Abstract
Protein misfolding and aggregation cause serious degenerative conditions such as Alzheimer’s, Parkinson, and prion diseases. Damage to membranes is thought to be one of the mechanisms underlying cellular toxicity of a range of amyloid assemblies. Previous studies have indicated that amyloid fibrils can cause membrane leakage and elicit cellular damage, and these effects are enhanced by fragmentation of the fibrils. Here we report direct 3D visualization of membrane damage by specific interactions of a lipid bilayer with amyloid-like fibrils formed in vitro from β2-microglobulin (β2m). Using cryoelectron tomography, we demonstrate that fragmented β2m amyloid fibrils interact strongly with liposomes and cause distortions to the membranes. The normally spherical liposomes form pointed teardrop-like shapes with the fibril ends seen in proximity to the pointed regions on the membranes. Moreover, the tomograms indicated that the fibrils extract lipid from the membranes at these points of distortion by removal or blebbing of the outer membrane leaflet. Tiny (15–25 nm) vesicles, presumably formed from the extracted lipids, were observed to be decorating the fibrils. The findings highlight a potential role of fibrils, and particularly fibril ends, in amyloid pathology, and report a previously undescribed class of lipid–protein interactions in membrane remodelling.
The failure of molecular chaperones to prevent the accumulation of misfolded proteins results in protein aggregation and amyloid formation, processes associated with severe human degenerative diseases (1, 2). Despite the attention focused on these problems during the century since these disorders were first identified (3–5) and advances in understanding the structure of the cross-β conformation of amyloid fibrils in atomic detail (6, 7), the basic pathological mechanisms of amyloidosis remain poorly understood and therapeutic intervention is lacking. The identity of the toxic species and the mechanisms of cytotoxicity remain major unsolved problems. In some systems, there is evidence suggesting that prefibrillar oligomers, rather than the fully formed fibrils, are the source of toxicity (8, 9). In these cases, cytotoxicity is thought to result from the formation of specific membrane pores (10, 11) although alternative models including membrane destabilization or membrane thinning have also been proposed (12–15). In other cases, toxicity may reside with the amyloid fibrils themselves. Evidence that toxicity correlates with fibrillar assemblies has been reported for yeast and mammalian prion proteins (16, 17), human lysozyme (18), Huntingtin exon 1, α-synuclein (19), and Amyloid-β (Aβ) (20, 21). Furthermore, Aβ plaques have been shown to form rapidly in vivo and to precede neuropathological changes in a mouse model (22). The end surfaces of fibrils (herein termed “fibril ends”) are unusually reactive entities: they play a key role in catalyzing recruitment and conformational conversion of amyloid-forming proteins (23, 24) and provide the sites for templated elongation of amyloid fibril growth (25, 26). Recently, Xue et al. (27) showed that short fibrils of β2-microglobulin (β2m), α-synuclein, and hen lysozyme, each prepared by fragmentation of longer fibrils, cause increased damage to membranes and disruption to cellular function compared with the initial long fibrils. Short and long fibril preparations differ in the number of fibril ends at a given protein concentration. Because these are known to be reactive sites, the above observations suggest a role for fibril ends in amyloid–lipid interaction and possibly in amyloid pathogenesis (23, 24, 27). Fragmented fibrils of all three proteins were also found to induce dye leakage from negatively charged liposomes, the most susceptible of which contain a mixture of the cellular lipids phosphatidylcholine (PC) and phosphatidylglycerol (PG), but liposomes with a variety of compositions were damaged by the fibrils in all cases (27).
Here, we use β2m amyloid fibrils formed in vitro as a model system to investigate the structural basis of membrane damage by amyloid fibrils (27, 28). Previous studies have shown that these fibrils possess a parallel in register cross-β structure (29, 30) assembled into multidomain filaments coiled together, described by cryo-EM (28). These fibrils bind amyloid-specific ligands such as serum amyloid P component with a similar affinity to their ex vivo counterparts (31). Using the conditions under which β2m amyloid fibrils induce dye release from liposomes (pH 7.4), we examined the effects of both long (∼1,400 nm) and fragmented (∼400 nm) β2m fibrils (27), as well as various control preparations, on the 3D structures of the liposomes by confocal microscopy, cryo-EM, and tomography. We found pronounced distortions in the liposomes, interruptions to the bilayer structure, and extraction of lipids that were induced by the presence of amyloid fibrils. The most severe distortions were seen in proximity to the fibril ends, which are enriched in the fragmented fibril samples. This type of membrane remodelling appears distinct from the actions of other previously described proteins that induce membrane breakage, as in the action of membrane pore-forming proteins (32). The results suggest a role of fibrils in membrane damage that could contribute to the cellular dysfunction associated with amyloid disease.
Results
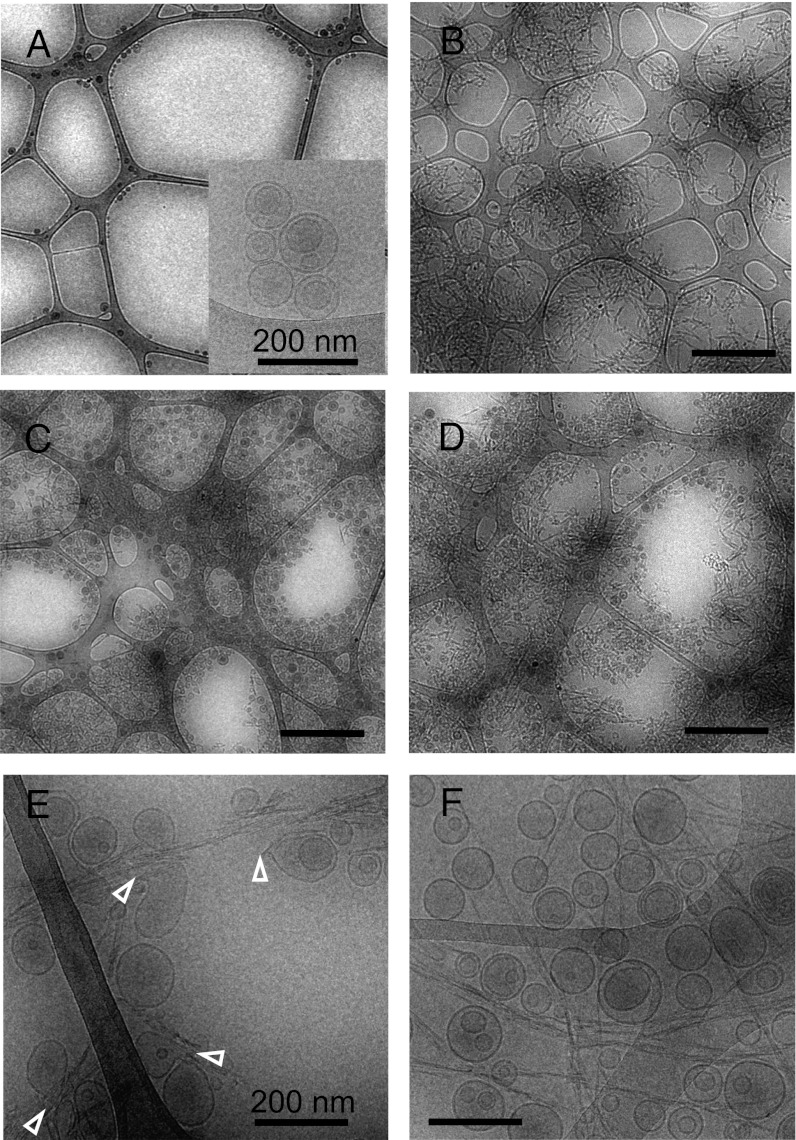
Large unilamellar vesicles (LUVs) formed from 80% egg PC and 20% egg PG, prepared by extrusion with a 100 nm filter, showed smoothly rounded shapes in cryo-EM images (size range 60–130 nm; Fig. 1A, Inset). The wide field view of the liposomes on a lacy carbon support film shows that the liposomes are sparsely distributed over the EM grid, mainly adhering to the carbon at the edges of holes (Fig. 1A). When β2m amyloid fibrils initially formed at pH 2.0 are transferred to pH 7.4, they associate into irregular bundles and aggregates that are distributed over the entire grid (Fig. 1B), consistent with our previous observations (28). Mixing liposomes with either short or long amyloid-like fibrils formed from β2m results in densely clustered liposomes entangled with the fibrils (Fig. 1 C and D). In contrast with the finding that the liposomes alone adhere poorly to EM grids and appear mainly at the carbon edges, the fibril–liposome mixtures are densely distributed across the entire grid, suggesting strong fibril–liposome interactions. Analysis of the liposome structure in the presence of short, fragmented fibrils revealed that some liposomes are distorted into extended, teardrop-like shapes (Fig. 1E, open arrowheads). In these samples, liposomes with altered shapes are readily observed, and the liposomes are permeabilized in dye release experiments under these conditions (27). Although the long fibrils also bind and concentrate the liposomes, in these samples many liposomes remain smoothly rounded, even when they contact large bundles of fibrils, consistent with the reduced ability of the long fibrils to cause dye leakage (27) (Fig. 1F). To quantitate the difference in frequency of liposomes with point distortions in samples incubated with short or long fibrils, we counted >5,000 liposomes. This revealed five times more point distortions in samples incubated with short fibrils compared with their longer counterparts (Table S1). Example images showing the criteria for classifying liposomes as pointed, smoothly rounded, or ambiguous are shown in Fig. S1. The ambiguous category was needed because half of the liposomes were partially obscured in the samples with short fibrils by contacts with fibril bundles and/or other liposomes (and one-third in the samples with long fibrils), preventing their reliable classification as either pointed or smooth.
Fig. 1.
Cryo-EM overviews of liposomes and β2m fibrils. Low-magnification images of (A) liposomes, (B) short β2m fibrils, (C) liposomes plus short fibrils, and (D) liposomes plus long fibrils. (Scale bar for A–D main images: 1 μm.) (E and F) Higher magnification views of liposomes with short (E) or long (F) fibrils. Examples of distorted liposomes are indicated by white arrowheads in E. The inset in A shows a higher magnification view of liposomes. (Scale bar for E and F: 200 nm.)
To determine whether the observations made are specific to the amyloid-like structure of the β2m fibrils, we examined the effect of a different type of β2m aggregate, known as “worm-like” (WL) fibrils, on the liposome preparations. These WL fibrils are formed under different conditions to the amyloid-like fibrils of β2m and do not contain a highly ordered cross-β structure, as revealed by EPR, magic angle spinning NMR, and FTIR (29, 30, 33). Moreover, they exhibit a reduced ability to disrupt cellular function and to induce dye release from liposomes compared with their amyloid-like counterparts (27). The WL fibrils also interact differently with egg PC/PG liposomes, showing only partial clustering of the liposomes as visualized by cryo-EM, with many liposomes remaining in regions free of fibrils (Fig. S2A). This suggests much weaker interaction of the WL fibrils with lipid compared to both the short and long amyloid-like fibrils, consistent with the liposome dye-release experiments described above. β2m monomers had no detectable effect on the distribution or appearance of the liposomes (Fig. S2B), nor did control filaments that lack a cross-β structure such as microtubules (Fig. S2C) or tobacco mosaic virus (TMV) (Fig. S2D).
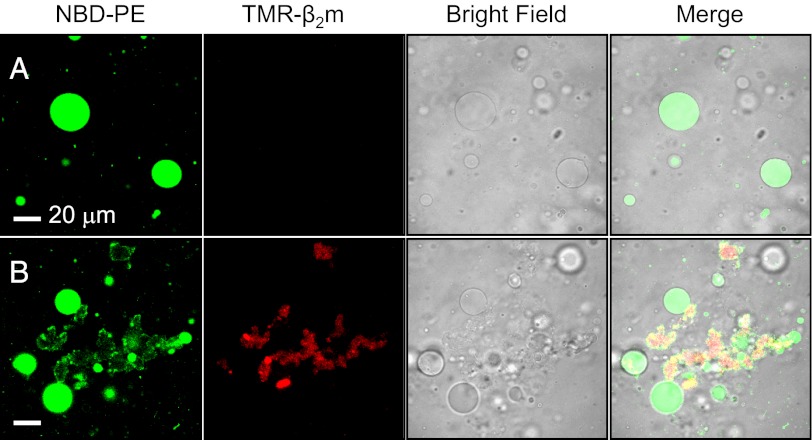
To examine the effect of fibrils on membrane integrity in solution, fluorescence microscopy was used to image giant vesicles (GVs) labeled with green fluorescent lipid, mixed with short β2m fibrils labeled with 5-(and 6-)carboxytetramethylrhodamine (TMR) (Fig. 2). Fluorescence and bright field images of these liposomes incubated with buffer alone showed stable, round structures that remained intact with no visible deformation even after 3 h of incubation (Fig. 2A and Fig. S3A). Similar results were obtained when the vesicles were incubated with TMR-labeled β2m monomers (Fig. S3B). Images of short or long β2m fibrils alone at pH 7.4 showed irregular aggregates, consistent with the bundling of fibrils observed by EM (Fig. S3 C and D), whereas small aggregates were observed for WL fibrils alone (Fig. S3E). Mixing the liposomes and short fibrils resulted in severe damage to the liposomes (Fig. 2 and Fig. S4A). Strikingly, the surfaces of the fibril aggregates colocalize with lipid (yellow regions), suggesting that lipid extraction from the liposome membranes had occurred upon mixing with the fibril sample. Less extensive damage occurred with long fibrils (Fig. S4B), whereas in the presence of WL fibrils many liposomes remained intact (Fig. S4C). These observations are consistent with the relative potencies of the different fibril types in perturbing liposome structure as indicated by cryo-EM (Fig. 1 and Fig. S2A), in dye-release experiments, and in causing cellular damage (short fibrils > long fibrils >> WL fibrils (27). However, these approaches cannot reveal the mechanism of membrane disruption, nor can they show the details of the fibril–liposome interaction.
Fig. 2.
Fluorescence microscopy showing liposome damage by short β2m fibrils. (A) Liposomes plus buffer and (B) liposomes plus short fibrils. Green, NBD-PE labeled giant vesicles; red, TMR-β2m fibrils. Yellow regions indicate colocalization of lipids and fibrils. In B, the vesicles are either completely disintegrated by fibrillar aggregates or show membrane damage with visible lipid extraction by the fibrils.
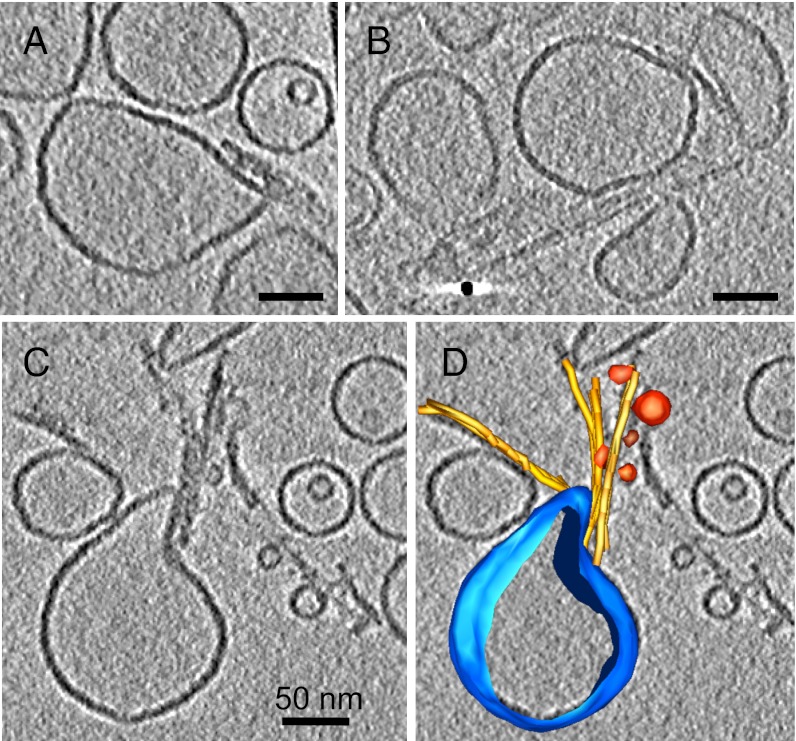
Although the cryo-EM images shown in Fig. 1 indicate a strong interaction of the amyloid fibrils with liposomes, it is not possible to determine from individual projection images whether apparent overlaps between fibrils and liposomes indeed indicate a contact in the same plane of the specimen. Therefore, we used cryo-electron tomography to examine the 3D structures at the sites of liposome interaction with short fibrils by reconstruction from a series of images recorded at different tilt angles. The results revealed examples of liposomes distorted into pointed, tear-drop-like shapes in every tomogram examined. These are shown in sections through representative 3D reconstructions (Fig. 3 A–C). The fibrils typically form lateral contacts to the lipid surfaces and examples were seen in which the fibrils terminate in close proximity to sharp discontinuities in the liposomes. The tomogram region in Fig. 3C was used to trace the structure of the 100 nm liposome with associated fibrils and small vesicles, and these are shown as rendered surfaces superposed on a tomogram section in Fig. 3D. Tiny vesicles, with diameters ranging from 15 to 25 nm, were observed in contact with the clusters of short fibrils in 80% of tomograms examined (Fig. 3D, red spheres). Although some vesicles in this size range are also observed inside larger ones, presumably formed during lipid extrusion, free vesicles of this size range were never observed in samples that had not been incubated with amyloid fibrils.
Fig. 3.
Cryoelectron tomography of liposome–fibril interactions. (A–C) Sections of tomograms showing liposomes clustered and distorted by short fibrils. (D) A rendered 3D model of a distorted liposome, surrounding fibrils and adjacent small vesicles from C. (Scale bar: 50 nm.)
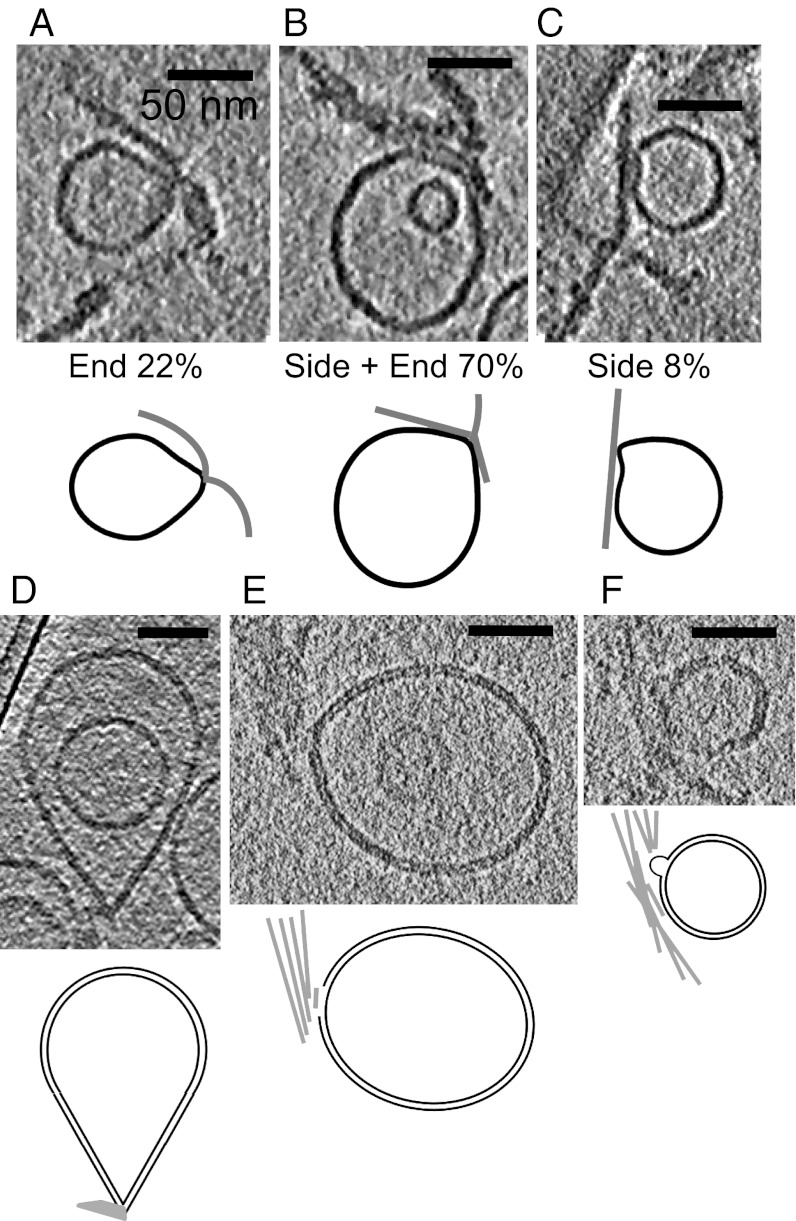
To quantify the nature of the observed fibril–vesicle interactions, we divided the contacts into three categories: fibrils binding the membrane surface by their sides, at their ends, and by a combination of both interactions. We counted the number of interactions in each category in a dataset of 244 clear examples (in 3D) of fibril–membrane interactions from four different tomograms collected from three different samples. The interactions typically involve a stretch of lateral contact between the liposome and the side of a fibril, terminating with a distortion or sharp discontinuity in the liposome shape at the end of the fibril. Of the 244 interactions, 172 interactions (70%) involved contacts of the vesicles with both sides and ends of the fibrils, 20 contacts (8%) involved only the sides of the fibrils, and 55 contacts (22%) were with fibril ends. An example of each of these types of interaction is shown in Fig. 4 A–C. In general, the membranes were flattened at sites of contact with other vesicles, edges of the carbon film and fibril sides, but showed sharper distortions in the vicinity of the fibril ends.
Fig. 4.
Distortions of liposomes in the vicinity of fibril ends. (A–C) Examples and cartoons of the different types of fibril–liposome interactions observed, with the percentage of each type measured by counting examples in four tomograms. (D–F) Sections of tomograms taken closer to focus, showing examples of the disruption of the lipid bilayer in the region of fibril ends, including (D) formation of a sharp point, (E) a break in the outer leaflet of the membrane, and (F) a bubble forming in the outer leaflet, with corresponding cartoons showing membranes as black lines and fibrils as gray lines. (Scale bar: 50 nm.)
The EM images suggest that the short fibrils bundle together more than their longer counterparts (Fig. 1 E and F). Bundling of fibrils decreases the lateral surface–volume ratio, and therefore the area of lateral surface available for interaction with liposomes. Nevertheless, the short fibrils, which have a higher proportion of exposed ends, increase dye permeability of liposomes (27), increase membrane disruption (Fig. 2), and cause more point distortions in the lipid bilayer in the vicinity of fibril ends (Figs. 3 and 4 and Movies S1 and S2). Together, these data suggest that the ends of the short fibrils are the most reactive species in membrane disruption, although the sides of the fibrils also play a role in membrane binding. Should fibril bundling be an important factor in the disruption of liposomes, the liposomes would be expected to form contacts to extended regions on the bundle surfaces—for example, the membrane might be expected to extend over, or wrap around, the bundle surface. However, such behavior was never observed in the tomograms. Instead, in all of the examples in which the liposome–fibril contact is clearly resolved (Figs. 3 and 4 and Movies S1 and S2), the membrane–fibril contacts are localized, so that the membrane appears to touch only one fibril in any given contact, even when that fibril is part of a bundle. These observations argue against a major effect of fibril bundling in the interaction with liposomes.
To better understand the details of membrane damage by the short β2m amyloid fibrils, we collected examples of these interactions in tomograms recorded with a lower defocus, so that the lipid bilayer was resolved, albeit at the expense of lower contrast. The examples shown in Fig. 4 D–F reveal the structural basis of how amyloid assemblies can extract lipid from membranes. Breaks and distortions appear in the outer leaflet of the bilayer, in regions adjacent to fibril ends, suggesting a specific interaction that leads to destabilization and eventually disintegration of the membrane.
Discussion
There is considerable evidence that amyloid assemblies interact with membranes, supporting the notion that these interactions may play a role in amyloid-associated cellular dysfunction (11, 13, 34). However, the physical basis for these interactions and the mechanisms of amyloid-induced dysfunction remain unresolved. Observations of ring-like structures formed from α-synuclein and prefibrillar assemblies of other amyloid-like systems, along with membrane leakage in the presence of prefibrillar oligomers, have led to the hypothesis of membrane pore formation, membrane thinning, and membrane destabilization by prefibrillar oligomers (8, 9, 12, 34, 35). In addition it has been shown that lipid-induced depolymerization of nontoxic Aβ fibrils leads to formation of so-called “reverse oligomers” that are cytotoxic, akin to the oligomers formed de novo during fibril assembly (36). Membrane-induced fibril depolymerization and membrane destabilization associated with fibril growth on lipid bilayers have also been proposed as alternative mechanisms of amyloid-mediated cytotoxicity (11, 13, 15). Using fluorescence methods, Reynolds et al. (12) observed fibril growth and lipid extraction/thinning by adding monomeric α-synuclein to membranes supported on a solid surface. In a similar study it was shown that addition of islet-associated polypeptide (IAPP) induces defects on supported lipid bilayers, accompanied by transfer of lipid vesicles onto growing amyloid fibrils (14). These studies provide a large body of evidence suggesting a significant involvement of lipid membranes in amyloid cytotoxicity.
Our results show that amyloid fibrils bind strongly to membrane surfaces, as shown by the marked clustering of liposomes in the presence of β2m fibrils. For many amyloidogenic proteins and amyloid fibrils, it has been shown that binding to membranes requires negatively charged lipids and positively charged residues or areas of high positive charge density on the protein assembly (12, 37–39). This is also the case for the interaction of β2m fibrils with liposomes: higher dye leakage was found for liposomes containing negatively charged lipids (27), suggesting that the fibril–membrane interaction has a strong electrostatic component. In the case of IAPP, binding of the monomeric protein to membranes is mainly driven by electrostatic interactions, but the membrane damage is dominated by hydrophobic interactions with IAPP oligomers, similar to the membrane-mediated toxicity of other amyloidogenic assemblies (11, 21, 39).
A key finding from the cryo-electron tomography images portrayed here is the sharp distortion of the membranes by the fibrils, suggesting that the fibrils make additional interactions that extract lipids from the outer membrane leaflet of the liposomes. Notably, initial studies of liposomes incubated with fragmented α-synuclein fibrils also showed examples of sharp distortions and interruptions to the bilayer at sites of contact with α-synuclein fibrils. The notion of lipid extraction by amyloid fibril ends is supported by the observation of tiny (15–25 nm) vesicles found near the areas of contact between β2m fibrils and distorted liposomes, and it is consistent with the colocalization of fibrils and disintegrated liposomes observed by fluorescence microscopy (Fig. 2B and Fig. S4 A and B). The observation that the sites of membrane disruption are mainly adjacent to fibril ends accords with the finding of the greatest liposome disruption by fibrils that were fragmented and therefore contained more ends, suggesting that the fibril ends have an enhanced ability (relative to the fibril shaft) to cause the sharp distortions and extract lipids from the membrane. It is likely that these distortions involve hydrophobic interactions in addition to the electrostatic component. Hydrophobic domains in proteins can induce dramatic distortions in lipid membranes. For example, the ability of epsin to induce membrane tubulation and vesiculation is attributed to distortion caused by the insertion of wedge-shaped hydrophobic domains into the membrane (40). The hydrophobicity of the fibril ends may be a property in common with prefibrillar or fibrillar oligomers, which are also reported to damage membranes and are considered as intermediates in amyloid fibril assembly and disassembly (11, 41–43).
Membrane distortion and breakage occur in many biological processes, such as viral fusion and pore formation, processes in which binding of extraneous proteins induces membrane curvature and breakage, resulting either in fusion with another membrane or in the formation of protein-lined transmembrane channels (32, 44). The fibril–liposome interaction appears to represent a new class of specific membrane distortion by a protein assembly, distinct from previously described mechanisms for distorting and disrupting lipid bilayers. Similar distortions have been seen by cryoelectron tomography of liposomes involved in a different process, the formation of viral fusion pores (45). In this case, formation of a pore and fusion of the membranes are directed by hemagglutinin spikes on the viral surface that bind and insert into the target liposome membrane. Binding occurs via the ends of the spikes, which undergo a low pH-induced conformational change to insert. In this case, a sharp point is also drawn out from the target membrane, as is observed with short β2m amyloid fibrils, but instead of destroying the liposome, the destabilized, extruded membrane region is fused to the viral membrane.
The cryoelectron tomography images of β2m fibril–membrane interactions presented here suggest that the cellular dysfunction associated with these and other fibrils or fibril-like assemblies (16–21, 27) involves direct bilayer disruption. This membrane damage might arise by direct interaction with the fibril ends and/or by the creation of new, toxic species by reaction of the fibril ends with the lipids. The ends of biological filaments differ dramatically in their structure and properties, with actin filament and microtubules being classic examples (46, 47). For amyloid fibrils, fibril ends are also distinct, being the sites of growth and disassembly, consistent with the notion that amyloid fibril ends have a specific role in membrane disruption. The results presented here suggest previously undescribed routes of amyloid-associated cellular dysfunction involving membrane disruption by fibril ends. These observations suggest, in turn, that inhibiting fibril–lipid interactions by capping fibril ends may provide a potential unique therapeutic strategy targeting amyloid pathogenesis in disease.
Methods
β2m Fibril Preparation.
Long straight fibrils were prepared from recombinant β2m in 10 mM NaH2PO4, 50 mM NaCl buffer pH 2.0 containing 0.1% (wt/wt) fibril seeds, and WL fibrillar aggregates were prepared in 25 mM NaH2PO4, 400 mM NaCl buffer pH 2.5 following previously described protocols (27, 48). Short fragmented fibril samples were generated by stirring preformed long straight fibril samples at 1,000 rpm for 48 h at 25 °C using a precision stirrer (custom-built by the workshop of the School of Physics and Astronomy, University of Leeds), as previously described (27). The protein monomer concentration was 120 μM for the straight fibrils and 45 μM for the WL aggregates. Fibril samples were stored at 25 °C and used within 3 mo. To prepare fibrils for confocal imaging, β2m monomers were labeled with TMR, as described in Porter et al. (49) and as detailed in SI Methods.
TMV and Microtubule Controls.
TMV (16 mg/mL) was supplied by J. W. M. van Lent (Laboratory of Virology, Wageningen, Netherlands). Microtubules prepared by polymerization of glycerol-free bovine brain tubulin (Cytoskeleton, Inc.), in 5 mM MgCl2, 1 mM EGTA, 80 mM Pipes, 1 mM GTP at pH 6.8, and with a tubulin monomer concentration of 50 μM, were provided by C. Moores (Birkbeck College, London, United Kingdom).
Liposome Preparation.
Liposome samples of LUVs of egg PC/PG were prepared as previously described (27, 50). Briefly, a stock solution of 62.5 mM lipids prepared from PC (Type XVI-E, chicken egg, Sigma Aldrich) and PG (chicken egg, Avanti Lipids) in chloroform to give a 4:1 molar ratio of PC–PG, and liposomes were formed by extrusion as detailed in SI Methods. GVs used in confocal microscopy experiments were prepared by a rapid evaporation method (51), as described in SI Methods. NBD-PE [1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2–1,3-benzoxadiazol-4-yl), ammonium salt, Avanti Lipids] was added to the egg PC–PG 4:1 lipid mixture at 0.04% (molar ratio) to make the vesicles fluorescent. Similar results were obtained with GVs prepared with a PC–PG ratio of 1:1.
Sample Preparation for EM.
Samples of fragmented β2m fibrils and liposomes were prepared by adding fragmented fibrils (60 μM) (all fibril concentrations are given in β2m monomer equivalent concentration) in 10 mM NaH2PO4, 50 mM NaCl (buffer pH 2.0) to stock solutions of PC–PG liposomes (1.3 or 2.6 mM lipids in 50 mM Hepes, 107 mM NaCl, 1 mM EDTA, pH 7.4), with a final concentration of 500–550 μM lipids and 13–18 μM β2m fibrils (30–40 molar excess of lipids). A typical cryo-EM sample contained a final concentration of 0.5 mM liposomes in 50 mM Hepes, 107 mM NaCl, 1 mM EDTA, and pH 7.4, which was vortexed for few seconds, mixed with 60 μM β2m fibril fragments in 10 mM NaH2PO4, 50 mM NaCl, and pH 2.0 to give a final concentration of 18 μM β2m fibrils. The mixture was vortexed for a few seconds and then incubated for 1–2 min at 25 °C. Three 5 μL aliquots of this solution were applied to negatively glow-discharged holey carbon grids (300–400 mesh Cu, Agar-Scientific), manually blotted, and plunge frozen in liquid ethane.
For cryotomography, an aliquot [12% (vol/vol) final concentration] of 10 nm gold beads (Protein A-gold conjugates, Aurion) was added to the sample immediately before grid preparation. For each sample of liposomes and fibrils, grids were prepared in batches of three, within ∼7 min after addition of fibrils to the liposomes. Liposome damage is complete within 10 min according to the dye release measurements (27).
Control samples containing only the PC–PG stock liposomes and samples of PC–PG liposomes with added TMV, microtubules, WL β2m fibrillar aggregates, long unfragmented β2m fibrils, or β2m monomer were prepared using the same protocol. Equivalent samples containing only fragmented, WL, or long (unfragmented) β2m fibrils were prepared in parallel. The final concentrations of TMV and microtubules in the samples containing PC–PG liposomes were 3.5 mg/mL and 7 μM (tubulin monomer equivalent), respectively. For each control sample, the effects of fragmented β2m fibrils were examined on the same stock of PC–PG liposomes to control for the inherent variability of extruded liposomes. Control samples of WL fibrillar aggregates and PC–PG liposomes prepared according to the above protocol contained a higher salt concentration (196 mM NaCl), as required for preparation of WL fibrillar aggregates (48). Therefore, samples of fragmented β2m fibrils and liposomes, as well as liposomes alone, were analyzed to verify that the higher salt did not affect the results obtained. Similarly, the pH 2.0 buffer used to assemble the amyloid-like fibrils of β2m had no effect on the liposome structure or integrity.
Cryo-EM, Tomography, and Image Processing.
Low-dose (15–20 e−/A2) images were recorded on a Gatan 4K × 4K charge coupled device (CCD) camera (Gatan) at a calibrated magnification of 29,000× (3.89 Å/pixel) and 2–3.5 μm underfocus with a Tecnai F20 FEG electron microscope operated at 200 kV and equipped with a Gatan cold stage. Overview images were recorded at 5,000×. Cryo-EM images collected from five different preparations of PC–PG liposomes showed that the liposomes were unilamellar, and more than 70% had diameters in the range 60–120 nm. No liposomes were observed with diameter less than ∼20 nm (Fig. S2).
Cryoelectron tomography was done on a Tecnai Polara electron microscope (FEI) operated at 300 keV with a calibrated magnification of 23,000× (5.1 Å/pixel). Tilt series covering an angular range of –60° to +60° at 2° increments and nominal underfocus of 2.5–5 μm and 70–90 e−/A2 per series were recorded on a Gatan Ultrascan 4000 4K × 4K CCD camera using FEI tomography software. A total of 33 tilt series were collected from four different samples of PG–PC liposomes and fragmented β2m fibrils. The images were binned to 10 Å/pixel and aligned with gold bead fiducial markers. Tomogram reconstructions were calculated by weighted back-projection in IMOD (52). Some tomograms were also reconstructed from unbinned images (5.1 Å/pixel) to resolve the lipid bilayer. Tomograms were denoised using nonlinear anisotropic diffusion (53) as implemented in IMOD (52). Images showing the interaction between fragmented fibrils and the lipid bilayer were obtained by averaging 10–30 slices of binned or unbinned, filtered tomograms, respectively.
Four representative, binned, filtered tomograms collected from three different grids at underfocus 2.5–5 μm were chosen for manual counting of contacts between fibril ends or sides with liposome surfaces. Contacts were divided into three categories: end interactions, side and end interactions, and side interactions only.
Fluorescence Microscopy.
TMR-labeled short β2m fibrils were 10-fold diluted into egg PC/PG/NBD-PE (4:1:2 × 10−3, molar ratio) giant vesicle suspension, yielding a 12 μM β2m monomer equivalent concentration and 1.8 mM total lipid concentration. The images were obtained following 15 min incubation of the fibrils with the vesicles on a Zeiss Axiovert 100M confocal laser scanning microscope using a Zeiss 63×/1.4 N.A. Plan Apochromat DIC oil immersion objective lens. The NBD-PE fluorescent probe was excited with the 488 nm line of an argon laser, whereas TMR was excited with argon-krypton laser at 568 nm. Long pass filters 505 and 580 were used for acquisition NBD and TMR fluorescence, respectively.
Supplementary Material
Acknowledgments
We thank Luchun Wang and Daniel Clare for help with electron microscopy and data analysis, Salvador Tomas for support in sample preparation and helpful discussions, and David Houldershaw and Richard Westlake for computing support, the Hewitt and Radford group members for helpful discussions and Giulia Zanetti for comments on the manuscript. This work was supported by Wellcome Trust Grants 08059 (to H.R.S. and S.E.R.), 075675 (to S.E.R.), and by a Wellcome Trust equipment Grant 079605 (to H.R.S.). Funding was also provided by the UK Biotechnology and Biological Sciences Research Council (BB/526502/1) (to S.E.R.) and the British Council (British Israel Research and Academic Exchange Award) (to S.E.R. and R.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206325109/-/DCSupplemental.
References
- 1.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 2.Pepys MB. Amyloidosis. Annu Rev Med. 2006;57:223–241. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- 3.Wadsworth JDF, Collinge J. Molecular pathology of human prion disease. Acta Neuropathol. 2011;121(1):69–77. doi: 10.1007/s00401-010-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 5.Buxbaum JN, Linke RP. A molecular history of the amyloidoses. J Mol Biol. 2012;421(2-3):142–159. doi: 10.1016/jmb.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 6.Eichner T, Radford SE. A diversity of assembly mechanisms of a generic amyloid fold. Mol Cell. 2011;43(1):8–18. doi: 10.1016/j.molcel.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Colletier JP, et al. Molecular basis for amyloid-beta polymorphism. Proc Natl Acad Sci USA. 2011;108(41):16938–16943. doi: 10.1073/pnas.1112600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lashuel HA, Lansbury PT., Jr Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q Rev Biophys. 2006;39(2):167–201. doi: 10.1017/S0033583506004422. [DOI] [PubMed] [Google Scholar]
- 9.Glabe CG. Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiol Aging. 2006;27(4):570–575. doi: 10.1016/j.neurobiolaging.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Demuro A, Smith M, Parker I. Single-channel Ca(2+) imaging implicates Aβ1-42 amyloid pores in Alzheimer’s disease pathology. J Cell Biol. 2011;195(3):515–524. doi: 10.1083/jcb.201104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butterfield SM, Lashuel HA. Amyloidogenic protein-membrane interactions: mechanistic insight from model systems. Angew Chem Int Ed Engl. 2010;49(33):5628–5654. doi: 10.1002/anie.200906670. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds NP, et al. Mechanism of membrane interaction and disruption by α-synuclein. J Am Chem Soc. 2011;133(48):19366–19375. doi: 10.1021/ja2029848. [DOI] [PubMed] [Google Scholar]
- 13.Stefani M. Biochemical and biophysical features of both oligomer/fibril and cell membrane in amyloid cytotoxicity. FEBS J. 2010;277(22):4602–4613. doi: 10.1111/j.1742-4658.2010.07889.x. [DOI] [PubMed] [Google Scholar]
- 14.Domanov YA, Kinnunen PKJ. Islet amyloid polypeptide forms rigid lipid-protein amyloid fibrils on supported phospholipid bilayers. J Mol Biol. 2008;376(1):42–54. doi: 10.1016/j.jmb.2007.11.077. [DOI] [PubMed] [Google Scholar]
- 15.Engel MFM, et al. Membrane damage by human islet amyloid polypeptide through fibril growth at the membrane. Proc Natl Acad Sci USA. 2008;105(16):6033–6038. doi: 10.1073/pnas.0708354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bucciantini M, et al. Toxic effects of amyloid fibrils on cell membranes: The importance of ganglioside GM1. FASEB J. 2012;26(2):818–831. doi: 10.1096/fj.11-189381. [DOI] [PubMed] [Google Scholar]
- 17.Novitskaya V, Bocharova OV, Bronstein I, Baskakov IV. Amyloid fibrils of mammalian prion protein are highly toxic to cultured cells and primary neurons. J Biol Chem. 2006;281(19):13828–13836. doi: 10.1074/jbc.M511174200. [DOI] [PubMed] [Google Scholar]
- 18.Mossuto MF, et al. The non-core regions of human lysozyme amyloid fibrils influence cytotoxicity. J Mol Biol. 2010;402(5):783–796. doi: 10.1016/j.jmb.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pieri L, Madiona K, Bousset L, Melki R. Fibrillar α-synuclein and huntingtin exon 1 assemblies are toxic to the cells. Biophys J. 2012;102(12):2894–2905. doi: 10.1016/j.bpj.2012.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomic JL, Pensalfini A, Head E, Glabe CG. Soluble fibrillar oligomer levels are elevated in Alzheimer’s disease brain and correlate with cognitive dysfunction. Neurobiol Dis. 2009;35(3):352–358. doi: 10.1016/j.nbd.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada T, Ikeda K, Wakabayashi M, Ogawa M, Matsuzaki K. Formation of toxic Abeta(1-40) fibrils on GM1 ganglioside-containing membranes mimicking lipid rafts: Polymorphisms in Abeta(1-40) fibrils. J Mol Biol. 2008;382(4):1066–1074. doi: 10.1016/j.jmb.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 22.Meyer-Luehmann M, et al. Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer’s disease. Nature. 2008;451(7179):720–724. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyedmers J, et al. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci USA. 2010;107(19):8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carulla N, et al. Molecular recycling within amyloid fibrils. Nature. 2005;436(7050):554–558. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- 25.Knowles TPJ, et al. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326(5959):1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 26.Xue WF, Homans SW, Radford SE. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc Natl Acad Sci USA. 2008;105(26):8926–8931. doi: 10.1073/pnas.0711664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue WF, et al. Fibril fragmentation enhances amyloid cytotoxicity. J Biol Chem. 2009;284(49):34272–34282. doi: 10.1074/jbc.M109.049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White HE, et al. Globular tetramers of β(2)-microglobulin assemble into elaborate amyloid fibrils. J Mol Biol. 2009;389(1):48–57. doi: 10.1016/j.jmb.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladner CL, et al. Stacked sets of parallel, in-register β-strands of β2-microglobulin in amyloid fibrils revealed by site-directed spin labeling and chemical labeling. J Biol Chem. 2010;285(22):17137–17147. doi: 10.1074/jbc.M110.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Debelouchina GT, Platt GW, Bayro MJ, Radford SE. Griffin RG Intermolecular alignment in beta(2)-microglobulin amyloid fibrils. J Am Chem Soc. 2010;132:17077–17079. doi: 10.1021/ja107987f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers SL, et al. A systematic study of the effect of physiological factors on β2-microglobulin amyloid formation at neutral pH. Biochemistry. 2006;45(7):2311–2321. doi: 10.1021/bi052434i. [DOI] [PubMed] [Google Scholar]
- 32.Tilley SJ, Saibil HR. The mechanism of pore formation by bacterial toxins. Curr Opin Struct Biol. 2006;16(2):230–236. doi: 10.1016/j.sbi.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Jahn TR, Tennent GA, Radford SE. A common β-sheet architecture underlies in vitro and in vivo β2-microglobulin amyloid fibrils. J Biol Chem. 2008;283(25):17279–17286. doi: 10.1074/jbc.M710351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hebda JA, Miranker AD. The interplay of catalysis and toxicity by amyloid intermediates on lipid bilayers: insights from type II diabetes. Annu Rev Biophys. 2009;38:125–152. doi: 10.1146/annurev.biophys.050708.133622. [DOI] [PubMed] [Google Scholar]
- 35.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416(6880):507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 36.Martins IC, et al. Lipids revert inert Abeta amyloid fibrils to neurotoxic protofibrils that affect learning in mice. EMBO J. 2008;27(1):224–233. doi: 10.1038/sj.emboj.7601953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasahara K, Hall D, Hamada D. Effect of lipid type on the binding of lipid vesicles to islet amyloid polypeptide amyloid fibrils. Biochemistry. 2010;49(14):3040–3048. doi: 10.1021/bi9019252. [DOI] [PubMed] [Google Scholar]
- 38.Yoshiike Y, Akagi T, Takashima A. Surface structure of amyloid-beta fibrils contributes to cytotoxicity. Biochemistry. 2007;46(34):9805–9812. doi: 10.1021/bi700455c. [DOI] [PubMed] [Google Scholar]
- 39.Knight JD, Miranker AD. Phospholipid catalysis of diabetic amyloid assembly. J Mol Biol. 2004;341(5):1175–1187. doi: 10.1016/j.jmb.2004.06.086. [DOI] [PubMed] [Google Scholar]
- 40.Boucrot E, et al. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149(1):124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campioni S, et al. A causative link between the structure of aberrant protein oligomers and their toxicity. Nat Chem Biol. 2010;6(2):140–147. doi: 10.1038/nchembio.283. [DOI] [PubMed] [Google Scholar]
- 42.Winner B, et al. In vivo demonstration that alpha-synuclein oligomers are toxic. Proc Natl Acad Sci USA. 2011;108(10):4194–4199. doi: 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cremades N, et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell. 2012;149(5):1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438(7068):590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 45.Lee K-K. Architecture of a nascent viral fusion pore. EMBO J. 2010;29(7):1299–1311. doi: 10.1038/emboj.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fourniol F, Moores CA. Kinesin motor activation: Microtubules pull the switches. Proc Natl Acad Sci USA. 2010;107(9):3949–3950. doi: 10.1073/pnas.0915158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433(7025):488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 48.Gosal WS, et al. Competing pathways determine fibril morphology in the self-assembly of β2-microglobulin into amyloid. J Mol Biol. 2005;351(4):850–864. doi: 10.1016/j.jmb.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 49.Porter MY, Routledge KE, Radford SE, Hewitt EW. Characterization of the response of primary cells relevant to dialysis-related amyloidosis to β2-microglobulin monomer and fibrils. PLoS ONE. 2011;6(11):e27353. doi: 10.1371/journal.pone.0027353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomas S, Milanesi L. Mutual modulation between membrane-embedded receptor clustering and ligand binding in lipid membranes. Nat Chem. 2010;2(12):1077–1083. doi: 10.1038/nchem.892. [DOI] [PubMed] [Google Scholar]
- 51.Moscho A, Orwar O, Chiu DT, Modi BP, Zare RN. Rapid preparation of giant unilamellar vesicles. Proc Natl Acad Sci USA. 1996;93(21):11443–11447. doi: 10.1073/pnas.93.21.11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 53.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135(3):239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.