Abstract
Impairment of ribosomal biogenesis can activate the p53 protein independently of DNA damage. The ability of ribosomal proteins L5, L11, L23, L26, or S7 to bind Mdm2 and inhibit its ubiquitin ligase activity has been suggested as a critical step in p53 activation under these conditions. Here, we report that L5 and L11 are particularly important for this response. Whereas several other newly synthesized ribosomal proteins are degraded by proteasomes upon inhibition of Pol I activity by actinomycin D, L5 and L11 accumulate in the ribosome-free fraction where they bind to Mdm2. This selective accumulation of free L5 and L11 is due to their mutual protection from proteasomal degradation. Furthermore, the endogenous, newly synthesized L5 and L11 continue to be imported into nucleoli even after nucleolar disruption and colocalize with Mdm2, p53, and promyelocytic leukemia protein. This suggests that the disrupted nucleoli may provide a platform for L5- and L11-dependent p53 activation, implying a role for the nucleolus in p53 activation by ribosomal biogenesis stress. These findings may have important implications with respect to understanding the pathogenesis of diseases caused by impaired ribosome biogenesis.
Keywords: proteasome, ribosomal stress
The exposure of cells to various stressors activates the p53 tumor suppressor, a transcription factor that regulates many coding and noncoding genes, with ensuing cellular outcomes such as cell cycle arrest, apoptosis, senescence, metabolic changes, and DNA repair (1, 2). Disruption of these functions promotes checkpoint defects, genomic instability, illegitimate survival, continuous proliferation, and evolution of stress-damaged cells (2). Given the fact that loss of wild-type p53 provides many selective advantages to cells, it is not surprising that ∼50% of all human cancers have mutations within the TP53 gene (3). In cancers retaining wild-type p53, the functions of p53 are likely inactivated by defects in its upstream regulators and downstream targets. Although DNA damage is common to most p53-activating stresses, evidence accumulated over the last decade suggests that perturbation of ribosome biogenesis triggers a p53-activating signaling pathway independently of DNA damage (2, 4–7). It was originally suggested that this pathway has a prominent role in preventing diseases by monitoring the fidelity of ribosome biogenesis (4). However, recent evidence suggests that p53 activation upon impairment of ribosome biogenesis might be responsible for specific pathological manifestations in mammals (8–11). Earlier work suggested that perturbation of ribosome biogenesis causes nucleolar disruption and translocation of a number of ectopically expressed ribosomal proteins (RPs), including L5, L11, L23, L26, and S7, from the nucleolus to the nucleoplasm, where they bind to Mdm2 and inhibit its ubiquitin ligase function toward p53, leading to p53 up-regulation (6, 12–21). An alternative model proposes that inhibition of ribosome biogenesis leads to specific up-regulation of L11 mRNA translation, resulting in the accumulation of ribosome-free L11 in the nucleoplasm where it associates with Mdm2 and blocks Mdm2-mediated p53 ubiquitination and degradation (22).
Here we demonstrate the central importance of L5 and L11 for p53 activation by pharmacological or siRNA-mediated inhibition of ribosome biogenesis and provide molecular insights into this mechanism.
Results
Specific Requirement for L5 and L11 in p53 Up-Regulation by Ribosomal Stressors.
A549 human lung carcinoma cells were transfected with siRNAs against the indicated RPs and treated with 5 nM actinomycin D (ActD), which selectively inhibits RNA polymerase I (Table S1). The effective silencing of each RP in all experiments was confirmed by quantitative RT-PCR (qRT-PCR) and immunoblotting (Fig. S1 A and B; Table S2).
Down-regulation of L5 or L11, alone or together, dramatically inhibited ActD-driven induction of p53 protein and of its targets p21WAF1 and Mdm2 in A549 cells (Fig. 1A and Fig. S2A). In contrast to L5 or L11, depletion of S7, L23, or L26 did not reduce p53 accumulation (Fig. 1A). While this article was in preparation, Fumagalli et al. reported a similar observation, except that in their experiments S7 or L23 depletion partially inhibited ActD-induced up-regulation of p53 (23). Moreover, depletion of L5 or L11, but not of S7, L23, or L26, largely rescued the entry of ActD-treated cells into the S phase of the cell cycle (Fig. S2B). Similar requirements for L5 and L11 were observed in ActD-treated U2OS cells (Fig. S2C), and a second set of siRNAs against the indicated RPs in A549 cells provided essentially analogous results (Fig. S2D). Together, these results provide strong evidence that under the conditions used L5 and L11, but not S7, L23, or L26, are required for ActD-mediated induction of p53 protein and its biological activities. Furthermore, the induction of p53 and its targets p21WAF1 and Mdm2 by 5-fluorouracil (5-FU), another pharmacological inhibitor of ribosome biogenesis, was dependent on L5 and L11, but not on L23, L26, and S7, in both A549 and U2OS cells (Fig. S2 E and F). Earlier studies have shown that down-regulation of S6 inhibits 40S ribosome biogenesis and consequently activates the p53 response in an L11-dependent manner (4, 9, 22). Here, we found that S6 down-regulation requires both L5 and L11, but not S7, L23, or L26, for the induction of p53, p21WAF1, and Mdm2 (Fig. S2G). Consistent with one recent published study (23) and in contrast to other studies (6), impairment of ribosome biogenesis by depletion of S7, L23, or L26 led to up-regulation of p53, p21WAF1, and Mdm2 protein levels (Fig. S2 H–J). These responses were specifically inhibited by L5 or L11 siRNAs, most likely by a mechanism similar to that operating upon down-regulation of S6 or ActD and 5-FU treatments (Fig. S2 H–J). Together, these results strongly point to a critical role of L5 and L11 in p53 up-regulation by impairement of ribosome biogenesis at various steps.
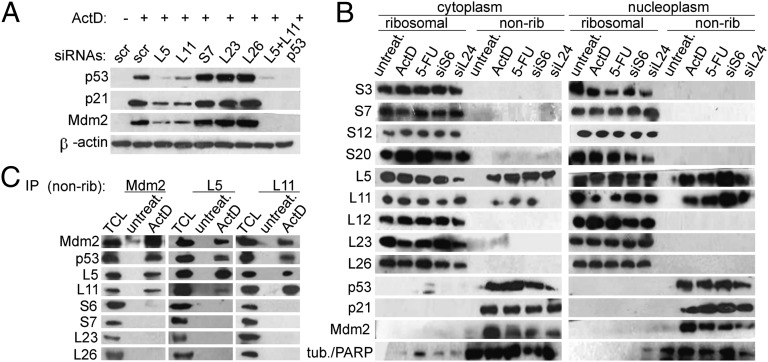
Fig. 1.
Specific requirement for L5 and L11 in p53 up-regulation upon inhibition of ribosome biogenesis in A549 cells. (A) Cells were transfected with the indicated siRNAs or scrambled (scr) negative control siRNA for 48 h and treated with ActD for 16 h. Expression of the indicated proteins was confirmed by immunoblotting. (B) Cells were transfected with the indicated siRNAs for 48 h or treated with ActD or 5-FU for 16 h. Ribosomal and nonribosomal (nonrib) cytoplasmic (Left) or ribosomal and nonribosomal nucleoplasmic fractions (Right) were immunoblotted with the indicated antibodies. The per-cell ratio of the amount of protein loaded onto a gel for cytoplasmic ribosomal, nucleoplasmic ribosomal, cytoplasmic nonribosomal, and nucleoplasmic nonribosomal fractions was 1:10:3:10. Tubulin (tub.) was used as a cytoplasmic marker and PARP as a nucleoplasmic marker. (C) Nonribosomal fractions from untreated and ActD-treated cells were immunoprecipitated with the indicated antibodies. These immunoprecipitates and the total cell lysate (TCL) from ActD-treated A549 cells were immunoblotted with the indicated antibodies.
Accumulation of Ribosome-Free L5 and L11 upon Ribosomal Stress.
It has been proposed that upon disruption of ribosome biogenesis several RPs, including L5, L11, S7, L23, and L26, accumulate outside of ribosomes and participate in p53 up-regulation (6). In light of our observations highlighting the centrality of L5 and L11 for p53 up-regulation in response to ribosomal stressors, we evaluated the expression of endogenous ribosome-free RPs in A549 cells by isolating ribosomal and nonribosomal cytoplasmic and nuclear fractions that were subjected to Western blot analysis with the indicated anti-RP antibodies developed in our laboratory (24) (Fig. 1B). Strikingly, whereas S3, S7, S20, L12, L23, and L26 remained exclusively associated with ribosomes after exposure to the indicated ribosomal stressors, we detected a significant increase in the amount of ribosome-free L5 and L11 in the cytoplasm (Fig. 1B, Left) and nucleoplasm under the same conditions (Fig.1B, Right), without a corresponding decrease in their amounts in the ribosome fractions. p53, Mdm2, and p21WAF1 were also present in the nonribosomal fraction. The observation that accumulation of ribosome-free L5 and L11 also occurred in mouse embryonic fibroblasts (MEFs) established from 13.5-d embryos upon ActD treatment suggests that this response has a wide biological significance (Fig. S3A). Overexpression experiments have shown that a number of RPs, including L5, L11, S7, L23, and L26, can bind to Mdm2 and inhibit its ubiquitin ligase activity, leading to p53 up-regulation (6, 12–21). We therefore evaluated the interactions of several endogenously expressed RPs with Mdm2 in total cell lysates from untreated and ActD-treated A549 cells. We were able to coimmunoprecipitate L5, L11, S7, L23, and L26 with Mdm2 in untreated cells, and these interactions were significantly increased by ActD treatment most likely due to the increased amounts of cellular Mdm2 (Fig. S3B). S6 was also bound to Mdm2 under these conditions (Fig. S3B). This, together with the observation that L5 and L11 are present in the nonribosomal fraction with Mdm2 upon ribosomal stresses (Fig. 1B), raised the question of the specificity of interactions of endogenous L23, L26, and S7 with Mdm2 in the total cell lysate. To address this issue, we homogenized untreated or ActD-treated A549 cells under conditions in which rRNA is protected from degradation, separated ribosomal and nonribosomal fractions, and retrieved endogenous Mdm2-RP complexes from the nonribosomal fraction (24) (Fig. 1C). As expected, Mdm2 levels were undetectable in untreated cells and were dramatically increased after ActD treatment. Mdm2 was associated with L5, L11, and p53 in this fraction. The specificity of these interactions was further supported by the observation that S6, S7, L23, or L26 did not form complexes with Mdm2 under the same conditions (Fig. 1C). Both L5 and L11 immunoprecipitates contained L5, L11, Mdm2, and p53 (Fig. 1C). The inefficient coimmunoprecipitation of L11 with L5 could reflect interference of these antibodies with formation of the L5-L11 complex. Altogether, these results further strengthen the critical role of L5 and L11 in p53 up-regulation by various defects in ribosome biogenesis (Fig. 1; Figs. S2 and S3).
De Novo Protein Synthesis Is Absolutely Required for the Accumulation of Ribosome-Free L5 and L11 upon Ribosomal Stress.
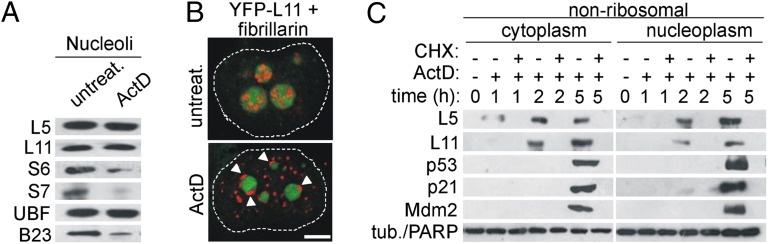
As mentioned above, two opposing models regarding the source of p53-activating ribosome-free RPs have been put forth. One proposes that perturbation of ribosome biogenesis causes nucleolar disruption and translocation of a number of RPs from the nucleolus into the nucleoplasm (6), and the other suggests that there is a selective up-regulation of L11 mRNA translation without disruption of nucleolar integrity (22). To identify the source of ribosomal stress-induced endogenous ribosome-free L5 and L11, we first focused on nucleoli. We showed that the production of 47S rRNA, a fundamental step in ribosome biogenesis (9), was inhibited by all tested p53-activating ribosomal stressors (Fig. S4A). It was shown that depletion of these RPs inhibits rRNA processing at various levels (4, 25). Despite interference with ribosome biogenesis, L23 and L26 depletion did not show any obvious effect on nucleolar structure visualized by fluorescence microscopy with antibodies against UBF, B23, and fibrillarin (Fig. S4B). On the other hand, depletion of either S6 or 5-FU treatment diminished nucleolar B23 staining and resulted in its diffused nucleoplasmic staining, whereas the localization of UBF and fibrillarin was not affected. S7 depletion specifically affected the localization of B23, which formed nuclear spot structures. ActD caused virtually complete nucleolar disruption, which is characterized by formation of nucleolar caps for both UBF and fibrillarin and translocation of B23 from the nucleolus to the nucleoplasm (Fig. S4B). The relative distribution of B23 between nucleolus and nucleoplasm upon exposure to various ribosomal stressors is shown in Fig. S4C. However, the observed morphological and functional changes in the nucleolus may not necessarily lead to the release of L5 and L11 from the nucleolus to the nucleoplasm. To clarify this issue, nucleoli from untreated and ActD-treated A549 cells were isolated, and the amount of endogenous nucleolar L5 and L11 was determined by Western blot analysis (26) (Fig. 2A). Surprisingly, ActD treatment did not substantially change the levels of nucleolar L5 and L11 (Fig. 2A), whereas it led to a loss of nucleolar S6 and S7. To provide further support for this finding, we used H1299 cells that were engineered to contain L11 fused to yellow fluorescent protein (YFP), expressed from its endogenous chromosomal location (27). Consistent with the results shown in Fig. 2A, confocal laser scanning microscopy (CLSM) demonstrated that the YFP-L11 signal was still present in the nucleolar remnants surrounded by fibrillarin cap structures in ActD-treated cells (Fig. 2B). These results imply that the nucleolus is not a significant source of endogenous ribosome-free L5 and L11 in response to ribosomal stress. Thus, experiments in which overexpressed RPs were used must be interpreted cautiously (6). Next, we focused on protein synthesis. To determine whether de novo protein synthesis is required for the accumulation of ribosome-free L5 and L11, A549 cells were treated for 1, 2, or 5 h with ActD (Fig. 2C) in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX). Treatment with CHX abrogated the accumulation of ribosome-free L5 and L11 in response to ActD (Fig. 2C) as well as the increases in p53, Mdm2, and p21WAF1 protein levels. RP mRNAs contain an oligopyrimidine tract at their 5′ end (5′ TOP), and the mTOR inhibitor rapamycin prevents the translation of 5′ TOP mRNAs (28). Similar to CHX, rapamycin prevented the accumulation of ribosome-free L5 and L11 as well as p53, Mdm2, and p21WAF1 by ActD (Fig. S4D). Together, these results demonstrate that new protein synthesis is absolutely required for the accumulation of ribosome-free L5 and L11 after inhibition of ribosome biogenesis.
Fig. 2.
De novo protein synthesis of L5 and L11 is required for their accumulation in the ribosome-free form. (A) A549 cells were left untreated or treated with ActD for 5 h. Purified nucleoli were immunoblotted with the indicated antibodies. (B) H1299 cells containing YFP-L11 (green) were treated with ActD for 5 h, stained with antifibrillarin antibodies (red), and analyzed by CLSM. Fibrillarin nucleolar caps are indicated by arrowheads. Nuclei are outlined with dashed lines. (Scale bar, 5 μm.) (C) A549 cells were treated with ActD in the presence or absence of CHX for 1, 2, or 5 h, and nonribosomal fractions were immunoblotted with the indicated antibodies.
Ribosome-Free L5 and L11 Are Not Degraded by Proteasomes upon ActD Treatment.
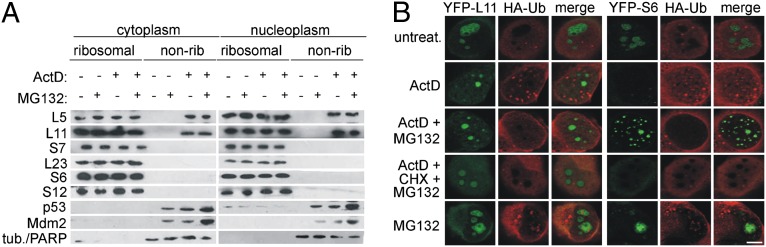
RPs continue to be synthesized upon inhibition of rRNA transcription by ActD and are rapidly degraded by proteasomes, probably to prevent deleterious effects of highly basic ribosome-free RPs on the cell (29–31). However, the observed accumulation of ribosome-free L5 and L11 in the cytoplasm and nucleoplasm upon exposure to various stressors that impair ribosome biogenesis suggests that these two RPs have a different fate than other RPs (Fig. 1B and Fig. S3A). To assess the stability of newly synthesized L5 and L11 upon ribosomal stress, A549 cells were treated with ActD for 5 h, then CHX was added for the indicated times, and the ribosome-free fraction was analyzed by immunoblotting with the indicated anti-RP antibodies (Fig. S5A). After up to 1 h of CHX treatment, L5 and L11 protein levels remained unchanged (Fig. S5A). By 2 h after addition of CHX, the amounts of ribosome-free L5 and L11 were partially decreased and became almost undetectable by the 4-h time point (Fig. S5A). On the basis of overexpression experiments, it has been suggested that Mdm2 protects L11 from degradation upon ActD treatment (32). To determine the extent to which this mechanism contributes to accumulation of ribosome-free L5 and L11 upon ActD treatment, we prevented Mdm2 expression by silencing its transcriptional activator, p53 (Fig. S5B). Endogenous ribosome-free L5 and L11 accumulated normally under such conditions, suggesting that Mdm2 does not play a role in their stabilization (32). In view of the relative stability of L5 and L11, we hypothesized that newly synthesized endogenous ribosome-free L5 and L11 are not degraded by the proteasome upon exposure to ActD, in contrast to other newly synthesized ribosome-free RPs. To test this, A549 cells were treated with ActD, and the fate of the indicated RPs was followed in the presence or absence of the proteasome inhibitor MG132 (Fig. 3A). MG132 treatment resulted in a significant elevation of ActD-induced p53 and Mdm2 protein levels. However, the treatment did not increase the amount of ribosome-free L5 and L11, consistent with the idea that they are not degraded by the proteasome. Intriguingly, under these conditions, L23, S7, S6, and S12 also did not accumulate in the ribosome-free fraction in the presence of MG132 and ActD (Fig. 3A), suggesting that they may be degraded through a proteasome-independent pathway. Alternatively, these RPs might be normally degraded by proteasomes upon ActD treatment, but they form insoluble aggregates when their proteosomal degradation is inhibited by MG132. To distinguish between these possibilities, we used H1299 cells containing endogenous L11 or S6 fused to YFP. Consistent with the results shown in Fig. 2A, ActD treatment led to a complete loss of nucleolar YFP-S6, but did not significantly decrease the total amount of nucleolar YFP-L11 (Fig. 3B). Although ActD treatment in combination with MG132 did not have a significant effect on the intensity and localization of the L11-YFP signal, it caused a dramatic accumulation of nuclear aggregates containing YFP-S6 (Fig. 3B). Furthermore, ActD treatment in combination with MG132 led to the nucleoplasmic accumulation of YFP-L4 aggregates (Fig. S6A). Notably, the protein synthesis inhibitor CHX prevented the formation of YFP-S6 and YFP-L4 nucleoplasmic aggregates (Fig. 3B and Fig. S6A). Despite the fact that proteasome inhibition by MG132 led to the nucleoplasmic accumulation of YFP-S6 and YFP-L4 aggregates in ActD-treated cells, we failed to observe their colocalization with HA-ubiquitin (HA-Ub) (Fig. 3B and Fig. S6A), suggesting that YFP-S6 and YFP-L4 are degraded through a proteasome-dependent, ubiquitin-independent pathway (33). In contrast, HA-Ub colocalized with p53, a known ubiquinated protein, upon proteasome inhibition in A549 cells (Fig. S6B) (1). Significantly, endogenous S6 and S12 also formed nucleoplasmic aggregates in the presence of ActD and MG132 (Fig. S6C). The most likely explanation of why endogenous S6 and S12 were not detected in the nonribosomal fraction under these conditions (Fig. 3A) is that S6 and S12 aggregates were removed by ultracentrifugation. Together, these results showed that newly synthesized L5 and L11 are not proteasomally degraded upon ActD treatment, in contrast to other tested RPs, and as a consequence they accumulate in free form.
Fig. 3.
Ribosome-free L5 and L11 are not degraded by proteasomes upon ActD treatment. (A) A549 cells were treated with ActD for 5 h in the presence or absence of MG132. Nonribosomal fractions were immunoblotted with the indicated antibodies. (B) H1299 cells expressing YFP-L11 or YFP-S6 were transfected with HA-Ub for 24 h and then treated as indicated for 6 h, followed by staining with anti-HA antibody (red) and analysis by CLSM. (Scale bar, 5 μm.)
Accumulation of Ribosome-Free L5 and L11 Is due to Their Mutual Protection from Proteasomal Degradation.
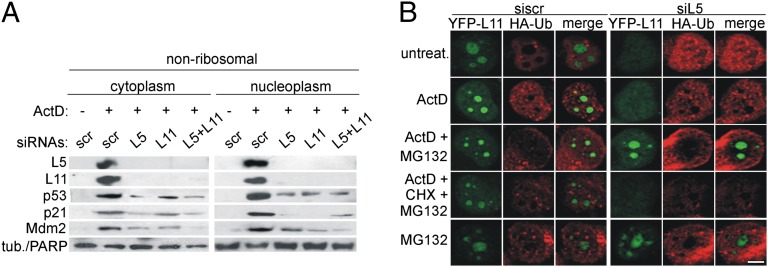
On the basis of overexpression experiments it was previously suggested that L5 and L11 can interact with Mdm2 independently of each other, and their cooperative binding to Mdm2 is needed for full p53 up-regulation (21). To gain further insight into the mutual dependence of L5 and L11 in p53 activation, we silenced either L5 or L11 and followed their expression in the nonribosomal fraction after ActD treatment (Fig. 4A). Surprisingly, L5 silencing abrogated the presence of L11 and vice versa (Fig. 4A). Therefore, we tested the possibility that L5 and L11 stabilize each other in ActD-treated cells. H1299 cells containing YFP-L11 were transfected with siL5 and treated with ActD in the presence or absence of MG132. Depletion of L5 resulted in the loss of nucleolar YFP-L11 staining in ActD-treated cells. MG132 not only prevented the YFP-L11 loss but also resulted in a net increase of nucleolar YFP-L11 in siL5- and ActD-treated cells (Fig. 4B). Notably, under these conditions HA-Ub did not colocalize with YFP-L11 (Fig. 4B). The rescue of YFP-L11 expression in siL5- and ActD-treated cells by MG132 was prevented by CHX (Fig. 4B). These results strongly indicate that ribosome-free L5 is required to prevent degradation of newly synthesized L11 upon ActD treatment through a proteasome-dependent, ubiquitin-independent pathway (Fig. 4 A and B) (33).
Fig. 4.
Accumulation of ribosome-free L5 and L11 upon ActD treatment is due to their mutual protection from proteasomal degradation. (A) A549 cells were transfected with the indicated siRNAs for 48 h and then treated with ActD for 5 h. Nonribosomal fractions were immunoblotted with the indicated antibodies. (B) H1299 cells expressing YFP-L11 (green) were transfected with L5 siRNA for 24 h and then transfected with HA-Ub for 24 h. Cells were then treated as indicated for 6 h, stained with anti-HA antibody (red), and analyzed by CLSM. (Scale bar, 5 μm.)
Newly Synthesized L5 and L11 Are Imported into Nucleoli upon ActD Treatment.
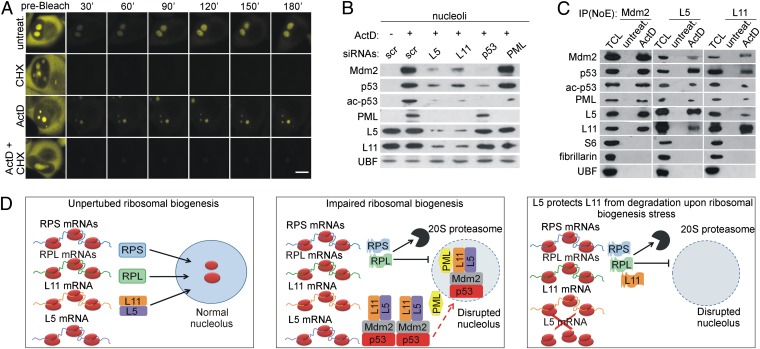
The fact that the amount of nucleolar L5 and L11 in ActD-treated A549 cells is significantly decreased by CHX indicates that newly synthesized L5 and L11 continue to accumulate in nucleoli upon ribosomal stress (Fig. S7A). To provide further support for this finding, we compared the dynamics of nucleoar recovery of YFP-L11 between untreated and ActD-treated H1299 cells by using fluorescence recovery after photobleaching (7, 27, 30). In the presence of ActD, we observed a significant recovery of fluorescent YFP-L11 in the nucleoli after photobleaching. Notably, CHX abolished that recovery (Fig. 5A and Fig. S7B), arguing that it was dependent on de novo protein synthesis rather than on import of preexisting protein. The newly synthesized YFP-L11 colocalized with fibrillarin and UBF in untreated cells (Fig. S8A). On the other hand, in ActD-treated cells, it localized in the nucleolar remnant (central body) in an RNA-dependent manner and was surrounded by fibrillarin and UBF cap structures (Fig. S8A).
Fig. 5.
Newly synthesized L5 and L11 colocalize with Mdm2, p53, and PML in the nucleolus upon ribosomal stress. (A) H1299 cells expressing YFP-L11 were treated with ActD for 5 h in the presence or absence of CHX and then photobleached using a 488-nm laser. Time-lapse laser scanning microscopy was used to monitor the recovery of the nucleolar fluorescence signal. (Scale bar, 5 μm.) (B) A549 cells were transfected with siRNAs against L5, L11, p53, or PML for 48 h and then left untreated or treated with ActD for 5 h. Purified nucleoli were immunoblotted with the indicated antibodies. (C) Nucleolar extracts (NoE) from untreated or ActD-treated (5 h) A549 cells were immunoprecipitated with antibodies against Mdm2, L5, or L11. These immunoprecipitates and total cell lysate (TCL) from ActD-treated A549 cells were immunoblotted with the indicated antibodies. (D) (Left) Under normal conditions, newly synthesized RPs of 40S (RPS) and 60S (RPL) ribosomal subunits are imported into the nucleolus. (Middle) Upon impairment of ribosomal biogenesis, the majority of RPL and RPS are synthesized, but they are degraded by nuclear 20S proteasomes. In contrast, L5 and L11 are not degraded, and they accumulate in the nonribosomal fraction, where they bind Mdm2. L5 and L11 colocalize with Mdm2, p53, and PML in the nucleolus after impairment of ribosomal biogenesis, where full p53 activation probably takes place. Less efficient import of newly synthesized L5 and L11 into the nucleolus upon inhibition of ribosomal biogenesis may also contribute to their accumulation in the nonribosomal fraction (indicated by dashed red arrow). (Right) In the absence of L5, ribosome-free L11 is degraded by proteasomes upon ribosomal biogenesis stress, suggesting that L5 and L11 protect each other from degradation and explaining their mutual requirement in p53 activation.
Newly Synthesized L5 and L11 Colocalize with p53, Mdm2, and Promyelocytic Leukemia in the Nucleolus upon Ribosomal Stress.
The observation that newly synthesized L5 and L11 accumulate in nucleoli upon ActD treatment suggests that they may play a role in p53 activation at that subnuclear location. To gain insight into this, we first tested the expression of endogenous p53 and Mdm2 in highly purified nucleoli from untreated and ActD-treated A549 cells by immunoblotting (Fig. 5B). Because the promyelocytic leukemia (PML) tumor suppressor forms nucleolar caps upon low-dose ActD treatment and regulates p53 acetylation at K382 (34, 35), we also tested the expression of nucleolar PML and p53 acetylated at K382 (ac-p53) under the same conditions (Fig. 5B). ActD treatment led to increased levels of nucleolar p53, ac-p53, Mdm2, and PML (Fig. 5B). CLSM confirmed localization of these proteins within the abberant nucleoli of ActD-treated A549 and H1299 cells (Fig. S8B). Under these conditions, a portion of Mdm2 and p53 colocalized with UBF, forming nucleolar cap-like structures that eventually engulfed the nucleolar remnants (Fig. S8B). Notably, p53 was also localized to the nucleolus following depletion of S6 in A549 cells (Fig. S8C). Coimmunoprecipitation experiments from the nucleolar lysates of ActD-treated A549 cells revealed that Mdm2 forms a complex with p53, L5, L11, and PML, but not with S6, fibrillarin, and UBF (Fig. 5C).
It was reported that L11 down-regulation impairs the ability of PML to localize to nucleoli upon doxorubicin treatment (36). Here, we determined the requirement of L5 and L11 for nucleolar localization of endogenous p53, Mdm2, and PML in response to ActD treatment of A549 cells by immunoblotting (Fig. 5B). Depletion of either L5 or L11 impaired PML, p53, ac-p53, and Mdm2 nucleolar accumulation (Fig. 5B). The expression levels of PML, L5, and L11 within nucleoli in ActD-treated A549 cells were independent of p53 and Mdm2 (Fig. 5B). Depletion of PML in these cells did not prevent association of p53, Mdm2, L5, and L11 with nucleoli, although it abolished accumulation of nucleolar ac-p53 (Fig. 5B). Thus, L5 and L11 are required for the recruitment of PML, p53, ac-p53, and Mdm2 to the nucleolus upon ActD treatment. Furthermore, our results show that L5- and L11-dependent recruitment of PML to nucleoli is independent of p53 and Mdm2. Interestingly, ac-p53 also accumulated in nonribosomal cytoplasmic and nucleoplasmic fractions in response to ActD, whereas PML depletion abrogated this ac-p53 accumulation without significantly affecting p53 levels (Fig. S8D).
Together, these results suggest that continuous nucleolar accumulation of newly synthesized L5 and L11 together with PML, Mdm2, and p53 have an important role in p53 activation upon ActD treatment.
Discussion
Here, we demonstrate that L5 and L11 are critical mediators of the activation of p53 by impaired ribosome biogenesis. Upon exposure of cells to various ribosomal stressors, L5 and L11 are unique among several RPs examined in being able to accumulate in the nonribosomal fraction where they specifically bind Mdm2. This further points to their specific role in activating p53. Importantly, the L5/L11-Mdm2-p53 pathway has been convincingly supported by an in vivo mouse model (37). RPs are among the most abundant proteins in mammalian cells and are highly basic proteins (29). If free from ribosomes, RPs can specifically or nonspecifically interact with various macromolecules, including proteins and RNA. Indeed, we were able to coimmunoprecipitate all tested RPs with Mdm2 from the total cell lysate. Our results are consistent with a recently published study in which a critical role of L5 and L11, but not of L23 and S7, in p53 activation by ActD and RP deficiencies was reported (23). However, our findings differ from previously published studies that showed that depletion of RPS7 (19–20), L23 (16–17), or L26 (18) compromises the induction of p53 after pharmacological inhibition of ribosome biogenesis. We do not know the basis for these differences, which could be due to time of treatment with the drug, siRNA sequences used, or cell-line differences. Given that these studies have convincingly shown that S7, L23, and L26 interact with Mdm2 and efficiently inhibit its ubiquitin ligase activity toward p53 (16–20), we cannot rule out the possibility that they can regulate p53 levels in certain cell types or upon exposure to specific ribosomal biogenesis stressors, or with kinetics different from L5 and L11. Additionally, these and possibly some other RPs can trigger p53 activation when they are made in excess even in the absence of “ribosomal biogenesis stress,” which may occur in certain situations, as, for example, when c-Myc becomes hyperactivated (7) or after transfection of specific RPs (6).
Considerable controversy exists regarding the source of ribosomal stress-induced ribosome-free “p53-activating” RPs (6, 22, 38). Here, we showed that ribosome-free L5 and L11 are synthesized upon ActD treatment. Although we do not detect a significant decrease in the levels of nucleolar L5 and L11 upon ActD treatment, we cannot exclude the possibility that a small portion of the newly synthesized nucleolar L5 and L11 may contribute to the accumulation of ribosome-free L5 and L11. We suggest that, in addition to new protein synthesis (Fig. 2C and Fig. S4D) (22, 38), the interdependent protection of ribosome-free L5 and L11 from proteasomal degradation may contribute to their accumulation in the nonribosomal fraction and p53 activation upon ribosomal stress (Fig. 5D and Fig. S7B).
It is known that RPs continue to be synthesized upon ActD treatment and that they are rapidly degraded by proteasomes (29–31). Here we showed that YFP-S6, YFP-L4, and endogenous S6 and S12 form nucleoplasmic aggregates in the presence of ActD and MG132, suggesting that they are degraded by proteasomes upon impairment of ribosome biogenesis. However, we failed to demonstrate colocalization of ubiquitin with these aggregates, implying that their degradation occurs through a proteasome-dependent, ubiquitin-independent pathway (33). Previous studies suggested that abnormal/misfolded proteins can be subjected to ubiquitin-independent proteasomal degradation upon cellular stress (33). It could be speculated that newly synthesized RPs, when not bound to rRNA, assume abnormal 3D structures that make them susceptible to degradation by the 20S proteasome, without a requirement for ubiquitin conjugation (Fig. 5D) (33). Such a mechanism would prevent potentially toxic accumulation of unbound, free RPs in the nucleoplasm (30). This raises the question as to why L5 and L11 are also not degraded upon ribosomal stress. The mutual protection of ribosome-free L5 and L11 from proteasomal degradation upon ribosomal stress may also offer an explanation for their codependence in p53 up-regulation (Fig. 5D) (21). It has been suggested that 5S rRNA forms a complex with Mdm2, L5, and L11 (21, 39). However, further studies will be required to fully understand a possible role of 5S rRNA in L5- and L11-dependent p53 activation.
What is the role of the nucleolus in p53 activation by ribosomal biogenesis stress? All tested ribosomal stressors caused functional and/or structural alterations in the nucleolus. Despite evident nucleolar disruption by ActD, we did not observe a significant decrease in the nucleolar levels of endogenous L5 and L11, which is in contrast with other studies in which the fate of exogenous RPs was followed (6). Surprisingly, endogenous L5 and L11 continue to accumulate in the nucleolar remnants where they colocalize with PML, Mdm2, and p53 under these conditions, suggesting that the disrupted nucleoli may provide a platform for PML-mediated posttranslational modifications and activation of p53 upon ribosomal stress (Fig. 5D) (34). An L11-dependent nucleolar localization of PML upon doxorubicin treatment has been demonstrated (36). Here, we showed that L5- and L11-mediated nucleolar localization of PML following ActD treatment is independent of Mdm2 and p53.
We speculate that activated p53 then relocalizes from nucleoli to nucleoplasm and cytoplasm. Future studies will be required to understand the relationship between nucleolar p53/Mdm2/PML/L5/L11 and accumulation of nucleoplasmic and cytoplasmic p53.
In summary, we demonstrate that L5 and L11 are required for p53 activation in response to various ribosomal biogenesis stressors. Whereas other newly synthesized RPs are degraded by proteasomes upon ActD treatment, L5 and L11 accumulate in the nonribosomal fraction where they interact with Mdm2. Importantly, we demonstrate that this selective accumulation of ribosome-free L5 and L11 is largely due to their mutual protection from proteasomal degradation. The observation that newly synthesized L5 and L11 continue to accumulate in nucleoli together with Mdm2, p53, and PML even after impairment of ribosome biogenesis suggests that the altered nucleoli may provide a site for L5- and L11-dependent p53 activation. A potential implication of our findings is that normal p53-activating functions of L5 and L11 might be compromised in various diseases (11).
Materials and Methods
Antibodies against RPs.
Rabbit polyclonal antibodies against L11 were produced previously (5). Here, we generated polyclonal antibodies against S7, S20, L5, L12, L23, and L26 and monoclonal antibodies against S3, S6, and S12 after immunization of rabbits or mice with specific His-tagged mouse RPs, respectively. Details of other methods used in this research are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Uri Alon for his kind gift of H1299 cells expressing YFP-RPs; Jon Ashwell and Nick Pullen for critical reading of the manuscript; and Ivan Dikic and Koraljka Husnjak for providing the HA-Ub plasmid. This work was supported by European Commission Seventh Framework Programme grants Inflammation and Cancer Research in Europe (INFLA-CARE) and Translational Medical Research in Rijeka (TransMedRi); the Unity through Knowledge Fund; a grant from the Ministry of Science, Education, and Sports of Croatia (to S.V.); and National Institutes of Health Grant CA58316 (to C.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218535109/-/DCSupplemental.
References
- 1.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1(5):a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol. 2010;2(3):a001016. doi: 10.1101/cshperspect.a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volarevic S, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288(5473):2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 5.Sulic S, et al. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 2005;19(24):3070–3082. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Lu H. Signaling to p53: Ribosomal proteins find their way. Cancer Cell. 2009;16(5):369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golomb L, et al. Importin 7 and exportin 1 link c-Myc and p53 to regulation of ribosomal biogenesis. Mol Cell. 2012;45(2):222–232. doi: 10.1016/j.molcel.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones NC, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14(2):125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barkić M, et al. The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol Cell Biol. 2009;29(10):2489–2504. doi: 10.1128/MCB.01588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellagatti A, et al. Induction of p53 and up-regulation of the p53 pathway in the human 5q- syndrome. Blood. 2010;115(13):2721–2723. doi: 10.1182/blood-2009-12-259705. [DOI] [PubMed] [Google Scholar]
- 11.Fumagalli S, Thomas G. The role of p53 in ribosomopathies. Semin Hematol. 2011;48(2):97–105. doi: 10.1053/j.seminhematol.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279(43):44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 13.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23(12):2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3(6):577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23(23):8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai MS, et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24(17):7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24(17):7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res. 2010;38(19):6544–6554. doi: 10.1093/nar/gkq536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: Binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26(35):5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Y, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35(3):316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27(44):5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- 22.Fumagalli S, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11(4):501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fumagalli S, Ivanenkov VV, Teng T, Thomas G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012;26(10):1028–1040. doi: 10.1101/gad.189951.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapik YR, Fernandes CJ, Lau LF, Pestov DG. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol Cell. 2004;15(1):17–29. doi: 10.1016/j.molcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 25.Robledo S, et al. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14(9):1918–1929. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen JS, et al. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12(1):1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 27.Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322(5907):1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 28.Volarević S, Thomas G. Role of S6 phosphorylation and S6 kinase in cell growth. Prog Nucleic Acid Res Mol Biol. 2001;65:101–127. doi: 10.1016/s0079-6603(00)65003-1. [DOI] [PubMed] [Google Scholar]
- 29.Warner JR. In the absence of ribosomal RNA synthesis, the ribosomal proteins of HeLa cells are synthesized normally and degraded rapidly. J Mol Biol. 1977;115(3):315–333. doi: 10.1016/0022-2836(77)90157-7. [DOI] [PubMed] [Google Scholar]
- 30.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17(9):749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen JS, et al. Nucleolar proteome dynamics. Nature. 2005;433(7021):77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 32.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10(10):1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nat Chem Biol. 2009;5(11):778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- 34.Condemine W, Takahashi Y, Le Bras M, de Thé H. A nucleolar targeting signal in PML-I addresses PML to nucleolar caps in stressed or senescent cells. J Cell Sci. 2007;120(Pt 18):3219–3227. doi: 10.1242/jcs.007492. [DOI] [PubMed] [Google Scholar]
- 35.Ito A, et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20(6):1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernardi R, et al. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6(7):665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 37.Macias E, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell. 2010;18(3):231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donati G, et al. The balance between rRNA and ribosomal protein synthesis up- and downregulates the tumour suppressor p53 in mammalian cells. Oncogene. 2011;30(29):3274–3288. doi: 10.1038/onc.2011.48. [DOI] [PubMed] [Google Scholar]
- 39.Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol. 1994;14(11):7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.