Abstract
In contrast to nearly all eukaryotes, the Old World Leishmania species L. infantum and L. major lack the bona fide RNAi machinery genes. Interestingly, both Leishmania genomes code for an atypical Argonaute-like protein that possesses a PIWI domain but lacks the PAZ domain found in Argonautes from RNAi proficient organisms. Using sub-cellular fractionation and confocal fluorescence microscopy, we show that unlike other eukaryotes, the PIWI-like protein is mainly localized in the single mitochondrion in Leishmania. To predict PIWI function, we generated a knockout mutant for the PIWI gene in both L. infantum (Lin) and L. major species by double-targeted gene replacement. Depletion of PIWI has no effect on the viability of insect promastigote forms but leads to an important growth defect of the mammalian amastigote lifestage in vitro and significantly delays disease pathology in mice, consistent with a higher expression of the PIWI transcript in amastigotes. Moreover, amastigotes lacking PIWI display a higher sensitivity to apoptosis inducing agents than wild type parasites, suggesting that PIWI may be a sensor for apoptotic stimuli. Furthermore, a whole-genome DNA microarray analysis revealed that loss of LinPIWI in Leishmania amastigotes affects mostly the expression of specific subsets of developmentally regulated genes. Several transcripts encoding surface and membrane-bound proteins were found downregulated in the LinPIWI(−/−) mutant whereas all histone transcripts were upregulated in the null mutant, supporting the possibility that PIWI plays a direct or indirect role in the stability of these transcripts. Although our data suggest that PIWI is not involved in the biogenesis or the stability of small noncoding RNAs, additional studies are required to gain further insights into the role of this protein on RNA regulation and amastigote development in Leishmania.
Introduction
The Argonaute (AGO) proteins represent a large family of proteins that bind to small RNAs like microRNAs (miRNAs), short interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs) [1]. They are central to many transcriptional and posttranscriptional gene silencing pathways [2] as well as involved in transposon control, which otherwise threaten the integrity of the genome [3]. Argonaute proteins are evolutionarily conserved and can be phylogenetically classified into three paralogous groups: the AGO subfamily proteins which are similar to Arabidopsis thaliana AGO1, the PIWI subfamily which is closely related to Drosophila melanogaster PIWI and the worm-specific WAGO subfamily which is related to the C. elegans specific group 3 Argonaute proteins [4]. The AGO-clade proteins have been found in nearly all eukaryotes, while the PIWI-clade proteins are restricted to animals and ciliates, organisms which undergo sexual reproduction [5]. Many prokaryotes and archaea also encode homologs of Argonaute proteins but their functions remain largely unknown [6]. Prokaryotic AGOs are predicted to function as key components of a new class of defense systems against mobile genetic elements [6]. AGO proteins share two main structural features – the ∼140 residue central PAZ domain and the C-terminal 350-residue PIWI domain [1]. The PAZ domain contains an OB (oligonucleotide/oligosaccharide binding) fold, which is a typical single-stranded nucleic acid binding motif that has been shown to bind the 3′ end of short RNAs [7]. The PIWI domain shows extensive structural similarity to RNase H enzymes and is the catalytic center for rendering some AGOs that retain conserved residues able to target cleavage of RNA molecules complementary to AGO-bound small RNAs [8].
In most organisms investigated so far, which include Drosophila, the zebrafish and the mouse, the PIWI subfamily proteins play diverse function in germline-specific events and gametogenesis, where they bind PIWI-interacting RNAs (piRNAs) [5]. There is considerable evidence that the PIWI subfamily proteins (e.g. PIWI, AUB and AGO3) and piRNAs regulate transposon activity in Drosophila [9], [10], [11] but also in vertebrates [12], [13]. PIWI-type proteins have also a role in epigenetic function. The murine PIWI homologs, MILI and MIW2, are needed for the methylation of transposon-encoding genome regions [12], [13]. Furthermore, an epigenetic role has been attributed to PIWI along with piRNAs in Drosophila where it associates with chromatin and interacts directly with the heterochromatin protein 1 [14]. PIWI is also involved in the epigenetic control in somatic cells of the ciliate Tetrahymena where it is vital for the conjugative replication of macronucleus by removing highly repetitive transposon-like sequences [15].
Leishmania is a unicellular parasite, which is responsible for leishmaniasis. The parasite has two life stages, the invertebrate promastigote stage and the mammalian amastigote stage within the phagolysosomes of macrophages. Leishmania and related kinetoplastids regulate gene expression mainly posttranscriptionally [16]. Old World Leishmania species such as L. infantum and L. major lack a functional RNAi pathway [17], [18], [19]. The absence of typical Argonaute proteins in these parasites suggests their inability to suppress gene expression by RNAi [17]. Unlike the Old World Leishmania species, the New World species L. braziliensis (subgenus Viannia) code for RNAi pathway homologs [20] and can downregulate target genes by RNAi [21]. Similarly, the related trypanosomatid protozoan Trypanosoma brucei possesses a functional RNAi machinery and codes for an Argonaute 1 protein (AGO1), which can function as slicer [22]. This suggests that the RNAi machinery genes must have been lost independently in Old World Leishmania during evolution of the trypanosomatid lineage. Interestingly, while the RNAi-negative L. major/L. infantum strains [19]; http://tritrypdb.org) and Trypanosoma cruzi [23] have lost their AGO genes, they encode an Argonaute/PIWI-like protein homolog.
Here, we characterized a PIWI-like protein homolog in Leishmania and evaluated its putative function in regulating gene expression in these parasites. We show that genetic deletion of PIWI in both L. infantum and L. major species leads to an important growth defect of amastigotes in vitro and significantly delays disease pathology in mice. Furthermore, we demonstrate that unlike other canonical PIWI proteins in eukaryotes, the Leishmania PIWI-like protein resides mainly in the mitochondrion. As determined by a whole-genome DNA microarray analysis, loss of PIWI in Leishmania amastigotes affects mostly the expression of specific subsets of developmentally regulated genes, suggesting an important role of this protein in posttranscriptional regulation.
Results
The RNAi-negative Old World Leishmania Species Encode an Argonaute/PIWI-like Protein Lacking the Central PAZ Domain
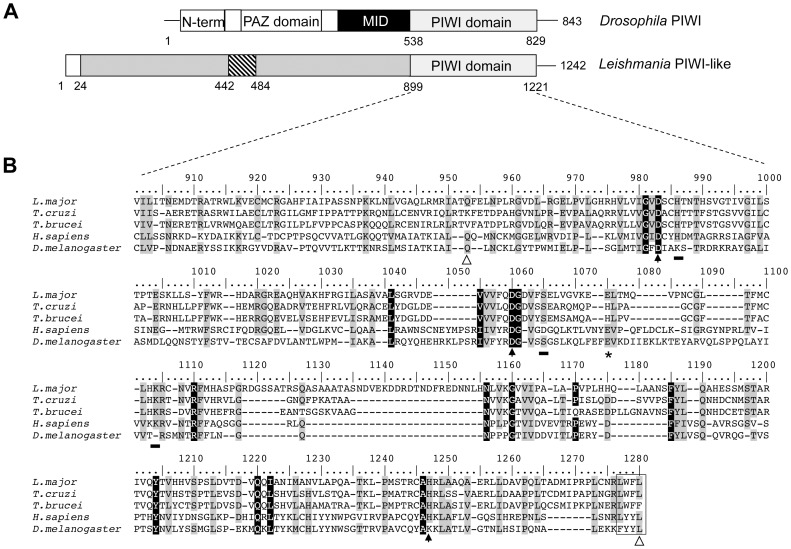
Argonaute (AGO) proteins found in eukaryotes are composed of four sections: N-terminal, PAZ domain, MID domain and PIWI domain [1], [24] (Figure 1A), while most archeal and prokaryotic AGOs do not contain the PAZ domain, and the domain architecture is variable [6]. The Argonaute/PIWI-like protein encoded by Leishmania infantum (LinJ.21.0470) and L. major (LmjF.21.0410) (http://tritrypdb.org) is a basic protein with a predicted MW of 133 kDa that is distinct from the canonical AGO/PIWI proteins in other eukaryotes. Indeed, the Leishmania PIWI-like homolog lacks the PAZ domain, which is typical of Argonaute proteins [4] but it has retained a PIWI-like C-terminal domain (from 899–1221 aa) (Figure 1A). Multiple sequence alignment of the L. major PIWI-like protein along with Trypanosoma cruzi (TcCLB.511367.240), T. brucei (Tb927.10.2220), and the H. sapiens (AAC97371) and Drosophila (AAF53043) PIWI homologs revealed a characteristic DDH motif composed of two aspartates and a histidine (Figure 1B), which constitutes a conserved catalytic core similar to the DDE (aspartate, aspartate, glutamate) catalytic triad of RNase H folding [25]. The Leishmania PIWI-like protein harbors also other invariable residues in all Argonaute proteins. These include conserved residues that define the PIWI and Argonaute subfamilies, the divalent cation-binding residues (Gln and Leu) for structural stability and catalytic activity and residues putatively involved in RNA binding [26], [27] (see Figure 1B).
Figure 1. The Leishmania PIWI-like protein homolog forms a distinct group of the Argonaute/PIWI subfamily.
(A) Structural domains (N-term, PAZ, MID, PIWI) of the Argonaute/PIWI proteins in higher eukaryotes (e.g. Drosophila) and in the unicellular parasitic protozoan Leishmania. In Leishmania only the C-terminal PIWI domain is found (residues 899–1221). The Leishmania PIWI-like homolog contains a small domain (residues 450–492, hatched box) with homology to the mitochondrial cytochrome C oxidase subunit VIIIb. A signal peptide of 24 residues has been predicted at the N-terminus of the Leishmania PIWI-like homolog. (B) Multiple sequence alignment of PIWI domains from L. major, the related trypanosomatids Trypanosoma cruzi and T. brucei, H. sapiens HIWI and the Drosophila melanogaster PIWI. The alignment was conducted using the Bioedit program [74]. Residues that are universally invariable among all eukaryotic Argonaute (AGO) proteins are shaded black. Residues that are highly conserved in all AGO proteins are shaded grey. Divalent anion-binding residues (Gln and Leu; residues 952 and 1242) are indicated by open arrows and the catalytic AGO triad (DDH; residues 981, 1048 and 1210) are indicated by black arrows. In the Drosophila PIWI homolog, the histidine (H) residue of DDH is not conserved but instead a lysine residue is present (DDK). The glutamate at position 1061, part of the RNase H-like DDE motif, is indicated by an asterisk. Residues putatively involved in RNA binding are underlined by a black rectangle. The C-terminus hydrophobic residues are boxed.
The L. major and L. infantum PIWI-like proteins share 67% identity with L. braziliensis 1–1200 aa), 43% identity with T. cruzi (mainly from 581–1200 aa) and 41% identity with T. brucei (mainly from residues 580–1200). Interestingly, unlike the T. cruzi TcPIWI homolog which comprises a full Argonaute architecture with an oligonucleotide/oligosaccharide binding (OB fold) motif at the N-terminus, a MID domain and a PIWI C-terminal domain [23], the Leishmania PIWI-like homolog has only retained the C-terminal PIWI domain. Unlike any other Argonaute protein, the Leishmania PIWI homolog contains a small domain at amino acid positions 450–492 with homology to the mitochondrial cytochrome C oxidase subunit VIIIb.
The Leishmania PIWI Transcript is Preferentially Expressed in the Amastigote Life Stage of the Parasite
It has been reported in various organisms that PIWI protein has specific functions during different stages of growth. For example, MILI, a mouse homolog of PIWI, was shown to be involved in spermatogenesis [28]. Similarly, PIWI modulates cell division rates in Drosophila germline stem cells [29]. To determine PIWI transcript expression in both life stages of L. infantum we carried out northern blot analysis on total RNA from both promastigote and amastigote life stages using the LinPIWI ORF as a probe. Northern blot hybridization revealed that the level of LinPIWI transcript was preferentially expressed in axenic amastigotes compared to log-phase and stationary promastigotes (Figure 2). Interestingly, L. major DNA microarray experiments comparing mouse lesion-derived amastigotes to promastigotes revealed a differential PIWI expression in the amastigote stage (Natalia S. Akopyants, et al., in preparation; http://tritrypdb.org).
Figure 2. The Leishmania PIWI transcript is preferentially expressed in the amastigote lifestage.
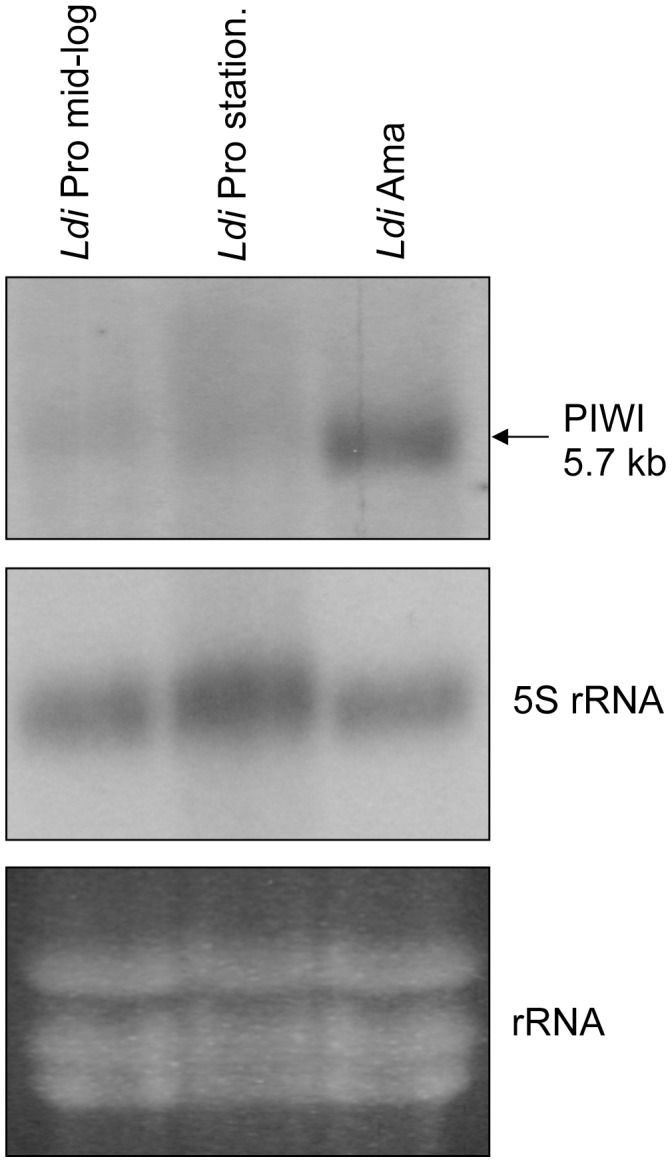
Northern blot hybridization to measure expression of the L. infantum LinPIWI transcript in exponentially- and stationary-grown promastigotes (Pro) and also in axenic amastigotes (Ama). The 5S rRNA probe was used for normalization of RNA loading. Northern blot experiments have been repeated two more times with identical results.
The Leishmania PIWI-like Protein Resides Mainly in the Mitochondrion
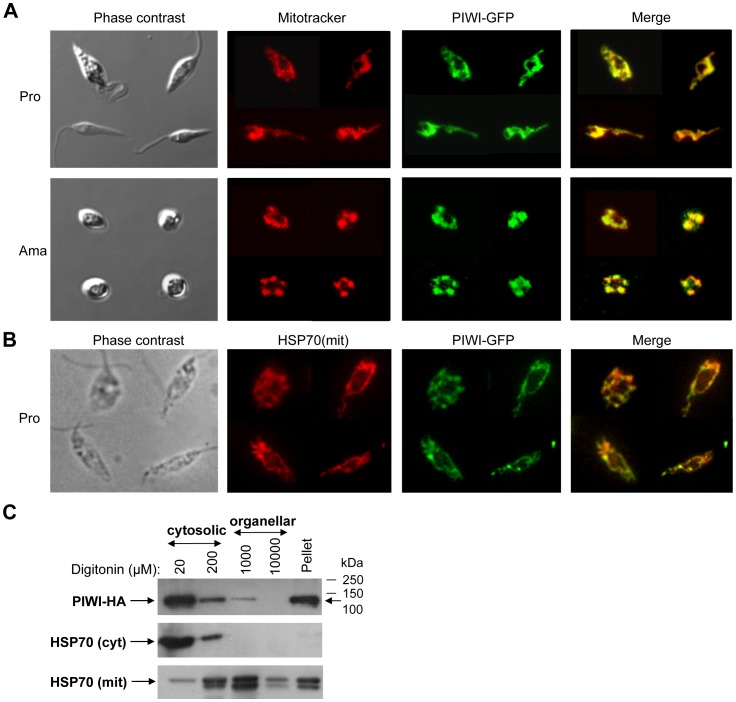
In higher eukaryotes, the PIWI subfamily proteins are localized in the cytosol or in the nucleus [5], [30]. Interestingly, the PIWI-like homolog in Leishmania possesses a putative mitochondrial targeting signal peptide at the N-terminus (1–24 aa) as suggested with the help of different servers that predict the presence and location of signal peptide cleavage sites in amino acid sequences from different organisms (e.g. SignalP 4.0, TargetP 1.1, PSORT and SOSUIsignal). Some of these softwares predict the sequence ARGECR (starting from amino acid 21) as the putative mitochondrial targeting signal peptide. MitoProt II-v1.101 analysis further asserted a possible targeting signal in the N-terminal 52 residues. To investigate whether the presence of a putative N-terminal mitochondrial targeting signal peptide could target the PIWI-like protein to the single mitochondrion of Leishmania, we fused the first 52 residues of the L. infantum PIWI-like protein to the GFP protein (designated as LinN52PIWI-GFP; Figure S1A) and then stably transfected this vector into L. infantum. Epifluorescence microscopy showed that the N-terminal 52 residues were able to direct the GFP protein into the mitochondrion both in promastigotes and in amastigotes (Figure S1B). To assess the sub-cellular localization of the full-length Leishmania PIWI-like protein, the PIWI homolog was fused with GFP at the C-terminus (LmjPIWI-GFP) and transfected into L. infantum. Confocal fluorescence microscopy showed that the PIWI-GFP protein was targeted specifically to the mitochondrion, both in promastigote and axenic amastigote forms (Figure 3A). To further confirm mitochondrial localization of the Leishmania PIWI-lke protein, we carried out co-localization studies with a known mitochondrial protein, HSP70. These studies clearly demonstrate that the PIWI-like protein largely co-localizes with the mitochondrial HSP70 protein (Figure 3B). Although most of the PIWI-lke protein in Leishmania seems to localize in the mitochondrion, there are some defined regions that did neither stain with Mitotracker nor co-localize with the mitochondrial HSP70 protein (Figure 3A–B), suggesting a partial cytosolic localization of this protein. Cellular compartmentalization by fractionation using various concentrations of digitonin (20 µM to 10 mM) confirms that part of the LinPIWI-HA tagged protein (134 kDa) is enriched in the organellar fraction but it also demonstrates an important enrichment in the cytosolic fraction and the membrane-bound fraction (Figure 3C and data not shown). The enrichment of the LinPIWI-HA protein in the pellet fraction, which represents total membrane fractions of the parasite, including the mitochondrial membrane, suggests that PIWI-like protein may be residing in the mitochondrial membrane. Interestingly, different softwares predicting the presence of membrane-spanning regions (e.g. TMpred, TopPred 0.01, PRED-TMR, HMMTOP, and SACS MEMSAT) revealed two short transmembrane helices in the L. infantum PIWI-like protein at positions 225–243 aa and 814–832 aa (data not shown), which may explain the membrane-association of this protein.
Figure 3. The Leishmania PIWI-like protein is mainly localized in the mitochondrion.
(A) Localization of PIWI-GFP protein was monitored in both L. infantum promastigote (Pro) and axenic amastigote (Ama) forms by confocal microscopy. Mitotracker (a red fluorscent dye) was used to stain the Leishmania single mitochondrion. The green fluorescence signal of GFP was mostly co-localized with that of the Mitotracker. (B) Immunolocalization studies using epifluorescence microscopy to confirm mitochondrial localization of the PIWI-like protein. PIWI largely co-localizes with a known mitochondrial protein, HSP70. (C) Digitonin fractionation was carried out in L. infantum LinHA-PIWI-HA recombinant promastigotes. Western blot of digitonin-fractionated samples (20 µM–10 mM) was probed with an anti-HA antibody to detect the PIWI protein and with anti-HSP70 (cytosolic) and anti-HSP70 (mitochondrial) antibodies used as controls for cytoplasmic and mitochondrial proteins, respectively. The 20 and 200 µM digitonin lanes correspond to cytosolic fractions and the 1 mM and 10 mM lanes correspond to organellar fractions. The pellet fraction contains membrane-associated proteins.
The Leishmania PIWI-like Protein is not Involved in the Biogenesis of Small Non-coding RNAs
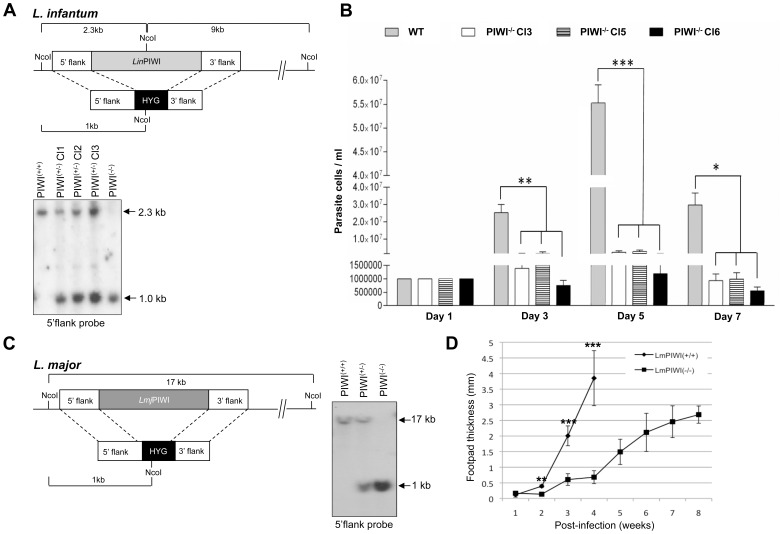
The evolutionarily conserved Argonaute/PIWI family of proteins are crucial for the biogenesis and function of the 24–32 nucleotide long piRNAs that are generated from long single-stranded RNA precursors often encoded by repetitive intergenic sequences in the genome [5]. As PIWI proteins do have the RNase H fold and some PIWI subfamily proteins have shown slicer activity, it is possible that they cleave the precursor into mature piRNAs [5]. The Leishmania genome is full of repetitive sequences within intergenic regions and 3′UTRs such as the highly structured SIDER retroposon elements [31], [32], which may be a substrate for PIWI. To assess the putative role of the Leishmania PIWI-like protein in the production of small non-coding RNA species, we engineered a L. infantum mutant lacking the PIWI gene (LinPIWI(−/−)) by gene replacement through homologous recombination. Both alleles of the PIWI single copy gene were replaced by the hygromycin (HYG) resistance marker gene by loss of heterozycocity as revealed by Southern blot analysis using the 5′-flank region of PIWI as a probe (Figure 4A). The integration of the HYG cassette into both LinPIWI alleles was confirmed by the presence of the ∼1 kb NcoI hybridizing band (Figure 4A upper panel and data not shown) and the absence of the 2.3 kb NcoI fragment corresponding to the wild type LinPIWI alleles (Figure 4A, bottom panel).
Figure 4. Genomic inactivation of the PIWI gene in L. infantum and L. major strains leads to a decrease in amastigote growth and disease pathology.
(A, upper panel) Strategy to inactivate the L. infantum PIWI gene (LinPIWI(−/−)) by genetic replacement. Both alleles of the LinPIWI single copy gene were replaced by the hygromycin phosphotransferase gene (HYG) by a loss of heterozygocity. (A, bottom panel) Southern blot hybridization of L. infantum genomic DNA digested with NcoI using the PIWI 5′ flank sequence as a probe. In LinPIWI(+/+), only a 2.3 kb band which corresponds to the wild type alleles was detected. In LinPIWI(+/−) clones (C1, C2 and C3), in addition to the wild type allele, one more band of 1.0 kb (for the HYG gene integration) was detected. In LinPIWI(−/−), only one band of 1.0 kb (for the HYG gene replacement) was detected but not the 2.3 kb band. (B) Growth curve of L. infantum axenic amastigotes for LinPIWI(+/+) and LinPIWI(−/−) independent clones 3, 5 and 6. The growth pattern of L. infantum WT and LinPIWI(−/−) clones on days 1, 3, 5 and 7 was analyzed by one-way ANOVA followed by a Tukey’s post-test using GraphPad Prism (version 3.03) software. Significant differences between the various groups are indicated (*, P<0.05; **, P<0.01; and ***, P<0.001). (C, left panel) Strategy to generate a PIWI null mutant in L. major (LmjPIWI(−/)). The LmjPIWI alleles were replaced by the HYG expression cassette by a single round of gene targeting. (C, right panel) Southern blot hybridization using the L. major PIWI 5′ flank sequences as a probe. The 17 kb band corresponds to the PIWI wild type alleles and the 1.0 kb band corresponds to the HYG gene integration into the PIWI genomic locus. (D) L. major infection rates in mice estimated by measuring the size of footpad lesions at different time points post-infection in BALB/c mice infected with either the wild type (LmjPIWI(+/+)) strain or the LmjPIWI(−/−) null mutant (clone 1). Control mice were sacrificed at 4 weeks post-infection due to the larger size of the lesions. Data shown here are the mean±SD of 6 mice per group and are representative of three independent experiments. The footpad thickness of L. infantum WT and LmjPIWI(−/−) null mutant on weeks 1, 2, 3 and 4 was analyzed by t-test (non parametric) using GraphPad Prism (version 3.03) software. Significant differences between WT and LmjPIWI(−/−) are indicated (**, P<0.01 and ***, P<0.001).
To evaluate if there is any change in the small RNA species population between the LinPIWI(−/−) mutant and wild type, we enriched for small RNAs (≤200 nt) (see Materials and Methods) that were labelled with [γ-32P]ATP and resolved on 15% urea acrylamide gel. This experiment revealed no differences between the LinPIWI(−/−) mutant and wild type in the pattern of small or microRNAs derived from either SIDER elements [31] and or other type of noncoding RNA species previously described in our laboratory [33] (Figure S2A and data not shown). It was previously reported that the mouse MIWI protein and piRNAs co-fractionate with polysomes, indicating a potential role of these components in translational control [34]. Therefore, small RNA species of the wild type and LinPIWI(−/−) mutant were enriched from RNPs, 40S and 60S subunits, 80S monosome and polysomal fractions following 15–45% sucrose gradient fractionation. As also seen with the total small RNA population, there were no notable differences in small RNA species associated with ribosomes between the wild type and LinPIWI(−/−) mutant (Figure S2B), hence suggesting that LinPIWI is most likely not involved in the biogenesis or the stability of small RNAs in Leishmania.
Mitochondrial localization of the Leishmania PIWI-like protein prompted us also to evaluate if PIWI through its association with small guide RNAs plays a role in site-specific edition of mitochondrial mRNAs, a posttranscriptional addition or deletion of uridine residues that takes place exclusively within the mitochondrion of trypanosomatid protozoa [35]. We therefore examined the cytochrome C oxidase subunit II (COXII) and cytochrome B (CYB) transcripts, known to be extensively edited in kinetoplastids [36]. The editing pattern of COXII (Figure S3A) and CYB (Figure S3B) transcripts in LinPIWI(−/−) was identical to that of wild type, suggesting that LinPIWI is not involved in RNA editing. Furthermore, we evaluated if LinPIWI plays any role in kinetoplastid DNA (kDNA) replication within the mitochondrion [37]. However, isolation of the kDNA from wild type and LinPIWI(−/−) strains showed no differences in kDNA pattern (Figure S4).
Genomic Depletion of the PIWI-like Protein Significantly Delays Leishmania Amastigote Growth and Disease Pathology in Mice
Next, we assessed the effect of PIWI depletion on parasite growth both in vitro and in vivo. The L. infantum PIWI(−/−) mutant did not show any growth defect when cultured as promastigotes (data not shown). However, growth of several independent clones lacking PIWI as axenic amastigotes was significantly delayed compared to the wild type (Figure 4B). To evaluate the role of PIWI in amastigote intracellular survival and disease development, we inactivated the PIWI gene in L. major (LmjPIWI(−/−)) (Figure 4C) as described above for L. infantum (Figure 4A). PCR analysis (data not shown) and Southern blot hybridization (Figure 4C) confirmed the inactivation of LmjPIWI gene, as determined by the absence of the ∼17 NcoI kb band corresponding to the wild type alleles and the detection of the ∼1 kb NcoI HYG integration band. An equal number of stationary-phase wild type (LmjPIWI(+/+)) and LmjPIWI(−/−) mutant (clone 1) were injected into the footpad of BALB/c mice and infection was monitored for up to 8 weeks by measuring the size of the footpad lesions. Three independent experiments confirmed a significant delay in lesion development in mice infected with LmjPIWI(−/−) in comparison to the wild type (LmjPIWI(+/+))-infected animals, even at 8 weeks post-infection (Figure 4D). Moreover, an independent experiment using a different LmjPIWI(−/−) clone (clone 3) showed a similar delay in lesion development than clone 1 (data not shown). All our attempts to complement the PIWI(−/−) null mutant phenotype failed as none of the episomally expressed PIWI epitope-tagged (e.g. HA, Myc, and GFP) proteins or even the original PIWI gene without any tag (Figure S5) transfected into LmjPIWI(−/−) or LinPIWI(−/−) mutant background could rescue efficiently for the loss of the endogenous PIWI protein (data not shown). The fact that we could only detect the PIWI-tagged proteins by western blot after enrichment or cell fractionation (Figure 3C and data not shown) suggests that the levels of the ectopically expressed PIWI protein might be too low or not properly regulated outside its genomic context. We cannot also exclude that the episomal vectors used for rescuing the mutant phenotype were lost in the absence of drug selection in mice. Nevertheless, it is important to highlight that we could observe a similar defect in the PIWI−/− mutant amastigote growth in two distinct Leishmania species and for different clones, which further supports the specificity of the role of PIWI protein in the amastigote growth of the parasite.
Loss of the L. infantum PIWI-like Protein Affects the Expression of Specific Subsets of Stage-regulated Transcripts
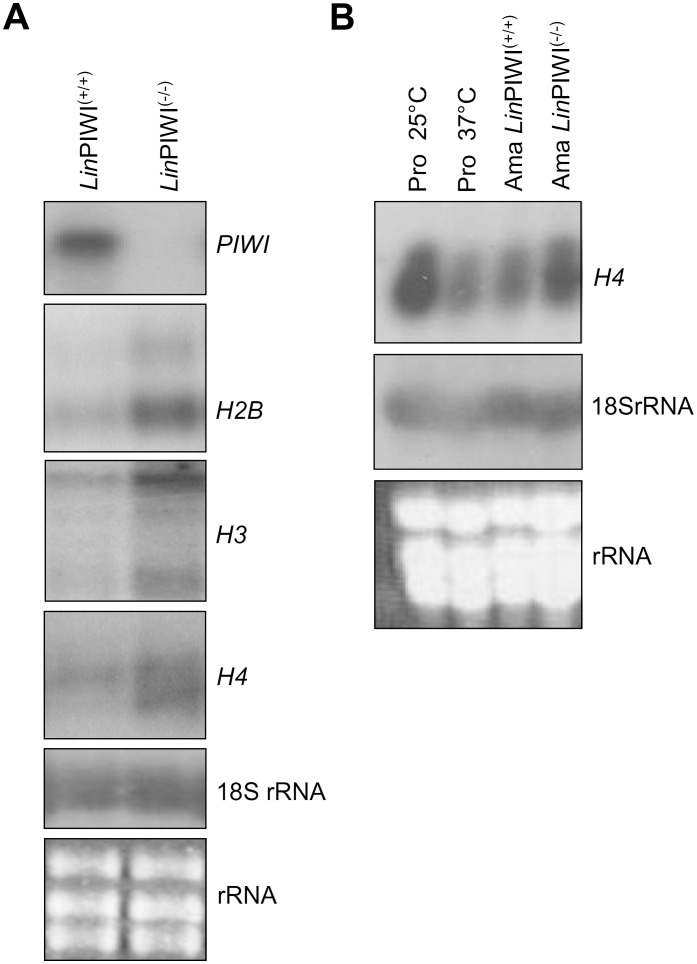
To determine whether PIWI depletion could affect expression levels of specific mRNA subsets, we performed a whole-genome DNA microarray analysis to compare gene expression profiles between L. infantum wild type (LinPIWI(+/+)) and LinPIWI(−/−) null mutant amastigotes. For this experiment, we used a previously described high-density 70-mer oligonucleotide Leishmania genome microarray [38]. After subtracting the background, the difference of 1.7-fold in the signal intensity between the experimental conditions used for a given gene was chosen as the cut-off, provided that the p value confidence was higher than 95%. The most significant differences in gene expression between L. infantum wild type and LinPIWI(−/−) null mutant are presented in Table 1. Several membrane-bound protein encoding transcripts seem to be downregulated in the LinPIWI(−/−) mutant, including the major surface protein GP63 leishmanolysin, the surface antigen-like protein, the membrane-bound acid phosphatase 2 and several transporters. The list of the downregulated genes includes also transcripts coding for protein kinases and hypothetical conserved proteins. Transcripts upregulated in the LinPIWI(−/−) mutant code mainly for histones. Histone H1, H2A, H2B, H3 and H4 transcripts were upregulated by 2-fold in average in the LinPIWI(−/−) mutant (Table 1). The accumulation of histone transcripts in LinPIWI(−/−) was further confirmed by northern blot analysis (Figure 5A). It was previously described [39], [40], [41], [42], [43] and shown also here that histone mRNAs in Leishmania are generally downregulated in the amastigote stage or when subjected to heat stress (Figure 5B).
Table 1. Genes differentially expressed between Leishmania infantum wild type and LinPIWI(−/−) axenic amastigotes as assessed by DNA microarray analysis.
| Accession number (TriTryp DB)/Biological processa | Gene description | Fold difference (Ax. Ama/Pro) | p value | Stage-specific gene expressionb |
| Downregulated genes | ||||
| Surface membrane proteins | ||||
| LinJ.04.0200 | surface antigen-like protein | 0.5824 | 4,32E−04 | Amastigote |
| LinJ.05.0900 | surface antigen-like protein | 0.5561 | 4,58E−04 | Amastigote |
| LinJ.36.2720 | membrane bound acid phosphatase 2, putative | 0.5375 | 2,59E−05 | Promastigote |
| LinJ.33.0310 | glucose transporter/membrane transporter D2, putative | 0.4638 | 4,20E−05 | Amastigote |
| LinJ.07.1340 | amino acid transporter, putative (AAT19) | 0.4731 | 1,22E−05 | Amastigotec |
| LinJ.10.0490, LinJ.10.0500, LinJ.10.0510, LinJ.10.0520, LinJ.10.0530 | GP63, leishmanolysin | 0.5641 | 4,20E−04 | Promastigote |
| LinJ.28.0600, LinJ.28.0610 | major surface protease GP63, putative, leishmanolysin, putative | 0.5409 | 2,59E−05 | Promastigote |
| LinJ.10.1450 | pteridine transporter, putative | 0.5859 | 2,87E−04 | Amastigote |
| LinJ.19.0870 | folate/biopterin transporter, putative | 0.4271 | 5,65E−05 | Promastigote |
| Lipid metabolic process | ||||
| LinJ.14.0710d, LinJ.14.0720d, LinJ.14.0730, LinJ.14.0740, LinJ.14.0760* | fatty acid elongase, putative | 0.3810 | 9,45E−06 | Amastigote |
| LinJ.23.1560 | lathosterol oxidase-like protein | 0.5687 | 1,50E−05 | Promastigote |
| Cell motility | ||||
| LinJ.16.0920, LinJ.16.0930 | flagellar calcium binding protein, putative | 0.5921 | 7,57E−04 | Amastigote |
| Cell communication | ||||
| LinJ.17.0130 | receptor-type adenylate cyclase, putative | 0.5439 | 4,32E−04 | Amastigote |
| Protein modification | ||||
| LinJ.08.0670* | protein kinase, putative | 0.5228 | 2,95E−05 | Amastigote |
| LinJ.04.1230 | protein kinase, putative, casein kinase I, putative | 0.5050 | 2,93E−04 | Amastigote |
| LinJ.36.2420 | protein kinase, putative, serine/threonine protein kinase, putative | 0.5683 | 2,48E−03 | Amastigote |
| LinJ.27.1680 | casein kinase I-like protein | 0.5586 | 8,69E−05 | Amastigote |
| Cell cycle | ||||
| LinJ.30.3690 | CYC2-like protein, putative | 0.5706 | 5,48E−04 | Amastigote |
| Unclassified | ||||
| LinJ.18.0080 | hypothetical protein, conserved | 0.4548 | 2,59E−05 | Amastigote |
| LinJ.18.0140* | hypothetical protein, conserved | 0.5865 | 9,24E−04 | Constitutive |
| LinJ.19.0560* | hypothetical protein, conserved | 0.4814 | 5,65E−05 | Promastigote |
| LinJ.19.0570* | hypothetical protein, conserved | 0.4814 | 5,65E−05 | Amastigote |
| LinJ.06.1360* | hypothetical protein, conserved | 0.4125 | 2,32E−04 | Amastigote |
| LinJ.10.0280* | hypothetical protein, conserved | 0.5471 | 4,25E−04 | Promastigote |
| LinJ.07.0850 | hypothetical protein, unknown function | 0.4690 | 1,69E−04 | Amastigote |
| LinJ.36.0630 | hypothetical protein, unknown function | 0.5452 | 6,11E−04 | Amastigotec |
| LinJ.23.0700* | hypothetical protein | 0.5694 | 1,78E−04 | Constitutive |
| LinJ.23.1190 | hypothetical protein, unknown function | 0.5340 | 3,33E−04 | Promastigote |
| LinJ.26.2710 | hypothetical protein, unknown function | 0.5522 | 8,69E−05 | Amastigote |
| LinJ.27.0960* | hypothetical protein, conserved | 0.5425 | 4,93E−04 | Promastigote |
| LinJ.29.2940 | hypothetical protein, conserved/RNA bndingprotein RBP6 | 0.3800 | 1,22E−05 | Promastigote |
| LinJ.31.0750* | hypothetical protein, conserved/Major facilitator superfamily | 0.4655 | 9,55E−05 | Amastigote |
| LinJ.36.4050* | similar to L. major 1411.4-like protein | 0.5921 | 2,93E−04 | Amastigote |
| LinJ.21.0470 | argonaute-like protein, putative, PIWI-like protein 1, putative | 0.3278 | 3,21E−03 | Amastigote |
| Upregulated genes | ||||
| Nucleosome assembly | ||||
| LinJ.27.1070 | histone H1, putative | 1.8331 | 2,97E−03 | Amastigote |
| LinJ.27.1120 | histone H1, putative | 1.8331 | 2,97E−03 | Amastigote |
| LinJ.21.1160, LinJ.21.1170 | histone H2A | 1.6954 | 9,88E−04 | Promastigote |
| LinJ.29.1850, LinJ.29.1860, LinJ.29.1870 | histone H2A, putative | 1.6954 | 9,88E−03 | Promastigote |
| LinJ.09.1410 | histone H2B | 2.2143 | 4,83E−04 | Promastigote |
| LinJ.17.1320 | histone H2B | 2.2143 | 4,83E−04 | Promastigote |
| LinJ.19.0030, LinJ.19.0040 | histone H2B | 2.2143 | 4,83E−04 | Promastigote |
| LinJ.10.0920 | histone H3 | 1.9454 | 6,30E−04 | Promastigote |
| LinJ.10.1050, LinJ.10.1070 | histone H3 | 1.9454 | 6,30E−04 | Promastigote |
| LinJ.16.0600, LinJ.16.0610 | histone H3, putative | 1.9454 | 6,30E−04 | Promastigote |
| LinJ.31.3320 | histone H4 | 2.2006 | 1,51E−04 | Promastigote |
| LinJ.15.0010 | histone H4 | 2.2006 | 1,51E−04 | Promastigote |
| LinJ.25.2560 | histone H4 | 2.2006 | 1,51E−04 | Promastigote |
| LinJ.06.0010 | histone H4 | 2.2006 | 1,51E−04 | Promastigote |
| LinJ.35.0020 | histone H4, putative, pseudogene | 2.2006 | 1,51E−04 | Promastigote |
| LinJ.35.1320 | histone H4 | 2.2006 | 1,51E−04 | Promastigote |
| LinJ.36.0020 | histone H4 | 2.2006 | 1,51E−04 | Promastigote |
| LinJ.21.0020 | histone H4 | 2.2006 | 1,51E−04 | Promastigote |
| GPI anchor biosynthetic process | ||||
| LinJ.12.0140* | Alg9-like mannosyltransferase, putative | 1.6925 | 1,51E−03 | Constitutive |
| Translation | ||||
| LinJ.26.0150, LinJ.26.0160 | 60S ribosomal protein L7, putative | 1.7545 | 1,85E−04 | Constitutive |
| Energy metabolism | ||||
| LinJ.26.0450* | ATPase subunit 9, putative | 1.8391 | 6,26E−03 | Constitutive |
| Unclassified | ||||
| LinJ.35.1470 | hypothetical protein, conserved | 1.9454 | 1,75E−04 | Constitutive |
| LinJ.06.0920 | hypothetical protein, conserved | 2.1498 | 1,43E−02 | Constitutive |
| LinJ.08.0440 | hypothetical protein, conserved | 2.5012 | 5,59E−03 | Amastigote |
| LinJ.16.0570 | hypothetical protein, conserved | 2.5899 | 2,67E−03 | Constitutive |
Gene functions are based on Gene Ontology (GO) annotation. The main categories are shown here.
Stage-specific gene expression of the modulated transcripts in the L. infantum LinPIWI(−/−) mutant is based on the data by [38], [44].
There is some discrepancy between the intracellular and axenic amastigote microarray data. These genes seem to be more expressed in promastigotes when comparing L. infantum intracellular amastigotes to promastigotes.
A probe recognizing LinJ.14.0710, LinJ.14.0720¸ LinJ.14.0730, LinJ.14.0740 and LinJ.14.0760 showed that theses genes were more expressed in the amastigote stage. Another probe recognizing LinJ.14.0710 and LinJ.14.0720 showed that theses genes were more expressed in the promastigote stage.
Genes encoding proteins with one to several transmembrane domains.
Figure 5. The histone mRNAs are upregulated in the L. infantum PIWI−/− null mutant.
(A) Northern blot hybridization of total RNA isolated from wild type LinPIWI(+/+) and LinPIWI(−/−) axenic amastigotes to evaluate changes in the accumulation of the histone H2B (LinJ.19.0030), H3 (LinJ.10.0920) and H4 (LinJ.21.0020) transcripts. Hybridization was carried out with radiolabeled PIWI ORF and histone ORF probes. There are two bands hybridizing with the H2B and H3 gene probes as reported previously [41], [43]. (B) Northern blot hybridization to confirm upregulation of the H4 transcript in LinPIWI(−/−) amastigotes or in heat-stressed promastigotes compared to unstressed parasites. The 18S RNA blot hybridization and EtBr-stained gels were used as loading controls.
Another intriging finding from the DNA microarray analysis is that the vast majority of the modulated transcripts in the LinPIWI(−/−) mutant has been reported previously to be differentially expressed in either lifestage of the parasite [44] (Table 1). Amongst the downregulated genes, the majority seems to be more expressed in amastigotes (24 amastigote-specific vs. 10 promastigote-specific) (Table 1). In the case of the upregulated genes, the large majority is more expressed in promastigotes (16 promastigote-specific vs. 3 amastigote-specific) (Table 1).
Leishmania Amastigotes Lacking PIWI Exhibit a Higher Sensitivity to Apoptosis-like Cell Death Inducing Agents
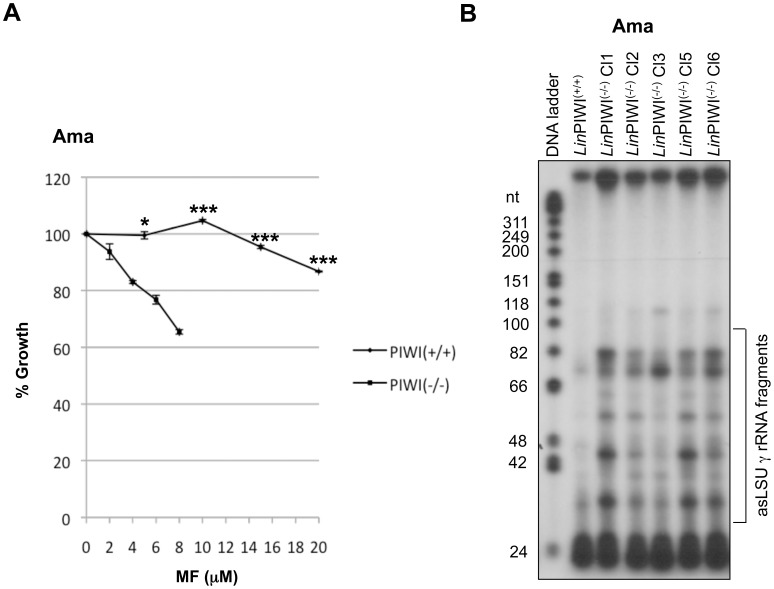
Leishmania undergoes various steps of apoptosis-like cell death following treatment with anti-leishmanial drugs such as trivalent antimony, miltefosine, and or exposure to H2O2 [45], [46], [47]. Since the Leishmania PIWI-like protein is mainly localized in the mitochondrion and mitochondria are known to be central for apoptosis-associated deregulation [48], we investigated whether sensitivity to apoptosis was altered in the LinPIWI(−/−) mutant. Indeed, exposure of the wild type and LinPIWI(−/−) mutant to increasing concentrations of miltefosine (MF) showed that parasites lacking PIWI were more sensitive to MF than the wild type cells (Figure 6A). We have reported recently that induction of apoptosis by MF triggers fragmentation of antisense (as) rRNA, which leads to rRNA degradation [49]. Since the LinPIWI(−/−) mutant is more sensitive to MF, we tested whether asrRNA fragmentation was increased in the PIWI null mutant as compared to the wild type cells. Primer extension analysis to specifically detect antisense RNA complementary to the large subunit gamma (LSU γ) rRNA, one of the six 28S rRNA processed fragments in Leishmania, revealed significantly higher fragmentation of the asLSU γ rRNA in the LinPIWI(−/−) mutant compared to the wild type (Figure 6B). Together, these results suggest a protective role of the Leishmania PIWI-like protein against apoptosis.
Figure 6. Leishmania amastigotes lacking PIWI are more sensitive to apoptosis-like cell death inducing agents.
(A) LinPIWI(−/−) was shown to be highly sensitive to miltefosine (MF). Equal number of L. infantum axenic amastigotes (Ama) of wild type and LinPIWI(−/−) mutant strains were treated with various concentrations of MF for a period of 8 hrs. Drug sensitivity was evaluated by measuring the OD at 600 nm in a 96 well plates. The results are expressed as mean ± SD of biological triplicates and the experiments were conducted twice with similar results. The comparison of growth percentage between L. infantum WT treated with 0, 5, 10, 15, 20 µM of MF and LinPIWI(−/−) treated with 0, 2, 4, 6 and 8 µM of MF, respectively (LinPIWI(−/−) is much more sensitive to MF than WT) was analyzed by t-test (non parametric) using GraphPad Prism (version 3.03) software. Significant differences between WT and LinPIWI(−/−) are indicated (*, P<0.05; ***, P<0.001). (B) Primer extension analysis of L. infantum wild type (LinPIWI(+/+)) and five independent clones (Cl 1–3, Cl 5 and Cl 6) of LinPIWI(−/−) null mutant grown as axenic amastigotes using a forward primer corresponding to nucleotides 101–118 of the sLSU γ rRNA to detect antisense (as) LSU γ rRNA fragmentation.
Discussion
Although Argonaute/PIWI proteins are conserved between species, they have undergone remarkable structural evolution and functional diversification. In this study, we show that unlike higher eukaryotes, Leishmania encodes a distinct homolog of AGO/PIWI-like proteins which possesses a PIWI domain harboring the conserved DDH catalytic triad but lacks the typical PAZ domain involved in the sequence-independent binding of ssRNA [4]. Thus, similarly to some prokaryotic AGO proteins [6], the N-terminal part of the Leishmania AGO/PIWI protein including PAZ domain was lost independently in several lineages. Interestingly, the recently characterized PIWI-like homolog from the related trypanosomatid T. cruzi exhibits a higher domain architecture similarity to the AGO/PIWI proteins of higher eukaryotes [23] than to the Leishmania PIWI-like protein. However, even, in T. cruzi, the PAZ domain is divergent from the canonical AGO proteins, suggesting that AGO/PIWI-like proteins in trypanosomatids devoided RNAi [22] may have other unknown functions unrelated to RNAi.
Functional analysis of the Leishmania PIWI-like protein revealed its important role in the disease-causing amastigote stage of the parasite. Genetically engineered L. infantum and L. major mutants lacking the PIWI gene demonstrated a significant growth defect as amastigotes both in vitro and in an experimental mouse system. These data are consistent with a higher expression of the PIWI transcript in amastigotes. The stage-specific expression of PIWI is well documented in higher organisms. In mammals, PIWI protein is specifically expressed in the testes of male during the germ cell development. In male mice, absence of PIWI homologs (MIWI, MILI and MIWI2) leads to the arrested spermatogenesis, subsequent apoptosis of the germ cells and eventual male sterility whereas in female, MIWI, MILI and MIWI2 knockout mice are fertile and produce fertile off springs [50], [51]. These piRNAs along with PIWI proteins are well known for their role in regulating transposon activity both in invertebrates (e.g. Drosophila and C. elegans) [52], [53] and in vertebrates (zebrafish) [54], thereby maintaining genomic integrity. The Leishmania genome is largely invaded by short interspersed degenerate retroposon elements (SIDERs), which are mainly located within 3′UTRs and intergenic regions and seem to play important roles in posttranscriptional regulation of gene expression [31], [32]. Therefore, we hypothesized that PIWI might be involved in the control of these widespread repetitive elements in the Leishmania genome. However, all our attempts to isolate small RNAs associated with PIWI derived either from the repetitive SIDER elements or from other type of noncoding RNA species identified previously in Leishmania [31], [33] failed, which excludes any major role of this protein in the biogenesis of small RNAs. This is in agreement with the absence of RNAi in Old World Leishmania species [17].
Here, we show that the Leishmania PIWI-like protein plays a role in the regulation of specific subsets of developmentally regulated transcripts in the two lifestages of the parasite. Indeed, most of the downregulated genes in the LinPIWI(−/−) mutant are preferentially expressed in amastigotes whereas the vast majority of the upregulated genes are differentially expressed in promastigotes. This suggests that LinPIWI is involved either directly or indirectly in the stabilization of subsets of amastigote-specific transcripts and destabilization of promastigote-specific transcript subsets. In Drosophila, it has been shown that the piRNA pathway is involved in the decay of maternal messenger RNAs and in translational repression [55]. How does LinPIWI protein modulate the expression of these specific subsets of mRNAs is not yet understood. It is really intriguing that many of the downregulated genes in the LinPIWI(−/−) mutant code for membrane-bound proteins and that the majority of the upregulated genes encode histones. Amongst the downregulated transcripts encoding surface or membrane-bound proteins some have been reported to contribute to parasite virulence. For example, the major surface protease GP63 plays a key role in the interaction of the parasite with its mammalian host and its survival within macrophages [56]. Glucose transporter deficient Leishmania exhibit higher sensitivity to oxidative stress and are not viable in the disease-causing amastigote stage of the parasite [57], [58]. Amino acid transporters have also been found to be important for intracellular amastigote growth [59] or to improve virulence [60]. Differences in the expression of surface proteins or essential transporters may impact on the parasite’s intracellular survival as corroborated by the decreased infectivity of the PIWI(−/−) mutant in mice. In contrast to transcripts encoding surface or membrane-bound proteins, the histone mRNAs were unregulated by ∼2-fold in the amastigote stage of the L. infantum PIWI (−/−) mutant. In Leishmania, H2A, H2B, H3 and H4 transcripts are generally expressed at higher levels in promastigotes than in amastigotes [39], [40], [41], [42], [43]. As histones are an integral part of nucleosomes, the basic unit of chromatin, it is possible that PIWI plays a role in epigenetic control in Leishmania, similarly to what has been reported in higher eukaryotes [5]. Indeed, a positive epigenetic role of PIWI along with its associated piRNAs has been attributed to the heterochromatic activation through histone modifications, although PIWI has a global function in silencing of heterochromatin [61]. To the best of our knowledge this is the first time that an inverse relationship between an Argonaute/PIWI protein and regulation of histone mRNAs is reported. It is interesting to mention that Leishmania amastigotes express higher levels of the PIWI transcript and lower levels of histone transcripts whereas promastigotes express low levels of PIWI transcript but higher levels of histones, suggesting that PIWI directly or indirectly negatively regulates stability of histone mRNAs.
Unlike other eukaryotes where PIWI proteins localize in the nucleus as epigenetic regulators and regulators of transposon activity [5], [11], [13], [28], [54], [62] or in the cytoplasm as regulators of RNA stability and translation [30], [34], [63], [64], the Leishmania PIWI-like protein is mainly localized in the single mitochondrion of the parasite. Targeting of the LinPIWI-like protein into the mitochondrion occurs most likely via its N-terminal signal targeting peptide. In contrast to Leishmania, the T. cruzi PIWI protein (TcPIWI) was shown to reside solely in the cytoplasm [23]. This difference may be attributed to the poor conservation of the N-terminal amino acid sequence of TcPIWI compared to LinPIWI. The enrichment of LinPIWI protein in the membrane-bound protein fraction and the presence of two predicted short transmembrane helices within the PIWI protein suggest localization in the mitochondrial outer membrane. This brings up the interesting possibility that LinPIWI might be exposed to the cytoplasmic side of the outer mitochondrial membrane, thus allowing it to interact with cytosolic proteins and to contribute to the stability or translation of specific subsets of mRNAs as shown by DNA microarray experiments. Despite its mitochondrial localization, the Leishmania PIWI-like protein does not seem to be involved in RNA editing [35] or in kinetoplast DNA (kDNA) replication [37], two processes that take place exclusively in the mitochondrion of these parasitic protozoa. Thus, the mitochondrial function of PIWI in Leishmania remains elusive.
Given that the Leishmania PIWI-like protein preferentially localizes into the mitochondrion and that mitochondria play a key role in cell viability, we also addressed the putative role of PIWI in apoptosis-like programmed cell death, which can be induced by various leishmanicidal agents. In higher organisms, PIWI is known to be involved in apoptosis. For example, in zebrafish and mice the loss of PIWI homologs triggers apoptosis in the germ cells [28], [54]. High levels of the PIWI family protein PIWI2 have been detected in various tumors where it acts like an oncogene that activates various signal transduction cascades that prevent apoptosis [65]. Thus, PIWI protein is guarding tumor cells from apoptosis. Interestingly, the LinPIWI(−/−) null mutant exhibits high sensitivity to the apoptosis inducing agent miltefosine. It is possible that in the absence of PIWI the mitochondrial membrane may be destabilized, rendering hence the parasite more prone to apoptosis inducing agents. Overall, our work demonstrates that the Leishmania PIWI-like protein homolog exhibits novel features that are distinct from other well-characterized members of the AGO/PIWI protein family in higher eukaryotes. Future studies will shed more light into the putative function of this divergent PIWI homolog on RNA regulation and amastigote development.
Materials and Methods
Leishmania Culture and Mouse Infection
L. infantum MHOM/MA/67/ITMAP-263 and L. major LV39 MRHO/SU/59/P strains and have been described previously [38]. Promastigotes were cultured at 25°C in SDM-79 medium pH 7.0 supplemented with 10% heat-inactivated FCS (Wisent) and 5 mg/ml of hemin. To differentiate L. infantum promastigotes into axenic amastigotes, late-stationary promastigotes were inoculated in MAA-20 medium supplemented with 20% FCS and grown at pH 5.6 and 37°C in a 5% CO2 atmosphere, as reported previously [66]. Fully differentiated amastigotes remained stable in culture and were used after 2–3 passages. For the in vivo studies, 1×107 L. major wild type and L. major PIWI(−/−) stationary promastigotes were used to infect female BALB/c mice subcutaneously in the footpad and cutaneous lesions were followed for up to 6–8 weeks post-infection. The infection protocol in mice has been approved by the animal protection committees of the CHUQ Research Centre (CPA-CHUQ) and Laval University (CPAUL) (authorization number: 2009148-3). L. infantum axenic amastigotes were subjected to various concentrations of miltefosine (Cayman Chemical) (0–20 µM for L. infantum wild type axenic amastigotes and 0–10 µM for the LinPIWI null mutant).
Plasmid Constructs and Targeting Cassettes for Gene Replacement and Transfections
The vector pSPBT1YNEOα-HALinPIWIHA was constructed as follows. The YNEOα fragment where Y is a 92 bp polypyrimidine stretch [67], NEO the neomycin phosphotransferase gene for resistance to G418, and α the intergenic region of the L. enriettii alpha-tubulin gene was amplified from vector pSPYNEOαLUC and inserted into NotI site of pSPBT1 [68]. The LinHA-PIWI-HA was amplified from L. infantum genomic DNA using the primers specified in Table S1 and cloned into the EcoRV site of pSPBT1YNEOα. To generate vector pGEMαNEOα-LmjPIWI-GFP, the L. major PIWI gene was amplified (see primers in Table S1) and cloned into the HindIII site of pGEMαNEOα-GFP vector. The LinN52PIWI-GFP construct was made as follows. A 156 bp fragment corresponding to the first 52 amino acids of the L. infantum PIWI protein was amplified from L. infantum genomic DNA and fused with GFP (primers are described in Table S1). The fused LinN52PIWI-GFP product was cloned into BamHI-XbaI sites of pSP72αNEOα. The purified plasmids were transfected into L. infantum promastigotes as described previously [68] and episomal expression of LinPIWI-GFP and LinN52PIWI-GFP recombinant strains were monitored by fluorescence. A targeted gene replacement strategy was used to inactivate the L. infantum (Lin) PIWI (LinJ.21.0470, http://tritrypdb.org) and L. major LmjPIWI (LmjF.21.0410, http://tritrypdb.org) genes. The 5′ and 3′ flanking regions of the PIWI gene (amplified from L. infantum and L. major genomic DNA independently) were fused to the hygromycin phosphotransferase (HYG) gene using a PCR fusion-based strategy. The 5′ flank, and 3′ flank regions were amplified from both L. infantum and L. major using primers described in Table S1. The HYG gene was amplified from the pSP72αHYGα Leishmania expression vector. The 5′-flank, HYG gene and 3′-flank regions were fused in a single PCR reaction using Phusion® High-Fidelity DNA Polymerase (NEB) and 5′ flank forward and 3′-flank reverse primers. The reverse primer of the 5′ flank region (5′ flank-reverse) has a tail that is complementary to the HYG forward primer. Similarly, the HYG reverse primer has a tail, which is complementary to the 3′ flank forward primer (Table S1). The resulting HYG cassettes corresponding to the L. infantum and L. major targeting constructs were transfected independently into both species. The two alleles of LinPIWI and LmjPIWI single copy genes were replaced by the HYG expression cassette following a single round of transfection by loss of heterozygocity using higher concentrations (200–600 µg/ml) of Hygromycin B (Sigma) as described previously [69].
DNA, RNA and Protein Blots
Genomic DNA and total RNA were isolated from L. major and L. infantum strains using the DNAzol and TRIzol reagents (Invitrogen), respectively following the manufacturer’s instructions. Southern and northern blot hybridizations were performed following standard procedures [70]. Double-stranded DNA probes were radiolabeled with [α-32P] dCTP using random oligonucleotides and Klenow fragment DNA polymerase I [(New England Biolabs). Enrichment of small (≤200 nt) RNA fraction was carried out using the mirVana™ miRNA Isolation Kit (Ambion) according the manufacturer’s procedure. The enriched small RNAs were labelled with [γ-32P]ATP and polynucleotide kinase (NEB) and resolved on a 15% Urea-polyacrylamide gel (National Diagnostics). The digitonin-fractionated proteins (see below) were resolved on SDS-PAGE and western blots were performed with different antibodies following standard procedures [70].
Primer Extension Analysis
Primer extension was performed with the SuperScript™ III RT kit (Invitrogen) as described previously [49]. The forward primer corresponding to nucleotides 101–118 of the sense LSU γ rRNA (see Table S1) was used to detect antisense (as) LSU γ rRNA fragmentation. Total RNA was isolated from L. infantum wild type and LinPIWI (−/−) mutant axenic amastigotes and used for reverse transcriptase reactions with [γ-32P] ATP-labeled forward primer to detect asLSU γ RNA cleavage products. Radiolabeled cDNA products were resolved on 10% Urea acrylamide gel (Sequagel, National Diagnostics) and visualized by autoradiography. A ΦX174 DNA/HinfI dephosphorylated DNA marker (Promega) was labeled with [γ-32P]ATP and PNK (New England Biolabs) according to the manufacturer’s recommendations.
DNA Microarray Analysis
Probes for DNA microarray hybridizations were prepared with 10 µg of total RNA for each condition. Purified cDNA from L. infantum axenic amastigotes, array pre-hybridization, hybridization and washings were carried out as described previously [38]. Four biological replicates of all hybridizations were performed to account for sample heterogeneity, variation between slides and variations due to hybridization. To prevent bias by preferential label incorporation into particular sequences, Alexa 555 and Alexa 647 dyes were swapped between the two RNA preparations. The fluorescence signal intensities of the slides hybridized with L. infantum wild type and LinPIWI−/− amastigote RNA were measured using the Perkin Elmer ScanArray Express Scanner. GenePix Pro 6.0 image analysis software (Axon Instruments, Union City, California, USA) was employed to measure the fluorescence signal intensities of the array features and local background. Data correction and normalization and statistical analyses were carried out as described [38]. Only genes statistically significant with an absolute log2 ratio greater than 0.75 were considered as differentially expressed. Gene ontology annotation was analyzed using the AmiGO website [71].
Protein Immunolocalization Studies
For immunolocalization studies of LinN52PIWI-GFP, 107 L. infantum promastigotes and axenic amastigotes were labeled with 20 nM MitoTracker Red (Invitrogen), a dye to stain mitochondria in live cells, in DMSO and incubated for 30 min. For LinPIWI-GFP, fluorescence images were acquired through a 60×1.4 NA objective (PlanApo, Olympus) and captured at serial optical sections at 1 µm intervals by an Olympus FLUOWVIEW FV300 laser scanning confocal microscope with a 488 nm Argon-ion laser for the green channel and a 543 nm He-Ne laser for the red channel at a resolution 1024×1024 pixel. Images were produced in Fluoview software (FV300 version 5). For immunolocalization studies of the mitochondrial HSP70 protein, the parasites were fixed with 2% paraformaldehyde, permeabilized with Triton X100 (Sigma) and reacted with an anti-rabbit HSP70 antibody as a first antibody (kindly provided by Dr Osvaldo de Melo Neto, Centro de Pesquisas Aggeu Magalhães, Fiocruz, Recife, Brazil; [72]) and an anti-rabbit Alexa 594 as the second antibody. The cells were washed twice with HEPES-NaCl (500 µl) and observed under a Nikon epifluorescence microscope.
Digitonin Fractionation
Exponentially growing L. infantum transfected with the LinHA-PIWI-HA construct were harvested and washed twice in 1XPBS buffer and subjected to digitonin fractionation as described previously [73]. Four fractions were obtained using increasing concentrations of digitonin (20 µM, 200 µM, 1 mM and 10 mM). Proteins were precipitated with equal volume of acetone at −20°C for 1 h and re-solubilized in 1X Laemmli buffer and separated by SDS-PAGE. Western blots were performed with antibodies against the HA epitope (1∶2500), the cytoplasmic (1/1000) HSP70 and the mitochondrial HSP70 (1/1000) proteins.
Supporting Information
The N-terminal signal peptide sequence of the Leishmania PIWI-like protein targets the PIWI protein to the mitochondrion. (A) Schematic representation of the LinN52PIWI-GFP construct used for subcellular localization studies in L. infantum promastigotes. The LinN52PIWI-GFP construct was made by fusing the N-terminal 52 amino acids of LinPIWI (harboring the mitochondrial signal peptide sequence as predicted by SignalP 4.0, TargetP 1.1, PSORT and SOSUIsignal) to the GFP protein and then cloned into pGEM-αNEOα-GFP vector and transfected into L. infantum promastigotes. (B) Immunolocalization studies of LinN52PIWI-GFP in L. infantum promastigotes (Pro) and amastigotes (Ama) using a Nikon epifluorescence microscope. Mitotracker (red) was used for defining the Leishmania single mitochondrion. The green fluorescence signal (GFP) was largely co-localized with that of the Mitotracker (red).
(TIF)
Small RNA profile in Leishmania wild type and the PIWI(−/−) null mutant. (A) Small RNAs (<200 nt) from L. infantum wild type (LinWT) and LinPIWI−/− null mutant were isolated using the mirVanaTM Kit (Ambion), 5′-labelled by polynucleotide kinase and loaded on denaturing 15% SDS-PAGE gel. More RNA fragments are seen in amastigotes (A) (this could also be due to degradation products) than in promastigotes (P) but there is no significant accumulation or decrease of small RNA species in the LinPIWI(−/−) mutant in comparison to wild type cells. The marker (M) corresponds to an end-labeled 20 nt DNA oligonucleotide. (B) Small RNAs isolated as in A and enriched by 15–45% sucrose gradient analysis to look for small RNAs associated or not with ribosomes. No significant differences in small RNA species between wild type and the LinPIWI(−/−) mutant were observed.
(TIF)
RNA editing is fully operational in the L. infantum PIWI(−/−) null mutant. The cDNAs were generated from total RNA of wild type and LinPIWI(−/−) axenic amastigotes by random hexamers. The cDNAs were used for PCR amplification for cytochrome C oxidase subunit II (COXII) (A, upper panel) using specific primers and sequenced. The edited sequence of COXII in which four uridines were added is indicated by red arrows (A, lower panel). The PCR fragment amplified from unedited mitochondrial DNA sequence was shown as control for this experiment (B, upper panel) cDNA PCR amplification for the cytochrome b (CYB) gene using specific primers. (B, lower panel) Nucleotide sequence of the CYB gene with the edited uracils underlined in red. There is no difference in the editing pattern of COXII (A) and CYB (B) transcripts between L. infantum wild type and LinPIWI(−/−) mutant. The experiment was repeated twice with similar results. The PCR framgment amplified from unedited mitochondrial DNA sequence was shown as control.
(TIF)
Kinetoplastid DNA pattern between the L. infantum wild type and Lin PIWI(−/−) null mutant. Ethidium bromide-stained gel of circular kinetoplastid DNA (kDNA) from both L. infantum wild type (LinPIWI(+/+)) and LinPIWI(−/−) null mutant. The circular kinetoplastid DNA was isolated by plasmid mini-preparation columns (Qiagen), digested with HindIII and EcoRI enzymes and run on a 1% agarose gel. No differences were observed in kDNA between the L. infantum wild type and LinPIWI(−/−) null mutant.
(TIF)
Expression vectors to rescue the Lin PIWI(−/−) null mutant. Schematic representation of the LinPIWI gene fused or not at either extremity with known epitope tag sequences (e.g. HA, MYC and GST) and cloned into the BamHI and XbaI sites of vector pSP72αNEOα (see Materials and Methods). These constructs were transfected into the L. infantum LinPIWI(−/−)strain to rescue the null mutant phenotype.
(TIF)
Primers used in this study.
(DOC)
Acknowledgments
We are thankful to Dr Osvaldo de Melo Neto for providing us with an antibody against the mitochondrial HSP70 protein. PP was a recipient of a postdoctoral fellowship from the Canadian Institutes of Health Research (CIHR) Strategic Training Program STP-53924 and the Centre for Host-Parasite Interactions (CHPI). M.J.S. is a Fonds de Recherche du Québec-Santé Junior 2 Scholar supported by CIHR. BP is a member of the Centre for Host-Parasite Interactions ‘Programme Regroupements Stratégiques’ of the Fonds de Québec pour la Recherche sur la Nature et les Technologies.
Funding Statement
This work was supported by CIHR operating grant MOP-12182 awarded to BP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hock J, Meister G (2008) The Argonaute protein family. Genome Biol 9: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524. [DOI] [PubMed] [Google Scholar]
- 3. Chung WJ, Okamura K, Martin R, Lai EC (2008) Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol 18: 795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32. [DOI] [PubMed] [Google Scholar]
- 5. Thomson T, Lin H (2009) The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 25: 355–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Makarova KS, Wolf YI, van der Oost J, Koonin EV (2009) Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct 4: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lingel A, Simon B, Izaurralde E, Sattler M (2003) Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465–469. [DOI] [PubMed] [Google Scholar]
- 8. Parker JS, Barford D (2006) Argonaute: A scaffold for the function of short regulatory RNAs. Trends Biochem Sci 31: 622–630. [DOI] [PubMed] [Google Scholar]
- 9. Girard A, Hannon GJ (2008) Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol 18: 136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, et al. (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. [DOI] [PubMed] [Google Scholar]
- 11. Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, et al. (2008) An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aravin AA, Hannon GJ, Brennecke J (2007) The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764. [DOI] [PubMed] [Google Scholar]
- 13. Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, et al. (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, et al. (2007) Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev 21: 2300–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA (2002) Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110: 689–699. [DOI] [PubMed] [Google Scholar]
- 16. Haile S, Papadopoulou B (2007) Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr Opin Microbiol 10: 569–577. [DOI] [PubMed] [Google Scholar]
- 17. Robinson KA, Beverley SM (2003) Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol 128: 217–228. [DOI] [PubMed] [Google Scholar]
- 18. DaRocha WD, Otsu K, Teixeira SM, Donelson JE (2004) Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol Biochem Parasitol 133: 175–186. [DOI] [PubMed] [Google Scholar]
- 19. Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, et al. (2005) The genome of the kinetoplastid parasite, Leishmania major. Science 309: 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, et al. (2007) Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lye LF, Owens K, Shi H, Murta SM, Vieira AC, et al. (2010) Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog 6: e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kolev NG, Tschudi C, Ullu E (2011) RNA interference in protozoan parasites: achievements and challenges. Eukaryot Cell 10: 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garcia Silva MR, Tosar JP, Frugier M, Pantano S, Bonilla B, et al. (2010) Cloning, characterization and subcellular localization of a Trypanosoma cruzi argonaute protein defining a new subfamily distinctive of trypanosomatids. Gene 466: 26–35. [DOI] [PubMed] [Google Scholar]
- 24. Wei KF, Wu LJ, Chen J, Chen YF, Xie DX (2012) Structural evolution and functional diversification analyses of argonaute protein. J Cell Biochem 113: 2576–2585. [DOI] [PubMed] [Google Scholar]
- 25. Tolia NH, Joshua-Tor L (2007) Slicer and the argonautes. Nat Chem Biol 3: 36–43. [DOI] [PubMed] [Google Scholar]
- 26. Parker JS, Roe SM, Barford D (2004) Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J 23: 4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nowotny M, Gaidamakov SA, Crouch RJ, Yang W (2005) Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121: 1005–1016. [DOI] [PubMed] [Google Scholar]
- 28. Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, et al. (2004) Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849. [DOI] [PubMed] [Google Scholar]
- 29. Cox DN, Chao A, Lin H (2000) piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514. [DOI] [PubMed] [Google Scholar]
- 30. Ohrt T, Muetze J, Svoboda P, Schwille P (2012) Intracellular localization and routing of miRNA and RNAi pathway components. Curr Top Med Chem 12: 79–88. [DOI] [PubMed] [Google Scholar]
- 31. Bringaud F, Muller M, Cerqueira GC, Smith M, Rochette A, et al. (2007) Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog 3: 1291–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muller M, Padmanabhan PK, Rochette A, Mukherjee D, Smith M, et al. (2010) Rapid decay of unstable Leishmania mRNAs bearing a conserved retroposon signature 3′-UTR motif is initiated by a site-specific endonucleolytic cleavage without prior deadenylation. Nucleic Acids Res 38: 5867–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dumas C, Chow C, Muller M, Papadopoulou B (2006) A novel class of developmentally regulated noncoding RNAs in Leishmania. Eukaryot Cell 5: 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grivna ST, Pyhtila B, Lin H (2006) MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A 103: 13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hajduk SL, Harris ME, Pollard VW (1993) RNA editing in kinetoplastid mitochondria. Faseb J 7: 54–63. [DOI] [PubMed] [Google Scholar]
- 36. Shaw JM, Campbell D, Simpson L (1989) Internal frameshifts within the mitochondrial genes for cytochrome oxidase subunit II and maxicircle unidentified reading frame 3 of Leishmania tarentolae are corrected by RNA editing: evidence for translation of the edited cytochrome oxidase subunit II mRNA. Proc Natl Acad Sci U S A 86: 6220–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT (2005) Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol 21: 363–369. [DOI] [PubMed] [Google Scholar]
- 38. Rochette A, Raymond F, Ubeda JM, Smith M, Messier N, et al. (2008) Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics 9: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Genske JE, Cairns BR, Stack SP, Landfear SM (1991) Structure and regulation of histone H2B mRNAs from Leishmania enriettii. Mol Cell Biol 11: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Noll TM, Desponds C, Belli SI, Glaser TA, Fasel NJ (1997) Histone H1 expression varies during the Leishmania major life cycle. Mol Biochem Parasitol 84: 215–227. [DOI] [PubMed] [Google Scholar]
- 41. Soto M, Requena JM, Quijada L, Alonso C (1996) Organization, transcription and regulation of the Leishmania infantum histone H3 genes. Biochem J 318 (Pt 3): 813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Soto M, Quijada L, Alonso C, Requena JM (1997) Molecular cloning and analysis of expression of the Leishmania infantum histone H4 genes. Mol Biochem Parasitol 90: 439–447. [DOI] [PubMed] [Google Scholar]
- 43. Soto M, Quijada L, Larreta R, Iborra S, Alonso C, et al. (2003) Leishmania infantum possesses a complex family of histone H2A genes: structural characterization and analysis of expression. Parasitology 127: 95–105. [DOI] [PubMed] [Google Scholar]
- 44. Rochette A, Raymond F, Corbeil J, Ouellette M, Papadopoulou B (2009) Whole-genome comparative RNA expression profiling of axenic and intracellular amastigote forms of Leishmania infantum. Mol Biochem Parasitol 165: 32–47. [DOI] [PubMed] [Google Scholar]
- 45. Sereno D, Holzmuller P, Mangot I, Cuny G, Ouaissi A, et al. (2001) Antimonial-mediated DNA fragmentation in Leishmania infantum amastigotes. Antimicrob Agents Chemother 45: 2064–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paris C, Loiseau PM, Bories C, Breard J (2004) Miltefosine induces apoptosis-like death in Leishmania donovani promastigotes. Antimicrob Agents Chemother 48: 852–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Das M, Mukherjee SB, Shaha C (2001) Hydrogen peroxide induces apoptosis-like death in Leishmania donovani promastigotes. J Cell Sci 114: 2461–2469. [DOI] [PubMed] [Google Scholar]
- 48. Zamzami N, Marchetti P, Castedo M, Decaudin D, Macho A, et al. (1995) Sequential reduction of mitochondrial transmembrane potential and generation of reactive oxygen species in early programmed cell death. J Exp Med 182: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Padmanabhan PK, Samant M, Cloutier S, Simard MJ, Papadopoulou B (2012) Apoptosis-like programmed cell death induces antisense ribosomal RNA (rRNA) fragmentation and rRNA degradation in Leishmania. Cell Death Differ 19: 1972–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, et al. (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514. [DOI] [PubMed] [Google Scholar]
- 51. Deng W, Lin H (2002) miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830. [DOI] [PubMed] [Google Scholar]
- 52.Simonelig M (2011) Developmental functions of piRNAs and transposable elements: A Drosophila point-of-view. RNA Biol 8. [DOI] [PMC free article] [PubMed]
- 53. Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, et al. (2012) Function, Targets, and Evolution of Caenorhabditis elegans piRNAs. Science 337: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, et al. (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82. [DOI] [PubMed] [Google Scholar]
- 55. Rouget C, Papin C, Boureux A, Meunier AC, Franco B, et al. (2010) Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467: 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olivier M, Atayde VD, Isnard A, Hassani K, Shio MT (2012) Leishmania virulence factors: Focus on the metalloprotease GP63. Microbes Infect. [DOI] [PubMed]
- 57. Rodriguez-Contreras D, Landfear SM (2006) Metabolic changes in glucose transporter-deficient Leishmania mexicana and parasite virulence. J Biol Chem 281: 20068–20076. [DOI] [PubMed] [Google Scholar]
- 58. Rodriguez-Contreras D, Feng X, Keeney KM, Bouwer HG, Landfear SM (2007) Phenotypic characterization of a glucose transporter null mutant in Leishmania mexicana. Mol Biochem Parasitol 153: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wanasen N, MacLeod CL, Ellies LG, Soong L (2007) L-arginine and cationic amino acid transporter 2B regulate growth and survival of Leishmania amazonensis amastigotes in macrophages. Infect Immun 75: 2802–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Darlyuk I, Goldman A, Roberts SC, Ullman B, Rentsch D, et al. (2009) Arginine homeostasis and transport in the human pathogen Leishmania donovani. J Biol Chem 284: 19800–19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yin H, Lin H (2007) An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450: 304–308. [DOI] [PubMed] [Google Scholar]
- 62. Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- 63. Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, et al. (2009) Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lim AK, Tao L, Kai T (2009) piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol 186: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lee JH, Schutte D, Wulf G, Fuzesi L, Radzun HJ, et al. (2006) Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet 15: 201–211. [DOI] [PubMed] [Google Scholar]
- 66. Sereno D, Roy G, Lemesre JL, Papadopoulou B, Ouellette M (2001) DNA transformation of Leishmania infantum axenic amastigotes and their use in drug screening. Antimicrob Agents Chemother 45: 1168–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Papadopoulou B, Roy G, Ouellette M (1994) Autonomous replication of bacterial DNA plasmid oligomers in Leishmania. Mol Biochem Parasitol 65: 39–49. [DOI] [PubMed] [Google Scholar]
- 68. McNicoll F, Muller M, Cloutier S, Boilard N, Rochette A, et al. (2005) Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J Biol Chem 280: 35238–35246. [DOI] [PubMed] [Google Scholar]
- 69. Gueiros-Filho FJ, Beverley SM (1996) Selection against the dihydrofolate reductase-thymidylate synthase (DHFR-TS) locus as a probe of genetic alterations in Leishmania major. Mol Cell Biol 16: 5655–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sambrook J, Russell D (2001) Molecular Cloning: A laboratory manual,. NEW YORK: Cold Spring Harbor Laboratory Press,.
- 71. Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, et al. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32: D258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Campos RM, Nascimento M, Ferraz JC, Pereira MM, Rocha PO, et al. (2008) Distinct mitochondrial HSP70 homologues conserved in various Leishmania species suggest novel biological functions. Mol Biochem Parasitol 160: 157–162. [DOI] [PubMed] [Google Scholar]
- 73. Foucher AL, Papadopoulou B, Ouellette M (2006) Prefractionation by digitonin extraction increases representation of the cytosolic and intracellular proteome of Leishmania infantum. J Proteome Res 5: 1741–1750. [DOI] [PubMed] [Google Scholar]
- 74. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series Vol. 41: 95–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The N-terminal signal peptide sequence of the Leishmania PIWI-like protein targets the PIWI protein to the mitochondrion. (A) Schematic representation of the LinN52PIWI-GFP construct used for subcellular localization studies in L. infantum promastigotes. The LinN52PIWI-GFP construct was made by fusing the N-terminal 52 amino acids of LinPIWI (harboring the mitochondrial signal peptide sequence as predicted by SignalP 4.0, TargetP 1.1, PSORT and SOSUIsignal) to the GFP protein and then cloned into pGEM-αNEOα-GFP vector and transfected into L. infantum promastigotes. (B) Immunolocalization studies of LinN52PIWI-GFP in L. infantum promastigotes (Pro) and amastigotes (Ama) using a Nikon epifluorescence microscope. Mitotracker (red) was used for defining the Leishmania single mitochondrion. The green fluorescence signal (GFP) was largely co-localized with that of the Mitotracker (red).
(TIF)
Small RNA profile in Leishmania wild type and the PIWI(−/−) null mutant. (A) Small RNAs (<200 nt) from L. infantum wild type (LinWT) and LinPIWI−/− null mutant were isolated using the mirVanaTM Kit (Ambion), 5′-labelled by polynucleotide kinase and loaded on denaturing 15% SDS-PAGE gel. More RNA fragments are seen in amastigotes (A) (this could also be due to degradation products) than in promastigotes (P) but there is no significant accumulation or decrease of small RNA species in the LinPIWI(−/−) mutant in comparison to wild type cells. The marker (M) corresponds to an end-labeled 20 nt DNA oligonucleotide. (B) Small RNAs isolated as in A and enriched by 15–45% sucrose gradient analysis to look for small RNAs associated or not with ribosomes. No significant differences in small RNA species between wild type and the LinPIWI(−/−) mutant were observed.
(TIF)
RNA editing is fully operational in the L. infantum PIWI(−/−) null mutant. The cDNAs were generated from total RNA of wild type and LinPIWI(−/−) axenic amastigotes by random hexamers. The cDNAs were used for PCR amplification for cytochrome C oxidase subunit II (COXII) (A, upper panel) using specific primers and sequenced. The edited sequence of COXII in which four uridines were added is indicated by red arrows (A, lower panel). The PCR fragment amplified from unedited mitochondrial DNA sequence was shown as control for this experiment (B, upper panel) cDNA PCR amplification for the cytochrome b (CYB) gene using specific primers. (B, lower panel) Nucleotide sequence of the CYB gene with the edited uracils underlined in red. There is no difference in the editing pattern of COXII (A) and CYB (B) transcripts between L. infantum wild type and LinPIWI(−/−) mutant. The experiment was repeated twice with similar results. The PCR framgment amplified from unedited mitochondrial DNA sequence was shown as control.
(TIF)
Kinetoplastid DNA pattern between the L. infantum wild type and Lin PIWI(−/−) null mutant. Ethidium bromide-stained gel of circular kinetoplastid DNA (kDNA) from both L. infantum wild type (LinPIWI(+/+)) and LinPIWI(−/−) null mutant. The circular kinetoplastid DNA was isolated by plasmid mini-preparation columns (Qiagen), digested with HindIII and EcoRI enzymes and run on a 1% agarose gel. No differences were observed in kDNA between the L. infantum wild type and LinPIWI(−/−) null mutant.
(TIF)
Expression vectors to rescue the Lin PIWI(−/−) null mutant. Schematic representation of the LinPIWI gene fused or not at either extremity with known epitope tag sequences (e.g. HA, MYC and GST) and cloned into the BamHI and XbaI sites of vector pSP72αNEOα (see Materials and Methods). These constructs were transfected into the L. infantum LinPIWI(−/−)strain to rescue the null mutant phenotype.
(TIF)
Primers used in this study.
(DOC)