Abstract
Background
The association between the methylenetetrahydrofolate reductase (MTHFR) C677T/A1298C polymorphisms and the susceptibility to cervical lesions was unclear. This study was designed to investigate their precise association using a large-scale meta-analysis.
Methods
The previous 16 studies were identified by searching PubMed, Embase and CBM databases. The crude odds ratios and their corresponding 95% confidence intervals (CIs) were used to estimate the association between the MTHFR C677T/A1298C polymorphisms and the susceptibility to the cervical lesions. The subgroup analyses were made on the following: pathological history, geographic region, ethnicity, source of controls and source of DNA for genotyping.
Results
Neither of the polymorphisms had a significant association with the susceptibility to the cervical lesions in all genetic models. Similar results were found in the subgroup analyses. No association was found between the MTHFR C677T polymorphism and the cervical lesions in the Asia or the America populations though a significant inverse association was found in the Europe population (additive model: P = 0.006, OR = 0.83, 95% CI = 0.72–0.95; CT vs. CC: P = 0.05, OR = 0.83, 95% CI = 0.69–1.00; TT vs. CC: P = 0.05, OR = 0.73, 95% CI = 0.53–1.00). Interestingly, women with the MTHFR A1298C polymorphisms had a marginally increased susceptibility to invasive cancer (ICC) when compared with no carriers but no statistically significant difference in the dominant model (P = 0.06, OR = 1.21, 95% CI = 0.99–1.49) and AC vs. AA (P = 0.09, OR = 1.21, 95% CI = 0.97–1.51).
Conclusions
The MTHFR C677T and A1298C polymorphisms may not increase the susceptibility to cervical lesions. However, the meta-analysis reveals a negative association between the MTHFR C677T polymorphisms and the cervical lesions, especially in the European populations. The marginal association between the MTHFR A1298C polymorphisms and the susceptibility to cervical cancer requires a further study.
Introduction
Cervical cancer is the third most frequently encountered cancer and the fourth leading cause of the women’s cancer death in the world, accounting for 9% (529,800) of the total newly-diagnosed cancer cases and 8% (275,100) of the total cancer deaths among females in 2008 [1]. However, cervical cancer is considered a preventable disease because of its relatively long period of precancerous lesions, including cervical intraepithelial neoplasia (CIN). The virological, molecular, clinical and epidemiological studies have provided evidence that cervical cancer is in fact a sequel to a long-term unresolved infection of certain genotypes of the Human Papilloma Virus (HPV) [2], [3]. High-risk HPVs are known to infect cervical epithelium, with a subset of these being associated with preneoblastic lesions that can progress to cervical cancer. Nevertheless, despite the extremely high rate of infection by these viruses, the rate of cervical cancer, even in the prescreening area, has been less than one tenth that of exposure [4], [5]. Thus, other factors are important for cervical lesion development and progression such as a long-term use of hormonal contraceptives, multiparty, smoking, and some nutritional factors [6]–[8].
Association between micronutrient depletion, particularly folate deficiency, and cervical lesions has been studied for a long time. Folate deficiency, as a potential risk for cervical cancer, was first reported by some cytopathologists in the 1960s, who had found that the cervical epithelial cells from folate-deficient women had some similarity to the dysplastic cervical cells in cytology [9]. Later on, Whitehead et al. demonstrated that macrocytic changes in the cervical cells of the oral contraceptive users could be reversed with folic acid supplementation [10]. However, conflicting results still existed in the conclusion of the association between the folate deficiency and the cervical dysplasia [11]–[13]. Furthermore, various clinical epidemiological studies have shown that low-level folate was not directly increase risk of cervical dysplasia but enhance HPV infection instead [14]–[16]. Therefore, despite the lack of a statistically significant association between folate status and cervical dysplasia, these trials indicated that folate may involve along with HPV to induce cervical carcinogenesis.
The apparent role of folate in carcinogenesis in cervical tissue has stimulated investigations of polymorphisms in the folate metabolizing enzymes. As we know, Methylenetetrahydrofolate reductase (MTHFR) is a crucial enzyme that can regulate the metabolism of folate and methionine, both of which are important in DNA methylation and synthesis [17]. This occurs through the conversion of 5, 10-methyltetrahydrofolate to 5-methyltetrahydrofolate (1-carbon metabolism), which is a dominant circulating form of folate. The MTHFR gene is located on the short arm of chromosome 1 (1p36.3) and has several well-described single nucleotide polymorphisms (SNPs). Two common SNPs are known to affect enzyme function and have been shown to have clinical significance. The most common mutation is a C-to-T transition at nucleotide 677 (rs1801133, C677T) in exon 4, resulting in a substitution of alanine with valine that affects the catalytic domain of the enzyme, leading to the enzyme activity reduction [18]. Another common variant is an A-to-C transversion at position 1298 in exon 7 (rs1801131, A1298C), resulting in a substitution of glutamate with alanine at codon 429. This polymorphism also reduces the enzyme activity to a lesser extent [19].
Several studies had been designed to evaluate associations between MTHFR genotypes and cervical lesions, including cervical cancer, but the results were inconsistent because of different stages of cervical lesions and the combinatorial effects of other risk factors. Precancerous cervical lesions are classified according to the degree of cellular abnormality. The lowest grade of abnormality is CIN1, and CIN2 and CIN3 describe the progressive epithelial dysplasia leading to invasive cancer. Preinvasive lesions have also been classified in terms of squamous intraepithelial lesions (SILs) included low-grade squamous intraepithelial lesions (LSIL, including HPV infection and CIN1) and high-grade squamous intraepithelial lesions (HSIL, including CIN2 and CIN3). The majority of the case-control genetic studies revealed no association between cervical lesions and MTHFR SNPs [20]–[25]. But some evidences indicated that the MTHFR variants are positively associated with the cervical cancer risk [26]–[31]; some other evidence indicated that the MTHFR variants are inversely associated with the cervical cancer risk [32]–[35]. For example, one study reported that the MTHFR variant genotype may increase CIN and cervical cancer risk in women who had low-level folate status [26]. Another study suggested women with MTHFR polymorphism and low riboflavin status were significantly less likely to have HSIL than women without the polymorphism and high riboflavin status [33].
These inconclusive results may due to limited sample size, because any single study may be underpowered to detect the precise effects. In addition, there also may be the causes of different characteristics among studies, such as ethnicity, pathological history, sources of controls, and source of DNA for genotyping. Therefore, we have done a meta-analysis on association between MTHFR polymorphisms and cervical lesions using data obtained from the published case-control genetic studies. Our aim was to identify whether the MTHFR polymorphisms affect the susceptibility to SIL or cervical cancer by means of a large-scale meta-analysis. Furthermore, we wanted to summarize the effect size of the polymorphism associated with the susceptibility to the cervical lesions.
Materials and Methods
Search Strategy and Selection Criteria
The computer-based search strategy was comprehensively used to find eligible studies for this meta-analysis. Two investigators (Long, Yang) searched in the PubMed and Emase independently from inception to July 22, 2012, for the studies on the association between the MTHFR C677T polymorphism (rs1801133) and A1289C polymorphism (rs1801131) and the cervical lesions. Following Medical Subject Heading (MeSH) terms and/or text words were used in our search, such as for methylenetetrahydrofolate reductase (“MTHFR” or “methylenetetrahydrofolate reductase” or Methylenetetrahydrofolate Reductase AND (NADPH2)) with terms for genetic variations (“polymorphism” or “variation” or “mutation” or “Single Nucleotide Polymorphism” or Polymorphism, Single Nucleotide” or “SNPs” ) and terms for cervical lesions(“Uterine Cervical Cancer” or “Neoplasms, Cervix” or “Neoplasms, Cervical” or “Cervix Neoplasms” or “Cervix Cancer” or “Cervical Neoplasms” or “Cancer of the Uterine Cervix” or “Cancer of the Cervix” or “Cancer of Cervix” or “Uterine Cervical Neoplasms” or “Uterine Cervical Neoplasms” or “Uterine Cervical Dysplasia” or “Neoplasia, Cervical Intraepithelia” or “Intraepithelial Neoplasia, Cervical” or “Cervical Intraepithelial Neoplasms” or “Cervical Intraepithelial Neoplasia” or “cin” ). Meanwhile, China Biological Medicine Database (CBM) was also searched for the eligible studies. Full articles published in English or Chinese were considered to be eligible for our study. In addition, reference list of the original research articles and reviews were also manually searched.
The eligible studies must meet the following inclusion criteria: (1) Exploration of associations between the MTHFR genetic polymorphisms (including C677T or A1298C or both) and the susceptibility to cervical cancer or SIL; (2) A case-control study; (3) Provision of information on genotype frequencies of the MTHFR C677T and/or A1298C polymorphism(s) or sufficient data for the calculation. The exclusion criteria were as follows: (1) A review, case report, editorial, or comment; (2) A duplicated study; (3) Laboratory molecular or animal studies. If studies contained overlapping cases and/or controls, the largest study with extractable data was preferred.
Because the data included in this study was taken from literatures, written consent given by the patients and ethical approval acquired by certain committee were not needed in our meta-analysis.
Data Extraction
According to the inclusion and exclusion criteria, extraction from each study was conducted independently by two authors (Long, Yang) and the consensus was achieved for all the data, which were as follows: the first author’s name, year of publication, source of controls, source of DNA for genotyping, country, ethnicity, goodness-in-fitness of Hardy-Weinberg Equilibrium (HWE) in the control group, histological stage of cervical lesions, numbers of cases/patients and controls, and distribution of genotypes in the case and control groups. The patients were recruited into the study regardless of whether they had a first-degree relative with cervical lesions. The controls were recruited regardless of whether they had other diseases, e.g., hysteromyoma. For studies with inadequate information, authors of those studies were contacted for further information by E-mail if possible.
Statistical Analysis
Meta-analysis was performed and reported as described previously [36], [37]. Crude ORs with 95% CIs were computed to assess the strength of the correlation between the MTHFR C677T/A1298C polymorphisms and the susceptibility to cervical lesions. The pooled ORs were performed for the dominant model (aa+Aa vs. AA), recessive model (aa vs. Aa+AA) and additive model (A vs. a). Moreover, the pooled estimates were also calculated for the pair-wise comparisons (allele Aa vs. AA, and allele aa vs. AA). The above-mentioned A and a represented the major and the minor allele respectively. Taking consideration of possible between-study heterogeneity, a statistical test for heterogeneity was performed by the χ2 test or Fisher exact test when appropriate. P<0.10 or I2>50% indicated an obvious of the between-study heterogeneity, and OR (95% CI) was calculated by the random-effects model using the DerSimonian and Laird method; otherwise, the fixed-effects model was used by the Mantel-Haenszel method [38], [39]. Subgroup analyses were mainly conducted using the corresponding pathological history (ICC, SIL), geographic region (Asia, Europe, United States), ethnicity (Asian, Caucasian, mixed), source of controls (healthy persons, patients), and source of DNA for genotyping (blood, cervical cells or tissue sample), all of which were used to explore and explain the heterogeneity between the different studies.
The allele frequencies, at which the MTHFR C677T/A1298C polymorphisms occurred in each respective study, were determined by the allele-counting method. A chi-square test was used to determine whether the observed frequencies of the genotypes in the controls conformed to Hardy Weinberg-Equilibrium (HWE) expectations if genotype data were available. Sensitivity analyses were performed on stability of the results, namely, one case-control study omitted each time to reflect the influence of the individual data set on the pooled OR. Several methods were used to detect any probable publication bias. Asymmetry of the funnel plot indicated the possible publication bias. In addition, the Egger and Begg quantitative tests were also used, and P<0.05 was considered a statistical significance [40], [41].
All analyses were performed using the RevMan 5.0 program (Cochrane Library, UK) and the STATA package version 11.0 program (Stata Corporation, College Station, Texas, USA). All P values were two-sided. To ensure the reliability of data, two reviewers (Long, Yang) independently performed the data analysis using the statistics programs for the same results.
Results
Characteristics of Eligible Studies
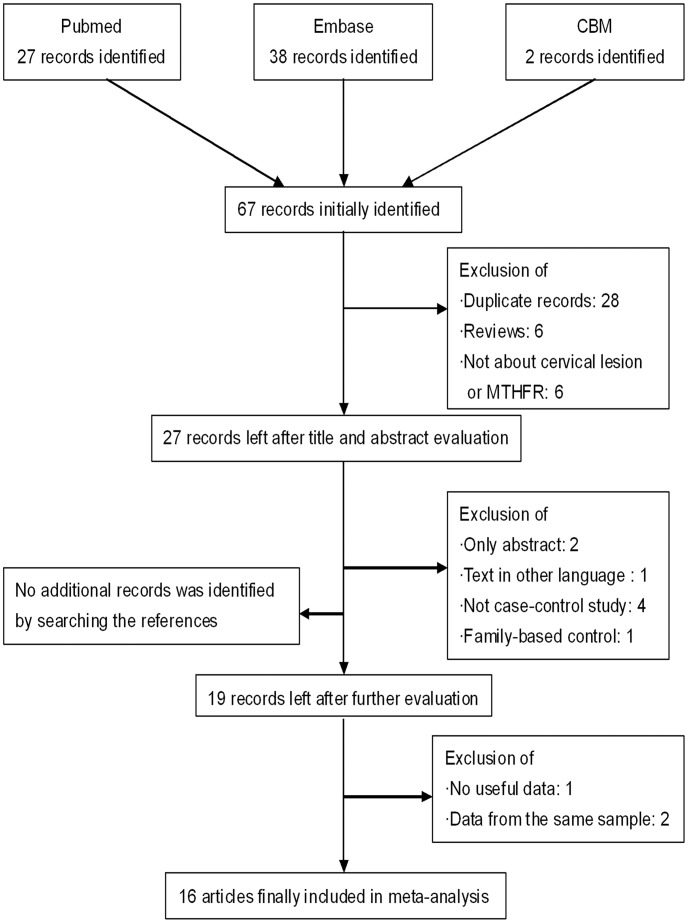
Detailed information for selecting eligible studies was showed in Figure 1. After comprehensively searching, 67 potentially-relevant publications were identified, and none of them were selected from the reference lists of the identified articles. After the careful selection, 16 eligible studies were finally included in our meta-analysis. Among them, 16 studies investigated the MTHFR C677T polymorphism with 3498 cases and 3594 controls and 5 studies investigated the MTHFR A1298C polymorphism with 1087 cases and 1202 controls. General characteristics of the included studies were evaluated for the association between variants and cervical lesions (Table 1, Table 2). For C677T, 11 studies recruited the controls from healthy persons; 1 study from hospital patients and 4 studies from both. 9 studies were performed in Asia; 4 studies performed in Europe; 3 studies performed in America. 5 studies talked about ICC; 3 studies talked about SIL and 8 studies talked about both. For A1298C, all 5 studies performed in Asian; 4 studies recruited controls from healthy persons and 1 study from both healthy persons and hospital patients. 1 study talked about ICC and 4 studies talked about both ICC and SIL. 14 of the studies presented NS (not significant) were conformed to Hardy Weinberg-Equilibrium (HWE) expectations (P>0.05). However, two of the studies [27], [35] presented NA (not available) were because we could not perform the HWE test for the subjects (either cases or controls) in those studies, for only the total number of the combined genotypes (CT/TT vs. CC or AC/CC vs. AA) were available. Therefore, this study was included in the analysis on the dominant model, not on other genetic models. Furthermore, the allele and genotype frequencies, at which the MTHFR C677T and the A1298C polymorphisms occurred in case and controls in each of the studies, were also summarized (Table 1, Table 2).
Figure 1. Flow diagram of the study selection process.
Table 1. Characteristics of the included case-control studies on the MTHFR C677T polymorphism in cervical lesions.
| Firstauthor[reference] | Year | Sourceofcontrol | SourceofDNA | Country | Ethnicity | HWE | Histology | Samplesize | Case | Control | |||||||||||
| case | control | C | T | CC | CT | TT | CT+TT | C | T | CC | CT | TT | CT+TT | ||||||||
| Prasad [20] | 2011 | Mixed | Blood | India | Asian | NS | ICC | 62 | 125 | 119 | 5 | 57 | 5 | 0 | 5 | 240 | 10 | 116 | 8 | 1 | 9 |
| Mostowska [21] | 2011 | Healthy persons | Blood | Poland | Caucasian | NS | ICC | 124 | 168 | 194 | 77 | 56 | 59 | 9 | 68 | 219 | 117 | 69 | 81 | 18 | 99 |
| Tong [26] | 2011 | Healthy persons | Blood | Korea | Asian | NS | LSIL | 159 | 427 | 186 | 132 | 52 | 82 | 25 | 107 | 502 | 352 | 152 | 198 | 77 | 275 |
| HSIL | 160 | 427 | 182 | 138 | 54 | 74 | 32 | 106 | 502 | 352 | 152 | 198 | 77 | 275 | |||||||
| ICC | 146 | 427 | 171 | 121 | 53 | 65 | 28 | 93 | 502 | 352 | 152 | 198 | 77 | 275 | |||||||
| Kohaar [22] | 2010 | Healthy persons | Tissue or cell | India | Asian | NS | HSIL | 39 | 231 | 67 | 11 | 28 | 11 | 0 | 11 | 387 | 75 | 161 | 65 | 5 | 70 |
| ICC | 164 | 231 | 273 | 55 | 113 | 47 | 4 | 51 | 387 | 75 | 161 | 65 | 5 | 70 | |||||||
| Shekari [32] | 2008 | Healthy persons | Blood | India | Asian | NS | ICC | 200 | 200 | 368 | 32 | 170 | 28 | 2 | 30 | 318 | 82 | 125 | 68 | 7 | 75 |
| Nandan [27] | 2008 | Healthy persons | Blood | India | Asian | NA | SIL | 80 | 77 | NA | NA | 34 | NA | NA | 46 | NA | NA | 53 | NA | NA | 24 |
| ICC | 62 | 77 | NA | NA | 36 | NA | NA | 26 | NA | NA | 53 | NA | NA | 24 | |||||||
| Piyathilake [33] | 2007 | Mixed | Blood | USA | Mixed | NS | HSIL | 80 | 355 | 134 | 26 | 59 | 16 | 5 | 21 | 562 | 148 | 223 | 116 | 16 | 132 |
| Zoodsma [34] | 2005 | Mixed | Blood | Netherlands | Caucasian | NS | HSIL | 264 | 592 | 362 | 166 | 121 | 120 | 23 | 143 | 808 | 376 | 273 | 262 | 57 | 319 |
| ICC | 636 | 592 | 944 | 328 | 357 | 230 | 49 | 279 | 808 | 376 | 273 | 262 | 57 | 319 | |||||||
| Kang [23] | 2005 | Healthy persons | Blood | Korean | Asian | NS | ICC | 79 | 74 | 86 | 72 | 27 | 32 | 20 | 52 | 92 | 56 | 30 | 32 | 12 | 44 |
| Sull [28] | 2004 | Healthy persons | Blood | Korean | Asian | NS | LSIL | 40 | 454 | 42 | 38 | 10 | 22 | 8 | 30 | 527 | 381 | 153 | 221 | 80 | 301 |
| HSIL | 176 | 454 | 190 | 162 | 50 | 90 | 36 | 126 | 527 | 381 | 153 | 221 | 80 | 301 | |||||||
| ICC | 246 | 454 | 261 | 231 | 73 | 115 | 58 | 173 | 527 | 381 | 153 | 221 | 80 | 301 | |||||||
| Lambropoulos [24] | 2003 | Healthy persons | Tissue or cell | Greece | Caucasian | NS | LSIL | 53 | 91 | 68 | 38 | 20 | 28 | 5 | 33 | 121 | 61 | 42 | 37 | 12 | 49 |
| HSIL | 64 | 91 | 83 | 45 | 27 | 29 | 8 | 37 | 121 | 61 | 42 | 37 | 12 | 49 | |||||||
| ICC | 21 | 91 | 30 | 12 | 11 | 8 | 2 | 10 | 121 | 61 | 42 | 37 | 12 | 49 | |||||||
| Goodman [29] | 2001 | Hospital patients | Blood | USA | Mixed | NS | SIL | 150 | 179 | 213 | 87 | 73 | 67 | 10 | 77 | 261 | 97 | 93 | 75 | 11 | 86 |
| Piyathilake [30] | 2000 | Healthy persons | Tissue or cell | USA | Mixed | NS | LSIL | 25 | 31 | 25 | 25 | 6 | 13 | 6 | 19 | 44 | 18 | 16 | 12 | 3 | 15 |
| HSIL | 39 | 31 | 45 | 33 | 11 | 23 | 5 | 28 | 44 | 18 | 16 | 12 | 3 | 15 | |||||||
| Agodi [35] | 2010 | Healthy persons | Cell | Italy | Caucasian | NA | SIL | 123 | 66 | NA | NA | 118 | NA | NA | 5 | NA | NA | 55 | NA | NA | 11 |
| Yang [25] | 2011 | Mixed | Blood | China | Asian | NS | SIL | 38 | 382 | 60 | 16 | 23 | 14 | 1 | 15 | 530 | 234 | 182 | 166 | 34 | 200 |
| ICC | 157 | 382 | 229 | 85 | 77 | 75 | 5 | 80 | 530 | 234 | 182 | 166 | 34 | 200 | |||||||
| Ma [31] | 2006 | Hospital patients | Blood | China | Asian | NS | ICC | 111 | 111 | 93 | 129 | 20 | 53 | 38 | 91 | 126 | 96 | 33 | 60 | 18 | 78 |
Abbreviations: HWE, Hardy-Weinberg Equilibrium; NA, not available; NS, not significant; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; ICC, invasive cervical cancer; SIL, squamous intra-epithelial lesion.
Table 2. Characteristics of the included case-control studies on the MTHFR A1298C polymorphism in cervical lesions.
| First author[reference] | Year | Source of control | Source of DNA | Country | Ethnicity | HWE | Histology | Sample size | Case | Control | |||||||||||
| case | control | A | C | AA | AC | CC | AC+CC | A | C | AA | AC | CC | AC+CC | ||||||||
| Tong [26] | 2011 | Healthy persons | Blood | Korea | Asian | NS | SIL | 160 | 428 | 260 | 60 | 107 | 46 | 7 | 53 | 688 | 168 | 278 | 132 | 18 | 150 |
| HSIL | 160 | 428 | 273 | 47 | 117 | 39 | 4 | 43 | 688 | 168 | 278 | 132 | 18 | 150 | |||||||
| ICC | 148 | 428 | 235 | 61 | 89 | 57 | 2 | 59 | 688 | 168 | 278 | 132 | 18 | 150 | |||||||
| Kohaar [22] | 2010 | Healthy persons | Tissue or cell | India | Asian | NS | HSIL | 39 | 231 | 50 | 28 | 15 | 20 | 4 | 24 | 289 | 173 | 85 | 119 | 27 | 146 |
| ICC | 164 | 231 | 199 | 129 | 58 | 83 | 23 | 106 | 289 | 173 | 85 | 119 | 27 | 146 | |||||||
| Nandan [27] | 2008 | Healthy persons | Blood | India | Asian | NA | SIL | 80 | 77 | NA | NA | 14 | NA | NA | 66 | NA | NA | 37 | NA | NA | 40 |
| ICC | 62 | 77 | NA | NA | 20 | NA | NA | 42 | NA | NA | 37 | NA | NA | 40 | |||||||
| Kang [23] | 2005 | Healthy persons | Blood | Korea | Asian | NS | ICC | 79 | 84 | 132 | 26 | 55 | 22 | 2 | 24 | 141 | 27 | 58 | 25 | 1 | 26 |
| Yang [25] | 2011 | Mixed | Blood | China | Asian | NS | SIL | 38 | 382 | 62 | 14 | 24 | 14 | 0 | 14 | 606 | 158 | 237 | 132 | 13 | 145 |
| ICC | 157 | 382 | 245 | 69 | 89 | 67 | 1 | 68 | 606 | 158 | 237 | 132 | 13 | 145 | |||||||
Abbreviations: HWE: Hardy-Weinberg Equilibrium; NA, not available; NS, not significant; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; ICC, invasive cervical cancer; SIL, squamous intra-epithelial lesion.
Quantitative Synthesis
Association between the MTHFR C677T polymorphisms and cervical lesions
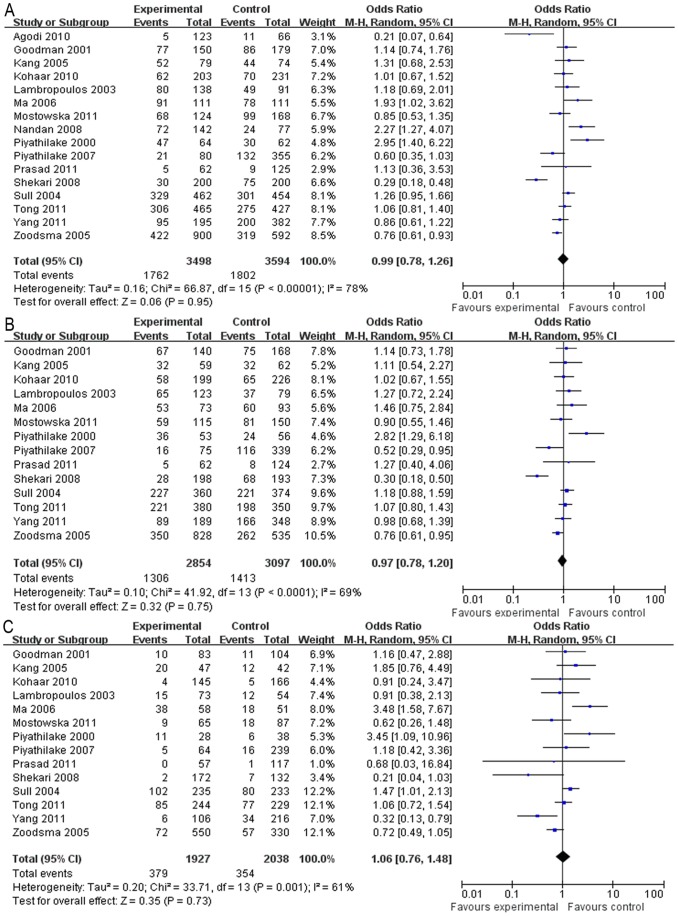
As for the C677T polymorphism, no association was found between the polymorphism and the susceptibility to cervical lesions in all the genetic models (Table 3, dominant model: OR = 0.99, 95% CI = 0.78–1.26, Figure 2A; recessive model: OR = 1.05, 95% CI = 0.80–1.38; additive model: OR = 0.97, 95% CI = 0.80–1.18,; CT vs. CC: OR = 0.97, 95% CI = 0.78–1.20, Figure 2B; TT vs. CC: OR = 1.06, 95% CI = 0.76–1.48, Figure 2C). The heterogeneity was significant in all the genetic models (P<0.05) and the random-effects model was used in the meta-analysis. The subgroup analysis of the C677T polymorphisms in the histological stages of the cervical lesions also revealed that the polymorphism was not associated with the risk of ICC or SIL in all the genetic models (Table 3). Although the subgroup analysis of C677T in the geographic regions revealed that no association was found between the C677T polymorphism and the cervical lesions in either the Asia or the America populations, the Europe population showed a significant inverse association in some genetic models (additive model: P = 0.006, OR = 0.83, 95% CI = 0.72–0.95; CT vs. CC: P = 0.05, OR = 0.83, 95% CI = 0.69–1.00; TT vs. CC: P = 0.05, OR = 0.73, 95% CI = 0.53–1.00). The heterogeneity was significantly reduced in the Europe populations in the recessive, additive, C/T vs. C/C, and T/T vs. C/C models.
Table 3. Pooled Analysis on Association between the MTHFR C677T polymorphism and the cervical lesion risk.
| Genetic model | Number of study | Sample Size | Analysis | I2 (%) | Ph | Test of Association | P(Publication bias test) | |||
| Case | Control | Model | P | OR(95%CI) | Begg’s test | Egger’s test | ||||
| Total | ||||||||||
| Dominant model | 16 | 3498 | 3594 | R | 78 | 0.00 | 0.95 | 0.99 [0.78, 1.26] | 0.558 | 0.626 |
| Recessive model | 14 | 3233 | 3451 | R | 51 | 0.01 | 0.75 | 1.05 [0.80, 1.38] | 0.827 | 0.956 |
| Additive model | 14 | 6177 | 6902 | R | 79 | 0.00 | 0.79 | 0.97 [0.80, 1.18] | 1.000 | 0.659 |
| CT vs. CC | 14 | 2854 | 3097 | R | 69 | 0.00 | 0.75 | 0.97 [0.78, 1.20] | 0.443 | 0.490 |
| TT vs. CC | 14 | 1927 | 2038 | R | 61 | 0.00 | 0.73 | 1.06 [0.76, 1.48] | 0.913 | 0.614 |
| Pathological type | ||||||||||
| ICC | ||||||||||
| Dominant model | 12 | 2008 | 2932 | R | 73 | 0.00 | 0.62 | 0.94 [0.72, 1.21] | ||
| Dominant model* | 11 | 1946 | 2855 | R | 73 | 0.00 | 0.44 | 0.90 [0.69, 1.18] | ||
| Recessive model | 11 | 1946 | 2855 | R | 59 | 0.00 | 0.96 | 1.01 [0.70, 1.45] | ||
| Additive model | 11 | 3915 | 5710 | R | 80 | 0.00 | 0.51 | 0.92 [0.73, 1.17] | ||
| CT vs. CC | 11 | 1731 | 2534 | R | 64 | 0.00 | 0.29 | 0.88 [0.69, 1.12] | ||
| TT vs. CC | 11 | 1229 | 1657 | R | 65 | 0.00 | 0.84 | 0.96 [0.62, 1.47] | ||
| SIL | ||||||||||
| Dominant model | 11 | 1490 | 2916 | R | 71 | 0.00 | 0.54 | 1.09 [0.82, 1.45] | ||
| Dominant modelˆ | 9 | 1287 | 2773 | R | 52 | 0.04 | 0.51 | 1.08 [0.86, 1.35] | ||
| Recessive model | 9 | 1287 | 2773 | F | 0 | 0.79 | 0.80 | 1.03 [0.83, 1.27] | ||
| Additive model | 9 | 2574 | 5546 | R | 43 | 0.08 | 0.59 | 1.04 [0.90, 1.21] | ||
| CT vs. CC | 9 | 1123 | 2475 | R | 47 | 0.06 | 0.27 | 1.09 [0.94, 1.26] | ||
| TT vs. CC | 9 | 698 | 1609 | F | 0 | 0.45 | 0.36 | 1.11 [0.88, 1.40] | ||
| Geographic area | ||||||||||
| Asian | ||||||||||
| Dominant model | 9 | 1919 | 2081 | R | 80 | 0.00 | 0.71 | 1.07 [0.76, 1.49] | ||
| Recessive model | 8 | 1777 | 2004 | R | 65 | 0.00 | 0.74 | 1.08 [0.70, 1.66] | ||
| Additive model | 8 | 3242 | 4008 | R | 83 | 0.00 | 0.82 | 0.97 [0.71, 1.31] | ||
| CT vs. CC | 8 | 1520 | 1770 | R | 72 | 0.00 | 0.72 | 0.95 [0.70, 1.28] | ||
| TT vs. CC | 8 | 1064 | 1186 | R | 69 | 0.00 | 0.77 | 1.08 [0.65, 1.80] | ||
| European | ||||||||||
| Dominant model | 4 | 1285 | 917 | R | 62 | 0.05 | 0.18 | 0.77 [0.52,1.13] | ||
| Recessive model | 3 | 1162 | 851 | F | 0 | 0.89 | 0.13 | 0.79 [0.58,1.07] | ||
| Additive model | 3 | 2347 | 1702 | F | 0 | 0.42 | 0.006 | 0.83 [0.72,0.95] | ||
| CT vs. CC | 3 | 1066 | 764 | F | 30 | 0.24 | 0.05 | 0.83 [0.69,1.00] | ||
| TT vs. CC | 3 | 688 | 471 | F | 0 | 0.82 | 0.05 | 0.73 [0.53,1.00] | ||
| USA | ||||||||||
| Dominant model | 3 | 294 | 596 | R | 83 | 0.00 | 0.62 | 1.22 [0.56, 2.65] | ||
| Recessive model | 3 | 294 | 596 | F | 0 | 0.72 | 0.25 | 1.39 [0.79, 2.45] | ||
| Additive model | 3 | 588 | 1192 | R | 76 | 0.02 | 0.57 | 1.16 [0.70, 1.93] | ||
| CT vs. CC | 3 | 268 | 563 | R | 83 | 0.00 | 0.74 | 1.15 [0.50, 2.63] | ||
| TT vs. CC | 3 | 175 | 381 | F | 20 | 0.29 | 0.13 | 1.56 [0.88, 2.77] | ||
Figure 2. Forest plot describing the association between the C677T polymorphism and the risk of cervical lesions.
(A) Meta-analysis in a random-effects model for CT+TT vs. CC (dominant model). (B) Meta-analysis in a random-effects model for CT vs. CC. (C) Meta-analysis in a random-effects model for TT vs. CC. Each study is shown by the point estimate of the OR (the size of the square is proportional to the weight of each study) and 95% CI for the OR (extending lines).
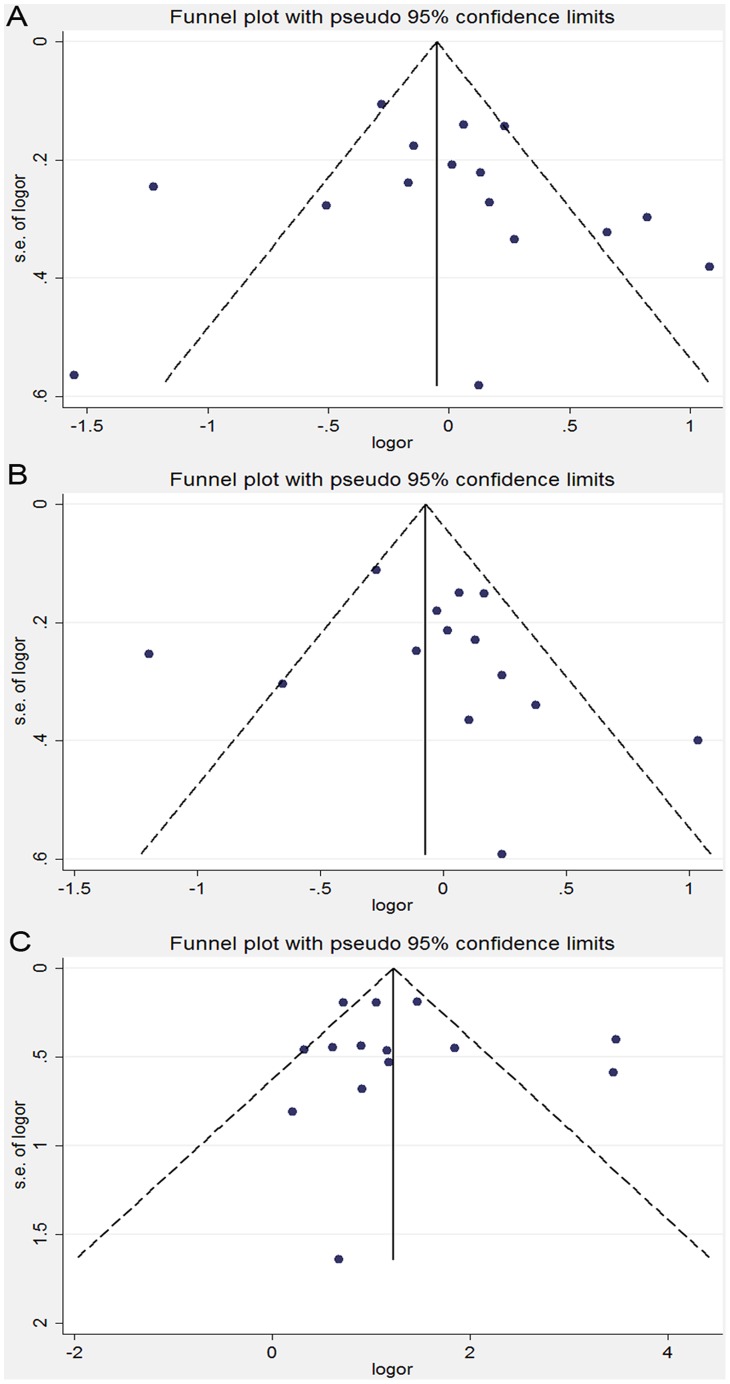
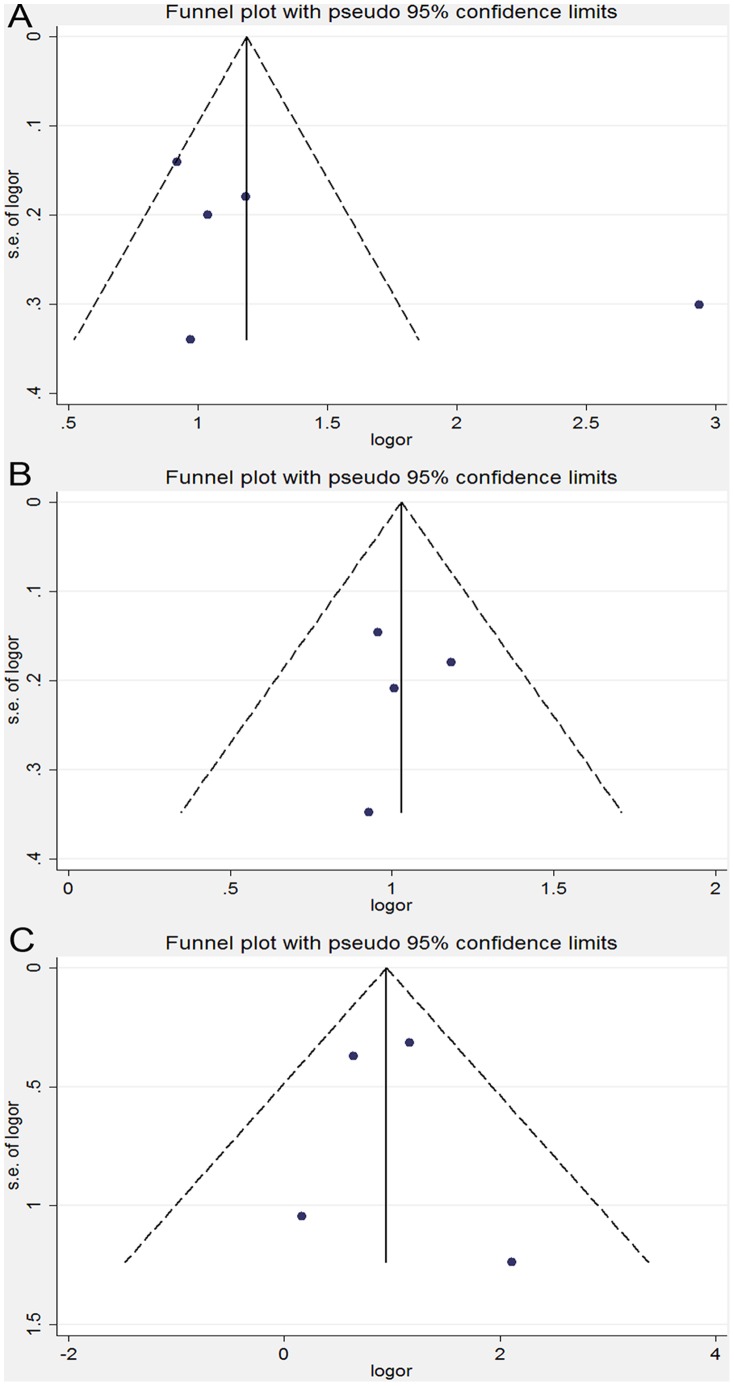
In the sensitivity analysis, the overall association between the MTHFR C677T genotype and the cervical lesions was unchanged after an exclusion of the individual study, including two studies [27], [35], which lacked enough data to calculate if it conformed to HWE among the control group. Similar results were found in the sensitivity analyses on the association between the MTHFR C677T genotype and ICC or SIL, indicating that our results were statistically robust. No obvious publication bias was detected according to the shapes of the funnel plots for the C677T polymorphism in all the genetic models (Figure 3). Consistent results of the Egger’s and the Begg’s tests were also obtained in all the genetic models (Table 3). Moreover, neither the funnel plots nor the Begg’s or Egger’s test detected any obvious evidence for the publication bias in the subgroup analyses on all the genetic models (data not shown).
Figure 3. Funnel plot analysis on the detection of the publication bias for the C677T polymorphism.
(A) Meta-analysis in a random-effects model for CT+TT vs. CC (dominant model). (B) Meta-analysis in a random-effects model for CT vs. CC. (C) Meta-analysis in a random-effects model for TT vs. CC. Each point represents an individual study for the indicated association. LogOR, natural logarithm of OR. Perpendicular line denotes the mean effect size.
Association between the MTHFR A1298C polymorphisms and cervical lesions
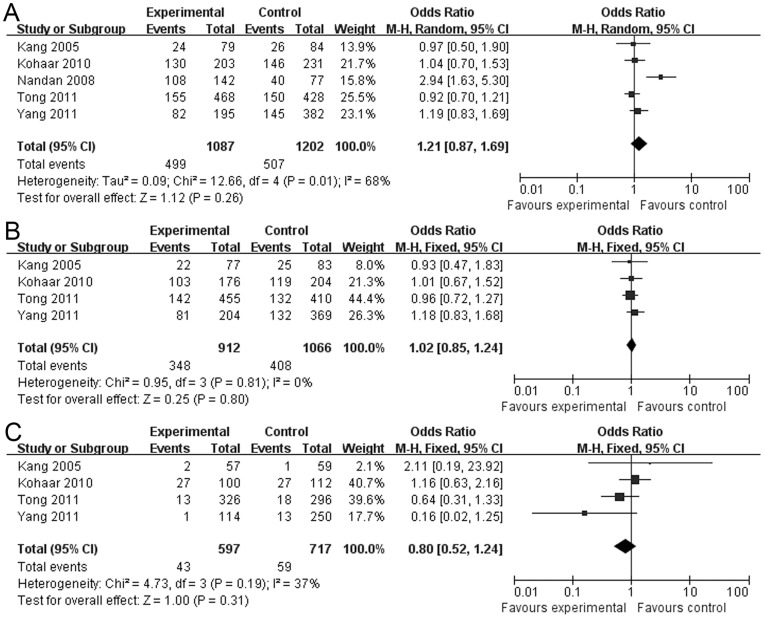
As for the A1298C polymorphism, no association was found between the polymorphism and the cervical lesions in all the genetic models (Table 4, dominant model: OR = 1.21, 95% CI = 0.87–1.690, Figure 4A; recessive model: OR = 0.81, 95% CI = 0.54–1.23; additive model: OR = 0.98, 95% CI = 0.85–1.14; AC vs. AA: OR = 1.02, 95% CI = 0.85–1.24, Figure 4B; CC vs. AA: OR = 0.80, 95% CI = 0.52–1.24, Figure 4C). The heterogeneity was significant in the dominant model (I2 = 68%, P = 0.01) and the random-effects model was performed. However, there was no significant heterogeneity for the comparison of other genetic models (P>0.1) and the fixed-effects method was performed for our investigation. In the subgroup analysis, no association was found between the A1298C polymorphism and SIL. Interestingly, the investigation on the women with A1298C polymorphisms vs. no carriers showed a marginally increased susceptibility to ICC but no statistically significant difference in dominant model (P = 0.06, OR = 1.21, 95% CI = 0.99–1.49) and AC vs. AA (P = 0.09, OR = 1.21, 95% CI = 0.97–1.51).
Table 4. Pooled Analysis on Association between the MTHFR A1298C polymorphism and the cervical lesion risk.
| Genetic model | Number of study | Sample Size | Analysis | I2 (%) | Ph | Test of Association | P(Publication bias test) | |||
| Case | Control | Model | P | OR(95%CI) | Begg’s test | Egger’s test | ||||
| Total | ||||||||||
| Dominant model | 5 | 1087 | 1202 | R | 68 | 0.01 | 0.26 | 1.21[0.87, 1.69] | 0.462 | 0.290 |
| Recessive model | 4 | 945 | 1125 | F | 42 | 0.16 | 0.33 | 0.81[0.54, 1.23] | 1.000 | 0.992 |
| Additive model | 4 | 1890 | 2250 | F | 0 | 0.81 | 0.82 | 0.98[0.85, 1.14] | 1.000 | 0.587 |
| AC vs. AA | 4 | 912 | 1066 | F | 0 | 0.81 | 0.80 | 1.02[0.85, 1.24] | 1.000 | 0.930 |
| CC vs. AA | 4 | 597 | 717 | F | 37 | 0.19 | 0.31 | 0.80[0.52, 1.24] | 1.000 | 0.971 |
| Pathological type | ||||||||||
| ICC | ||||||||||
| Dominant model | 5 | 610 | 1202 | F | 0 | 0.63 | 0.06 | 1.21[0.99, 1.49] | ||
| Recessive model | 4 | 548 | 1125 | R | 51 | 0.10 | 0.46 | 0.67[0.24, 1.93] | ||
| Additive model | 4 | 1096 | 2250 | F | 0 | 1.00 | 0.43 | 1.07[0.90, 1.27] | ||
| AC vs. AA | 4 | 520 | 1066 | F | 0 | 0.62 | 0.09 | 1.21[0.97, 1.51] | ||
| CC vs. AA | 4 | 319 | 717 | F | 43 | 0.15 | 0.46 | 0.82[0.49, 1.38] | ||
| SIL | ||||||||||
| Dominant model | 4 | 477 | 1118 | R | 83 | 0.00 | 0.49 | 1.28[0.63, 2.60] | ||
| Recessive model | 3 | 397 | 1041 | F | 0 | 0.85 | 0.43 | 0.78[0.42, 1.44] | ||
| Additive model | 3 | 794 | 2082 | F | 0 | 0.90 | 0.14 | 0.85[0.68, 1.06] | ||
| AC vs. AA | 3 | 382 | 983 | F | 0 | 0.75 | 0.25 | 0.85[0.65, 1.12] | ||
| CC vs. AA | 3 | 278 | 658 | F | 0 | 0.86 | 0.34 | 0.74[0.40, 1.38] | ||
Dominant model: CC+AC vs. AA; Recessive model: CC vs. AC+AA; Additive model: C vs. A; R, Random-effects model; F, fixed-effects model; ICC, invasive cervical cancer; ICC: invasive cervical cancer; SIL, squamous intra-epithelial lesion.
Figure 4. Forest plot describing the association between the A1298C polymorphism and the risk of cervical lesions.
(A) Meta-analysis in a random-effects model for AC+CC vs. AA (dominant model). (B) Meta-analysis in a random-effects model for AC vs. AA. (C) Meta-analysis in a random-effects model for CC vs. AA. Each study is shown by the point estimate of the OR (the size of the square is proportional to the weight of each study) and 95% CI for the OR (extending lines).
In the sensitivity analyses, the overall association between the MTHFR A1298C genotype and the cervical lesions was changed after an exclusion of one study [27] which lacked enough data to calculate if it conformed to HWE among the control group. However, the results of the sensitivity analysis on the cervical lesions were virtually unchanged after an exclusion of any other individual study (Figure 5). The shape of the funnel plots was symmetrical, which showed that no evidence was found for the publication bias among the studies (Figure 6). No publication bias was also detected according to the results of Egger’s and Begg’s tests (Table 4). Furthermore, neither the funnel plots nor the Begg’s and Egger’s tests found any obvious evidence for the publication bias in the subgroup analysis on all genetic models (data not shown).
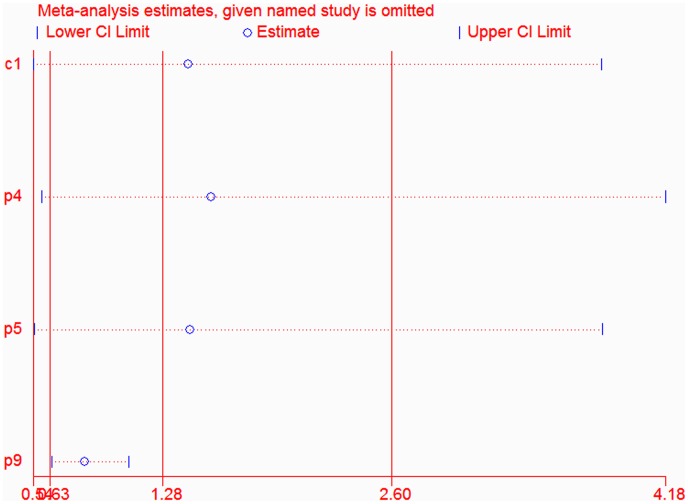
Figure 5. Influence analysis of the summary odds ratio coefficients on the association between the A1298C polymorphism and cervical cancer in dominant model.
The results were computed by omitting each study (left column) in turn. Bars, 95% CIs.
Figure 6. Funnel plot analysis on the detection of the publication bias for the A1298C polymorphism.
(A) Meta-analysis in a random-effects model for AC+CC vs. AA (dominant model). (B) Meta-analysis in a random-effects model for AC vs. AA. (C) Meta-analysis in a random-effects model for CC vs. AA. Each point represents an individual study for the indicated association. LogOR, natural logarithm of OR. Perpendicular line denotes the mean effect size.
Discussion
As we know, HPV infection may be necessary but is not sufficient to cause cervical cancer. Other factors may play some important roles in this cancer development. For example, the nutritional factors may affect the persistence of HPV infection and thereby influence progression of early precancerous lesions to invasive cancer. Specifically, folate plays a key role in DNA synthesis, repair, and methylation, and this forms the basis of mechanistic explanations for a putative role for folate in cancer prevention. However, the effect of folate in these processes may be modulated by the genotype for the common C677T or A1298C variants of MTHFR, the homozygosity of which is associated with a lower level of the enzyme activity, lower plasma and red blood cell folate, and elevated plasma homocysteine [42], [43]. Several studies investigated the association between the MTHFR polymorphisms and the preinvasive cervical lesions or cervical cancer, but the results were not consistent. Thus, our meta-analysis could better evaluate association between the MTHFR C677T/A1298C polymorphisms and the susceptibility to cervical lesions. Our findings demonstrate that there was no association between them. To our knowledge, this is the first meta-analysis on association between MTHFR C677T/A1298C polymorphisms and susceptibility to cervical lesions, and the largest-scale meta-analysis examining the risk of cervical cancer.
As for the MTHFR C677T, most evidence points to decrease in the susceptibility to colorectal cancer and an increase in the susceptibility to esophagus and gastric cancer [44]–[48], but the effect on the cervical cancer susceptibility was not consistent. In our meta-analysis, no statistically significant difference was found in the frequency of the MTHFR C677T polymorphism in the patients with cervical lesions when compared with the controls. This finding was consistent with that of one previous meta-analysis [49]. However, 9 new studies [20]–[22], [25]–[27], [32], [33], [35] have been published since 2006 and all recruited in our study dramatically increased the case number of cervical lesion and controls with genetic information, which indicated that our results could be more reliable. In addition, multiple subgroup analyses made our meta-analysis more convincing too. We meta-analyzed the eligible case-control studies for C677T by geographic regions. No association was found between the C677T polymorphism and the cervical lesions in either in the Asian or in the American populations. However, a significant inverse association was found in the European population. Different genetic backgrounds or environmental conditions could explain the discrepancy. The meta-analysis also stratified by histological stages of cervical lesions showed that there was no association between the MTHFR C677T variants and cervical lesion development. To assess the effect of individual study on the overall meta-analysis estimate, we excluded one study at a time, and the exclusion of any single report did not change the significance of the final conclusion, which indicated that the outcomes were robust. Taken together, we could make a conclusion that cervical lesion were not primarily caused by genetically-determined enzymatic defects in the folate metabolic pathway, which might be different from the pathways supposed for colorectal or gastric carcinogenesis. The effect of those polymorphisms on the cervical cancer susceptibility seems to be further modulated by other cofactors such as infection with the HPV and smoking.
As for MTHFR A1298C, some studies reported a positive association with cervical lesions, which had only borderline significance [25]. More recent studies have revealed no association between the MTHFR A1298C and the cervical lesions [22], [23], [26], [27]. Our meta-analysis confirmed that there is no association between the A1298C polymorphism and cervical lesions, similar to that found by the subgroup analysis on the ethnic groups and the histological stages of cervical lesions. No association was found between the A1298C polymorphism and SIL, but the ICC showed a marginally positive association though with no statistically significant difference. This result suggested that a probably higher risk for cervical cancer was linked to the A1298C variants, implying their important role in later stages of cervical carcinogenesis but not in SILs. Sensitivity analyses revealed that the overall association between the MTHFR A1298C genotype and cervical lesions could be changed after excluding one study [27] which lacked sufficient data to calculate whether it conformed to HWE among or not in the control group. In contrast, the results were virtually unchanged after the exclusion of any other individual study. To sum up, it is possibly indicated that the study by Nandan et al. could be the main source of the observed heterogeneity across the studies in this meta-analysis. Alternatively, the study may had limitations or because of other unknown factors.
To some extent, several limitations of this meta-analysis should be addressed. One limitation of the present study was that the sample size of A1298C mutation involved is not big enough. We neen more original researches to make our conclusions more reliable and accurate. The studies on the A1298C variant had reported only 5 articles, and their participants were entirely Asians with no population variation in minor allele frequency. So, the subgroup meta-analysis on this gene polymorphism was not possible by race. Another limitation was that significant heterogeneity in the studies was mainly present in overall analyses and subgroup analyses. Though several possible sources of the between-study heterogeneity were investigated, including pathological history, geographic region, ethnicity, source of controls, and source of DNA for genotyping ethnicity (data not shown), none of them could sufficiently explain the heterogeneity. The effect estimates might depend on some unidentified sources of heterogeneity. Besides, part of the exposure information was still lacking in the available studies, E.g., HPV infection status, smoking status or nutritional status (particularly folate intake or level). Therefore, effects of environment exposure or lifestyle on association between MTHFR variants and cervical lesions could not be determined by this meta-analysis.
In summary, despite the above-mentioned limitations, the present study provides evidence that the MTHFR C677T and A1298C polymorphisms may not increase the susceptibility to cervical cancer development. However, our meta-analysis reveals a negative association between the MTHFR C677T mutations and cervical lesions, especially in the European populations. The marginal association between the MTHFR A1298C polymorphisms and the susceptibility for cervical cancer need to be further studied.
Supporting Information
PRISMA checklist.
(DOC)
Funding Statement
The authors have no support or funding to report.
References
- 1. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Bosch F, Lorincz A, Munoz N, Meijer C, Shah K (2002) The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 55: 244–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walboomers JMM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, et al. (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189: 12–19. [DOI] [PubMed] [Google Scholar]
- 4. Elfgren K, Kalantari M, Moberger B, Hagmar B, Dillner J (2000) A population-based five-year follow-up study of cervical human papillomavirus infection. Am J Obstet Gynecol 183: 561–567. [DOI] [PubMed] [Google Scholar]
- 5. Insinga RP, Dasbach EJ, Elbasha EH (2009) Epidemiologic natural history and clinical management of Human Papillomavirus (HPV) disease: a critical and systematic review of the literature in the development of an HPV dynamic transmission model. BMC Infect Dis 9: 119–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castellsagué X, Muñoz N (2003) Cofactors in human papillomavirus carcinogenesis–role of parity, oral contraceptives, and tobacco smoking. JNCI Monographs 2003: 20–28. [PubMed] [Google Scholar]
- 7. Castellsague X, Bosch FX, Munoz N (2002) Environmental co-factors in HPV carcinogenesis. Virus Res 89: 191–199. [DOI] [PubMed] [Google Scholar]
- 8. García-Closas R, Castellsagué X, Bosch X, González CA (2005) The role of diet and nutrition in cervical carcinogenesis: a review of recent evidence. Int J Cancer 117: 629–637. [DOI] [PubMed] [Google Scholar]
- 9. Van Niekerk W (1966) Cervical cytological abnormalities caused by folic acid deficiency. Acta Cytol 10: 67–73. [Google Scholar]
- 10. Whitehead N, Reyner F, Lindenbaum J (1973) Megaloblastic changes in the cervical epithelium: association with oral contraceptive therapy and reversal with folic acid. JAMA 226: 1421–1424. [PubMed] [Google Scholar]
- 11. VanEenwyk J, Davis FG, Colman N (1992) Folate, vitamin C, and cervical intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev 1: 119–124. [PubMed] [Google Scholar]
- 12. Butterworth Jr C, Hatch KD, Macaluso M, Cole P, Sauberlich HE, et al. (1992) Folate deficiency and cervical dysplasia. JAMA 267: 528–533. [PubMed] [Google Scholar]
- 13. Potischman N, Brinton LA, Laiming VA, Reeves WC, Brenes MM, et al. (1991) A case-control study of serum folate levels and invasive cervical cancer. Cancer Res 51: 4785–4789. [PubMed] [Google Scholar]
- 14. Sedjo RL, Inserra P, Abrahamsen M, Harris RB, Roe DJ, et al. (2002) Human papillomavirus persistence and nutrients involved in the methylation pathway among a cohort of young women. Cancer Epidemiol Biomarkers Prev 11: 353–359. [PubMed] [Google Scholar]
- 15. Piyathilake CJ, Henao OL, Macaluso M, Cornwell PE, Meleth S, et al. (2004) Folate is associated with the natural history of high-risk human papillomaviruses. Cancer Res 64: 8788–8793. [DOI] [PubMed] [Google Scholar]
- 16. Pillai M, Chacko P, Kesari L, Jayaprakash P, Jayaram H, et al. (2003) Expression of folate receptors and heterogeneous nuclear ribonucleoprotein E1 in women with human papillomavirus mediated transformation of cervical tissue to cancer. J Clin Pathol 56: 569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, et al. (1997) Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA 94: 3290–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamada K, Chen Z, Rozen R, Matthews RG (2001) Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc Nati Acad Sci USA 98: 14853–14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van der Put NMJ, Gabreëls F, Stevens E, Smeitink JAM, Trijbels FJM, et al. (1998) A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet 62: 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prasad V, Wilkhoo H (2011) Association of the Functional Polymorphism C677T in the Methylenetetrahydrofolate Reductase Gene with Colorectal, Thyroid, Breast, Ovarian, and Cervical Cancers. Onkologie 34: 422–426. [DOI] [PubMed] [Google Scholar]
- 21. Mostowska A, Myka M, Lianeri M, Roszak A, Jagodzinski PP (2011) Folate and choline metabolism gene variants and development of uterine cervical carcinoma. Clin Biochem 44: 596–600. [DOI] [PubMed] [Google Scholar]
- 22. Kohaar I, Kumar J, Thakur N, Hussain S, Niyaz MK, et al. (2010) Homocysteine levels are associated with cervical cancer independent of methylene tetrahydrofolate reductase gene (MTHFR) polymorphisms in Indian population. Biomarkers 15: 61–68. [DOI] [PubMed] [Google Scholar]
- 23. Kang S, Kim JW, Kang GH, Park NH, Song YS, et al. (2005) Polymorphism in folate-and methionine-metabolizing enzyme and aberrant CpG island hypermethylation in uterine cervical cancer. Gynecol Oncol 96: 173–180. [DOI] [PubMed] [Google Scholar]
- 24. Lambropoulos A, Agorastos T, Foka Z, Chrisafi S, Constantinidis T, et al. (2003) Methylenetetrahydrofolate reductase polymorphism C677T is not associated to the risk of cervical dysplasia. Cancer Lett 191: 187–191. [DOI] [PubMed] [Google Scholar]
- 25. Yang F, Zhou Y, Jiang Y, Fan Y, Li J (2010) Study on the correlation between polymorphism of MTHFR gene and the pathogenesis of cervical cancer. J China Maternal Child Health 23: 4122–4124. [Google Scholar]
- 26. Tong S, Kim MK, Lee JK, Lee JM, Choi SW, et al. (2011) Common polymorphisms in methylenetetrahydrofolate reductase gene are associated with risks of cervical intraepithelial neoplasia and cervical cancer in women with low serum folate and vitamin B12. Cancer Causes Control 22: 63–72. [DOI] [PubMed] [Google Scholar]
- 27. Nandan NK, Wajid S, Biswas S, Juneja SS, Rizvi M, et al. (2008) Allelic variations in 5, 10-methylenetetrahydrofolate reductase gene and susceptibility to cervical cancer in Indian women. Drug Metabolism Lett 2: 18–22. [DOI] [PubMed] [Google Scholar]
- 28. Sull JW, Jee SH, Yi S, Lee JE, Park JS, et al. (2004) The effect of methylenetetrahydrofolate reductase polymorphism C677T on cervical cancer in Korean women. Gynecol Oncol 95: 557–563. [DOI] [PubMed] [Google Scholar]
- 29. Goodman MT, McDuffie K, Hernandez B, Wilkens LR, Bertram CC, et al. (2001) Association of methylenetetrahydrofolate reductase polymorphism C677T and dietary folate with the risk of cervical dysplasia. Cancer Epidemiol Biomarkers Prev 10: 1275–1280. [PubMed] [Google Scholar]
- 30. Piyathilake C, Macaluso M, Johanning G, Whiteside M, Heimburger D, et al. (2000) Methylenetetrahydrofolate reductase (MTHFR) polymorphism increases the risk of cervical intraepithelial neoplasia. Anticancer Res 20: 1751–1757. [PubMed] [Google Scholar]
- 31. Ma X, Wang J, Zhou Q, Ding L, Cheng Y, et al. (2006) Relationship between Methylenetetrahydrofolate reductase polymorphism and cervical cancer susceptibility. China Public Health 22: 1247–1248. [Google Scholar]
- 32. Shekari M, Sobti RC, Kordi Tamandani DM, Suri V (2008) Impact of methylenetetrahydrofolate reductase (MTHFR) codon (677) and methionine synthase (MS) codon (2756) on risk of cervical carcinogenesis in North Indian population. Arch Gynecol Obstet 278: 517–524. [DOI] [PubMed] [Google Scholar]
- 33. Piyathilake CJ, Azrad M, Macaluso M, Johanning GL, Cornwell PE, et al. (2007) Protective association of MTHFR polymorphism on cervical intraepithelial neoplasia is modified by riboflavin status. Nutrition 23: 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zoodsma M, Nolte IM, Schipper M, Oosterom E, van der Steege G, et al. (2005) Methylenetetrahydrofolate reductase (MTHFR) and susceptibility for (pre)neoplastic cervical disease. Hum Genet 116: 247–254. [DOI] [PubMed] [Google Scholar]
- 35. Agodi A, Barchitta M, Cipresso R, Marzagalli R, La Rosa N, et al. (2010) Distribution of p53, GST, and MTHFR polymorphisms and risk of cervical intraepithelial lesions in sicily. Int J Gynecol Cancer 20: 141–146. [DOI] [PubMed] [Google Scholar]
- 36. Collin SM, Metcalfe C, Zuccolo L, Lewis SJ, Chen L, et al. (2009) Association of folate-pathway gene polymorphisms with the risk of prostate cancer: a population-based nested case-control study, systematic review, and meta-analysis. Cancer Epidemiol Biomarkers Prev 18: 2528–2539. [DOI] [PubMed] [Google Scholar]
- 37. Moher D, Liberati A, Tetzlaff J, Altman DG (2009) The PRISMA Group (2009) Preferred reporting items for systematic reviews and meta-analyses: The PRISMA Statement. PLoS Med 6(6): e1000097 doi:10.1371/journal.pmed1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Higgins J, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 39. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clini Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 40. Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101. [PubMed] [Google Scholar]
- 41. Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jacques PF, Bostom AG, Williams RR, Ellison RC, Eckfeldt JH, et al. (1996) Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation 93: 7–9. [DOI] [PubMed] [Google Scholar]
- 43. Ashfield-Watt PAL, Pullin CH, Whiting JM, Clark ZE, Moat SJ, et al. (2002) Methylenetetrahydrofolate reductase 677C>T genotype modulates homocysteine responses to a folate-rich diet or a low-dose folic acid supplement: a randomized controlled trial. Am J Clin Nutrition 76: 180–186. [DOI] [PubMed] [Google Scholar]
- 44. Hubner RA, Houlston RS (2007) MTHFR C677T and colorectal cancer risk: A meta-analysis of 25 populations. Int J Cancer 120: 1027–1035. [DOI] [PubMed] [Google Scholar]
- 45. Huang Y, Han S, Li Y, Mao Y, Xie Y (2007) Different roles of MTHFR C677T and A1298C polymorphisms in colorectal adenoma and colorectal cancer: a meta-analysis. J Hum Genet 52: 73–85. [DOI] [PubMed] [Google Scholar]
- 46. Larsson SC, Giovannucci E, Wolk A (2006) Folate Intake, MTHFR Polymorphisms, and Risk of Esophageal, Gastric, and Pancreatic Cancer: A Meta-analysis. Gastroenterology 131: 1271–1283. [DOI] [PubMed] [Google Scholar]
- 47. Langevin S, Lin D, Matsuo K, Gao C, Takezaki T, et al. (2009) Review and pooled analysis of studies on MTHFR C677T polymorphism and esophageal cancer. Toxicol Lett 184: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zintzaras E (2006) Association of methylenetetrahydrofolate reductase (MTHFR) polymorphisms with genetic susceptibility to gastric cancer: a meta-analysis. J Hum Genet 51: 618–624. [DOI] [PubMed] [Google Scholar]
- 49. Zacho J, Yazdanyar S, Bojesen SE, Tybjærg-Hansen A, Nordestgaard BG (2011) Hyperhomocysteinemia, methylenetetrahydrofolate reductase 677C>T polymorphism and risk of cancer: Cross-sectional and prospective studies and meta-analyses of 75,000 cases and 93,000 controls. Int J Cancer 128: 644–652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA checklist.
(DOC)