Abstract
Multidrug resistance in Gram-negative bacteria, to which multidrug efflux pumps such as the AcrB transporter makes a major contribution, is becoming a major public health problem. Unfortunately only a few compounds have been cocrystallized with AcrB, and thus computational approaches are essential in elucidating the interaction between diverse ligands and the pump protein. We used molecular dynamics simulation to examine the binding of nine substrates, two inhibitors, and two nonsubstrates to the distal binding pocket of AcrB, identified earlier by X-ray crystallography. This approach gave us more realistic views of the binding than the previously used docking approach, as the explicit water molecules contributed to the process and the flexible binding site was often seen to undergo large structural changes. We analyzed the interaction in detail in terms of the binding energy, hydrophobic surface-matching, and the residues involved in the process. We found that all substrates tested bound to the pocket, whereas the binding to this site was not preferred for the nonsubstrates. Interestingly, both inhibitors [Phe-Arg-β-naphthylamide and 1-(1-naphtylmethyl)-piperazine] tended to move out of the pocket at least partially, getting into contact with a glycine-rich loop that separates the distal pocket from the more proximal region of the protein and is thought to control the access of substrates to the distal pocket.
Keywords: bacterial drug efflux pump, drug-binding pocket, efflux inhibitors
The past decades have seen the return of bacterial resistance as a major problem in public health (1, 2). Of particular concern is the appearance of multidrug-resistant (MDR) Gram-negative bacteria (1, 2), where the efflux pumps of the resistance nodulation division (RND) family make a major contribution to the MDR phenotype (3, 4). Substrates of these pumps include antibiotics and biocides with very different structural features, although they all seem to have a significant lipophilic portion (5). RND pumps are tripartite complexes (5, 6). In the major MDR efflux pump in Escherichia coli, AcrAB-TolC, the RND secondary transporter AcrB, driven by the proton-motive force (7), determines the substrate specificity.
The structure of AcrB, a homotrimer, has been solved in a threefold symmetric form (8) and, later, in an asymmetric conformation (9–11), where each protomer assumes a different conformation: access, binding, and extrusion (9). Furthermore, minocycline and doxorubicin were cocrystallized within a distal binding pocket located in the periplasmic domain of the binding conformer (9) (Fig. 1). On the basis of these findings, a functional rotation hypothesis was suggested, in which each protomer assumes successively one of the aforementioned conformations (9–11). This idea has been supported by several biochemical studies (12–14), as well as by molecular simulation (15, 16). It thus seems that this distal binding pocket (Fig. 1), which collapses in the extrusion protomer, plays a major role in the binding and selection of substrates by AcrB. Indeed site-directed mutagenesis and real-time efflux experiments confirmed this hypothesis (17–21), although very recently some antibiotics have been cocrystallized bound to a more proximal binding pocket in the access protomer (22, 23), which presumably represents an earlier stage in the drug efflux process, and it is consistent with earlier cocrystallization and biochemical studies (24, 25).
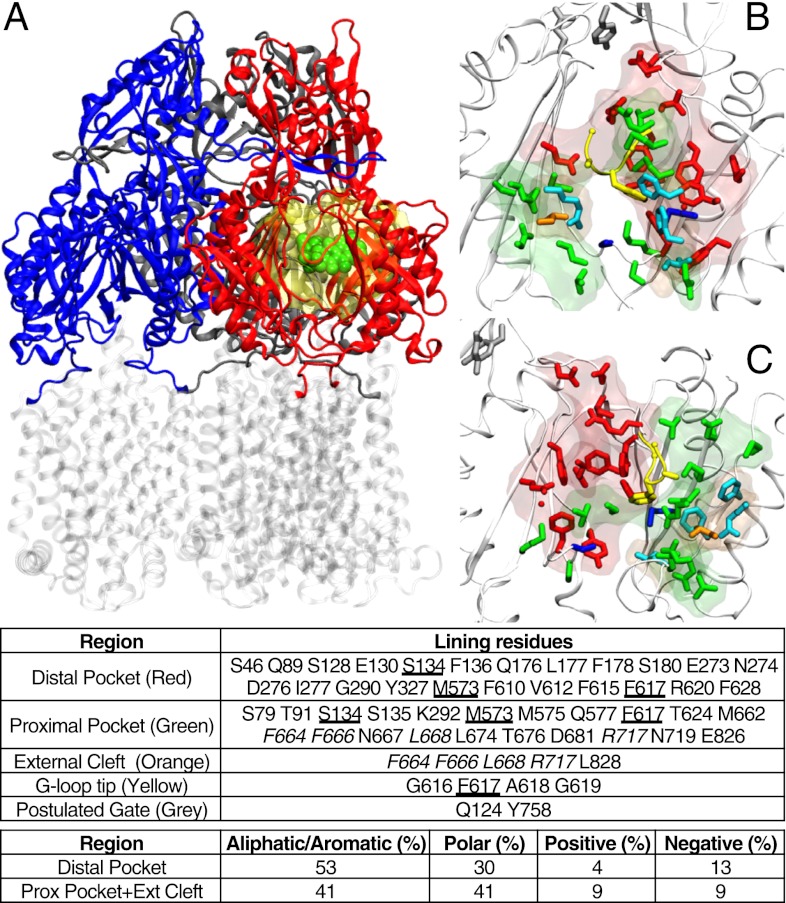
Fig. 1.
(A) Reduced model of AcrB used in this work. The transmembrane domain (in transparent gray) was cut off from the protein, and only the periplasmic domain (blue, red, and gray for access, binding, and extrusion monomer, respectively; residues 33–335 and 565–871 of the intact protein) was kept. The substrate is shown in spheres (green), and the molecular surface of the residues not restrained during the partially restrained MD is shown as a transparent yellow surface. (B and C) Front (B) and side (C) views of the regions of AcrB interacting with substrates (binding monomer; PDB ID code 4DX5). The entrance of the cleft, the proximal, and the distal pockets are shown as transparent surfaces, and the side chains of residues lining them are shown as sticks (orange, green, and red, respectively). Some residues (in addition to those belonging to the G-loop) are shared between the distal and proximal pocket (blue sticks), and four residues of the cleft entrance are also defined as belonging to the proximal cleft (cyan sticks). In addition, the residues of the tip of G-loop and of the gate are shown with yellow and gray sticks respectively. In the table identifying the residues belonging to each region (Lower), those common to the cleft and the proximal pocket are italicized, and those shared between the proximal and the distal pockets are underlined.
The distal binding pocket in the binding protomer is extensive (surrounded by more than 20 residues) and contains many hydrophobic residues, as well as several charged and polar ones (Fig. 1). These features fit with the extreme variety of substrates recognized by AcrB (6), ranging from basic dyes to most antibiotics, detergents, and even solvents (26, 27). It is of crucial importance, therefore, to understand the molecular mechanism by which such a diverse range of substrates are recognized by AcrB; this can help in the design of antimicrobial agents that would not be pumped out quickly, and of better inhibitors of these pumps. However, only two drugs have been cocrystallized within the distal pocket (9, 23), and we must use computational methods to predict the interaction of other ligands with this pocket. An initial attempt was made by using a computer docking program (28). Docking calculations, however, are subject to serious limitations, such as the lack of consideration of protein backbone flexibility (29), the absence of explicit water molecules, and the fact that the scoring functions are derived mostly from the complexes whose binding pockets exclude water (30). These issues are especially serious with AcrB, which is very flexible (15, 21, 31) and features a direct connection between the distal binding pocket and a very large, presumably water-filled, tunnel (28).
In this work we reconsider the binding of a dozen compounds to the distal pocket of AcrB by means of all-atom molecular dynamics (MD) simulations. The compounds include good substrates as well as poor substrates and inhibitors. The results not only deepened our understanding of the ligand-binding process in AcrB but also allowed us to speculate on the possible action mechanism of inhibitors.
Results
The binding of the following compounds to the distal binding pocket of AcrB was characterized through MD simulations, and included known substrates of the pump: minocycline (MIN), taurocholic acid (TAU), nitrocefin (NCF), chloramphenicol (CHL), ethidium (ETH), oxacillin (OXA), ciprofloxacin (CIN), cephalothin (CEF), and also erythromycin (ERY), which was shown to bind preferentially to a proximal site (22). The inhibitors 1-(1-naphtylmethyl)-piperazine (NMP) and phenylalanyl-arginine-β-naphthylamide (PAβN) were also examined. Finally, we evaluated the binding of two nonsubstrates, kanamycin A (KAN) and glucose (GLC) (Fig. S1; Table 1).
Table 1.
Free energy of binding and the surface-matching coefficients for different compounds evaluated in this study
| % of ΔGb |
Surface matching |
|||||||
| Cpd* | ΔGb value (kcal/mol)† | Distal pocket‡ | Proximal pocket | G-loop | Interface | External cleft | SML§ | SMH§ |
| MIN | −29.3 ± 4.7 (−22.7)¶ | 40 | — | — | — | — | 0.81 (0.80)¶ | 0.75 (0.67) |
| TAU | −28.4 ± 4.1 (−18.1) | 61 | — | — | — | — | 0.78 (0.69) | 0.50 (0.79) |
| ERY | −43.5 ± 5.3 (−10.0) | 24 | — | — | — | — | 0.45 (0.42) | 0.60 (0.66) |
| ERYA | −54.4 ± 5.8 (−7.7) | — | 12 | 4 | — | 17 | 0.66 (0.71) | 0.50 (0.67) |
| NCF | −42.5 ± 3.9 (−18.9) | 41 | — | — | — | — | 0.62 (0.54) | 0.29 (0.27) |
| CHL | −23.3 ± 4.6 (−20.3) | 52 | — | — | — | — | 0.78 (0.50) | 0.59 (0.20) |
| ETH | −31.0 ± 2.1 (−31.2) | 41 | — | — | — | — | 0.90 (0.84) | 0.00 (0.67) |
| OXA | −23.2 ± 4.2 (−20.9) | 52 | — | — | — | — | 0.70 (0.74) | 0.33 (0.60) |
| CIN | −22.9 ± 5.0 (−22.4) | 43 | — | — | 14 | — | 0.69 (0.55) | 0.48 (0.10) |
| CEF | −34.6 ± 3.3 (−28.8) | 32 | — | — | — | — | 0.77 (0.68) | 0.89 (0.84) |
| CEFA | −28.1 ± 5.0 (−20.5) | — | — | 9 | — | 48 | 0.51 (0.38) | 0.33 (0.78) |
| NMP | −22.3 ± 2.4 (−21.8) | 20 | — | 6 | 3 | 3 | 0.77 (0.67) | 0.48 (0.10) |
| NMP′ | −20.4 ± 2.9 (−11.6) | — | 3 | 7 | — | 14 | 0.63 (0.77) | 0.61 (0.38) |
| PAβN | −30.1 ± 5.3 (−2.4) | 29 | — | — | — | — | 0.68 (0.58) | 0.62 (0.19) |
| GLC | −22.4 ± 3.7 (−22.2) | 24 | — | — | — | — | 0.00 (0.00) | 0.84 (0.97) |
| KAN | −32.6 ± 6.7 (18.0) | 29 | — | — | — | — | 0.00 (0.00) | 0.64 (0.82) |
The calculations for all compounds refer to the drugs bound to the distal pocket of the binding protomer, except ERYA and CEFA, which deal with the drugs bound to the proximal pocket of the access protomer. NMP and NMP′ represent two independent runs of NMP from the distal binding site.
The contribution of the configurational entropy of the solute has not been included (SI Materials and Methods). Concerning the contributions of different regions (defined in Fig. 1) to ΔGb, only those larger than 2% are listed.
Residues within the various regions are listed in Fig. 1.
Calculated on the conformation of the complex with the lower rmsd from the average extracted from the unbiased MD simulations. SML and SMH refer to the lipophilic and hydrophilic-surface matching coefficients. See SI Materials and Methods for further details.
Values in parentheses are those for starting poses. ΔGb was calculated after 1,000 steps of structural optimization with restraints on heavy atoms of both protein and the ligand, to avoid the artifacts of high positive values created by the differences in bond lengths, angles, etc. specified by the docking program and the AMBER force field.
Validation of the Reduced Model of AcrB.
Because the number of ligands examined was large, we used a reduced model of AcrB, which did not contain the transmembrane domains (Fig. 1). An extensive validation of this model is reported in SI Materials and Methods (Figs. S2–S4).
Time Course of the MD Simulation.
In the initial phase of simulation, lasting ∼20 ns in most cases (Table S1), a partially restrained simulation was carried out by applying harmonic restraints (k = 1 kcal⋅mol−1⋅Å-2) on all Cα atoms except those near the ligand (see SI Materials and Methods for details). Then, unrestrained simulation of 48–83 ns was carried out (Table S1). In all cases, after a few nanoseconds of unbiased MD, the protein entered into a state of oscillation around an average conformation with very little drifts in the Cα-rmsd (black curves in Fig. S5). This finding is consistent with recent computational studies of the full model of AcrB (31, 32). With such compounds as OXA and NMP, which showed a rather unstable behavior (Fig. S5), multiple simulations were performed (Table S1).
Major Improvements Generated by MD Simulation.
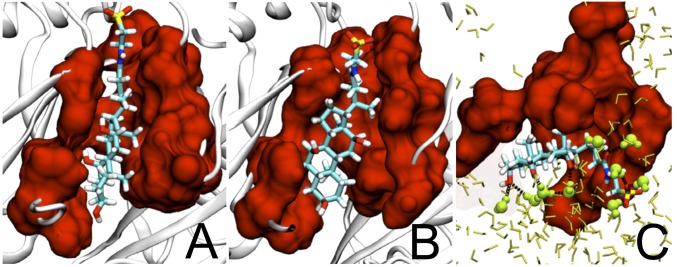
In contrast to docking, MD simulation introduces dynamics and an aqueous medium, which is relevant because the deep binding pocket of AcrB faces a large, presumably water-filled channel (figure 2C in ref. 28). As an example, we can examine the binding of TAU (Fig. 2). Bile salts are unusual detergents containing hydrophobic and hydrophilic groups on the different sides or faces of the planar structure (33). In the structure obtained by docking, the two sides of TAU are both facing the walls of the groove (28) of the binding pocket, lined by residues 178, 277, 279, 280, 285, 610, 612, and 615. In contrast, the plane of the molecule turned by ∼90° after 10 ns of partially biased MD (see SI Materials and Methods for details), so that its hydrophobic side faced the hydrophobic surface of the protein, and its hydrophilic side, with its three hydroxyl groups, faced outward to the water-filled channel, a pose clearly more likely to occur in the real AcrB protein. Similarly strong interaction with water molecules occurs with most other substrates and inhibitors (Table S2). Thus, PAβN, which is predicted to bind tightly to the distal pocket by docking (28), interacted strongly with water molecules in the channel in the MD simulation, with its Arg side-chain and the N-terminal amino group now sticking out into the channel (Fig. 3). In addition, H-bonds to groups on the binding pocket are also optimized, and the details with all of the compounds can be seen in Fig. S6.
Fig. 2.
Binding of TAU to the distal binding pocket. The binding pocket residues, defined in ref. 28, are shown as a red surface, and TAU is shown in a stick model with carbon atoms in cyan. (A) The binding pose found by a docking program, AutoDock Vina. (B) The binding pose after optimization with unrestrained MD simulation. (C) The view from the top of the binding complex shown in B, with water molecules. Those in H-bonding interaction with the ligand are shown as yellow-green spheres. In all views, structures in front of those shown were clipped off.
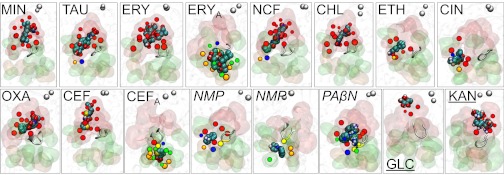
Fig. 3.
Average positions of the examined ligands (in spheres colored according to atom types, with nonpolar hydrogens removed) at the final phase of MD simulation, with respect to the distal (transparent red surface) and proximal (green surface) pockets, and the cleft (orange surface). The tip of the G-loop is shown as a gray cartoon. Residues that are within 3.5 Å from the ligand are shown as beads (red, green, orange, and yellow for those of distal and proximal pockets, cleft, and G-loop, respectively). Those shared by the two pockets are colored blue. For orientation, residues in the gate area (far away from the ligand) are shown as gray beads.
Another major improvement obtained by MD simulation is the optimization of ligand/protein contacts achieved by the movement of binding-site residues. This optimization is evident in the case of TAU, whose tail lost contact with the narrow groove identified earlier (28), due to conformational changes in this region of AcrB (Fig. 3). The 3.4 ± 0.3 Å rmsd of the groove, calculated with respect to the X-ray structure 2J8S, was significantly larger than that for MIN (2.5 ± 0.2 Å), where no large conformational alteration occurred in this area. Similarly, the groove conformation was altered more by the binding of CEF and OXA (rmsds of 3.5 ± 0.2 Å and 3.3 ± 0.5 Å, respectively) than of CHL and NCF (rmsds of 2.3 ± 0.2 Å and 2.4 ± 0.2 Å, respectively). The flexible conformation of the binding pocket, determined by the nature of the bound ligand, apparently helps AcrB in accommodating substrates of very different properties.
Binding of Substrates.
MD simulation confirmed the conclusion from the previous docking studies (28) in its broadest outline, because all of the noninhibitor substrates remained bound in the distal binding site (Fig. 3; Fig. S6). In most cases the ligand moved only a few angstroms from the position predicted by the docking program (Figs. S5 and S6). [Exceptions (CHL, OXA, and CIN) are discussed below]. In contrast to these substrates that remained in the pocket, the nonsubstrates GLC and KAN were predicted to bind outside the distal pocket by the docking program searching a 20 × 20 × 20 Å grid centered on the binding pocket, and remained outside the pocket even after long simulation runs (Fig. 3).
When we examined the docking of substrates earlier by using the binding protomer of AcrB model 2DRD (9), we saw that some, including MIN and NCF, bound to the narrow groove of the distal pocket, whereas others, including CHL, bound to a more open, cave-like area at the bottom half of the pocket (28), containing residues 136, 139, 327, 617, 626, and 628. This distinction, however, did not hold up in the present study. In fact, docking with the model 2J8S (11), the model used in this work, predicted CHL to bind to the groove area (Fig. S6), and MD simulation did not much alter its overall location despite its vertical flipping mentioned below (Fig. 3; Fig. S6). It appears that the narrower groove in the model 2DRD prevented the binding of CHL to this area. The implication of these results is examined in Discussion.
We estimated the strength of ligand binding in two ways. In the first approach, binding free energies were calculated by molecular mechanics–generalized Born surface area (MM-GBSA) method (34) as detailed in SI Materials and Methods. Table 1 shows that most of these substrates bound to the distal site with a higher affinity, or larger negative values of ΔGb, after MD simulation compared with the energy-minimized docked poses. Residues in the distal binding pocket contributed in general more than 40% of ΔGb for substrates (except with ERY and CEF, discussed below). In contrast, with the nonsubstrates GLC and KAN, the residues in the distal binding pocket contributed only small fractions of ΔGb, as predicted.
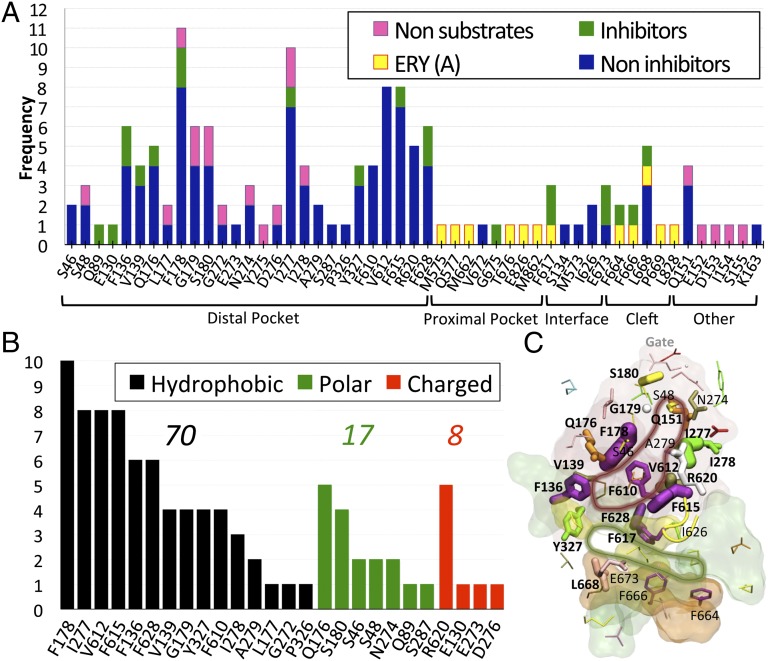
The contribution of various residues to binding of all ligands examined is summarized in Fig. 4. Residues F178, I277, V612, and F615 contributed to the stabilization of almost all substrates. Furthermore, in addition to 15 hydrophobic residues, 11 polar or charged amino acid residues were found to contribute to the binding. Such a wide spectrum of interaction types is consistent with the multidrug recognition capacity of AcrB.
Fig. 4.
(A) Frequency of contribution to the binding free energy from different residues belonging to the key regions defined in Fig. 1 and from additional residues. Only residues contributing with more than kT (∼0.6 kcal/mol at 310 K) are reported. Amino acids shared among different regions are reported only in one. The residues belong to the binding monomer for all complexes but ERYA, where they belong to the access monomer. (B) Frequency of contribution to the binding free energy of substrates (thus excluding GLC and KAN) by hydrophobic (black bars), polar (green), and charged (red) residues. Above each group is the sum of all frequencies. (C) Residues are shown with sticks, whose width is proportional to the frequency of binding contacts to ligands. In red, green, orange, and yellow transparent surfaces are shown the distal and proximal pockets, the entrance cleft, and the distal/proximal interface, respectively. The tip of the G-loop is also shown in yellow cartoon. The residues having frequency of contribution to binding larger than three (considering substrates and nonsubstrates of the transporter) are also labeled. Bold labels refer to residues whose frequency of contribution to substrates (inhibitors or not) is larger than three. The red and green lines highlight the distal and proximal pockets, respectively, drawn according to this analysis.
We also used a second approach, implemented in the software PLATINUM (35) (SI Materials and Methods), which correlates the affinity to the matching between the hydrophobic and hydrophilic surfaces of the ligand and the protein. This analysis (Table 1) also showed that the lipophilic surface matching was improved in all cases in comparison with the starting, docked structure, confirming that MD simulation resulted in a more realistic fit between the ligand and the binding site. In contrast, the matching between hydrophilic surface patches was not usually improved.
Details of the interactions of the ligand with protein residues can be examined by using MIN as an example. First, the pose predicted by the MD simulation was in very good agreement with the crystallographic structures (9, 23) (Fig. S4), and MIN interacted with the key residues identified there. With respect to docking, most of the initial contacts were conserved along the MD trajectory, and the new stable interactions were formed with residues S48 and S180 (Fig. S6). Importantly, polar contacts contributed significantly to the stabilization of the MIN–AcrB complex (Table S2), although hydrophobic residues (F178, I277, V612, and F615) had the highest impact. The basic residue R620 also contributed to stabilize the complex, whereas D276 (the only acidic residue within 3.5 Å from the substrate) did not. The details for other compounds are shown in Fig. S6.
During MD simulation, significant rearrangements occurred for CHL, OXA, and CIN in comparison with their original docked poses (Figs. S5 and S6). CHL especially underwent a vertical flipping so that the nitrophenyl group, which was at the top after docking, now was at the bottom (Fig. S6; Movie S1). However, even with these substrates the center of the mass of ligand molecule was moved only by several (≤5) angstroms during MD simulation (Fig. S5, Bottom). This forms a marked contrast with the movement of inhibitors described below.
ERY and CEF.
ERY was unusual among substrates in that its docked structure had only small values of both ΔGb and lipophilic surface matching coefficient (Table 1). Although MD simulation improved the value of ΔGb, the residues in the distal pocket contributed only 24% to it (Table 1), and the lipophilic matching coefficient remained low. Most importantly, we found that ERY bound much better to the proximal site (ERYA in Table 1 and Fig. 3), a result consistent with the recent report (22) of the crystal structure of AcrB with bound ERY in the proximal site.
With the binding of CEF to the distal site, again the residues of this pocket contributed only 32% to the binding energy (Table 1), although the lipophilic matching coefficient was comparable to that of other substrates. Thus, we examined the binding of CEF to the proximal site (CEFA in Table 1). In contrast to ERY, its affinity to the proximal site was weaker than that to the distal site.
Binding of Inhibitors.
The outcome of MD simulation was different for efflux inhibitors tested. Indeed, both PAβN and NMP moved quite far away (∼9–16 Å) from the initial docked positions in the distal pocket (Fig. 3; Figs. S5 and S6). With PAβN, docking predicted tight binding of most of this molecule to the deep groove (28) (see above) of the distal site, whereas MD simulation moved the positively charged Arg side-chain and the N-terminal amino group out of the binding site so that they could interact with water molecules in the channel. Even more importantly, the binding-site residues moved significantly to eliminate, essentially, the crevice of the original structure, pushing PAβN in a direction closer to the proximal site (Fig. 3; Fig. S6). This large movement of PAβN caused it to straddle the “G-loop” structure [called also F617 loop (22) or switch loop (23)], which separates the distal binding site from the proximal site. Consistently, the residues of the distal site were less involved in the stabilization of the complex than with substrates (Table S2).
The different behavior of inhibitors with respect to substrates was even more evident with NMP. Not only did this compound moved toward the G-loop, but in one simulation it was able to reach parts of the proximal pocket (Fig. 3, NMP′; Movie S2). In view of this behavior, we performed three additional simulations (in total two for each of the two score-equivalent poses found by AutoDock Vina; Figs. S6 and S7). In all simulations there was a significant (≥9 Å) displacement of NMP toward the proximal pocket. The thermodynamic stabilities and the average final conformations of NMP were similar, although its orientation was different (Table 1; see legend to Fig. S7). Moreover, in all simulations, NMP straddled the tip of the G-loop (Fig. 3; Fig. S7), which contributed to its stability (Table 1). When NMP crossed the G-loop in one simulation, its stabilization came from interaction of its naphthyl ring with F617, F664, and F666, and its hydrophilic groups interacted with waters and D681. When staying mainly on the side of the distal pocket, the ring was stacked between F136, F615, and F617, whereas the amine interacted with waters and Q89 and R620.
The behavior of these inhibitors forms a contrast with that of substrates. Even when they transiently moved away from the pocket, they came back eventually to the pocket, as seen with ETH (Movie S3).
Discussion
Compared with docking, MD simulations allowed us to assess the contribution of various residues in ligand binding and produced a much more realistic picture of the interaction of various ligands with AcrB; this occurred mainly because of two factors: the presence of explicit water and the malleability of the binding site, which improves the “fit” with a wide range of ligands. However, the inclusion of these additional degrees of freedom creates an enormous number of potential configurations, so that insufficient sampling could become an issue (36), in particular for systems of the size considered here, as discussed recently (31). In addition, one has to consider the inaccuracies in the model. For example, many of the substrates and inhibitors of AcrB contain weakly acidic or basic groups, and their charge states are likely to become altered in a low dielectric constant environment of the binding site; this is not taken into account in the standard MD simulation procedures currently in use (37). Finally, our system may be incomplete because AcrA appears to be needed for the activity, and thus for the active conformation, of AcrB (6).
With the advantages and limitations of our approach in mind, it would be desirable to compare the results of MD simulation with the known properties of the AcrB pump. Overall, there is a perfect fit, because all of the noninhibitor substrates remained bound within the distal binding pocket, presumably ready to be extruded by the conformational change in the protein, whereas both of the noninhibitors tested, GLC and KAN, remained outside the pocket. However, it was difficult to compare the details of binding of various substrates as determined by MD simulation with their experimentally determined binding affinities. Unfortunately, a simple ligand binding assay (38) does not indicate the affinity to the distal binding pocket, because the binding could have occurred at any other accessible sites on the giant AcrB protein (6). So far, only the export of β-lactams by AcrB has been analyzed in detail (39, 40), but KM values are obviously not directly correlated with the KD values of the substrates. In addition, the comparison is made even more tenuous because in most cases the efflux kinetics shows positive cooperativity, and KM values cannot be calculated. If we take the substrate concentrations where half-maximal rates are obtained, i.e., K0.5 values, those for NCF and CEF were 5.0 and 91 µM (40), which is consistent with the trend in the calculated ΔGb, −43.5 and −34.6 kcal/mol, respectively, for these compounds (Table 1). However, OXA with a K0.5 of 1.0 µM (39), is estimated to bind to the distal pocket only with a lower affinity (ΔGb of −23.2 kcal/mol). This discrepancy might be indicative of difficulty in comparing cephalosporins (NCF and CEF) with a penicillin (OXA), or more generally in comparing K0.5 (or KM) with affinity to the binding site; alternatively, it may have been caused by the inaccuracy of our ΔGb values, which did not include the contribution from conformational entropy.
Despite these limitations, we were able to assess the importance of the various residues lining the binding pocket for the stability of ligand–AcrB complex (Fig. 4). A hydrophobic core made of residues F178, I277, V612, and F615 was identified (Fig. 4C), and the role of polar and charged residues pinpointed.
It is expected that the binding to the distal pocket, the subject of the present study, does not explain the entire transport process; the other phases of the transport process, such as the binding to the proximal pocket, the subsequent movement of the substrate to the distal pocket, and finally the detachment of the substrate from the pocket and its extrusion, may exert a major influence on the overall kinetics of export. Biased MD simulation will be a useful approach in such studies, and indeed has already shown that the poor drug efflux activity of F610A mutant AcrB is largely due to the slowing down of the drug extrusion process, rather than to the lowered binding affinity to the pocket (21).
In the previous docking study with the model 2DRD (28), we found that NCF efflux was inhibited by MIN, but not by CHL. Because NCF and MIN were found in the groove area of the binding site, whereas CHL was in the cave area, these different locations seemed to explain the results of NCF efflux assay (28). However, the present study, starting from the model 2J8S, suggests that all three substrates above bind to an overlapping site (Fig. S6), and the presence or absence of efflux inhibition must now be explained in other ways, perhaps those involving different phase(s) of export process, rather than the simple binding to the distal pocket.
Although inhibitors of AcrB efflux are known, it is unknown how the inhibition occurs. PAβN is known to act also as a substrate of AcrB homologs (41), whereas there is no evidence that NMP is a substrate, because NMP susceptibility of E. coli did not change detectably by the deletion of acrAB genes (42). However, this criterion is not good for deciding if any drug is pumped out at a significant rate (39). A priori, inhibitors can bind very tightly to the binding pocket so that either the subsequent binding of substrate drugs is prevented, or a conformational change in the protein is hindered. Docking predicts that PAβN and NMP bind fairly well to the distal pocket with the estimated energy of −9.4 and −9.0 kcal/mol, but these are not the extremely strong binding expected from this hypothesis, because some substrates—e.g., NCF—bind even more tightly (in this case, −10.2 kcal/mol). We can draw the same conclusion from the ΔGb values in Table 1. So how do the inhibitors work? MD simulations gave us a hint on this question. All of the nine noninhibitor substrates stayed well within the distal binding pocket throughout the MD runs (Fig. 3). In a striking contrast, both inhibitors PAβN and NMP moved at least partially out of the pocket so that they would now straddle the G-loop structure (Fig. 3), which forms a boundary between the proximal and distal binding sites and is proposed to control the movement of substrates between these two pockets (22, 23). It appears possible that this predicted interaction with the G-loop, likely reducing its flexibility, explains at least some of the mechanisms of inhibition caused by PAβN and NMP. This hypothesis is also consistent with the recent MD simulation study (32) of the G616P and G619P AcrB mutants, where mutations in the G-loop impair the export of drugs (22, 23).
Materials and Methods
MD (43) simulations were performed using the program NAMD 2.8 (44). They usually started from the docked structure obtained with AutoDock Vina (30). The details of the simulation procedure and the analysis of data are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Lawrencium cluster staff at Lawrence Berkeley National Laboratory for their help in running simulations, and computer time from CINECA, Italian SuperComputer Resource Allocation-A Grant HP10ARSZ26. This study was supported in part by Public Health Service Grant AI-009644 and “Regione Autonoma della Sardegna” research Fellowship PO Sardegna FSE 2007-2013, L.R.7/2007 Promozione Della Ricerca Scientifica e dell’Innovazione Tecnologica in Sardegna (to A.V.V.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218348109/-/DCSupplemental.
References
- 1.Tam VH, et al. Prevalence, resistance mechanisms, and susceptibility of multidrug-resistant bloodstream isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54(3):1160–1164. doi: 10.1128/AAC.01446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: An update. Drugs. 2009;69(12):1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikaido H, Pagès JM. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev. 2012;36(2):340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178(20):5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikaido H. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol. 2011;77:1–60. doi: 10.1002/9780470920541.ch1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma D, et al. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175(19):6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419(6907):587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 9.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443(7108):173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 10.Seeger MA, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313(5791):1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 11.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5(1):e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takatsuka Y, Nikaido H. Site-directed disulfide cross-linking shows that cleft flexibility in the periplasmic domain is needed for the multidrug efflux pump AcrB of Escherichia coli. J Bacteriol. 2007;189(23):8677–8684. doi: 10.1128/JB.01127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takatsuka Y, Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J Bacteriol. 2009;191(6):1729–1737. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeger MA, et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15(2):199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 15.Schulz R, Vargiu AV, Collu F, Kleinekathöfer U, Ruggerone P. Functional rotation of the transporter AcrB: Insights into drug extrusion from simulations. PLOS Comput Biol. 2010;6(6):e1000806. doi: 10.1371/journal.pcbi.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao XQ, Kenzaki H, Murakami S, Takada S. Drug export and allosteric coupling in a multidrug transporter revealed by molecular simulations. Nat Commun. 2010;1:117. doi: 10.1038/ncomms1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohnert JA, Schuster S, Fähnrich E, Trittler R, Kern WV. Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF) J Antimicrob Chemother. 2007;59(6):1216–1222. doi: 10.1093/jac/dkl426. [DOI] [PubMed] [Google Scholar]
- 18.Bohnert JA, et al. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J Bacteriol. 2008;190(24):8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohnert JA, Karamian B, Nikaido H. Optimized Nile Red efflux assay of AcrAB-TolC multidrug efflux system shows competition between substrates. Antimicrob Agents Chemother. 2010;54(9):3770–3775. doi: 10.1128/AAC.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wehmeier C, Schuster S, Fähnrich E, Kern WV, Bohnert JA. Site-directed mutagenesis reveals amino acid residues in the Escherichia coli RND efflux pump AcrB that confer macrolide resistance. Antimicrob Agents Chemother. 2009;53(1):329–330. doi: 10.1128/AAC.00921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vargiu AV, et al. Effect of the F610A mutation on substrate extrusion in the AcrB transporter: Explanation and rationale by molecular dynamics simulations. J Am Chem Soc. 2011;133(28):10704–10707. doi: 10.1021/ja202666x. [DOI] [PubMed] [Google Scholar]
- 22.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480(7378):565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 23.Eicher T, et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci USA. 2012;109(15):5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: A crystallographic and site-directed mutagenesis study. J Bacteriol. 2005;187(19):6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Husain F, Nikaido H. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol Microbiol. 2010;78(2):320–330. doi: 10.1111/j.1365-2958.2010.07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukagoshi N, Aono R. Entry into and release of solvents by Escherichia coli in an organic-aqueous two-liquid-phase system and substrate specificity of the AcrAB-TolC solvent-extruding pump. J Bacteriol. 2000;182(17):4803–4810. doi: 10.1128/jb.182.17.4803-4810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White DG, Goldman JD, Demple B, Levy SB. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179(19):6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takatsuka Y, Chen C, Nikaido H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci USA. 2010;107(15):6559–6565. doi: 10.1073/pnas.1001460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Totrov M, Abagyan R. Flexible ligand docking to multiple receptor conformations: A practical alternative. Curr Opin Struct Biol. 2008;18(2):178–184. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer N, Kandt C. Three ways in, one way out: Water dynamics in the trans-membrane domains of the inner membrane translocase AcrB. Proteins. 2011;79(10):2871–2885. doi: 10.1002/prot.23122. [DOI] [PubMed] [Google Scholar]
- 32.Feng Z, Hou T, Li Y. Unidirectional peristaltic movement in multisite drug binding pockets of AcrB from molecular dynamics simulations. Mol Biosyst. 2012;8(10):2699–2709. doi: 10.1039/c2mb25184a. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong MJ, Carey MC. The hydrophobic-hydrophilic balance of bile salts. Inverse correlation between reverse-phase high performance liquid chromatographic mobilities and micellar cholesterol-solubilizing capacities. J Lipid Res. 1982;23(1):70–80. [PubMed] [Google Scholar]
- 34.Kollman PA, et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 35.Pyrkov TV, Chugunov AO, Krylov NA, Nolde DE, Efremov RG. PLATINUM: A web tool for analysis of hydrophobic/hydrophilic organization of biomolecular complexes. Bioinformatics. 2009;25(9):1201–1202. doi: 10.1093/bioinformatics/btp111. [DOI] [PubMed] [Google Scholar]
- 36.Grossfield A, Zuckerman DM. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu Rep Comput Chem. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopes PEM, Roux B, Mackerell AD., Jr Molecular modeling and dynamics studies with explicit inclusion of electronic polarizability. Theory and applications. Theor Chem Acc. 2009;124(1-2):11–28. doi: 10.1007/s00214-009-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su CC, Yu EW. Ligand-transporter interaction in the AcrB multidrug efflux pump determined by fluorescence polarization assay. FEBS Lett. 2007;581(25):4972–4976. doi: 10.1016/j.febslet.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim SP, Nikaido H. Kinetic parameters of efflux of penicillins by the multidrug efflux transporter AcrAB-TolC of Escherichia coli. Antimicrob Agents Chemother. 2010;54(5):1800–1806. doi: 10.1128/AAC.01714-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci USA. 2009;106(14):5854–5858. doi: 10.1073/pnas.0901695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lomovskaya O, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45(1):105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bohnert JA, Kern WV. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob Agents Chemother. 2005;49(2):849–852. doi: 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlitter J, Engels M, Kruger P, Jacoby E, Wollmer A. Targeted molecular-dynamics simulation of conformational change-application to the T→R transition in insulin. Mol Simul. 1993;10(2–6):291–308. [Google Scholar]
- 44.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.