Abstract
Temporally restricted feeding (RF) can phase reset the circadian clocks in numerous tissues in mammals, contributing to altered timing of behavioral and physiological rhythms. However, little is known regarding the underlying molecular mechanism. Here we demonstrate a role for the gamma isotype of protein kinase C (PKCγ) in food-mediated entrainment of behavior and the molecular clock. We found that daytime RF reduced late-night activity in wild-type mice but not mice homozygous for a null mutation of PKCγ (PKCγ−/−). Molecular analysis revealed that PKCγ exhibited RF-induced changes in activation patterns in the cerebral cortex and that RF failed to substantially phase shift the oscillation of clock gene transcripts in the absence of PKCγ. PKCγ exerts effects on the clock, at least in part, by stabilizing the core clock component brain and muscle aryl hydrocarbon receptor nuclear translocator like 1 (BMAL1) and reducing its ubiquitylation in a deubiquitination-dependent manner. Taken together, these results suggest that PKCγ plays a role in food entrainment by regulating BMAL1 stability.
Many organisms exhibit 24-h or circadian rhythms in a vast repertoire of cellular, physiological, and behavioral processes. Circadian rhythms are believed to arise from a molecular clock consisting of a series of transcriptional/posttranscriptional feedback loops with Clock and Bmal1 at the center of the loops (1). CLOCK–BMAL1 dimers activate the transcription of three Period genes (Per1, Per2, and Per3) and two Cryptochrome genes (Cry1 and Cry2). PER and CRY heterodimerize and translocate into the nucleus, inhibiting the transcriptional activity of CLOCK–BMAL1. In a second loop, CLOCK–BMAL1 activates the transcription of the retinoic acid-related orphan receptors Rev-erbα and Rorα. The former inhibits, whereas the latter activates, transcription of Bmal1. In the forebrain, neuronal Per/Arnt/Sim domain protein 2 (NPAS2) functions as a CLOCK analog (2). In addition to transcriptional control, posttranscriptional modifications (e.g., phosphorylation) also play a critical role in setting the speed of the clock.
Circadian rhythms are primarily entrained by light, but other cues, such as timed food signals (or temporally restricted feeding; RF), can also serve as potent entrainment signals that trigger behavioral and physiological rhythms expressed in the circadian range (3). These food-entrained rhythms are accompanied by phase resetting of the molecular clock in the brain (4, 5) and various peripheral tissues (6), but not in the light-entrainable pacemaker, the suprachiasmatic nucleus (SCN) (4, 5). In the brain, a network of structures such as the cerebral cortex, hippocampus, dorsal medial hypothalamic nucleus, and cerebellum are entrained by food and may participate in regulating RF-dependent biological rhythms (4, 7, 8). Because the molecular oscillations in various brain structures but not the SCN are phase reset by RF, this can lead to desynchrony between the SCN clock and clocks in other brain regions. Similarly, eating at the “wrong time” of the day for humans (such as night-eating syndrome and shift work) may also result in desynchrony between the SCN clock and extra-SCN clocks in the brain. Desynchrony between the molecular oscillations of the SCN and extra-SCN brain structures may contribute to altered mood and cognitive functions. Indeed, individuals with night-eating syndrome and shift workers tend to exhibit depressed mood and impairment in cognitive function (9, 10). However, the mechanism of how RF phase resets the molecular clocks in extra-SCN brain regions is not yet understood.
The mammalian protein kinase C (PKC) family has been shown to be involved in phase resetting of the circadian clock (11, 12). PKCα is ubiquitously expressed in both central and peripheral tissues (13), including the SCN (14), and has recently been identified as a core clock component (15). PKCα also mediates light entrainment by stabilizing PER2 in the SCN (11), whereas both PKCα and PKCγ may phosphorylate and activate CLOCK in response to serum stimulation (12). Unlike PKCα, PKCγ is neuron-specific, with the most abundant expression in the cerebral cortex, hippocampus, and cerebellum (16), all of which are regions that can be entrained by food (4, 7, 8), whereas there is little or no expression in the SCN (14). Here we set out to investigate whether PKCγ plays a role in food entrainment to further reveal the molecular mechanism regarding the resetting of the circadian clock by RF. We find that RF reduces late-night activity in wild-type (WT) but not PKCγ knockout mutant mice (PKCγ−/−). Moreover, the activation status of PKCγ is altered by RF compared with food ad libitum (AL) in the cerebral cortex. RF substantially phase advances the oscillation of several core clock genes in WT but not PKCγ−/− mice. PKCγ may regulate the clock by reducing BMAL1 ubiquitylation, thus stabilizing BMAL1, which could in turn lead to phase resetting.
Results
Restricted Feeding Reduces Late-Night Activity in Wild-Type but Not PKCγ−/− Mice.
To examine the expression pattern of PKCγ, we performed immunostaining with a PKCγ antibody. Consistent with previous results (16) and the Allen Brain Atlas (www.brain-map.org), we observed the most intense labeling in the cerebral cortex, hippocampus, and cerebellum (Fig. S1). The levels of PKCγ did not exhibit substantial oscillation throughout the day (Fig. S1).
Due to the importance of PKCγ in the neuronal functions of the food-entrainable brain regions (16), we examined whether PKCγ is involved in regulating behavior during RF. PKCγ−/− mice adapted their daily food intake to RF (food was provided only for 4 h during the light phase) more slowly than WT (Fig. S2A). Both genotypes exhibited a sharp decrease in the amount of food intake at the onset of RF compared with AL, but the food intake amount of WT had returned to baseline levels by RF day 4, whereas that of PKCγ−/− did not until RF day 8. This may reflect defects in the ability to entrain to food in PKCγ−/−. Both WT and PKCγ−/− mice exhibited food-anticipatory activity that was not significantly different (Fig. S2B).
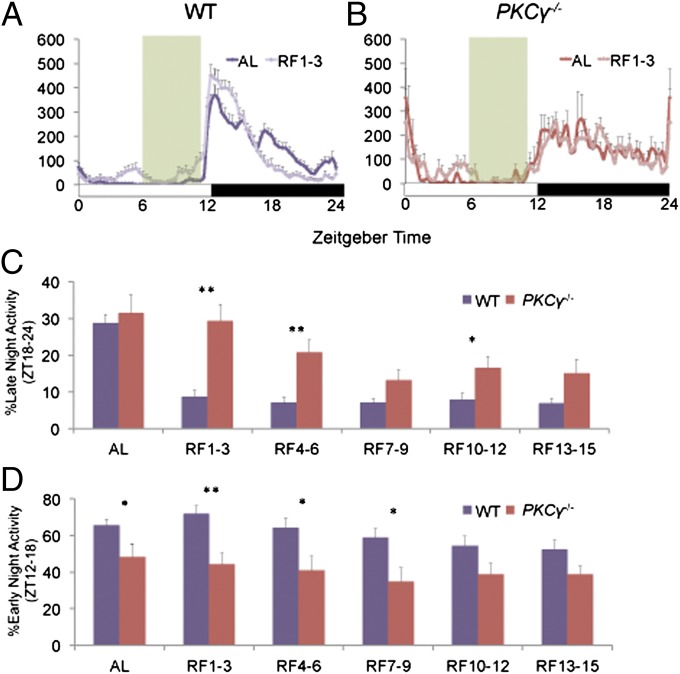
Under AL conditions, WT mice showed substantially higher activity during the early night than the late night, whereas PKCγ−/− showed relatively constant activity levels throughout the entire dark phase (Fig. 1 A and B). Under RF conditions, WT mice demonstrated increased activity in the early night and reduced activity in the late night compared with AL conditions (Fig. 1A). This change was not observed in PKCγ−/− mice under RF, which had activity patterns similar to the AL condition (Fig. 1B). This resulted in significantly higher late-night [Zeitgeber time (ZT)18–24] activity in PKCγ−/− relative to WT throughout the entire duration of RF, but the difference was most prominent during RF days 1–3 (Fig. 1C). On the other hand, PKCγ−/− mice exhibited significantly lower early-night (ZT12–18) activity during AL and for the first ∼10 d of RF (Fig. 1D).
Fig. 1.
Restricted feeding reduces late-night activity of wild-type but not PKCγ−/− mice. (A and B) Normalized wheel-running activity profiles of WT (A) and PKCγ−/− (B) mice averaged over 3 d of 12L12D conditions during AL (darker shade) and RF (lighter shade) days 1–3. The white and black boxes indicate light regime, whereas food availability during RF is indicated by the green shading. (C and D) The percentage of late-night (C) and early-night (D) activity out of total daily activity in WT (purple) and PKCγ−/− (red) mice during AL and between days 1–3, 4–6, 7–9, 10–12, and 13–15 of RF. Error bars represent SEM (n = 10–16). Asterisks mark significant differences between the two genotypes (Student’s t test, *P < 0.05, **P < 0.01).
PKCγ Activation Status Is Differentially Regulated During Food ad Libitum Versus Restricted Feeding.
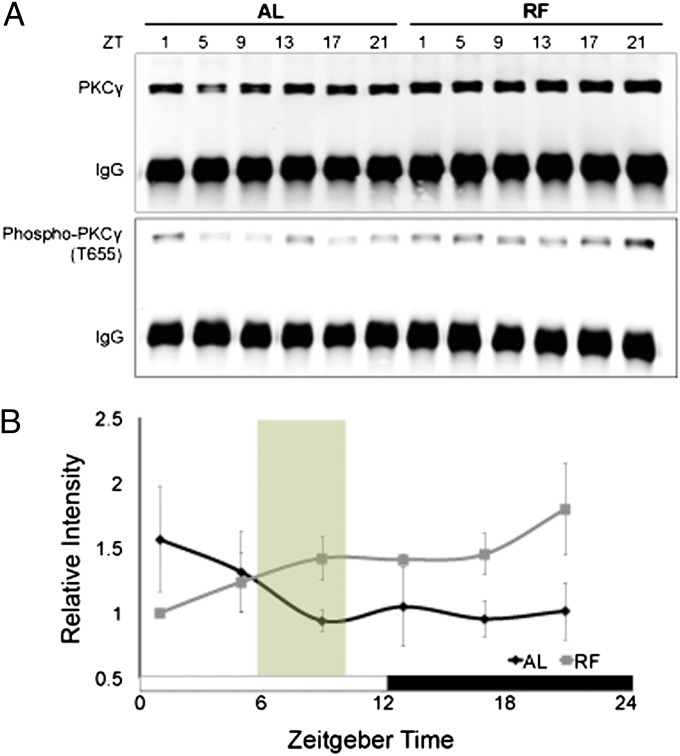
To characterize how PKCγ regulates RF-induced changes in nighttime behavior, we compared the activation status of PKCγ during RF vs. AL using an antibody that labels PKCγ phosphorylated at threonine 655 (PKCγ-phosphoT655). Phosphorylation at this site indicates a catalytically mature kinase (17). To prevent cross-reaction of the PKCγ-phosphoT655 antibody with other PKC isotypes, we first performed immunoprecipitation with the PKCγ antibody to specifically pull down PKCγ from cerebral cortex lysates of WT mice (Fig. 2A, Upper), and then probed with the phosphoT655 antibody (Fig. 2A, Lower). We found that the temporal pattern of PKCγ activation status is significantly different throughout the day during RF vs. AL, with a tendency toward reduced phospho-PKCγ levels at ZT1 (i.e., 1 h after lights-on) and increased phospho-PKCγ levels from ZT9 to ZT21 under RF conditions relative to AL (Fig. 2B). This suggests that RF modulates PKCγ activity.
Fig. 2.
PKCγ activation status is altered during restricted feeding compared with food ad libitum. (A) PKCγ was immunoprecipitated from cerebral cortex protein extracts of WT mice collected during AL or on RF day 2. PKCγ and PKCγ-phosphoT655 were detected in the immunoprecipitates by Western blotting. (B) Quantification of PKCγ-phosphoT655 levels vs. ZT. PKCγ-phosphoT655 levels were first normalized to IgG and then to total PKCγ. The white and black boxes indicate light regime, whereas food availability during RF is indicated by the green shading. Error bars represent SEM (n = 3). Significant treatment x ZT interaction (two-way repeated measures ANOVA, P = 0.0094).
Restricted Feeding Substantially Phase Advances the Cerebral Cortex Clock of Wild-Type but Not PKCγ−/− Mice.
The RF-dependent changes in PKCγ activity prompted us to investigate whether PKCγ plays a role in the phase resetting of the molecular clock by RF. We found that there were significant differences in the diurnal expression of several clock gene transcripts in the cerebral cortex of WT mice during RF vs. AL, including Per2, Per3, Cry1, Bmal1, and Rev-erbα (Fig. S3). RF appeared to elicit a phase advance in the oscillations of most of these genes (namely Per2, Per3, Cry1, and Rev-erbα). In PKCγ−/−, on the other hand, the molecular oscillations did not exhibit a significant phase shift. These results suggest that PKCγ is involved in the phase resetting of the cerebral circadian clock by RF.
PKCγ Interacts with BMAL1.
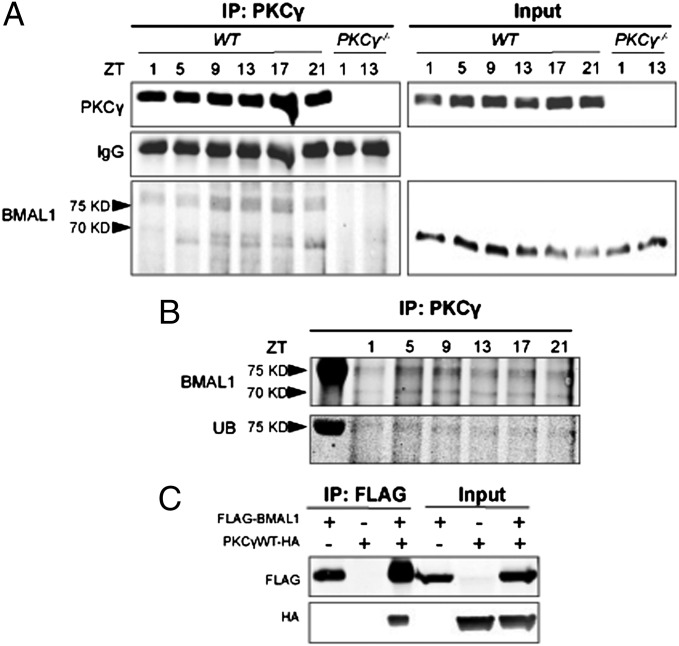
To identify the target of PKCγ in the molecular clock, we performed immunoprecipitation from cerebral cortex lysates with a PKCγ antibody and probed the precipitates for core clock components, including the different PERs, CRYs, BMAL1, CLOCK, and NPAS2. Among the clock components examined, BMAL1 was the only one detected in the precipitates of WT lysates. We found BMAL1 in two forms, one ∼69 kDa, which corresponds to the native size, and a more prominent form of ∼77 kDa, which we suspect represents a fraction of BMAL1 that had undergone posttranslational modification (Fig. 3A). Indeed, this form of BMAL1 could be colabeled with an anti-ubiquitin antibody (Fig. 3B). Based on the size of the protein, we predicted that it was a monoubiquitylated form of BMAL1. To further confirm the interaction between BMAL1 and PKCγ, we performed immunoprecipitation by using FLAG-tagged BMAL1 and HA-tagged PKCγ in cell culture. FLAG-BMAL1 was immunoprecipitated with a FLAG antibody and PKCγ-HA was detected in the precipitates (Fig. 3C), which is consistent with what we observed in vivo. These results suggest that PKCγ and BMAL1 physically interact, thus providing a potential mechanism for PKCγ to exert its effect on the manifested molecular oscillation changes.
Fig. 3.
PKCγ interacts with BMAL1. (A) PKCγ was immunoprecipitated from cerebral cortex protein extracts of WT and PKCγ−/− mice collected on RF day 2. (Left) PKCγ and BMAL1 were detected in the immunoprecipitates (IP) by Western blotting. (Right) Protein input control. (B) PKCγ was immunoprecipitated from cerebral cortex protein extracts of WT mice collected on RF day 2; BMAL1 and ubiquitin (UB) were detected in the same Western blot in the immunoprecipitates. The leftmost lane is the marker. (C) FLAG-BMAL1 and PKCγWT-HA were coexpressed in HEK293T cells in the combinations noted in the headers. The left half of each panel depicts proteins that were immunoprecipitated with anti-FLAG followed by Western blotting with anti-FLAG (Upper) or anti-HA (Lower).
PKCγ Stabilizes BMAL1.
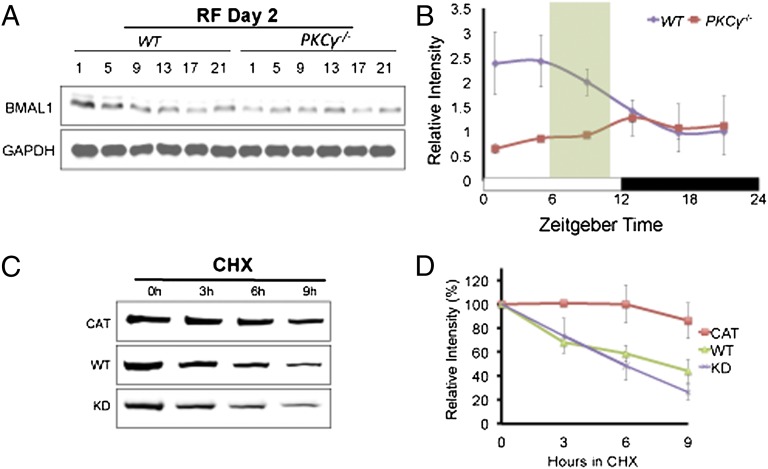
Given that PKCγ targets BMAL1, we examined the diurnal expression pattern of BMAL1 during AL and RF in the cerebral cortex of WT and PKCγ−/− animals (Fig. 4A). The temporal expression of BMAL1 throughout the day is significantly different between WT and PKCγ−/− during RF (Fig. 4B), and this difference is due to reduced BMAL1 levels in PKCγ−/− animals in the light phase under RF conditions.
Fig. 4.
PKCγ stabilizes BMAL1. (A) Representative Western blots of BMAL1 and GAPDH from cerebral cortex protein extracts of WT and PKCγ−/− mice on day 2 of RF. Mice were maintained under 12L12D conditions with lights-on at ZT0 and lights-off at ZT12. Food was given at ZT6 and removed at ZT10. (B) Line graph showing quantification of Western blots in A (n = 3–4). BMAL1 levels were normalized to GAPDH, and for each time series the value of a certain time point of WT was set to 1. Error bars represent SEM. Significant interaction of genotype X ZT during RF day 2 (two-way repeated measures ANOVA, P = 0.0498). (C) FLAG-BMAL1 and different forms of PKCγ (CAT, WT, or KD) were coexpressed in HEK293T cells. Western blots show the rate of FLAG-BMAL1 degradation from HEK293T cell extracts. Duration of CHX treatment is indicated. (D) Line graph showing quantification of Western blots in C (n = 3–4). FLAG-BMAL1 levels were normalized to GAPDH and the amount of FLAG-BMAL1 present at the onset of CHX treatment (0 h) was set to 100%. Error bars represent SEM. Significant interaction of mutation X hours in CHX for CAT vs. KD (two-way repeated measures ANOVA, P = 0.0115).
Because RF leads to decreased BMAL1 levels in PKCγ−/−, we examined whether PKCγ affects the stability of BMAL1. We coexpressed BMAL1 with a constitutively active PKCγ (PKCγCAT), a wild-type PKCγ (PKCγWT), or a kinase-dead PKCγ (PKCγKD) in HEK293T cells (18). The transfected cells were treated with cycloheximide (CHX) to block de novo protein synthesis and BMAL1 levels were assayed (Fig. 4 C and D). Relative BMAL1 levels in the 9-h period after CHX treatment were significantly higher in cells transfected with PKCγCAT vs. that of PKCγKD (Fig. 4D), indicating that PKCγ functions to stabilize BMAL1 in a kinase-dependent manner.
PKCγ Reduces BMAL1 Ubiquitylation.
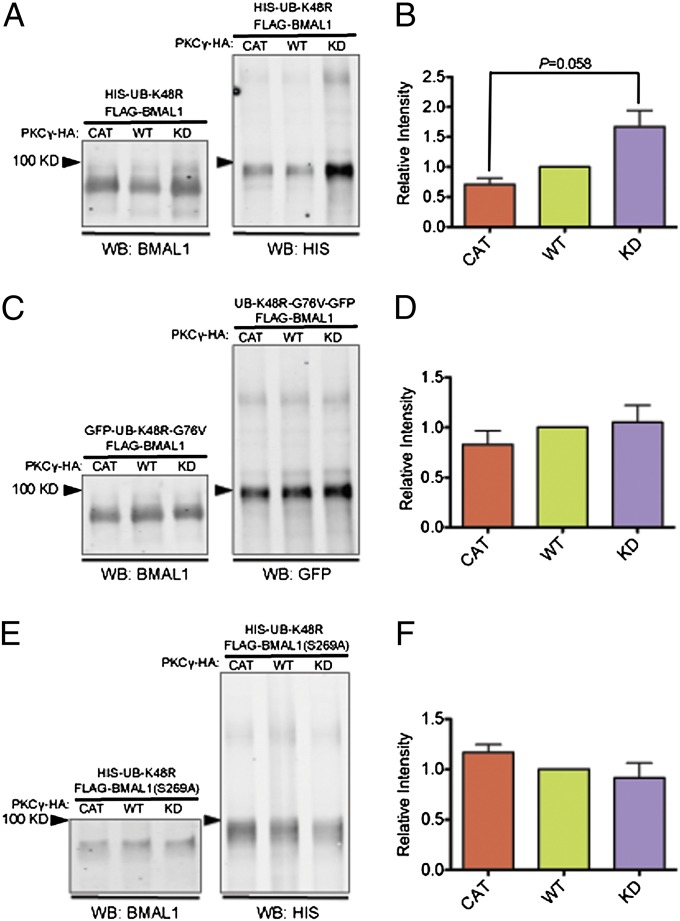
Because PKCγ is associated with ubiquitylated BMAL1 in vivo (Fig. 3B) and exerts a stabilizing effect on BMAL1, we investigated whether PKCγ affects the ubiquitylation of BMAL1. We coexpressed FLAG-BMAL1 with PKCγCAT, PKCγWT, or PKCγKD, and a GFP-tagged ubiquitin with all of its lysines mutated to arginines (GFP-UB-KO) (19). Ubiquitylation can progress until a mutant ubiquitin is incorporated, thus preventing further extension of the ubiquitin chain and allowing for accumulation of ubiquitylated substrate (Fig. S4 A and B). FLAG-BMAL1 was immunoprecipitated and a GFP antibody was used to assay the amount of ubiquitylated BMAL1 in the precipitates. We found that cells expressing PKCγCAT exhibited a reduction in the amount of ubiquitylated BMAL1 compared with cells expressing PKCγKD (Fig. S5). A well-studied ubiquitin-mediated pathway involved in protein degradation is one in which polyubiquitin chains are assembled through lysine 48 (K48) and targets the substrate to the 26S proteasome for degradation (20). We coexpressed FLAG-BMAL1 in cell culture with PKCγCAT, PKCγWT, or PKCγKD, and a mutated ubiquitin carrying a lysine-to-arginine mutation at K48 (UB-K48R) (21). This K-to-R mutation leads to accumulation of ubiquitylated substrate in K48-dependent pathways (Fig. S4 A and C). We found that cells expressing PKCγCAT exhibited substantially lower levels of BMAL1 conjugated with UB-K48R vs. cells expressing PKCγKD (Fig. 5 A and B). These results demonstrate that PKCγ interferes with K48-mediated ubiquitylation of BMAL1, which results in less degradation by the 26S proteasome, thus stabilizing BMAL1. Moreover, when we expressed UB-K48R with glycine 76 (G76) mutated to valine (UB-K48R-G76V), which blocks ubiquitin cleavage from BMAL1 (Fig. S4 A and D) (22), we no longer observed differences in the levels of ubiquitylated BMAL1 in cells expressing PKCγCAT, PKCγWT, or PKCγKD (Fig. 5 C and D). Collectively, these data suggest that PKCγ facilitates ubiquitin cleavage and prevents the formation of polyubiquitylated chains on BMAL1, which then leads to a stabilization of BMAL1. Last, we tested another well-characterized ubiquitin-mediated pathway that is primarily involved in cell signaling (23). In this pathway, polyubiquitin chains are assembled via lysine 63 (K63), and thus we used a mutated ubiquitin carrying a lysine-to-arginine mutation at K63 (UB-K63R) that leads to accumulation of ubiquitylated substrate in the K63-dependent pathway (21) (Fig. S4 A and E). The quantity of BMAL1 conjugated with UB-K63R was not significantly different among PKCγCAT-, PKCγWT-, and PKCγKD-expressing cells (Fig. S6), indicating that PKCγ specifically impinges on K48- but not K63-mediated ubiquitylation of BMAL1.
Fig. 5.
PKCγ reduces K48-mediated ubiquitylation of BMAL1. (A) FLAG-BMAL1, HIS-UB-K48R, and different forms of PKCγ (CAT, WT, or KD) were coexpressed in HEK293T cells. FLAG-BMAL1 was immunoprecipitated from cell lysates; BMAL1 and HIS-UB-K48R were detected in the same Western blot (WB) in the immunoprecipitated proteins with antibodies as specified. (B) Bar graph showing quantification of Western blots in A (n = 3). HIS-UB-K48R levels were normalized to the amount of FLAG-BMAL1 pulled down, and the amount of HIS-UB-K48R in cells transfected with PKCγWT was set to 1. Error bars represent SEM. Student’s t test was applied. (C) FLAG-BMAL1, UB-K48R-G76V-GFP, and different forms of PKCγ (CAT, WT, or KD) were coexpressed in HEK293T cells. FLAG-BMAL1 was immunoprecipitated from cell lysates; BMAL1 and UB-K48R-G76V-GFP were detected on the same Western blot in the immunoprecipitated proteins with antibodies as specified. (D) Bar graph showing quantification of Western blots in C (n = 4). HIS-UB-K48R-G76V levels were normalized to the amount of FLAG-BMAL1 pulled down, and the amount of HIS-UB-K48R-G76V in cells transfected with PKCγWT was set to 1. Error bars represent SEM. (E) FLAG-BMAL1-S269A, HIS-UB-K48R, and different forms of PKCγ (CAT, WT, or KD) were coexpressed in HEK293T cells. FLAG-BMAL1-S269A was immunoprecipitated from cell lysates; BMAL1 and HIS-UB-K48R were detected in the same Western blot in the immunoprecipitated proteins with antibodies as specified. (F) Bar graph showing quantification of Western blots in E (n = 2). HIS-UB-K48R levels were normalized to the amount of FLAG-BMAL1-S269A pulled down, and the amount of HIS-UB-K48R in cells transfected with PKCγWT was set to 1. Error bars represent SEM.
Considering that PKCγ reduces BMAL1 ubiquitylation in a kinase-dependent manner, we investigated whether PKCγ phosphorylates certain sites on BMAL1 to regulate its ubiquitylation. To determine what sites on BMAL1 are important for PKCγ to modulate its ubiquitylation, we performed phosphorylation prediction analysis and found that serine 256 (S256) and serine 269 (S269) exhibit the highest probability of being phosphorylated by classical PKC isotypes, including PKCγ. Thus, we mutated these two sites to alanine. We coexpressed FLAG-BMAL1-S256A or FLAG-BMAL1-S269A in cell culture with PKCγCAT, PKCγWT, or PKCγKD, and UB-K48R. In the presence of the S269A mutation, PKCγ failed to reduce BMAL1 ubiquitylation, and cells expressing PKCγCAT or PKCγWT did not show lower levels of ubiquitylated BMAL1 compared with cells expressing PKCγKD (Fig. 5 E and F). This implies that phosphorylation at S269 is necessary for PKCγ to reduce BMAL1 ubiquitylation. On the other hand, the S256A mutation did not eliminate the effects of PKCγ on BMAL1 ubiquitylation, as we observed significantly lower levels of ubiquitylated BMAL1 in cells expressing PKCγCAT vs. PKCγKD (Fig. S7).
Discussion
In the current study, we identify PKCγ as a potential feeding-dependent regulator of behavior and the circadian clock in the cerebral cortex. Although PKCγ−/− mice exhibit food-anticipatory activity comparable to that of WT, they do not reduce their late-night activity or adapt their food intake amount to RF as efficiently as WT animals. This implies that PKCγ is involved in regulating behavior in response to RF, but it does not appear to be important for the food-entrainable oscillator, which drives food-anticipatory activity. This also suggests that nighttime activity and food-anticipatory activity are differentially regulated. During AL, PKCγ−/− mice show reduced early-night activity and increased activity to lights-on relative to WT. The latter may reflect RF-independent functions of PKCγ, as PKCγ−/− mice appear more easily startled when presented with external stimulus such as handling or feeding, which may explain the exaggerated response to lights-on.
We examined the activation status of PKCγ in the cerebral cortex using levels of PKCγ-phosphoT655 as an indicator and found that RF altered the diurnal activation pattern of PKCγ relative to AL, implying that RF induced a change in the diurnal pattern of PKCγ activity. This means that PKCγ may function as an instructive signal during RF to regulate behavior and the circadian clock. The altered diurnal pattern of PKCγ activity in RF vs. AL could result in increased PKCγ function at certain times of the day during RF compared with AL when its targets (such as BMAL1) are responsive to its effects, thus conferring RF-specific effects. However, PKCγ activity does not perfectly coincide with BMAL1 protein levels. During RF, PKCγ activity is highest late at night, whereas BMAL1 levels are highest early in the day. We reason that there are other factors besides PKCγ that are involved in determining BMAL1 levels, some of which may counteract the stabilizing effects of PKCγ, resulting in a time lag between PKCγ activity and BMAL1 levels. Another possibility is that the stabilizing effects of PKCγ on BMAL1 are not immediate.
During AL, the SCN, which is entrained by light, synchronizes the extra-SCN clocks, including that of the cortex (24). Although RF is a stronger entrainment signal than the light/dark cycle for the cerebral cortex clock and can override the influence of the SCN by phase shifting the cerebral cortex clock, feeding signals may also exert some modest effects on the clock even under AL conditions. This could explain the small differences observed in the diurnal expression of clock gene transcripts between WT and PKCγ−/− during AL; these feeding-dependent effects become more prominent under RF conditions, and thus greater differences in the molecular oscillations are observed. These results suggest that PKCγ plays a role in connecting feeding signals to the circadian clock in the cerebral cortex.
PKCγ and BMAL1 coprecipitate both in vivo and in cell culture, implicating a potential physical association between the two proteins. We were not able to detect the other core clock components in the precipitates in vivo, but this may be due to the sensitivity of the assay and does not necessarily mean that BMAL1 is the only target of PKCγ in the clock. PKCγ appears to be constitutively associated with BMAL1 throughout the day based on our immunoprecipitation results but, as aforementioned, there are differences in PKCγ activity level throughout the day and PKCγ activity may be effective on BMAL1 in a time-dependent manner due to the presence of other factors that could counteract the influence of PKCγ. Therefore, the functional outcome of PKCγ activity on BMAL1 and the clock may be evident only at certain times of the day.
BMAL1 protein levels are reduced earlier in the day in PKCγ−/− mice relative to WT during RF. This could contribute to the lack of an RF-induced phase advance of clock gene transcript oscillations in PKCγ−/− mice. Instead of a phase shift, the cycling of Per2 and Cry1 mRNAs are damped in PKCγ−/− with reduced peak levels compared with WT, which is consistent with reduced BMAL1 levels. We hypothesize that PKCγ stabilizes BMAL1, resulting in changes in BMAL1-mediated transcription. These effects on BMAL1 and the molecular clock could contribute to RF-induced behavioral changes such as reduction of late-night activity, but not food-anticipatory activity, which is intact in PKCγ−/− mice. This is consistent with previous studies demonstrating that food-anticipatory activity does not require the known clock components (25, 26).
Interestingly, the ubiquitously expressed PKCα is believed to function as an integral part of the clock to suppress BMAL1 transcriptional activity (15), which may be one of the factors that mask the influence of PKCγ on the clock, particularly during AL when PKCγ appears dispensable (PKCγ−/− did not exhibit substantial molecular phenotypes during AL). Whereas PKCα is rhythmically activated by internal processes (15), PKCγ may be involved in relaying external stimuli (such as feeding signals) to the clock. Two candidate upstream regulators of PKCγ are the μ-opioid receptor and melanocortin-3 receptor. Both genes have been shown to be involved in food entrainment (27, 28) and physically associate with PKCγ (29, 30).
Cell-culture experiments reveal that PKCγCAT significantly stabilizes BMAL1, which may be a result of the inhibitory effects that PKCγ exerts on BMAL1 ubiquitylation, thus preventing BMAL1 degradation by the 26S proteasome. Moreover, PKCγ acts to reduce BMAL1 ubiquitylation in a deubiquitination-dependent manner. Taken together, these results suggest that PKCγ stabilizes BMAL1 by promoting ubiquitin cleavage from BMAL1. This is consistent with in vivo data demonstrating that PKCγ primarily coprecipitates with a ubiquitylated form of BMAL1. We have also shown that a serine-to-alanine mutation at a potential PKCγ phosphorylation site on BMAL1 (S269A) eliminates the effect of PKCγ on BMAL1 ubiquitylation. Given the physical interaction observed between PKCγ and BMAL1, these data suggest that phosphorylation by PKCγ of BMAL1 at S269 is required for PKCγ to promote ubiquitin cleavage from BMAL1. Interestingly, a recent study demonstrated that GSK3β phosphorylates BMAL1 at serine 17 and threonine 21 and primes it for ubiquitylation (31), providing a potential mechanism that counteracts the effects of PKCγ during AL and perhaps RF as well, contributing to the time lag between PKCγ activity and BMAL1 levels.
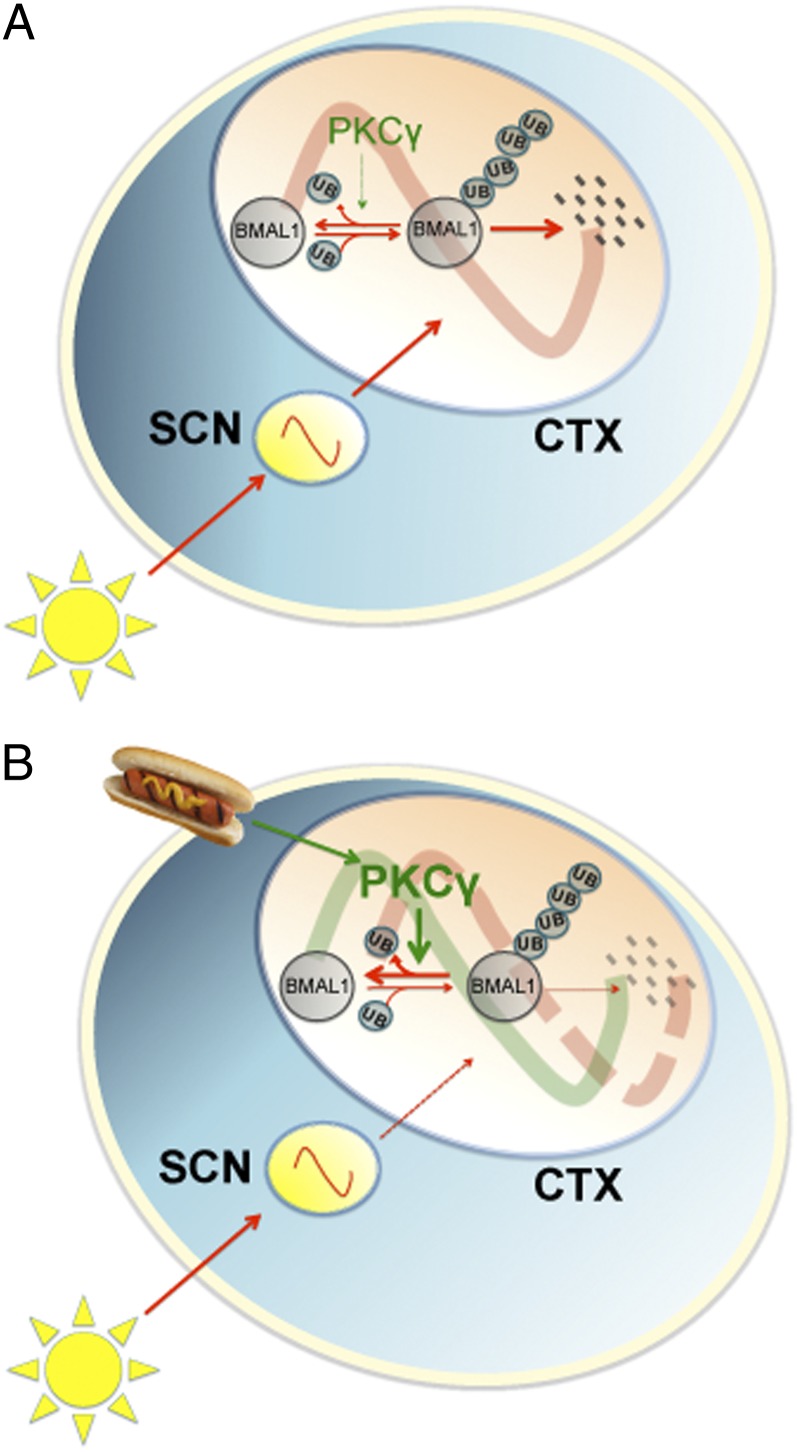
Based on our results, we hypothesize that timed food signals recruit PKCγ to phase reset the clock in food-entrainable regions of the brain (in particular the cerebral cortex) by stabilizing BMAL1. This contributes to the phase-resetting effects of RF on behavior. During AL, light is the strongest entrainment factor. Light synchronizes the SCN, which in turn synchronizes the cerebral cortex clock via signaling pathways that override or mask the effects of PKCγ (Fig. 6A). During temporally RF, on the other hand, the PKCγ activity pattern is altered, enabling it to override the influences from the SCN by enhancing ubiquitin cleavage from BMAL1, thus suppressing its degradation, resulting in phase resetting of the cerebral cortex clock (Fig. 6B). This provides a mechanism for eating during the rest phase (e.g., night-eating syndrome, shift work, or jet lag) to phase reset the clock in food-entrainable regions of the brain (e.g., cerebral cortex) and result in desynchrony between the SCN and other brain regions.
Fig. 6.
Model of the mechanism of PKCγ in regulating the circadian clock during temporally restricted feeding. (A) Under light/dark conditions with food ad libitum, light is the most prominent entrainment factor. Light entrains the SCN, which synchronizes the cerebral cortex clock. PKCγ is dispensable under this condition, and BMAL1 is degraded via a ubiquitin-dependent pathway. (B) During RF, timed food signals modulate the activity of PKCγ. PKCγ functions to stabilize BMAL1 by facilitating ubiquitin cleavage from BMAL1 and therefore contributes to a phase shift of the cerebral cortex clock. Red arrows represent light entrainment-regulated signaling pathways and green arrows represent food entrainment-regulated signaling pathways. CTX, cerebral cortex.
Both night-eating syndrome and shift work may be associated with obesity and metabolic syndromes (9, 32, 33), which could result in susceptibility to various diseases such as cardiovascular diseases and cancer. Psychiatric symptoms are associated with night-eating syndrome, shift work, and jet lag conditions (9, 10, 34). Desynchrony between the molecular oscillations of the SCN and food-entrainable regions (as induced by RF described here) may serve as part of the pathophysiology underlying these disorders/diseases. Therefore, understanding the molecular mechanism of how eating at the “wrong” time of the day desynchronizes the SCN clock from the clock of food-entrainable regions can facilitate the development of better treatments for disorders associated with night-eating syndrome, shift work, and jet lag.
Experimental Procedures
Animals and Behavioral Analysis.
Animal studies were approved by the Animal Care and Use Committee at the University of California, San Francisco. For all experiments, male WT and PKCγ−/− mice (35) on a C57BL6 × 129SvJ background and 2–6 mo of age were used. Mouse housing, handling, and wheel-running activity monitoring were performed as described earlier (36). Wheel revolutions were normalized to the average number of revolutions for each mouse. Each data point represents the average wheel-running activity across 3 d for a single 20-min bin. For daytime restricted feeding, mice were entrained to 12-h light/12-h dark (12L12D) for at least 2 wk with ad libitum access to food. On the last day of AL, food was removed at ZT10 (10 h after lights-on). Food restriction was started by a stepwise reduction in food access from 8 h (ZT2–10) to 6 h (ZT4–10) to 4 h (ZT6–10) per day over a 3-d period and then kept at 4 h/d for a total of 15 d.
Immunoprecipitation and Western Blot.
Immunoprecipitation was performed using an immunoprecipitation kit (Roche) and rabbit anti-PKCγ C-19 (Santa Cruz) or EZview Red Anti-FLAG M2 Affinity Gel (Sigma). For Western blot, tissues and cells were homogenized in extraction buffer [20 mM Hepes, pH 7.5, 100 mM KCl, 5% (vol/vol) glycerol, 20 mM β-glycerophosphate, 100 μM sodium orthovanadate, 10 mM EDTA, 0.1% Triton X-100, 1 mM DTT, protease inhibitor mixture (Roche), phosphatase inhibitor mixture (Active Motif)]. After centrifugation at 16,000 × g for 10 min, supernatants were boiled for 5 min in SDS sample buffer and separated by SDS/PAGE. After transfer to nitrocellulose membranes and blocking with Odyssey blocking buffer (LI-COR), proteins were detected with the following antibodies: rabbit anti-PKCγ C-19 (Santa Cruz), 1:1,000–1:2,000; rabbit anti-BMAL1, 1:1,000; mouse anti-Ub (Biomol), 1:1,000; goat anti-BMAL1 N-20 (Santa Cruz), 1:1,000; mouse anti-GAPDH (Millipore), 1:5,000; mouse anti-FLAG M2 (Sigma), 1:5,000; rat anti-HA (Roche), 1:1,000; mouse anti-HIS (Calbiochem), 1:500; secondary donkey anti-rabbit/mouse/goat antibodies (LI-COR), 1:10,000. Signal was scanned and quantified using an Odyssey-2415 (LI-COR).
Plasmid Construction.
Syrian hamster Bmal1 was amplified by PCR using pcDNA3.1-Bmal1 (37) as template and was subsequently ligated into a pCMV-3Tag vector (Stratagene). pCMV-3Tag-Bmal1-S256A and pCMV-3Tag-Bmal1-S269A were generated by PCR-based site-directed mutagenesis using pCMV-3Tag-Bmal1 as template. All constructs were confirmed by sequencing.
Transient Transfection, Protein Stability, and Ubiquitylation Assays.
HEK293T cells were plated in six-well plates or 10-cm plates and transfected with FuGENE HD transfection reagent (Roche) or X-tremeGENE 9 transfection reagent (Roche). DNA constructs used for transfections were as follows: pCMV-3Tag-shBmal1, pHACE-PKCγCAT (18), pHACE-PKCγWT (18), pHACE-PKCγKD (18), pEGFP-C1-Ub-KO (19), 6XHis-Ub-K48R-EGFP (21), 6XHis-Ub-K63R-EGFP (21), and pEGFP-N1-Ub-K48R-G76V (22). pHACE is a mammalian expression vector containing a CMV promoter, Kozak translational initiation sequence, ATG start codon, EcoRI cloning site, C-terminal HA epitope tag and stop codon. Approximately 40 h later, cycloheximide (100 μg/mL; Sigma) or MG132 (25 μM; Calbiochem) was added and cells were harvested 3, 6, or 9 h after cycloheximide treatment or 5 h after MG132 treatment. Kinase phosphorylation prediction was performed using Motif Scan (http://scansite.mit.edu).
Supplementary Material
Acknowledgments
We thank D. A. Gray for His-Ub-K48R and His-Ub-K63R constructs, and N. P. Dantuma for Ub-K48R-G76V-GFP construct. This work is supported by National Institutes of Health Grants GM079180 and HL059596 (to Y.-H.F. and L.J.P.) and the Sandler Neurogenetics Fund. L.J.P. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218699110/-/DCSupplemental.
References
- 1.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec no 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 2.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: An analog of clock operative in the mammalian forebrain. Science. 2001;293(5529):506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 3.Carneiro BT, Araujo JF. The food-entrainable oscillator: A network of interconnected brain structures entrained by humoral signals? Chronobiol Int. 2009;26(7):1273–1289. doi: 10.3109/07420520903404480. [DOI] [PubMed] [Google Scholar]
- 4.Verwey M, Amir S. Food-entrainable circadian oscillators in the brain. Eur J Neurosci. 2009;30(9):1650–1657. doi: 10.1111/j.1460-9568.2009.06960.x. [DOI] [PubMed] [Google Scholar]
- 5.Challet E, Mendoza J. Metabolic and reward feeding synchronises the rhythmic brain. Cell Tissue Res. 2010;341(1):1–11. doi: 10.1007/s00441-010-1001-9. [DOI] [PubMed] [Google Scholar]
- 6.Escobar C, Cailotto C, Angeles-Castellanos M, Delgado RS, Buijs RM. Peripheral oscillators: The driving force for food-anticipatory activity. Eur J Neurosci. 2009;30(9):1665–1675. doi: 10.1111/j.1460-9568.2009.06972.x. [DOI] [PubMed] [Google Scholar]
- 7.Wakamatsu H, et al. Restricted-feeding-induced anticipatory activity rhythm is associated with a phase-shift of the expression of mPer1 and mPer2 mRNA in the cerebral cortex and hippocampus but not in the suprachiasmatic nucleus of mice. Eur J Neurosci. 2001;13(6):1190–1196. doi: 10.1046/j.0953-816x.2001.01483.x. [DOI] [PubMed] [Google Scholar]
- 8.Mendoza J, Pévet P, Felder-Schmittbuhl MP, Bailly Y, Challet E. The cerebellum harbors a circadian oscillator involved in food anticipation. J Neurosci. 2010;30(5):1894–1904. doi: 10.1523/JNEUROSCI.5855-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallant AR, Lundgren J, Drapeau V. The night-eating syndrome and obesity. Obes Rev. 2012;13(6):528–536. doi: 10.1111/j.1467-789X.2011.00975.x. [DOI] [PubMed] [Google Scholar]
- 10.Zee PC, Goldstein CA. Treatment of shift work disorder and jet lag. Curr Treat Options Neurol. 2010;12(5):396–411. doi: 10.1007/s11940-010-0090-9. [DOI] [PubMed] [Google Scholar]
- 11.Jakubcakova V, et al. Light entrainment of the mammalian circadian clock by a PRKCA-dependent posttranslational mechanism. Neuron. 2007;54(5):831–843. doi: 10.1016/j.neuron.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Shim HS, et al. Rapid activation of CLOCK by Ca2+-dependent protein kinase C mediates resetting of the mammalian circadian clock. EMBO Rep. 2007;8(4):366–371. doi: 10.1038/sj.embor.7400920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konopatskaya O, Poole AW. Protein kinase Calpha: Disease regulator and therapeutic target. Trends Pharmacol Sci. 2010;31(1):8–14. doi: 10.1016/j.tips.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Zee EA, Bult A. Distribution of AVP and Ca(2+)-dependent PKC-isozymes in the suprachiasmatic nucleus of the mouse and rabbit. Brain Res. 1995;701(1-2):99–107. doi: 10.1016/0006-8993(95)00968-1. [DOI] [PubMed] [Google Scholar]
- 15.Robles MS, Boyault C, Knutti D, Padmanabhan K, Weitz CJ. Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science. 2010;327(5964):463–466. doi: 10.1126/science.1180067. [DOI] [PubMed] [Google Scholar]
- 16.Saito N, Shirai Y. Protein kinase C gamma (PKC gamma): Function of neuron specific isotype. J Biochem. 2002;132(5):683–687. doi: 10.1093/oxfordjournals.jbchem.a003274. [DOI] [PubMed] [Google Scholar]
- 17.Newton AC. Protein kinase C: Structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev. 2001;101(8):2353–2364. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- 18.Soh JW, Weinstein IB. Roles of specific isoforms of protein kinase C in the transcriptional control of cyclin D1 and related genes. J Biol Chem. 2003;278(36):34709–34716. doi: 10.1074/jbc.M302016200. [DOI] [PubMed] [Google Scholar]
- 19.Bergink S, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20(10):1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickart CM. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11(13):1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 21.Tsirigotis M, Zhang M, Chiu RK, Wouters BG, Gray DA. Sensitivity of mammalian cells expressing mutant ubiquitin to protein-damaging agents. J Biol Chem. 2001;276(49):46073–46078. doi: 10.1074/jbc.M109023200. [DOI] [PubMed] [Google Scholar]
- 22.Lindsten K, et al. Mutant ubiquitin found in neurodegenerative disorders is a ubiquitin fusion degradation substrate that blocks proteasomal degradation. J Cell Biol. 2002;157(3):417–427. doi: 10.1083/jcb.200111034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33(3):275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Inouye ST, Kawamura H. Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus. Proc Natl Acad Sci USA. 1979;76(11):5962–5966. doi: 10.1073/pnas.76.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pendergast JS, et al. Robust food anticipatory activity in BMAL1-deficient mice. PLoS ONE. 2009;4(3):e4860. doi: 10.1371/journal.pone.0004860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storch KF, Weitz CJ. Daily rhythms of food-anticipatory behavioral activity do not require the known circadian clock. Proc Natl Acad Sci USA. 2009;106(16):6808–6813. doi: 10.1073/pnas.0902063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kas MJ, et al. Mu-opioid receptor knockout mice show diminished food-anticipatory activity. Eur J Neurosci. 2004;20(6):1624–1632. doi: 10.1111/j.1460-9568.2004.03581.x. [DOI] [PubMed] [Google Scholar]
- 28.Sutton GM, et al. The melanocortin-3 receptor is required for entrainment to meal intake. J Neurosci. 2008;28(48):12946–12955. doi: 10.1523/JNEUROSCI.3615-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodríguez-Muñoz M, de la Torre-Madrid E, Sánchez-Blázquez P, Wang JB, Garzón J. NMDAR-nNOS generated zinc recruits PKCgamma to the HINT1-RGS17 complex bound to the C terminus of mu-opioid receptors. Cell Signal. 2008;20(10):1855–1864. doi: 10.1016/j.cellsig.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 30.Wachira SJ, Hughes-Darden CA, Taylor CV, Ochillo R, Robinson TJ. Evidence for the interaction of protein kinase C and melanocortin 3-receptor signaling pathways. Neuropeptides. 2003;37(4):201–210. doi: 10.1016/s0143-4179(03)00026-x. [DOI] [PubMed] [Google Scholar]
- 31.Sahar S, Zocchi L, Kinoshita C, Borrelli E, Sassone-Corsi P. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS ONE. 2010;5(1):e8561. doi: 10.1371/journal.pone.0008561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup Environ Med. 2001;58(11):747–752. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Lorenzo L, et al. Effect of shift work on body mass index: Results of a study performed in 319 glucose-tolerant men working in a southern Italian industry. Int J Obes Relat Metab Disord. 2003;27(11):1353–1358. doi: 10.1038/sj.ijo.0802419. [DOI] [PubMed] [Google Scholar]
- 34.Katz G. Jet lag and psychotic disorders. Curr Psychiatry Rep. 2011;13(3):187–192. doi: 10.1007/s11920-011-0192-4. [DOI] [PubMed] [Google Scholar]
- 35.Abeliovich A, et al. Modified hippocampal long-term potentiation in PKC gamma-mutant mice. Cell. 1993;75(7):1253–1262. doi: 10.1016/0092-8674(93)90613-u. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434(7033):640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 37.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.