Abstract
Higher-order structures that originate from the specific and reversible DNA-directed self-assembly of microscopic building blocks hold great promise for future technologies. Here, we functionalized biotinylated soft colloid oil-in-water emulsion droplets with biotinylated single-stranded DNA oligonucleotides using streptavidin as an intermediary linker. We show the components of this modular linking system to be stable and to induce sequence-specific aggregation of binary mixtures of emulsion droplets. Three length scales were thereby involved: nanoscale DNA base pairing linking microscopic building blocks resulted in macroscopic aggregates visible to the naked eye. The aggregation process was reversible by changing the temperature and electrolyte concentration and by the addition of competing oligonucleotides. The system was reset and reused by subsequent refunctionalization of the emulsion droplets. DNA-directed self-assembly of oil-in-water emulsion droplets, therefore, offers a solid basis for programmable and recyclable soft materials that undergo structural rearrangements on demand and that range in application from information technology to medicine.
Keywords: diethyl phthalate, functionalization, lipid emulsion, POPC, smart material
Complex higher-order structures that spontaneously arise from the specific and reversible self-assembly of simple building blocks hold great promise to become the foundation of tomorrow’s technology (1). In the context of DNA-directed self-assembly of hard colloids and artificial vesicles, linear DNA oligonucleotides were shown to be adhesive elements that allowed the self-assembly process not only to be specific but also to be reversible by a variety of external stimuli such as temperature (2–4), changes in the electrolyte concentration (4, 5), addition of competing oligonucleotides (6), and exploitation of the secondary structure of DNA oligonucleotides (7).
Oil-in-water (o/w) emulsion droplets (EDs) offer a robust, configurable, and recyclable soft-colloid system with interesting properties for the fabrication of new delivery systems and advanced synthetic materials with adaptable properties (8). By exploiting the well-characterized ligand–receptor pair biotin–streptavidin, EDs were immobilized on glass surfaces (9) and actively transported along microtubules (10). In the context of self-assembly, both polymer-mediated (11–13) and biotin–streptavidin-mediated aggregation of EDs was shown (14). Despite the advantages a combination of EDs as building blocks and DNA oligonucleotides as linking agent offers, a protocol for DNA-directed self-assembly of EDs is currently missing.
To close this gap, here we prepared an o/w emulsion composed of heavier-than-water diethyl phthalate (DEP) dispersed in an aqueous medium, stabilized by phospholipids, and functionalized with single-stranded DNA (ssDNA) oligonucleotides. Palmitoyloleoyl phosphatidylcholine (POPC) and distearoylphosphatidyl ethanolamine (DSPE) in a 9:1 molar ratio were used to stabilize the EDs. The DSPE was grafted with flexible poly(ethylene glycol) (PEG) tethers with a molecular weight of 2,000 (DSPE-PEG). The PEG tethers are known to prevent nonspecific aggregation of lipid surfaces driven by attractive van der Waals and other less pronounced aggregating potentials such as polymer bridging and the hydrophobic effect (15). Ten mole percent of the PEG tethers were additionally functionalized with terminal biotin-moieties (DSPE-PEG-btn). This allowed the ED surface to be functionalized in a highly modular way with biotin–ssDNA (btn-ssDNA) oligonucleotides using streptavidin as a connecting element (Figs. 1A and 2A). Exploiting the noncovalent biotin–streptavidin binding to functionalize the ED surface instead of incorporating DNA oligonucleotides covalently attached to a large hydrophobic group (16) eases the reuse of the components involved (17).
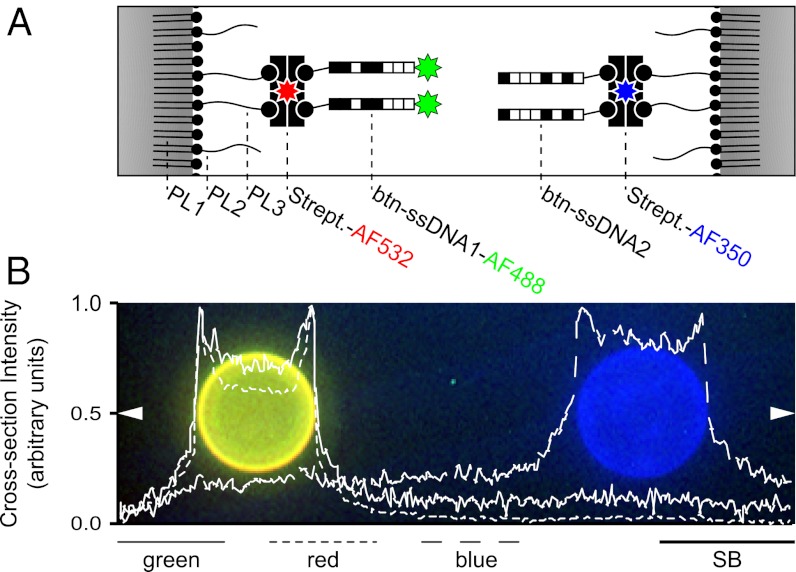
Fig. 1.
Surface functionalization and stability. (A) Scheme of the surface functionalization of a binary mixture of emulsion droplet (ED) populations functionalized with noncomplementary biotinylated single-stranded DNA (btn-ssDNA) oligonucleotides. ED1 (Left) and ED2 (Right) were both stabilized by a ternary mixture of phospholipids POPC (PL1), DSPE-PEG (PL2), and DSPE-PEG-btn (PL3) in a molar ratio of 90:9:1. The surface of the EDs was functionalized with btn-ssDNA oligonucleotides (btn-ssDNA1-AF488 and btn-ssDNA2) using streptavidin (Strept.) as a connecting element. Both Strept. populations and one btn-ssDNA population were fluorescently modified by Alexa Fluor (AF), resulting in ED1 labeled green (AF488) and red (AF532) and ED2 labeled blue (AF350). (B) Representative fluorescence micrograph of a binary mixture of ED1 and ED2. The cross-section intensities of the two EDs were acquired along the centerline of the micrograph (arrows) and show no interdroplet exchange of fluorescence signal after 1 wk of coincubation. Each fluorescent marker was measured across the micrograph and superimposed on the image as intensity. For full names of phospholipids see text. [Scale bar (SB): 10 μm.]
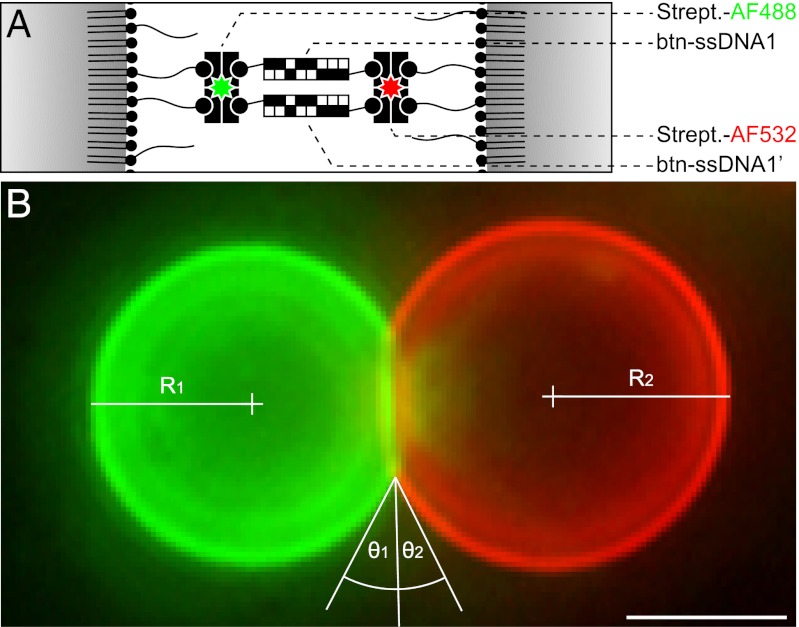
Fig. 2.
Specific detail of emulsion droplet aggregation. (A) Scheme of the surface functionalization of a binary mixture of emulsion droplet (ED) populations functionalized with complementary biotinylated single-stranded DNA (btn-ssDNA) oligonucleotides. The surface of the EDs was functionalized with btn-ssDNA oligonucleotides using streptavidin (Strept.) as a connecting element. Both Strept. populations were fluorescently modified by Alexa Fluor (AF), resulting in ED3 (Left) labeled green (AF488) and ED3′ (Right) labeled red (AF532). (B) Representative fluorescence micrograph of a binary mixture of ED3 and ED3′ functionalized with complementary btn-ssDNA oligonucleotides. θ1 and θ2, angles of contact; R1 and R2, radii of EDs. For numerical values see text. (Scale bar: 5 μm.)
Results
Surface Tension, Stability of the Surface Functionalization, and Size Distribution.
Certain basic characteristics of the ED system were characterized before assembly. The surface tension of the DEP–water interface was found to be 16.24 ± 0.05 mN/m (SD) for pure DEP and 4.08 ± 0.04 mN/m (SD) for DEP stabilized by phospholipids, indicating the formation of a phospholipid surface surrounding the EDs. To assess the stability of the ED surface functionalization over time, a binary mixture of ED populations functionalized with noncomplementary btn-ssDNA oligonucleotides (ED1:ED2) was incubated for 1 wk. Because both streptavidin molecules and one of the btn-ssDNA oligonucleotide populations differed in the covalently attached fluorescence label (Fig. 1A), the location of each component was readily accessible by fluorescent signal. The absence of a significant interdroplet exchange of streptavidin or btn-ssDNA oligonucleotides at the end of the incubation period (Fig. 1B) reveals the stability of the ED surface functionalization. The size distributions of two functionalized ED populations (ED3 and ED3′) used in subsequent assembly assays were skewed to the right (Fig. S1 B and C) with mean diameters of 10.29 ± 4.47 μm (SD) for ED3 and 10.97 ± 5.14 μm (SD) for ED3′, medians of 9.59 μm (ED3) and 10.15 μm (ED3′), and modes of 6.75 μm (ED3) and 6.89 μm (ED3′).
Specificity of the Self-Assembly Process.
The specificity of the DNA-directed self-assembly of ED aggregates was analyzed both on individual and on population levels by comparing binary mixtures of ED populations functionalized either with complementary (ED3:ED3′) or noncomplementary (ED3:ED4) btn-ssDNA oligonucleotides. On both levels, aggregates were found exclusively if the ssDNA oligonucleotides were of complementary sequence (Figs. 2B and 3 C and D), whereas no aggregates were found otherwise (Figs. 1B and 3 E and F). Under the conditions demonstrated here, the majority of the EDs participated in the aggregation, resulting in very large aggregates visible on the macroscopic scale.
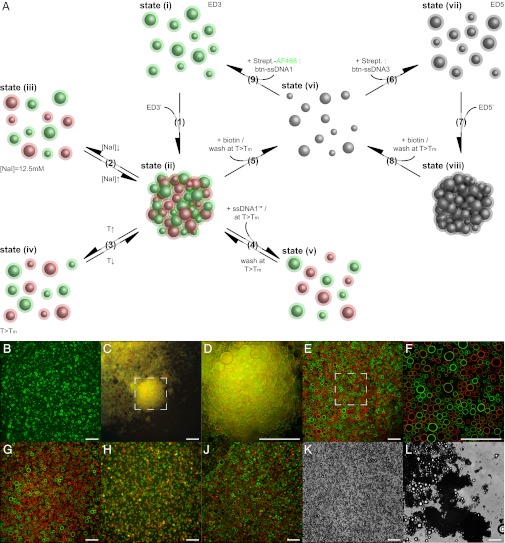
Fig. 3.
Specificity and reversibility of the self-assembly process and recyclability of emulsion droplets. (A) Scheme of the steps (Arabic numerals) and states (Roman numerals) for the experimental procedure. For all states the temperature was at room temperature, [i.e., below the melting temperature (Tm) of the dsDNA oligonucleotides] and the sodium iodide concentration was 25 mM unless stated otherwise. Representative fluorescence (B–J) and transmission (K and L) micrographs. (B) State i, emulsion droplet population 3 (ED3) after surface functionalization with fluorescently labeled streptavidin (green) and biotinylated single-stranded DNA oligonucleotides of sequence 1 (btn-ssDNA1). (C and D) State ii, binary aggregates of complementary ED3 and ED3′ (red). (E and F) Aggregates were absent in binary control mixtures of ED populations functionalized with noncomplementary btn-ssDNA oligonucleotides. (D and F) Zoomed-in displays of regions indicated in C and E. Disassembled states after (G) decreasing the electrolyte concentration (state iii) and (H) increasing the temperature (state iv). (J) Preserved disassembled state after addition of competing nonbiotinylated ssDNA (ssDNA1′*) complementary to btn-ssDNA1 (state v). (K) ED5 after surface reset and refunctionalization (state vii). (L) Binary aggregates of complementary ED5 and ED5′ (state viii). For experimental details see text. (Scale bars: 250 μm.)
Reversibility of the Self-Assembly Process.
The reversibility of the assembly process and the recyclability of the EDs both were analyzed by a variety of manipulations on the population level, summarized in Fig. 3A. Starting from extensive ED3:ED3′ aggregates (Fig. 3A, state ii), decreasing the sodium iodide concentration from 25 mM to 12.5 mM (Fig. 3A, step 2) resulted in the disassembly of the aggregates (Fig. 3G). Reestablishing the original electrolyte concentration induced the reassembly of the ED aggregates (Fig. S2H). In step 3 (Fig. 3A), the ED aggregates were disassembled by an incubation above the dsDNA melting temperature (Tm). The disassembled state (Fig. 3H) was found to be stable only if the temperature was kept above the dsDNA Tm. A decrease in temperature below the dsDNA Tm resulted in the spontaneous reassembly of the ED aggregates. To prevent the EDs from reassembling, in step 4 (Fig. 3A) an excess of competing nonbiotinylated (*) ssDNA* oligonucleotides of complementary sequence was added before decreasing the temperature below the dsDNA Tm. When cooled below dsDNA Tm, this blocked the rehybridization of the anchored ssDNA oligonucleotides (Fig. S3C, state v.1,2) and preserved the disassembled state (Fig. 3J). Addition of excess soluble ssDNA* oligonucleotides of noncomplementary sequence as a control did not block the reassembly (Fig. S2F). Repeated washing above the dsDNA Tm removed the soluble ssDNA* oligonucleotides and left behind solely the anchored complementary ssDNA oligonucleotides still bound to the surface of the two ED populations. This restored the ability of the EDs to reassemble through dsDNA when cooled below the dsDNA Tm (Fig. S2N). A subsequent temperature-induced disassembled state was then again preserved by a second addition of an excess of competing ssDNA* oligonucleotides of complementary sequence (Fig. S2P).
Recyclability of EDs.
The first three disassembly methods exploited properties of the DNA hybridization. In step 5 (Fig. 3A), the biotin–streptavidin linkage was broken by the addition of soluble biotin and an incubation above the dsDNA Tm; this resulted not only in a disassembly of the ED aggregates but also in a removal of the biotinylated ssDNA oligonucleotides and/or of the streptavidin from the biotinylated ED surface. Consequently, the two ED populations lost their identity previously defined by their distinct surface functionalization as the EDs were reset to their initial state, that is, to the biotinylated ED surface without DNA oligonucleotides (Fig. S2K). Neither addition of water and washing above the dsDNA Tm (Fig. S2L) nor addition of biotin but washing below the dsDNA Tm (Fig. S2M) successfully removed the streptavidin and the biotinylated ssDNA oligonucleotides from the biotinylated ED surface in two independent control experiments. Consequently, aggregation was not prevented. The reset of the surface functionalization to biotinylation without DNA oligonucleotides (Fig. S2A) unified the two previously distinct ED populations and enabled a refunctionalization of the EDs in step 6 (Fig. 3A) using a different btn-ssDNA oligonucleotide–streptavidin combination. We used nonfluorescent streptavidin for the refunctionalization to be able to qualitatively evaluate the removal of the fluorescently active streptavidin in step 5. As indicated by the slight fluorescence activity of the resulting ED5 (Fig. S2O), the replacement of the fluorescently labeled streptavidin of the first functionalization operation by nonfluorescent streptavidin molecules of the refunctionalization operation was not complete. However, no ED aggregates were found in state vii (Fig. 3K), indicating that the removal of linking agents was sufficient to prevent reassembly induced by ssDNA oligonucleotides still linked to the ED surface (Fig. S2 L and M). In step 7 (Fig. 3A), binary mixtures of ED populations functionalized with complementary btn-ssDNA oligonucleotides (ED5:ED5′) resulted in reassembled ED aggregates (Fig. 3L). No reassembly was observed in the control experiment with ED populations functionalized with noncomplementary (ED5:ED6) btn-ssDNA oligonucleotides (Fig. S2R). The results on the reset of the EDs in step 5 indicate that EDs could optionally be rereset in step 8 (Fig. 3A) and rerefunctionalized in step 9 (Fig. 3A) with the original functionalization for a fully recursive assembly procedure.
Discussion
In this study, we showed the surface functionalization of EDs to be stable and the DNA-directed self-assembly of EDs to be highly specific and reversible by several distinct external triggers. The higher-than-water density of the DEP eased the removal of the supernatant during the decoration, refunctionalization, and reassembly procedures and also increased the encounter probability of EDs due to a pellet formation at the bottom of the incubation vessel as the EDs spontaneously sedimented. The large diameter of the EDs made them readily available for facile imaging with optical microscopy. The EDs were shown to maintain their structural integrity during numerous manipulation steps: repeated freeze-thawing; long-term storage up to several months at −20 °C, increasing the reproducibility and comparability by using the same batch of EDs for all experiments; several washing steps (i.e., removal of the supernatant); and repeated cycles of assembly, disassembly, reassembly, and refunctionalization operations—up to more than 20 different manipulations for some ED populations. The positive skew of the size distribution of ED3 and ED3′ (Fig. S1 B and C) may be a consequence of a positive selection for larger particles. Because the sedimentation speed depends linearly on the reference area of an object, smaller EDs had an increased probability of being removed when the supernatant was replaced during the wash procedure.
To estimate the DNA surface coverage of an ED, we estimate the average lipid head-group area as 65.8 Å2 and the average packing area per PEG tether Γ as 658 Å2 using an area per lipid head group of 68.3 Å2 for POPC (18) and 43 Å2 for DSPE (19) and a 9:1 molar ratio of POPC to DSPE. Because only 10 mol % of all PEG tethers were biotinylated, the average packing area of the PEG-btn tethers is Γbtn = 6,580 Å2. We assume the four biotin binding sites of a streptavidin molecule to be occupied by two btn-DNA oligonucleotides on average. This assumption is based on the 2:1 molar ratio of the btn-ssDNA oligonucleotides to streptavidin we have chosen during their coincubation. This leads to a 1:1 ratio of DSPE-PEG-btn to btn-ssDNA oligonucleotides and consequently to a DNA surface coverage of about 15,200 strands/μm2. However, for btn-DNA coupled via streptavidin to a biotin-doped supported phospholipid bilayer, Larsson et al. (20) showed that the intuitive assumption of a 2:1 molar ratio of btn-ssDNA oligonucleotides may be too simplistic and that the combination of electrostatic repulsion and steric hindrance may prevent efficient coupling of two btn-DNA oligonucleotides per streptavidin. In this case, only one btn-DNA oligonucleotide would be coupled per streptavidin; consequently, the DNA surface coverage would be halved.
In the next step, we estimate the force acting on the linkers composed of the anchoring DSPE-PEG-btn, streptavidin, and dsDNA. Following Chiruvolu et al. (21), the interdroplet adhesion force F0 for a contact angle θ is given by
for the ED radius R and surface tension σ0. For the two aggregated EDs shown in Fig. 2B with contact angles θ1 = 29° and θ2 = 26°, and radii R1 = 5.03 μm and R2 = 5.53 μm, and the measured surface tension σ0 = 4.08 ± 0.04 mN/m (SD) for DEP stabilized by phospholipids, the adhesion force is between 28.7 and 32.3 nN. The contact area is 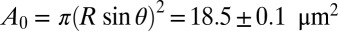 (SD). With Γbtn calculated above, the number n of linkers is given by
(SD). With Γbtn calculated above, the number n of linkers is given by
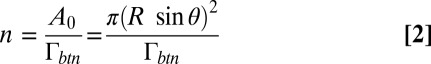 |
and resolves to 280,000. This number results in a force per linker F = 110 fN. Consequently, the force acting on the linkers is between 100 and 1,000 times lower than the force needed to separate the 15-mer dsDNA (22), for the biotin–streptavidin linkage to fail (23), and to extract the lipid anchor from the membrane (24). Thus, the force will result only in a deformation of the PEG moiety even if the DNA surface coverage is halved (see above). To estimate d, the average extension of the PEG-btn tether at the adhesion site, the force per PEG-btn tether can be expressed in relation to d by the phenomenological model of Alexander (25) and De Gennes (26, 27) as
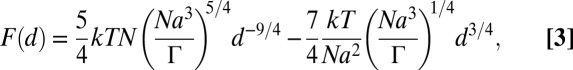 |
where n = 45 is the number of PEG monomer units and a = 3.5 Å is the length of a PEG monomer ([–CH2CH2O–]). We express d by relating Eqs. 1–3;
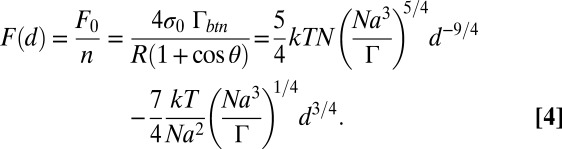 |
Numerically solving this equation for d yields 36.8 Å, which is in good agreement with RF = 35 Å for 2,000-Mr PEG tethers. These calculations ignore a potential accumulation of linkers at the site of contact that was reported for aggregated vesicles (28). However, assuming that all ssDNA oligonucleotides in the contact area undergo hybridization may overestimate the number of bridging linkers. To what extent these opposing effects cancel out or affect the ED self-assembly is not clear.
Interestingly, we found no ED aggregates at room temperature and a sodium iodide concentration of 12.5 mM (Fig. 3G), although a salt-adjusted estimation of Tm for the DNA strands in solution (i.e., 0.25 μM) using OligoCalc (29) reveals a Tm of 30.6 °C. Although calculations of the melting temperature and melting curve are routine tools for DNA strands in solution, existing models for DNA-induced particle interactions typically overpredict the melting temperature (30). Dreyfus et al. (31) showed that the formation of interparticle bonds is accompanied by a significant configurational entropy loss due to binding and confinement of the tethered DNA oligonucleotides between neighboring particles. This entropy loss is not present for the hybridization of DNA in bulk solution and weakens the interaction of colloids, shifting the dissociation curve downward. Only recently, a model for DNA-induced interactions between pairs of polystyrene hard colloids was reported that quantitatively captures the temperature-dependent strength of these interactions, without empirical corrections (32). However, Mognetti et al. (33) soon after challenged this model and pointed out that “to obtain more reliable predictions of experimental results on DNA-coated colloids, we must go beyond the current level of description of DNA-mediated interactions.” This clearly shows that predicting DNA-mediated colloidal-pair interactions is subject to ongoing debates even for hard colloids. To what extent such models are applicable to the more complex soft, malleable colloids is currently unknown. The influence of the mechanical properties of the system of interest becomes apparent when the current study is compared with a previously reported study by the authors. There, DNA-directed aggregates of artificial giant unilamellar vesicles (GUVs) were still found at a sodium iodide concentration of 12.5 mM (34). These GUVs were not only of comparable size but also functionalized in the same way as the EDs in this study. Evans and Parsegian (35) recognized that extensive adherent contacts can only be formed if the adhering bodies can deform. In addition, the linkage-induced accumulation of linkers we showed for GUVs (34) is reported to increase the binding strength significantly (36). One can therefore speculate that the relatively larger adherent contacts of aggregated GUVs not only compensate the weakening of the DNA hybridization because of the reduction in the Na+ concentration, but also impede the disassembly of GUV aggregates by elevated temperatures, as has been reported by Beales and Vanderlick (4).
Another interesting finding is the need for elevated temperatures for soluble biotin to induce disassembly of ED aggregates (Fig. S2M). Due to its higher binding affinity for streptavidin than grafted biotin (37), soluble biotin was reported to be efficient at room temperature to spontaneously and immediately reverse the streptavidin-induced aggregation process of building blocks at the nanoscale at room temperature [e.g., 50-nm vesicles (21) and 12-nm nanoparticles (38)]. For colloid systems at the microscale, mechanical disturbance, for example, shaking and induced flow, was applied to reverse the streptavidin-induced aggregation process of cholesterol-PEG-biotin–modified lymphoblastoid cells of an average diameter of 10 µm (39) as well as the binding of 2-µm GUVs (40) at room temperature. Although the dependency of soluble biotin on the mechanical agitation was not analyzed in these studies, we speculate that the elevated temperatures per se did not considerably affect the strength of the biotin–streptavidin binding, but induced a convective mixing. The resulting flow of fluids both allowed the soluble biotin to penetrate the densely packed ED pellets faster than at room temperature and carried unbound binding elements (i.e., btn-ssDNA oligonucleotides and streptavidin) off the binding site, thereby reducing their local concentration and thus the probability of a reattachment to the surface of the EDs.
Regarding the handling and the technical potential of EDs, EDs rank between soft artificial vesicles, for which the aqueous internal compartment is widely used to encapsulate chemical cargo but for which the delicate lipid bilayer limits the versatility, and hard colloids, which are stable but lack both an internal compartment and properties inherent to lipid membranes (e.g., dynamic rearrangements and controlled transport across membranes). One potential advantage of using EDs is that the anhydrous interior is ideal for hosting hydrophobic substances and therapeutics, such as amphotericin B (41). Pontani et al. (14) recently reported on the exploitation of the biomimetic characteristics of ED membranes when they used protein-mediated self-assembly of EDs as mimics of cell–cell adhesion. In addition, the internal compartments of EDs are reported to possess fluid dynamic properties leading to self-division (42–44) and self-propelled motion (45). Aggregates of EDs, therefore, offer compartments for embedded (bio-)chemical information processing that act cooperatively to configure and reconfigure themselves into set structures to trigger functions and may be used, for example, in the generation of novel biocompatible, biologically inspired, and recyclable programmable smart materials. The use of DNA oligonucleotides as adhesive elements may provide a common “programming language” for the assembly of hybrid structures composed of artificial vesicles, hard colloids, and natural cells (46). In this context, we intentionally chose the solution to host the EDs to contain glucose and sodium iodide, which the authors already reported to successfully host DNA-directed assemblies of GUVs (34) and to affect the vesicle stability the least (47). This will ease, for example, the development of a hybrid system of specifically assembled vesicles and EDs that offers carriers for combination therapies of hydrophilic and hydrophobic therapeutic or diagnostic substances. The specific and reversible DNA-directed self-assembly of EDs may therefore bring about a universe of customizable, scalable new materials for use in various sectors from smart materials to medicine.
Materials and Methods
Preparation of EDs.
The phospholipids POPC, DSPE-PEG, and DSPE-PEG-btn, all provided in chloroform by Avanti Polar Lipids, were mixed in a molar ratio of 90:9:1. After chloroform was removed (under vacuum, 60 min), DEP (1 mL, Sigma-Aldrich) was added, resulting in a 2-mM final concentration of phospholipids. The phospholipid solution (PS) was then sonicated (30 min, 50 °C) using a Sonorex Digitec DT 156 BH (Bandelin GmbH), followed by an overnight incubation at room temperature. For the emulsification, PS (20 μL) was placed in an aqueous medium (450 μL), called the hosting solution (HS). The HS1 contained 25 mM sodium iodide (Sigma-Aldrich), 475 mM glucose (Sigma-Aldrich), and 2 mM potassium phosphate buffer. The mixture was emulsified by mechanical agitation. For the disassembly and reassembly experiments involving changes in the electrolyte concentration, two additional HS solutions were prepared: HS2 lacking sodium iodide and HS3 with an increased sodium iodide concentration of 37.5 mM. High-quality water (Milli-Q; Millipore) was used throughout the experiments.
Tensiometry.
Interfacial tension of the oil–water interface was determined using a PAT1D tensiometer (Sinterface Technologies) by the pendent drop method using the Sinterface software. The samples were prepared with either DEP or PS as the internal phase and an external aqueous phase containing 25 mM sodium iodide without glucose. Values used in the analysis (100–600 s for DEP; 200–400 s for PS) were taken after the interfacial tension values reached a plateau. For PS, no further change in tension was observed up to 1,200 s.
Functionalization of EDs.
Unlabeled streptavidin (Strept.) from Sigma-Aldrich and streptavidin Alexa Fluor 350, 488, and 532 conjugates (Strept.-AF350, Strept.-AF488, and Strept.-AF532) from Invitrogen were dissolved to a final concentration of 1 mg/mL. 15-mer ssDNA oligonucleotides (ssDNA1–ssDNA4) were synthesized, modified, purified by HPLC, and dissolved to a final concentration of 100 μM by the supplier (Sigma-Genosys); the sequences and the modifications of the DNA oligonucleotides are shown in Fig. S3A. For the surface functionalization of ED1, equal volumes of ED solution and a 1.35:0.5:98.15 (volume ratio) mixture of preincubated (30 min at room temperature) Strept.-AF532: btn-ssDNA1-AF488: HS1 were mixed by aspiration. This procedure was repeated for combinations of Strept.-AF350: btn-ssDNA2 (ED2), Strept.-AF488: btn-ssDNA1 (ED3), Strept.-AF532: btn-ssDNA1′ (ED3′), Strept.-AF532: btn-ssDNA2 (ED4), Strept.: btn-ssDNA3 (ED5), Strept.: btn-ssDNA3′ (ED5′), and Strept.: btn-ssDNA4 (ED6). A schematic representation of the surface functionalization of all ED populations is shown in Fig. S3A. After a 1-h incubation, the supernatant was removed and replaced by fresh HS1. This washing procedure was repeated four times. The functionalized EDs were frozen in liquid nitrogen and stored at −20 °C until use.
Stability of the Surface Functionalization.
Equal volumes of ED1 and ED2 were mixed by aspiration and incubated for 7 d at 4 °C. Analysis of the distribution of fluorescence was done by fluorescence microscopy, described below. Fig. S3B provides a schematic representation of the hybridization state of the DNA oligonucleotides.
Size Distribution.
The data acquisition for the size distribution (Fig. S1) was done by using a polydimethylsiloxane microfluidic chip with channels 130 µm in diameter. The particles were transferred to a carrying fluid containing sucrose (1,700 g/L) to avoid on-chip sedimentation and sequentially pumped through the channels at a flow rate of 0.5 µL/min. Transmission micrographs were taken as described below. A custom Matlab algorithm (Matlab R2012a 7.14.0.739; Mathworks) was used to estimate and subtract the background for contrast enhancement, to threshold the images for black-and-white conversion by blackening pixels of intensity lower than three SDs above the intensity mean, to discard unfocused and nonspherical (overlapping) particles, and to automatically measure particles of ED3 and ED3′. The accuracy of the method was tested in two independent control experiments using polystyrene beads (1.0 µm; Duke Scientific Corporation) and silica beads (5.0 µm; Bangs Laboratories) (Fig. S1A). One thousand (controls) and 4,000 (ED3 and ED3′) measurements, respectively, were randomly picked from the original datasets. These processed datasets were subjected to the Grubbs outlier test with a P-value cutoff of 0.05, resulting in 893 (1.0 µm beads), 963 (5.0 µm beads), 3,966 (ED3), and 3,990 (ED3′) measurements plotted in Fig. S1. The histograms of ED3 and ED3′ were constructed using the Freedman-Diaconis rule. Because the histograms of ED3 and ED3′ were found to be skewed to the right, log-normal distributions were fitted to these histograms (Fig. S1 B and C). Normal distributions were fitted to the histograms of the two controls (Fig. S1A). Matlab was used for data processing, plotting, and fitting operations.
Specificity and Reversibility of the Self-Assembly Process and Recyclability of EDs.
A detailed representation of the steps mentioned hereafter is provided in Fig. S2. Fig. S3C in addition provides a schematic representation of the hybridization state of the DNA oligonucleotides. In steps 1, 1c, 7, and 7c, binary mixtures of ED3:ED3′, ED3:ED4, ED5:ED5′, and ED5:ED6 were incubated for 60 min at room temperature. For assessing the specificity of the DNA-directed self-assembly on the level of individual EDs (Fig. 2B), the ED solutions were diluted 1:10 before incubation. In step 2f, the hosting medium was 1:1 (vol/vol) diluted with HS2 (HS3 for step 2r), mixed by aspiration, and incubated for 60 min. For the transition to state iv (i.e., step 3), the ED solution was incubated at 60 °C for 30 min using a standard analog dry-block heater (VWR Scientific) equipped with VWR modular heating blocks for microcentrifuge tubes. In step 4f, the sample was heated to 60 °C, ssDNA1′* (ssDNA2* for step 4c) was added in a 10-fold excess, and the sample was mixed by aspiration. After incubation for 30 min, the temperature was subsequently lowered to room temperature. In step 4r, the ED solution was washed five times (see above) with HS1 heated to 60 °C. During the washing procedure the temperature of the ED solution was carefully kept at 60 °C. In step 5, the addition of soluble biotin in a 1,000-fold excess, or of water as control (step 5c1), was followed by an incubation at 60 °C for 24 h and the abovementioned washing procedure at 60 °C or by an incubation and washing at room temperature as control (step 5c2). The soluble biotin (Sigma-Aldrich) was dissolved to a final concentration of 0.2 mg/mL. In step 6, refunctionalization of EDs took place by using the abovementioned functionalization protocol with a combination of Strept.: btn-ssDNA3, resulting in ED5.
Microscopy and Image Acquisition.
The ED solutions were mixed by aspiration before transfer to a coverslip. The coverslip was equipped with a spacer of about 1 mm in height before covering with a microscope slide. For the temperature-induced disassembly experiments (Fig. 3A, step 3), a glass Petri dish was filled with water heated to 60 °C and placed on top of the microscope slide to maintain the temperature above the dsDNA Tm. The temperature was continuously monitored using an infrared thermometer (IR001; Ryobi Technologies, Inc.) and samples were discarded when cooled below 50 °C. All samples were evaluated using an inverted light and fluorescence microscope Nikon Eclipse TE2000-S with a Nikon Intensilight light source. Images were captured with a Photometrics Cascade II 512 camera and Micro-Manager open-source microscopy software (version 1.4) for the size distribution, and in-house software otherwise. A 40× air objective (Nikon) was used for the transmission micrograph acquisition for the size distribution. A 100× oil-immersion objective (Nikon) was used for the micrographs of Figs. 1B and 2B. A 4× air objective (Nikon) was used otherwise, except for Fig. 3 D and F, for which a 10× air objective (Nikon) was used. The fluorescence micrographs of Figs. 1B and 2B were rotated and cropped. The transmission and fluorescence micrographs of Figs. 1–3 and Fig. S2 were automatically and independently for each channel contrast-adjusted (equally across the entire image). The cross-section intensity of Fig. 1B was normalized to 1 after contrast adjustment. For all manipulations and data acquisition except size distribution measurements Adobe Photoshop (CS5, version 12.0.4) was used. All figures were prepared using Adobe Illustrator (CS5, version 15.0.2).
Supplementary Material
Acknowledgments
M.H. was supported by the Swiss National Science Foundation; E.B. by the Japanese research funding program Exploratory Research for Advanced Technology; and M.M.H. by the Danish National Research Foundation under The Center for Fundamental Living Technology. The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under Grant Agreement 249032 (MATCHIT).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214386109/-/DCSupplemental.
References
- 1.Whitesides GM, Grzybowski B. Self-assembly at all scales. Science. 2002;295(5564):2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382(6592):607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 3.Valignat MP, Theodoly O, Crocker JC, Russel WB, Chaikin PM. Reversible self-assembly and directed assembly of DNA-linked micrometer-sized colloids. Proc Natl Acad Sci USA. 2005;102(12):4225–4229. doi: 10.1073/pnas.0500507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beales PA, Vanderlick TK. Specific binding of different vesicle populations by the hybridization of membrane-anchored DNA. J Phys Chem A. 2007;111(49):12372–12380. doi: 10.1021/jp075792z. [DOI] [PubMed] [Google Scholar]
- 5.Biancaniello PL, Crocker JC, Hammer DA, Milam VT. DNA-mediated phase behavior of microsphere suspensions. Langmuir. 2007;23(5):2688–2693. doi: 10.1021/la062885j. [DOI] [PubMed] [Google Scholar]
- 6.Tison CK, Milam VT. Reversing DNA-mediated adhesion at a fixed temperature. Langmuir. 2007;23(19):9728–9736. doi: 10.1021/la700601j. [DOI] [PubMed] [Google Scholar]
- 7.Leunissen ME, et al. Switchable self-protected attractions in DNA-functionalized colloids. Nat Mater. 2009;8(7):590–595. doi: 10.1038/nmat2471. [DOI] [PubMed] [Google Scholar]
- 8.Bibette J, Calderon FL, Poulin P. Emulsions: Basic principles. Rep Prog Phys. 1999;62(6):969–1033. [Google Scholar]
- 9.Fattaccioli J, Baudry J, Henry N, Brochard-Wyart F, Bibette J. Specific wetting probed with biomimetic emulsion droplets. Soft Matter. 2008;4(12):2434–2440. [Google Scholar]
- 10.Bottier C, et al. Active transport of oil droplets along oriented microtubules by kinesin molecular motors. Lab Chip. 2009;9(12):1694–1700. doi: 10.1039/b822519b. [DOI] [PubMed] [Google Scholar]
- 11.Weaver JVM, Rannard SP, Cooper AI. Polymer-mediated hierarchical and reversible emulsion droplet assembly. Angew Chem Int Ed Engl. 2009;48(12):2131–2134. doi: 10.1002/anie.200805448. [DOI] [PubMed] [Google Scholar]
- 12.Woodward RT, et al. Controlling responsive emulsion properties via polymer design. Chem Commun (Camb) 2009;(24):3554–3556. doi: 10.1039/b904320a. [DOI] [PubMed] [Google Scholar]
- 13.Woodward RT, Weaver JVM. The role of responsive branched copolymer composition in controlling pH-triggered aggregation of “engineered” emulsion droplets: Towards selective droplet assembly. Polym. Chem. 2011;2(2):403–410. [Google Scholar]
- 14.Pontani LL, Jorjadze I, Viasnoff V, Brujic J. Biomimetic emulsions reveal the effect of mechanical forces on cell-cell adhesion. Proc Natl Acad Sci USA. 2012;109(25):9839–9844. doi: 10.1073/pnas.1201499109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Needham D, Kim DH. PEG-covered lipid surfaces: Bilayers and monolayers. Colloids Surf B Biointerfaces. 2000;18(3–4):183–195. doi: 10.1016/s0927-7765(99)00147-2. [DOI] [PubMed] [Google Scholar]
- 16.Xu C, Taylor P, Fletcher PDI, Paunov VN. Adsorption and hybridisation of DNA-surfactants at fluid surfaces and lipid bilayers. J Mater Chem. 2005;15(3):394–402. [Google Scholar]
- 17.Holmberg A, et al. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005;26(3):501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 18.Kucerka N, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J Membr Biol. 2005;208(3):193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 19.Kuhl TL, Leckband DE, Lasic DD, Israelachvili JN. Modulation of interaction forces between bilayers exposing short-chained ethylene oxide headgroups. Biophys J. 1994;66(5):1479–1488. doi: 10.1016/S0006-3495(94)80938-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsson C, Rodahl M, Höök F. Characterization of DNA immobilization and subsequent hybridization on a 2D arrangement of streptavidin on a biotin-modified lipid bilayer supported on SiO2. Anal Chem. 2003;75(19):5080–5087. doi: 10.1021/ac034269n. [DOI] [PubMed] [Google Scholar]
- 21.Chiruvolu S, et al. Higher order self-assembly of vesicles by site-specific binding. Science. 1994;264(5166):1753–1756. doi: 10.1126/science.8209255. [DOI] [PubMed] [Google Scholar]
- 22.Hatch K, Danilowicz C, Coljee V, Prentiss M. Demonstration that the shear force required to separate short double-stranded DNA does not increase significantly with sequence length for sequences longer than 25 base pairs. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78(1 Pt 1):011920. doi: 10.1103/PhysRevE.78.011920. [DOI] [PubMed] [Google Scholar]
- 23.Florin EL, Moy VT, Gaub HE. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264(5157):415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 24.Uster PS, et al. Insertion of poly(ethylene glycol) derivatized phospholipid into pre-formed liposomes results in prolonged in vivo circulation time. FEBS Lett. 1996;386(2-3):243–246. doi: 10.1016/0014-5793(96)00452-8. [DOI] [PubMed] [Google Scholar]
- 25.Alexander S. Adsorption of chain molecules with a polar head: A scaling description. J Phys. 1977;38(8):983–987. [Google Scholar]
- 26.De Gennes PG. Polymers at an interface: A simplified view. Adv Colloid Interface Sci. 1987;27(3-4):189–209. [Google Scholar]
- 27.De Gennes PG. Polymer-solutions near an interface: 1. Adsorption and depletion layers. Macromolecules. 1981;14(6):1637–1644. [Google Scholar]
- 28.Noppl-Simson DA, Needham D. Avidin-biotin interactions at vesicle surfaces: Adsorption and binding, cross-bridge formation, and lateral interactions. Biophys J. 1996;70(3):1391–1401. doi: 10.1016/S0006-3495(96)79697-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kibbe WA. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35(Web Server issue):W43–46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vainrub A, Pettitt BM. Coulomb blockage of hybridization in two-dimensional DNA arrays. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;66(4 Pt 1):041905. doi: 10.1103/PhysRevE.66.041905. [DOI] [PubMed] [Google Scholar]
- 31.Dreyfus R, et al. Simple quantitative model for the reversible association of DNA coated colloids. Phys Rev Lett. 2009;102(4):048301. doi: 10.1103/PhysRevLett.102.048301. [DOI] [PubMed] [Google Scholar]
- 32.Rogers WB, Crocker JC. Direct measurements of DNA-mediated colloidal interactions and their quantitative modeling. Proc Natl Acad Sci USA. 2011;108(38):15687–15692. doi: 10.1073/pnas.1109853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mognetti BM, et al. Predicting DNA-mediated colloidal pair interactions. Proc Natl Acad Sci USA. 2012;109(7):E378–E379, author reply E380. doi: 10.1073/pnas.1119991109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadorn M, Eggenberger Hotz P. DNA-mediated self-assembly of artificial vesicles. PLoS ONE. 2010;5(3):e9886. doi: 10.1371/journal.pone.0009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans EA, Parsegian VA. Energetics of membrane deformation and adhesion in cell and vesicle aggregation. Ann NY Acad Sci. 1983;416:13–33. doi: 10.1111/j.1749-6632.1983.tb35176.x. [DOI] [PubMed] [Google Scholar]
- 36.Smith AS, Sengupta K, Goennenwein S, Seifert U, Sackmann E. Force-induced growth of adhesion domains is controlled by receptor mobility. Proc Natl Acad Sci USA. 2008;105(19):6906–6911. doi: 10.1073/pnas.0801706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers DD, Willard BL, Carbonell RG, Kilpatrick PK. Affinity precipitation of proteins by surfactant-solubilized, ligand-modified phospholipids. Biotechnol Prog. 1992;8(5):436–453. doi: 10.1021/bp00017a011. [DOI] [PubMed] [Google Scholar]
- 38.Li M, Wong KKW, Mann S. Organization of inorganic nanoparticles using biotin-streptavidin connectors. Chem Mater. 1999;11(1):23–26. [Google Scholar]
- 39.Meier W. Reversible cell aggregation induced by specific ligand-receptor coupling. Langmuir. 2000;16(3):1457–1459. [Google Scholar]
- 40.Kuhl TL, et al. A neutron reflectivity study of polymer-modified phospholipid monolayers at the solid-solution interface: Polyethylene glycol-lipids on silane-modified substrates. Biophys J. 1998;75(5):2352–2362. doi: 10.1016/S0006-3495(98)77679-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boswell GW, Buell D, Bekersky I. AmBisome (liposomal amphotericin B): A comparative review. J Clin Pharmacol. 1998;38(7):583–592. doi: 10.1002/j.1552-4604.1998.tb04464.x. [DOI] [PubMed] [Google Scholar]
- 42.Sumino Y, Kitahata H, Seto H, Yoshikawa K. Blebbing dynamics in an oil-water-surfactant system through the generation and destruction of a gel-like structure. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76(5 Pt 2):055202. doi: 10.1103/PhysRevE.76.055202. [DOI] [PubMed] [Google Scholar]
- 43.Sumino Y, Kitahata H, Seto H, Nakata S, Yoshikawa K. Spontaneous deformation of an oil droplet induced by the cooperative transport of cationic and anionic surfactants through the interface. J Phys Chem B. 2009;113(48):15709–15714. doi: 10.1021/jp9037733. [DOI] [PubMed] [Google Scholar]
- 44.Browne KP, Walker DA, Bishop KJM, Grzybowski BA. Self-division of macroscopic droplets: Partitioning of nanosized cargo into nanoscale micelles. Angew Chem Int Ed Engl. 2010;49(38):6756–6759. doi: 10.1002/anie.201002551. [DOI] [PubMed] [Google Scholar]
- 45.Hanczyc MM, Toyota T, Ikegami T, Packard N, Sugawara T. Fatty acid chemistry at the oil-water interface: Self-propelled oil droplets. J Am Chem Soc. 2007;129(30):9386–9391. doi: 10.1021/ja0706955. [DOI] [PubMed] [Google Scholar]
- 46.Gartner ZJ, Bertozzi CR. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc Natl Acad Sci USA. 2009;106(12):4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hadorn M, Boenzli E, Eggenberger Hotz P. A quantitative analytical method to test for salt effects on giant unilamellar vesicles. Sci Rep. 2011;1:168. doi: 10.1038/srep00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.