (5). In addition, a fraction of stromal cells in the bone marrow are sometimes called erythroblast nurse cells because they absorb erythroblast nuclei and assist their differentiation into red blood cells (6). Thus, nurse cells have been identified in various biological systems as cells that play an essential role in nurturing the survival and differentiation of neighboring cells or organisms.
The “thymic nurse cell” (TNC), which was first reported more than 30 y ago, is a large epithelial cell that completely envelops many viable lymphoid cells within its intracellular vesicles and is isolated by protease digestion of mouse and rat thymus tissues (7–10). TNC complexes are found not only in rodent but also in many vertebrate species, including human, bird, and fish (9, 11–13). It was hypothesized that TNCs provide a microenvironment that is necessary for lymphocyte proliferation and differentiation and that the intra-TNC differentiation is an essential step in intrathymic T-cell development (7, 8, 14, 15). It was further hypothesized that the TNC complex is a site for the positive and negative selection of T cells (16–18). However, how TNCs are involved in T-cell development, and selection has not been established (19). Whether TNCs are abundant in the thymic cortex (10) or derived from all parts of the thymus including the medulla (20) has not been clarified as well. It has even been questioned whether TNC complexes indeed represent the structures that are present in the thymus in vivo or are artificially generated during cell isolation procedures in vitro (10, 20).
In the present study, we examined the characteristics of thymic cortical epithelial cells (cTECs) that express the recently identified β5t. β5t is a cTEC-specific component of the thymoproteasome, which is essential for the positive selection of functionally competent CD8+ T cells (21–23). We found that in the postnatal mouse thymus, a majority of β5t-expressing cTECs, but not thymic medullary epithelial cells (mTECs), are tightly associated with thymocytes. Approximately 10% of β5t-expressing cTECs in the adult mouse thymus represent previously reported TNC complexes that completely enclose CD4+CD8+ cortical thymocytes. These cTEC–thymocyte complexes, including TNCs, are detected in the thymic cortex intravitally. Interestingly, we noted that TNC complexes appear late during ontogeny but are not detected in the adult thymus of various T-cell receptor (TCR)-transgenic mouse lines in that the majority of thymocytes can be positively selected (referred to as “positive-selector” TCR-transgenic mice), indicating that the formation of the TNC complex is not an absolute requirement for T-cell development or positive selection. Rather, our data show that the TNC complex represents a persistent interaction between adhesive cTECs and long-lived CD4+CD8+ thymocytes that undergo secondary TCRα rearrangement. Thus, this study reveals that TNCs represent a subpopulation of β5t+ cTECs that provide a microenvironment for the optimization of TCR selection by supporting the secondary TCR-Vα rearrangement in long-lived CD4+CD8+ thymocytes.
Results
Fraction of β5t-Expressing cTECs Form Multicellular TNC Complexes.
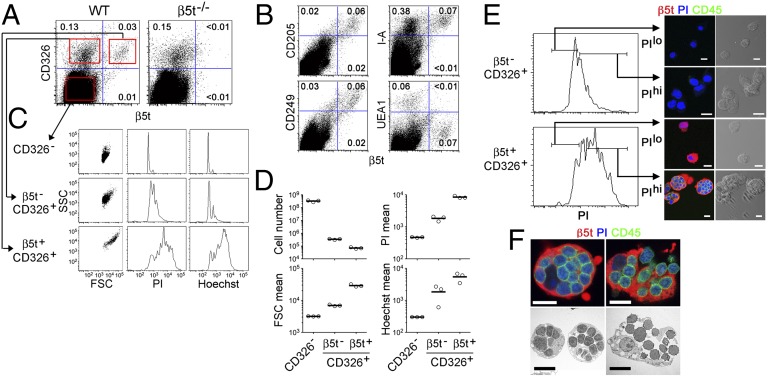
To analyze the characteristics of β5t-expressing cells in the thymus, unfractionated cell suspensions of collagenase-digested postnatal mouse thymus were stained for various cell surface molecules and for intracellular β5t. β5t was expressed in ∼0.03–0.06% of total unfractionated cells from the collagenase-digested postnatal mouse thymus (corresponding to ∼0.5–1.5 × 105 cells per mouse) (Fig. 1A). The detection was specific for β5t, as shown by the lack of detection in the thymuses of β5t-deficient mice (Fig. 1A). Most β5t+ cells were confined in CD326 (EpCAM)+ I-A class II MHC+ TECs and CD205+CD249 (Ly51)+ cTECs but not in Ulex europaeus agglutinin (UEA)-1+ mTECs (Fig. 1 A and B). Thus, β5t+ cells in the postnatal mouse thymus represent the majority of CD205+CD249+CD326+ cTECs, whereas most β5t−CD326+ TECs represent UEA-1+CD205−CD249− mTECs. In comparison with β5t−CD326+ mTECs and β5t−CD326− thymic cells (mostly thymocytes), β5t+ CD326+ cTECs were large, as defined by forward- and side-scatter intensities (Fig. 1 C and D). Unlike other thymic cells, most cTECs gave multinuclear signals, as measured by propidium iodide (PI) and Hoechst 33342 staining of cellular DNA (Fig. 1 C and D). Microscopic observation indicated that most PIhigh β5t+ CD326+ cTECs represented multicellular complexes between one β5t+ cTEC and many CD45+ thymocytes (Fig. 1 E and F). A fraction of β5t−CD326+ mTECs also exhibited multinuclear signals (Fig. 1 C and D) that mostly reflected the aggregates of CD45− stromal cells (Fig. 1E). Flow-cytometric measurements indicated that one β5t+ cTEC–thymocyte complex was associated with ∼15 thymocytes on average (Fig. 1 D and E). Confocal microscopy and transmission-electron microscopy indicated that the large cTEC–thymocyte complexes were heterogeneous; some complexes were β5t+ cTECs that completely enwrapped many CD45+ thymocytes (closed complexes), whereas other complexes represented an open association between β5t+ cTECs and multiple CD45+ thymocytes (open complexes) (Fig. 1F). Electron microscopy indicated that the closed complexes corresponded to previously described TNCs that completely enwrapped thymocytes (7, 8). The TEC–thymocyte complexes, including the TNC complexes, were detected specifically in β5t+ cTECs (Fig. 1E). These results indicate that the previously reported TNCs represent a fraction of β5t+ cTECs in the postnatal mouse thymus.
Fig. 1.
Association of β5t+ cTECs with CD45+ thymocytes. (A) Collagenase-digested cell suspensions of thymuses from wild-type (WT) (C57BL/6) mice and β5t−/− mice were analyzed by flow cytometry for intracellular β5t and cell surface CD326. Numbers indicate frequencies of cells within boxes. Representative profiles of three independent measurements are shown. (B) Flow-cytometric profiles of collagenase-digested cell suspension of WT thymus for intracellular β5t and indicated cell surface molecules. Representative profiles of three independent measurements are shown. (C) Thymic cell populations, gated as shown in red boxes in A, were analyzed for forward-scatter intensity (FSC), side scatter intensity (SSC), PI staining, and Hoechst 33342 (Hoechst) staining. All of the parameters are plotted in the logarithmic scale. Representative profiles of three independent measurements are shown. (D) Cell number per mouse and geometric mean values of FSC intensity, PI-staining intensity, and Hoechst-staining intensity of indicated cell populations. Open circles indicate values for individual mice and bars indicate means (n = 3). (E) β5t−CD326+ and β5t+ CD326+ thymic cells were sorted into PIlow or PIhigh cells, as defined in the left graphs. Sorted cells were further stained for CD45 and analyzed by confocal microscopy. Representative results of three-color fluorescence images and transmitted light images are shown (n = 2). (F) Isolated β5t+ CD326+ thymic cells further stained for CD45 and PI (Upper) were analyzed by transmission electron microscopy (Lower). Representative images of closed (Left) and open (Right) complexes are shown (confocal microscopic data from five independent experiments and electron microscopy data from two independent experiments). (Scale bars: 10 μm.)
A two-color staining profile for extracellular CD45 without saponin permeabilization (eCD45) and intracellular CD45 after saponin permeabilization (iCD45) revealed three populations of β5t+ CD326+ cTECs (Fig. S1A). Multicolor confocal microscopy of the three populations isolated by a cell sorter indicated that the eCD45high population (P1) represented the open association between cTECs and thymocytes, the eCD45low iCD45high population (P2) represented the closed TNC complexes between cTECs and thymocytes, and the eCD45low iCD45low population (P3) was associated with no or a few thymocytes (Fig. S1 A and B); these three populations comprised ∼70%, 10%, and 20% of β5t+ CD326+ cTECs, respectively (Fig. S1A). Thus, TNCs that form closed complexes with thymocytes, as defined in P2, comprise ∼10% of cTECs in the adult mouse thymus.
In the present study, cTECs that are openly associated with thymocytes in P1 are not defined as TNCs. However, the detection of open association between thymocytes and the majority of cTECs (P1) suggests that the depletion of CD45+ thymocytes during conventional procedures for the analysis and isolation of thymic stromal cells (24–26) would remove the majority of cTECs. Indeed, magnetic fractionation of collagenase-digested thymic cells for CD45− cells removed the majority of β5t+CD326+ cTECs (Fig. S1 C and D). The loss of cTECs was essentially attributable to the loss of P1 open complexes rather than P2 TNCs or P3 cells (Fig. S1 C and D). It is, thus, important to note that conventional methods for the preparation of CD45− thymic stromal cells deplete the majority of cTECs. It should also be noted that the conventional methods for the preparation of TNCs involving trypsin digestion of thymic cells and 1 × g sedimentation (7, 8) reasonably enriched P2 TNC complexes without significant loss (P > 0.05) (Fig. S1 A and C).
Intravital Imaging of cTEC–Thymocyte Complexes.
To further characterize the cTEC–thymocyte complexes, we next examined whether P1 open complexes and P2 TNC complexes were derived from cellular complexes that existed in the thymic cortex in vivo or were artificially formed in vitro during cell preparation after collagenase digestion, as previously suggested (10, 19, 20). Collagenase-digested thymic cells from C57BL/6 (B6) mice were mixed 1:1 with thymocytes prepared without enzyme digestion from B6.SJL-Ptprca (B6-Ly5.1) congenic mice (Fig. S2A). More than 95% of β5t+ cTECs in this mixture were derived from the collagenase-digested B6 thymus (Fig. S2A). P1 and P2 cTEC–thymocyte complexes were isolated by flow cytometry, and thymocytes associated with these complexes were released and further stained for CD45.1 and CD45.2, which specifically detected B6-Ly5.1 and B6 thymocytes, respectively. We found that the thymocytes isolated from P2 TNC complexes were exclusively derived from CD45.2+ B6 thymocytes (Fig. S2B). Further experiments of mixing intact thymic lobes from β5tVenus/+ B6 mice with those from B6-Ly5.1 mice before collagenase digestion demonstrated that most of the thymocytes contained within β5tVenus+ TNC complexes were CD45.2+ (Fig. S3). These results indicate that the P2 TNC complexes are not artificially formed during cell preparation before or after collagenase digestion but are derived from cellular complexes endogenously formed in the thymus. Indeed, P2 TNCs predominantly contained CD4+CD8+TCRlowCD69low thymocytes (Figs. S2B and S4), indicating that the P2 TNC complexes represent cellular complexes between β5t+ cTECs and CD4+CD8+ thymocytes formed within the thymic cortex in situ.
On the other hand, thymocytes released from P1 open cTEC–thymocyte complexes were a mixture of B6 and B6-Ly5.1 thymocytes (Fig. S2B and S3B), indicating that the P1 open complexes contain thymocytes that are artificially associated during cell preparation. Indeed, the P1 open complexes contained many CD4+CD8− and CD4−CD8+ medullary thymocytes (Fig. S2B and S4), in agreement with the possibility that cTECs isolated from the cortex are highly adhesive to lymphoid cells, including medulla-derived mature CD4+CD8− and CD4−CD8+ thymocytes and form P1 open complexes during cell preparation in vitro.
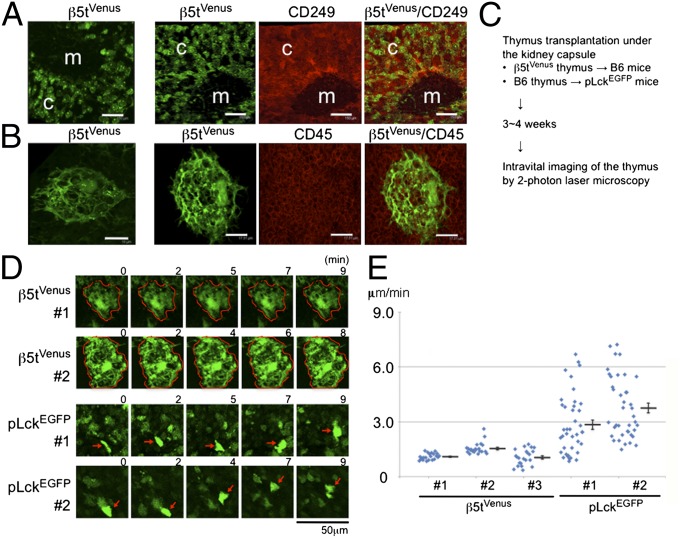
Confocal microscopy of fluorescence signals in 60-μm-thick sections of β5tVenus/+ postnatal mouse thymus revealed that most β5tVenus+ cTECs were detected in the cortex rather than the medulla and that many of the β5tVenus+ cTECs formed globular structures (Fig. 2A and B). β5tVenus+ cTECs, including the globular structures, were distributed throughout the cortex from the subcapsular area to the deep cortical area (Fig. 2A). Costaining for CD249 and CD45 indicated that the globular β5tVenus+ cells were included in CD249+ cTECs and contained CD45+ thymocytes (Fig. 2B). For intravital imaging of β5tVenus+ cTECs, neonatal thymus lobes from β5tVenus/+ mice were transplanted under the kidney capsules of wild-type mice. Lymphoid progenitor cells from host mice reconstitute the transplanted thymus lobes to undergo T-cell development and form the thymic microenvironments of the cortex and the medulla (27). After the reconstitution, the transplanted thymus lobes of anesthetized mice were time-lapse visualized intravitally by two-photon microscopy (28) (Fig. 2C). Globular cTEC structures were detected intravitally in situ, and most of the structures were stable for at least 10 min (Fig. 2 D and E and Movies S1 and S2). Parallel visualization of pLckEGFP-expressing thymocytes intravitally in the wild-type thymus lobes transplanted under the kidney capsules of pLckEGFP-transgenic mice revealed that the thymocytes exhibited heterogeneous motility (Fig. 2 D and E and Movies S3 and S4), as previously reported for ex vivo–cultured mouse thymuses (29, 30) and intravitally visualized fish thymuses (31). These results indicate that the globular structures of β5t+ cTECs are intravitally detected in the postnatal thymic cortex in situ.
Fig. 2.
In situ detection of cTEC–thymocyte complexes. (A and B) Confocal microscopy of thymuses from β5tVenus/+ mice. Venus fluorescence (green) and indicated antibody-stained fluorescence (red) are shown. Cortical region (c) and medullary region (m) are indicated in A. (Scale bars: A, 150 μm; B, 18 μm.) Representative images of four independent measurements are shown. (C–E) Intravital imaging of β5tVenus+ cTECs and pLckEGFP+ thymocytes. Time-lapse images obtained from the experiments briefly described in C are shown in Movies S1, S2, S3, and S4. Representative images of β5tVenus+ cTECs and pLckEGFP+ thymocytes are shown in D. Red arrows indicate the tracing of individual cells in a set of time-lapse images (D). Average velocities of individual cells (n = 22 for cTECs; n = 40 for thymocytes) and means and SEs of measurements of individual mice (E) show that cTECs are dormant in comparison with thymocytes, which exhibit heterogeneous cellular movement as reported previously (29–31).
The globular structures of β5t+ cTECs detected in vivo could contain P1 open complexes and P2 TNC complexes. Flow-cytometric analysis showed that the ratio of P1 to P2 complexes in isolated cTECs was ∼7:1 (Fig. S1), suggesting that the frequency of P2 TNC complexes in thymocyte-associated cTECs was ∼12% (∼1 in 8). However, cell mixture experiments demonstrated that P1 but not P2 complexes in isolated cTECs contained in vitro artifacts (Fig. S2–S4). Therefore, the homogeneously dormant and globular β5t+ cTEC structures detected in the postnatal thymic cortex in situ (>100 structures traced in >5 individual experiments) must contain closed TNC complexes at a frequency of higher than 12% (more than 1 in 8).
T-Cell Differentiation, Including Positive Selection, Occurs Without TNC Complexes.
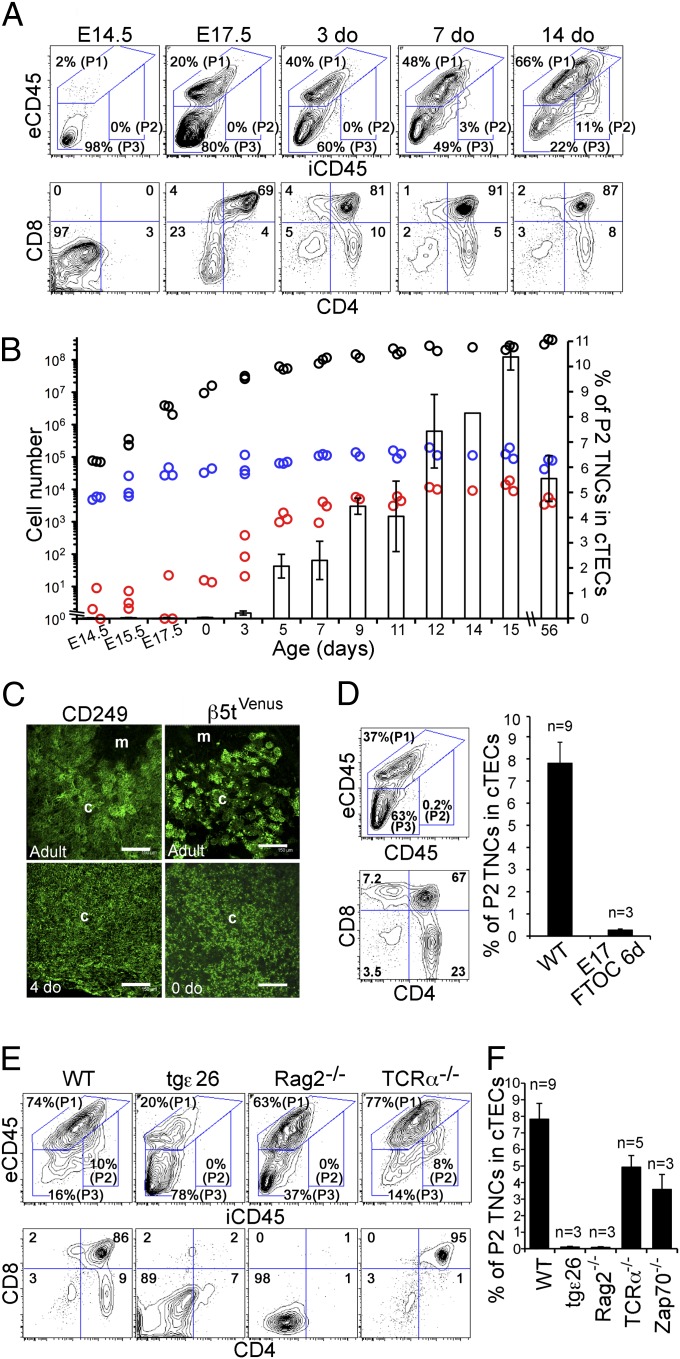
We next examined TNC complex development during ontogeny. β5t+ cTECs in mouse embryos are generated as early as embryonic day (E)12.5 (23). Unlike the postnatal thymus, however, the embryonic thymus at E14.5 formed neither P1 open complexes nor P2 TNC complexes (Fig. 3A). P1 open complexes were detected by E17.5, which coincided with CD4+CD8+ thymocyte development (Fig. 3A). On the other hand, P2 TNC complexes were negligible throughout the embryogenesis and the perinatal period until 3 or 4 d old (<1% of β5t+ cTECs) and became detectable only at 5 or 6 d old (Fig. 3 A and B). The frequency of the P2 TNC complexes within β5t+ cTECs increased thereafter and reached a plateau at ∼8–12% by 2 wk of age (Fig. 3 A and B). Section analysis indicated that the thymic cortices of newborn mice rarely contained the globular structures unlike the thymic cortices of adult mice (Fig. 3C). It should be noted that T-cell development in the thymus, including positive selection and export to the circulation, occurs even before 3 or 4 d old in mice (32) (Fig. 3A). Moreover, no P2 TNC complexes were detected in fetal thymus organ culture, in which CD4+CD8− and CD4−CD8+ mature thymocytes were generated (Fig. 3D). These results indicate that T-cell development in embryonic and perinatal thymuses occurs independent of the formation of closed TNC complexes.
Fig. 3.
Ontogeny and formation of cTEC–thymocyte complexes in T-cell–deficient mice. (A and B) Collagenase-digested cells of B6 thymus at the indicated age were analyzed as shown in Fig. S1. Representative flow-cytometric profiles of extracellular CD45 (eCD45) and intracellular CD45 (iCD45) in β5t+ CD326+ cTECs (Upper) and of cell surface CD4 and CD8 in total cells (Lower) are shown in A. Numbers indicate frequencies of cells within indicated areas (A). Numbers per mouse of total thymic cells (black circles), total β5t+ CD326+ cTECs (blue circles), and P2 TNC subpopulation of cTECs (red circles) are plotted in B. Symbols in B indicate measurement results from individual mice. The frequency (means and SEs) of P2 TNCs in cTECs is also plotted (B). (C) Confocal microscopy of anti-CD249 antibody–stained thymus sections from B6 mice (Left) and unstained thymus sections from β5tVenus/+ mice (Right) at the indicated age. Representative images of fluorescence signals from three independent measurements are shown. Cortical region (c) and medullary region (m) are indicated. (Scale bars: 150 μm.) (D) Cells were obtained from 6-d organ-cultured E17.5 B6 fetal thymus lobes. Representative flow-cytometric profiles of eCD45 and iCD45 in β5t+ CD326+ cTECs (Upper) and of cell surface CD4 and CD8 in total cells (Lower) are shown. Numbers indicate frequencies of cells within indicated areas. The frequency (means, SEs, and number of measurements) of P2 TNCs in cTECs is also plotted. (E and F) Representative flow-cytometric profiles of eCD45 and iCD45 in β5t+ CD326+ cTECs (Upper) and of cell surface CD4 and CD8 in total cells (Lower) in indicated mice are shown. Numbers indicate frequencies of cells within indicated areas. The frequency (means, SEs, and number of measurements) of P2 TNCs in cTECs is plotted in F.
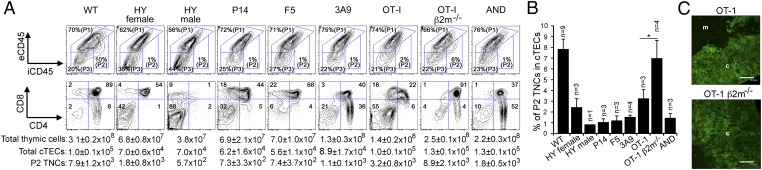
We further found that the P2 TNC complexes were undetectable in the adult thymuses of human CD3ε-transgenic tgε26 mice and Rag2-deficient mice (Fig. 3 E and F), in which T-cell development is arrested in immature CD4−CD8− thymocytes (33, 34). On the other hand, the P2 TNC complexes were detected in both adult TCRα-deficient mice and ZAP70-deficient mice (Fig. 3 E and F), in which T-cell development is arrested in CD4+CD8+ thymocytes by the lack of TCR-mediated positive- and negative-selection signals (35, 36). These results indicate that the postnatal P2 TNC complex formation coincides with CD4+CD8+ thymocyte development but requires neither positive nor negative selection of CD4+CD8+ thymocytes. In contrast, we found that the P2 TNC complexes were underrepresented in the adult thymuses of various positive-selector TCR-transgenic mice, including class I MHC–restricted TCR-transgenic mice (HY female, P14, F5, and OT-I) and class II MHC–restricted TCR-transgenic mice (AND and 3A9), as well as the “negative-selector” HY-TCR–transgenic male mice (Fig. 4 A and B). Thus, postnatal T-cell development including positive and negative selection in TCR-transgenic mice can occur without the closed TNC complexes.
Fig. 4.
Formation of cTEC–thymocyte complexes in TCR-transgenic mice. (A and B) Representative flow-cytometric profiles of eCD45 and iCD45 in β5t+ CD326+ cTECs (Upper) and of cell surface CD4 and CD8 in total cells (Lower) in indicated mice are shown. Numbers indicate frequencies of cells within indicated areas. The number (means and SEs) of total thymic cells, total cTECs, and P2 TNCs are also indicated (A, Lower). The frequency (means, SEs, and number of measurements) of P2 TNCs in cTECs is plotted in B. (C) Confocal microscopy of anti-CD249 antibody–stained thymus sections from indicated mice. Representative images of fluorescence signals from four independent measurements are shown. Cortical region (c) and medullary region (m) are indicated. (Scale bars: 150 μm.)
On the other hand, the formation of P1 open complexes was detected in the adult thymuses of all of the mutant mice, including tgε26 mice and Rag2-deficient mice, and all of the TCR-transgenic mice examined in this study (Figs. 3 E and F and 4 A and B). Thus, the adhesiveness of cTECs to thymocytes is postnatally formed even in the absence of T-cell development beyond CD4−CD8− thymocytes.
Persistent Interaction of cTECs with Long-Lived Thymocytes Leads to TNC Complex Formation for Secondary TCRα Rearrangement.
Finally, we addressed the role of TNC complexes in T-cell development. We found that the thymuses of “null-selector” β2-microglobulin–deficient OT-I TCR-transgenic mice contained a significantly (P < 0.05) larger number of P2 TNCs than those of positive-selector OT-I TCR-transgenic mice (Fig. 4 A and B). Both the absolute number and frequency of P2 TNCs within cTECs were approximately two times larger in the null-selector OT-I TCR-transgenic mice than in the positive-selector ones (Fig. 4 A and B). Accordingly, the globular cTEC structures were more apparent in the thymuses of the null-selector OT-I TCR-transgenic mice than in those of the positive-selector ones (Fig. 4C). Thus, the failure of positive selection appears to promote the formation of P2 TNC complexes.
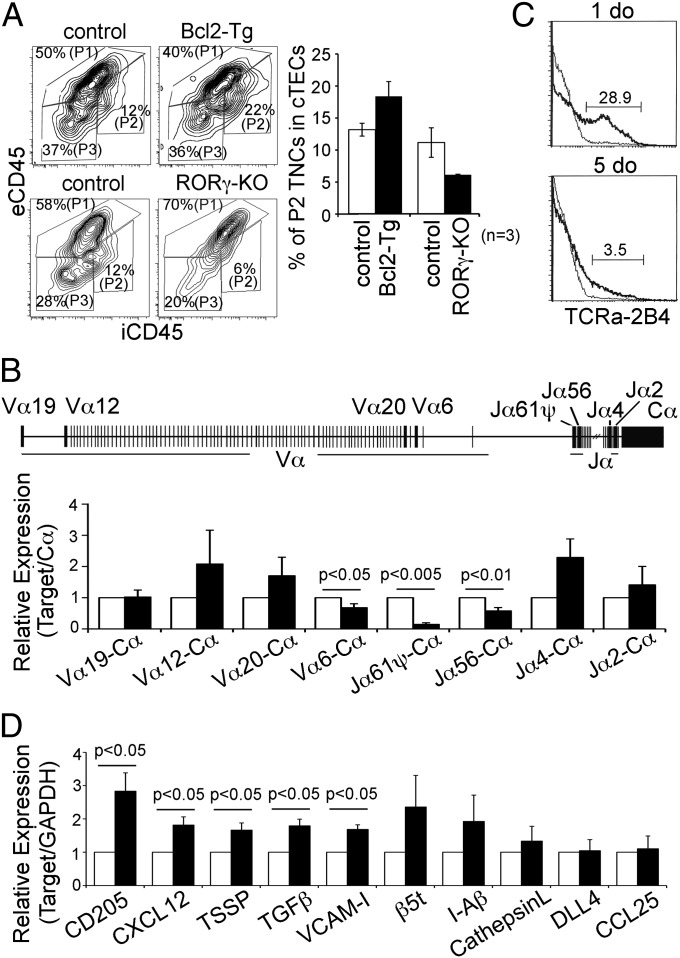
We further found that the number of P2 TNC complexes was elevated in Bcl2-transgenic mice and reduced in RORγ-deficient mice (Fig. 5A), in which the lifespan of CD4+CD8+ thymocytes is extended and shortened, respectively (37, 38). Thus, the lifespan of cortical thymocytes affects TNC complex formation. These results indicate that the persistent interaction of cTECs with long-lived CD4+CD8+ thymocytes, which are accumulated because of the failure to undergo efficient positive selection, promotes closed TNC complex formation.
Fig. 5.
Persistent interaction of cTECs with long-lived thymocytes leads to TNC complex formation for secondary TCRα rearrangement. (A) Representative flow-cytometric profiles of eCD45 and iCD45 in β5t+ CD326+ cTECs of indicated mice are shown. Numbers indicate frequencies of cells within indicated areas. The frequency (means, SEs, and number of measurements) of P2 TNCs in cTECs is plotted (Right). (B) Schematic map of mouse TCRα locus shows the locations of Vα and Jα loci analyzed for expression in CD4+CD8+ thymocytes. CD4+CD8+ thymocytes enclosed in TNCs (closed bars) were isolated from eCD45− cells mechanically released from purified CD205+CD326+ cTECs of B6 mice as shown in Fig. S2. Total CD4+CD8+ thymocytes were isolated as control (open bars). Rearranged Vα-Cα and Jα-Cα mRNA expression levels were measured by quantitative RT-PCR analysis and normalized to total Cα mRNA expression levels. Bar graphs show means ± SEs of three independent measurements. (C) CD4+CD8+ thymocytes of heterozygous knock-in (KI) mice carrying the rearranged 2B4 TCR VαJα gene to replace the genomic Jα50-Jα47 segment were isolated at 1 d old (1 do) or 5 d old (5do) and stained for cell surface 2B4 TCRα (solid lines) or for control profiles by normal IgG (gray lines). Numbers indicate frequencies of cells within indicated areas. Representative results of three independent measurements are shown. (D) eCD45−CD205+CD326+ TNC complexes were isolated from B6 mice and mechanically separated from enwrapping CD4+CD8+ thymocytes. mRNA expression levels of indicated genes in TNC cTECs in comparison with total cTECs was measured by quantitative RT-PCR analysis. The separation of TNC cTECs from CD4+CD8+ thymocytes was confirmed by the reduction of CD4 mRNA expression level (less than 14% of the expression levels in comparison with total thymocytes). Means ± SEs of three independent measurements are shown.
It was reported previously that the survival window of CD4+CD8+ thymocytes regulates TCR repertoire by the secondary TCRα rearrangement to revise TCR-recognition specificity for multiple opportunities of positive selection (39). It was also shown that TCRα rearrangement is initiated by rearranging a 3′ Vα segment and a 5′ Jα segment and then proceeds by using upstream Vα and downstream Jα segments until it is terminated by successful positive selection (40). We, thus, examined the use of Vα and Jα in CD4+CD8+ thymocytes confined in closed TNC complexes. As shown in Fig. 5B, the use of the most 3′ Vα segment, Vα6, and the most 5′ Jα segments, Jα61ψ and Jα56, was underrepresented in CD4+CD8+ thymocytes isolated from P2 TNC complexes compared with total CD4+CD8+ thymocytes. These results indicate that most of the CD4+CD8+ thymocytes confined in closed TNC complexes have undergone the secondary TCRα rearrangement.
By contrast, secondary TCRα rearrangement measured in mice that carried a rearranged 2B4 TCR VαJα gene to replace the genomic Jα50-Jα47 segment (41) was much less prominent in CD4+CD8+ thymocytes at 1 d old (Fig. 5C), at which P2 TNC complexes were still underdeveloped (Fig. 3 A and B), than in CD4+CD8+ thymocytes at 5 d old (Fig. 5C), at which P2 TNC complexes were generated (Fig. 3 A and B). Thus, the generation of closed TNC complexes coincides with the onset of secondary TCRα rearrangement in ontogeny.
cTECs that were isolated from closed TNC complexes exhibited significant (P < 0.05) elevations in CD205, CXCL12, TGF-β, TSSP, and VCAM-1 mRNA expression levels in comparison with total cTECs (Fig. 5D). β5t, I-Aβ, cathepsin L, CCL25, and DLL4 mRNA expression levels were not significantly different between TNC cTECs and total cTECs (Fig. 5D). These results indicate that cTECs that form TNC complexes have gene expression profiles distinct from those of the majority of cTECs, suggesting molecular heterogeneity in the cTEC subpopulations.
Discussion
The parenchyma of the thymus features multiple microenvironments that play individual roles in supporting various aspects of T-cell development and repertoire selection. The thymic cortex possesses microenvironments that promote the differentiation of lymphoid progenitor cells into T-lineage cells and positively select a diverse and functional repertoire of newly generated T cells, whereas the thymic medulla offers microenvironments that verify self-tolerance by deleting self-reactive T cells and generating regulatory T cells (42, 43). It is well appreciated that multilateral communication among thymic stromal cells and hematopoietic cells critically contributes to the formation of the medullary region and, thereby, the development and tolerance of T cells (44, 45). However, the developmental origin and the physiological functions of many other thymic microenvironments are still unknown. One of the vaguely understood microenvironments in the thymus is TNC. TNC is defined as a large epithelial cell that completely envelops many viable lymphoid cells within its intracellular vesicles (7–10). Despite the initial suggestion that TNC may be important for T-cell development and selection, its in vivo existence has been questioned and its function has remained unclear (10, 19, 20). The present results, including the analysis of the intravital imaging results of β5t+ cTECs, reveal that basket-like globular cTECs, including closed TNC complexes, are indeed present in the thymic cortex in situ and are not merely artificial structures formed during cell preparation. We found that TNCs represent a subpopulation of cTECs that express β5t-containing thymoproteasomes and carry a distinctive gene expression profile. Thymocytes that are enclosed in TNCs are enriched with CD4+CD8+ thymocytes that undergo the secondary TCRα rearrangement for the alteration of TCR-recognition specificity. Thus, this study reveals heterogeneity in the thymic microenvironments formed by cTECs and that TNC provides a microenvironment that optimizes T-cell selection through the secondary TCRα rearrangement in cortical CD4+CD8+ thymocytes.
Our results show that CD4+CD8+ thymocytes isolated from TNC complexes are enriched with cells that have undergone the secondary TCRα rearrangement. It was reported previously that cortical thymocytes that undergo the secondary TCRα rearrangement exhibit an alteration of the TCRα variable region and, thereby, the TCR-recognition specificity (39–41). It was also shown that optimal T-cell development relies on the ability of developing thymocytes to change their TCRα genes (46, 47). In addition, TNCs are known to express class I and class II MHC molecules on the surface that enwraps thymocytes (15, 48). Collectively, we think that TNC complexes provide a thymic microenvironment for the secondary TCRα rearrangement in cortical thymocytes, which crucially optimizes the efficiency of TCR-mediated repertoire selection.
Our results also demonstrate that the formation of TNC complexes is enhanced in the thymic cortex where TCR-induced positive and negative selection does not proceed efficiently in CD4+CD8+ thymocytes. On the other hand, various positive-selector TCR-transgenic mice in which positive and negative selection proceeds efficiently exhibit reduced formation of TNC complexes. TNC complexes are postnatally generated in ontogeny and are not detected in embryonic and perinatal thymuses where the initial cohorts of developing thymocytes undergo positive and negative selection. In the postnatal thymus, TNC complex formation is correlated with the survival window of cortical thymocytes, as noted in Bcl2-trangenic mice and RORγ-deficient mice. These results suggest that persistent interactions between adhesive cTECs and long-lived cortical thymocytes that fail to be efficiently selected by the initially expressed TCRs contribute to the formation of TNC complexes. Nonetheless, these results indicate that TNCs are neither needed for embryonic T-cell development, including positive and negative selection, in the thymus nor needed for postnatal T-cell development in many TCR-transgenic mice. Thus, it is important to note that TNC is not a nurse cell for all T cells.
In conclusion, the present findings demonstrate that TNCs represent a subpopulation of β5t+ cTECs and reveal the function of TNCs as a thymic microenvironment that supports the secondary TCRα rearrangement in cortical thymocytes. This study also reveals the heterogeneity of the microenvironments formed by cTECs. We propose that the term “TNC” be renamed to a developmentally and functionally relevant term, such as “cTEC for secondary TCR rearrangement (CSR).” Additional discussion is available in SI Discussion.
Methods
Mice, antibodies, thymus transplantation, intravital imaging, fetal thymus organ culture, preparation of thymic stromal cells, flow cytometry, confocal microscopy, electron microscopy, and quantitative mRNA analysis are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Drs. Dan Littman, Sidonia Fagarasan, Jos Domen, and Sho Yamasaki for providing RORγ-deficient mice and Bcl2-transgenic mice and Sachiko Nitta and Masako Ino for technical help. This study was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Japan Society for the Promotion of Science (JSPS) Grants 23249025 and 24111004, as well as Astellas, Naito, Uehara, Takeda, and Inamori Foundations.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.A.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213069109/-/DCSupplemental.
References
- 1.Bacci G. [Histochemical data on oocytes and nurse cells of Ophryotrocha puerilis.] Boll Soc Ital Biol Sper. 1952;28(6):1293–1295. [PubMed] [Google Scholar]
- 2.Mische S, Li M, Serr M, Hays TS. Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol Biol Cell. 2007;18(6):2254–2263. doi: 10.1091/mbc.E06-10-0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritz IB. Reflections on the evolution of the regulation of spermatogenesis. Prog Clin Biol Res. 1986;226:371–388. [PubMed] [Google Scholar]
- 4.Rao MK, et al. Tissue-specific RNAi reveals that WT1 expression in nurse cells controls germ cell survival and spermatogenesis. Genes Dev. 2006;20(2):147–152. doi: 10.1101/gad1367806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Despommier DD. Trichinella spiralis and the concept of niche. J Parasitol. 1993;79(4):472–482. [PubMed] [Google Scholar]
- 6.Manwani D, Bieker JJ. The erythroblastic island. Curr Top Dev Biol. 2008;82:23–53. doi: 10.1016/S0070-2153(07)00002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wekerle H, Ketelsen UP. Thymic nurse cells—Ia-bearing epithelium involved in T-lymphocyte differentiation? Nature. 1980;283(5745):402–404. doi: 10.1038/283402a0. [DOI] [PubMed] [Google Scholar]
- 8.Wekerle H, Ketelsen UP, Ernst M. Thymic nurse cells. Lymphoepithelial cell complexes in murine thymuses: Morphological and serological characterization. J Exp Med. 1980;151(4):925–944. doi: 10.1084/jem.151.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritter MA, Sauvage CA, Cotmore SF. The human thymus microenvironment: In vivo identification of thymic nurse cells and other antigenically-distinct subpopulations of epithelial cells. Immunology. 1981;44(3):439–446. [PMC free article] [PubMed] [Google Scholar]
- 10.Kyewski BA, Kaplan HS. Lymphoepithelial interactions in the mouse thymus: Phenotypic and kinetic studies on thymic nurse cells. J Immunol. 1982;128(5):2287–2294. [PubMed] [Google Scholar]
- 11.van de Wijngaert FP, Rademakers LH, Schuurman HJ, de Weger RA, Kater L. Identification and in situ localization of the “thymic nurse cell” in man. J Immunol. 1983;130(5):2348–2351. [PubMed] [Google Scholar]
- 12.Rieker T, Penninger J, Romani N, Wick G. Chicken thymic nurse cells: An overview. Dev Comp Immunol. 1995;19(4):281–289. doi: 10.1016/0145-305x(95)00008-h. [DOI] [PubMed] [Google Scholar]
- 13.Flaño E, et al. In vitro and in situ characterization of fish thymic nurse cells. Dev Immunol. 1996;5(1):17–24. doi: 10.1155/1996/14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shortman K, Scollay R, Andrews P, Boyd R. Development of T lymphocytes within the thymus and within thymic nurse cells. Curr Top Microbiol Immunol. 1986;126:5–18. doi: 10.1007/978-3-642-71152-7_2. [DOI] [PubMed] [Google Scholar]
- 15.de Waal Malefijt R, Leene W, Roholl PJ, Wormmeester J, Hoeben KA. T cell differentiation within thymic nurse cells. Lab Invest. 1986;55(1):25–34. [PubMed] [Google Scholar]
- 16.Wick G, Rieker T, Penninger J. Thymic nurse cells: A site for positive selection and differentiation of T cells. Curr Top Microbiol Immunol. 1991;173:99–105. doi: 10.1007/978-3-642-76492-9_14. [DOI] [PubMed] [Google Scholar]
- 17.Aguilar LK, Aguilar-Cordova E, Cartwright J, Jr, Belmont JW. Thymic nurse cells are sites of thymocyte apoptosis. J Immunol. 1994;152(6):2645–2651. [PubMed] [Google Scholar]
- 18.Guyden JC, Pezzano M. Thymic nurse cells: A microenvironment for thymocyte development and selection. Int Rev Cytol. 2003;223:1–37. doi: 10.1016/s0074-7696(05)23001-2. [DOI] [PubMed] [Google Scholar]
- 19.Pezzano M, Samms M, Martinez M, Guyden J. Questionable thymic nurse cell. Microbiol Mol Biol Rev. 2001;65(3):390–403. doi: 10.1128/MMBR.65.3.390-403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toussaint-Demylle D, Scheiff JM, Haumont S. Thymic nurse cells: Morphological study during their isolation from murine thymus. Cell Tissue Res. 1990;261(1):115–123. doi: 10.1007/BF00329444. [DOI] [PubMed] [Google Scholar]
- 21.Murata S, et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316(5829):1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 22.Nitta T, et al. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity. 2010;32(1):29–40. doi: 10.1016/j.immuni.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Ripen AM, Nitta T, Murata S, Tanaka K, Takahama Y. Ontogeny of thymic cortical epithelial cells expressing the thymoproteasome subunit β5t. Eur J Immunol. 2011;41(5):1278–1287. doi: 10.1002/eji.201041375. [DOI] [PubMed] [Google Scholar]
- 24.Gray DH, Chidgey AP, Boyd RL. Analysis of thymic stromal cell populations using flow cytometry. J Immunol Methods. 2002;260(1-2):15–28. doi: 10.1016/s0022-1759(01)00493-8. [DOI] [PubMed] [Google Scholar]
- 25.Gray DH, et al. Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood. 2006;108(12):3777–3785. doi: 10.1182/blood-2006-02-004531. [DOI] [PubMed] [Google Scholar]
- 26.Williams KM, et al. Single cell analysis of complex thymus stromal cell populations: Rapid thymic epithelia preparation characterizes radiation injury. Clin Transl Sci. 2009;2(4):279–285. doi: 10.1111/j.1752-8062.2009.00128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandel T, Russell PJ. Differentation of foetal mouse thymus. Ultrastructure of organ cultures and of subcapsular grafts. Immunology. 1971;21(4):659–674. [PMC free article] [PubMed] [Google Scholar]
- 28.Caetano SS, Teixeira T, Tadokoro CE. Intravital imaging of the mouse thymus using 2-photon microscopy. J Vis Exp. 2012;59(59):e3504. doi: 10.3791/3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bousso P, Bhakta NR, Lewis RS, Robey E. Dynamics of thymocyte-stromal cell interactions visualized by two-photon microscopy. Science. 2002;296(5574):1876–1880. doi: 10.1126/science.1070945. [DOI] [PubMed] [Google Scholar]
- 30.Witt CM, Raychaudhuri S, Schaefer B, Chakraborty AK, Robey EA. Directed migration of positively selected thymocytes visualized in real time. PLoS Biol. 2005;3(6):e160. doi: 10.1371/journal.pbio.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Iwanami N, Hoa VQ, Furutani-Seiki M, Takahama Y. Noninvasive intravital imaging of thymocyte dynamics in medaka. J Immunol. 2007;179(3):1605–1615. doi: 10.4049/jimmunol.179.3.1605. [DOI] [PubMed] [Google Scholar]
- 32.Fowlkes BJ, Pardoll DM. Molecular and cellular events of T cell development. Adv Immunol. 1989;44:207–264. doi: 10.1016/s0065-2776(08)60643-4. [DOI] [PubMed] [Google Scholar]
- 33.Wang B, et al. A block in both early T lymphocyte and natural killer cell development in transgenic mice with high-copy numbers of the human CD3E gene. Proc Natl Acad Sci USA. 1994;91(20):9402–9406. doi: 10.1073/pnas.91.20.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shinkai Y, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68(5):855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 35.Mombaerts P, et al. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature. 1992;360(6401):225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 36.Negishi I, et al. Essential role for ZAP-70 in both positive and negative selection of thymocytes. Nature. 1995;376(6539):435–438. doi: 10.1038/376435a0. [DOI] [PubMed] [Google Scholar]
- 37.Domen J, Gandy KL, Weissman IL. Systemic overexpression of BCL-2 in the hematopoietic system protects transgenic mice from the consequences of lethal irradiation. Blood. 1998;91(7):2272–2282. [PubMed] [Google Scholar]
- 38.Sun Z, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288(5475):2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 39.Guo J, et al. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol. 2002;3(5):469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 40.Petrie HT, et al. Multiple rearrangements in T cell receptor α chain genes maximize the production of useful thymocytes. J Exp Med. 1993;178(2):615–622. doi: 10.1084/jem.178.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, Huang CY, Kanagawa O. Rapid deletion of rearranged T cell antigen receptor (TCR) Valpha-Jalpha segment by secondary rearrangement in the thymus: Role of continuous rearrangement of TCR α chain gene and positive selection in the T cell repertoire formation. Proc Natl Acad Sci USA. 1998;95(20):11834–11839. doi: 10.1073/pnas.95.20.11834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9(12):833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 43.Anderson G, Takahama Y. Thymic epithelial cells: Working class heroes for T cell development and repertoire selection. Trends Immunol. 2012;33(6):256–263. doi: 10.1016/j.it.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 44.van Ewijk W, Shores EW, Singer A. Crosstalk in the mouse thymus. Immunol Today. 1994;15(5):214–217. doi: 10.1016/0167-5699(94)90246-1. [DOI] [PubMed] [Google Scholar]
- 45.Nitta T, Ohigashi I, Nakagawa Y, Takahama Y. Cytokine crosstalk for thymic medulla formation. Curr Opin Immunol. 2011;23(2):190–197. doi: 10.1016/j.coi.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Huang C, Kanagawa O. Ordered and coordinated rearrangement of the TCR α locus: Role of secondary rearrangement in thymic selection. J Immunol. 2001;166(4):2597–2601. doi: 10.4049/jimmunol.166.4.2597. [DOI] [PubMed] [Google Scholar]
- 47.Huang CY, Sleckman BP, Kanagawa O. Revision of T cell receptor α chain genes is required for normal T lymphocyte development. Proc Natl Acad Sci USA. 2005;102(40):14356–14361. doi: 10.1073/pnas.0505564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCormack JE, Wade T, Morales H, Kappler J, Marrack P. Analysis of class II MHC structure in thymic nurse cells. Cell Immunol. 1991;138(2):413–422. doi: 10.1016/0008-8749(91)90165-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.