Abstract
In 2011, a novel strain of O104:H4 Escherichia coli caused a serious outbreak of foodborne hemolytic uremic syndrome and bloody diarrhea in Germany. Antibiotics were of questionable use and 54 deaths occurred. Candidate therapeutic bacteriophages that efficiently lyse the E. coli O104:H4 outbreak strain could be selected rather easily from a phage bank or isolated from the environment. It is argued that phage therapy should be more considered as a potential armament against the growing threat of (resistant) bacterial infections.
Introduction
From May to July 2011, Germany was struck by the largest outbreak of hemolytic uremic syndrome (HUS) and bloody diarrhea caused by Escherichia coli ever reported [1]. A total of 3,816 cases were reported, 845 (22%) of which involved HUS. This high rate of HUS was the first indicator that the bacterial cause of illness was not a typical enterohaemorrhagic E. coli (EHEC) strain [2]. It was shown to be a highly pathogenic enteroaggregative E. coli (EAEC) strain, which in addition carried the EHEC genes for the classical HUS-associated Shiga toxin 2 [3]–[4]. Notwithstanding the timeliness of the German surveillance system (reporting occurred faster than required by law) for HUS and Shiga toxin-producing E. coli notifiable diseases [5], no less than 54 patients died. Eventually, sprouts were identified as the most likely outbreak vehicle [6]. The outbreak strain was resistant to beta-lactam antibiotics and third-generation cephalosporins and was partially resistant to fluoroquinolones, but sensitive to carbapenems and ciprofloxacin [1]. However, a Cochrane database review analyzing seven randomized controlled clinical trials of pediatric HUS associated with EHEC infections found no benefit of antibiotic treatment over supportive therapy alone [7]. Moreover, the use of antibiotics to treat Shiga toxin producing E. coli infections has been discouraged because it has been shown to release toxins. The genes for the Shiga toxin are actually not bacterial genes, but (bacterio)phage genes. When an E. coli bacterium gets infected with a temperate phage harboring a Shiga toxin gene, upon integration of the phage genome (as prophage) into the bacterial genome (lysogeny), it can be expressed by the bacterium, which then becomes pathogenic. Some types of antibiotics have been shown to induce the so-called SOS response in bacteria, i.e. a ubiquitous response to DNA damage, which induces phage replication and lytic cycle [8]. As such, the use of antibiotics may be helping Shiga toxin genes to spread. Phage-mediated transfer of bacterial virulence, fitness and antibiotic resistance is a negative (from the human point of view) consequence of bacterial phage coevolution.
Paradoxically, phages from a very different, constitutionally lytic genus could also help fight EHEC and EAEC. Phages do play a major role in controlling bacterial densities in the biosphere, including humans, which is the basis of sustainable phage therapy [9]. For example, phages appear to be key players in ending cholera epidemics. Faruque et al. [10] observed that seasonal epidemics of cholera inversely correlated with the prevalence of environmental cholera phages. Phages could be used therapeutically, as an additional tool or in combination with antibiotics, to treat bacterial infections that do not respond to conventional antibiotics. Importantly, they can be chosen to be harmless to the commensal bacteria, such as those of the gut microflora. The oral application of phages to humans is likely to be very safe [11] and several phage based preparations were given regulatory approval and are commercially available for the decontamination of foods [12]. Muniesa et al. [13], in a review on experimental treatment and prevention options for HUS, reported phages to be an effective tool for disinfection of various vegetables and meat products contaminated by E. coli strain O157. Steers that received phage by the rectal route showed a 100-fold fecal titer decrease of O157 compared to controls. In sheep, oral T4-like coliphage reduced the O157 counts in the caecum and rectum. Recently, French scientists isolated three morphologically different types of tailed phages that infect E. coli O104:H4. The cocktail of all three phages showed sustained replication in the intestines of mice, but without decreasing the intestinal titer of O104:H4 cells [14].
Here we present the results of an ad hoc multidisciplinary working group, including institutions involved in infectious diseases surveillance, phage therapy and molecular biology research, which set out to isolate, select and characterize candidate therapeutic phages active against the E. coli O104:H4 outbreak strain.
Materials and Methods
Isolation and selection of E. coli O104:H4 phages were performed according to the methods described by Merabishvili et al. [15]. Considering laboratory safety in the handling of EHEC, all manipulations involving the O104:H4 outbreak strain were performed in a dedicated biosafetylevel 3 facility. A set of 16 phages known to infect E. coli was screened against the outbreak strain. The set included the well-known T4 phage, nine phages from the therapeutic phage library of the Eliava Institute of Bacteriophage, Microbiology and Virology in Tbilisi (Georgia) and six phages that were recently isolated from wastewater of the Queen Astrid Military Hospital in Brussels.
The lytic potential of the 16 phages was tested on a collection of 92 E. coli strains using the horizontal bacterial strips method on agar plates [15]. The tested collection included ten different pathogenic E. coli serotypes (Table 1) and 82 clinical and non-clinical E. coli strains.
Table 1. Susceptibility of ten pathogenic E. coli serotypes to infection with phage GEC-3S.
| Serotype | ||||||||||
| O26:K60 | O55:K59 | O86:K61 | O104:H4 | O111:K58 | O111:K60 | O125:K70 | O128:K67 | O142:K86 | O157:H7 | |
| GEC-3S | − | − | − | cl | − | − | cl | − | – | – |
−, no lysis; cl, confluent lysis.
Phage genome sequence was determined by pyrosequencing (454 technology) according to Kropinski et al. [16] and analyzed according to Vandersteegen et al. [17]. Open reading frames were predicted, and their functions defined, by ORF finder (http://www.ncbi.nlm.nih.gov/projects/gorf/), GeneMark™ [18] and BLASTp [19] programs, followed by manual confirmation. EMBOSS stretcher [20], [21] was used to compare DNA homology between phages.
Results and Discussion
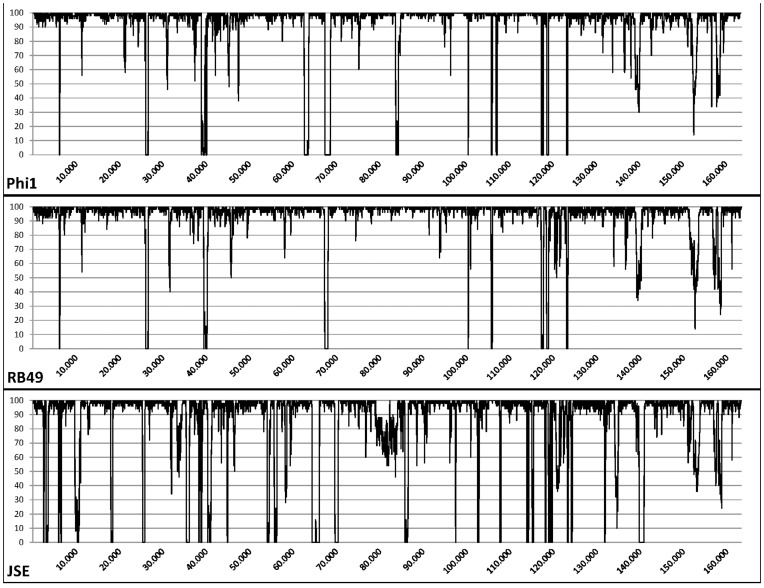
Only one out of 16 phages, myovirus GEC-3S (Fig. 1) from the Eliava collection, showed activity against the outbreak strain. In addition, two myoviruses, QAMH-FFP.1 and QAMH-NES.1, were newly isolated from the wastewater of the Brussels Military Hospital using the culture enrichment method with the original outbreak strain as the host. In contrast to phages QAMH-FFP.1 and QAMH-NES.1, phage GEC-3S could be propagated in E. coli strain K12 and this without significant decrease of titer or activity against strain O104:H4, which is not negligible in the context of an eventual production of therapeutic or prophylactic phage preparations. The non-pathogenic K12 strain is commonly used in Good Manufacturing Practices (GMP) and clean-room compliant preparation of medicinal products, whereas the handling of the O104:H4 strain requires strict biocontainment precautions. Phages QAMH-FFP.1 and QAMH-NES.1 showed no activity against the K12 strain. Of the ten tested pathogenic E. coli serotypes, only two – O104:H4 and O125:K70– were susceptible to phage GEC-3S (Table 1). The overall activity of the phage against 92 pathogenic and non-pathogenic E. coli strains proved to be 20.7% (19/92). Complete genome sequencing (Accession number HE978309) revealed that phage GEC-3S belongs to the genus of T4-like viruses with high similarity to the representatives of the RB49 group. Two hundred and seventy-six open reading frames were predicted and their functions were defined. At the nucleotide level, GEC-3S showed high homology to phi1, RB49 and JSE phages, respectively 94.6, 93.6 and 88.0%. The shared differences between GEC-3S and the other phages are spread throughout the whole genome and in particular in the regions between 39,000–40,000, 67,000–68,000, 100,000–110,000 and 115,000–125,000 bp, loci which mostly comprise ORFs coding for small hypothetical proteins and homing endonucleases (Fig. 2). Phage GEC-3S harbors six unique genes that are not present in the genomes of the other representatives of the RB49 group (phi1p263, RB49p263, EpJSE_00266) (Table 2). Four of these genes are ORFans, while the other two are coding homing endonucleases, extremely widespread enzymes in T-even-like phages according to available genomic data [22]. The only gene that is typical for the other three phages from this group, but is not present in GEC-3S is a hypothetical protein of 51 amino acids length (Table S1).
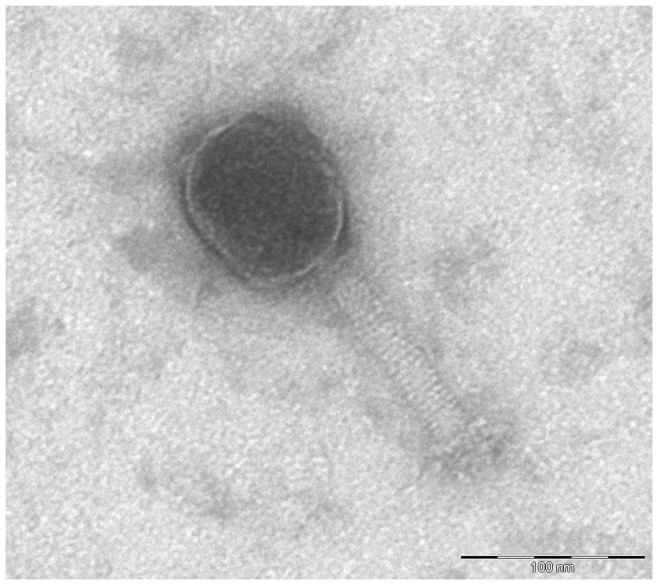
Figure 1. Transmission electron micrographs of myovirus GEC-3S.
Phage particles were analyzed by transmission electron microscopy as described by Imberechts et al. [27]. The head is somewhat elongated, appears hexagonal in outline and has a mean diameter of 102 nm (SEM = 3 nm). The head is separated from the tail by a neck. The tail is a rigid, contractile tube with a mean length of 109 nm (SEM = 1 nm). It appears cross-banded, suggesting helical symmetry. The tail ends in short and long fibers. The latter do not show up very well on this micrograph.
Figure 2. Pairwise DNA homology of GEC-3S and phi1, GEC-3S and RB49, GEC-3S and JSE, compared using a sliding window of 50 bp.
Table 2. ORFs of phage GEC-3S not presented in any other representative of the RB49 group.
| N | Amino acid length | E value | Putative functions/Known Homologs* |
| ORF 63 | 251 | 5×10−16 | putative homing endonuclease RB16 6 (Enterobacteria phage RB16) |
| ORF 137 | 246 | 1×10−9 | putative H-N-H-endonuclease P-TflX (Enterobacteria phage T5) |
| ORF 163 | 32 | – | ORFan |
| ORF 171 | 44 | – | ORFan |
| ORF 265 | 80 | – | ORFan |
| ORF 266 | 66 | – | ORFan |
Putative functions and known homologs of ORFs were identified using BlastP against the non-redundant database.
Being a T4-like phage (of the RB49 group), much is known about the properties and safety of phage GEC-3S, further strengthening its appropriateness for therapeutic applications. In general, temperate phages are considered inappropriate for use as therapeutic phages because they can transduce unwanted genes. Sequence similarity searches against the non redundant NCBI database confirmed the lytic (non temperate) character of phage GEC-3S and the absence of genes known to encode virulence or pathogenicity associated, or potentially allergenic, proteins. This is important, as the E. coli outbreak strain itself is a good illustration of phage-mediated increased pathogenicity.
Because sequencing of the O104:H4 outbreak strain failed to provide clues about its origin and evolution, it is unclear whether we may expect similar upcoming outbreaks to occur recurrently or spontaneously in the future [23]. Even though the epidemic has been declared finished, the health community should explore new (prophylactic) therapies. Theoretically, phage GEC-3S could have been used to help control (in humans, animals, the environment or food) the O104:H4 outbreak that caused the death of more than 50 patients. A cocktail of purified and fully defined and safe phages selected against the most problematic EHEC and EAEC strains could be produced and stored as an additional antimicrobial when an outbreak similar to the one witnessed in Germany occurs in the future. The high bacterial strain specificity of phages make it necessary to provide cocktails for treatment of disease. If required, new phages could be added to the cocktail. The isolation and selection process of the here presented phages took only 3 days to complete. A useful strategy would be to set up an international bank by isolating a large number of phages from E. coli-pathogen-rich sites using strain K12, checking their ability to grow efficiently on all the important pathogenic E. coli strains available (e.g. O104:H4), and from these choosing a set that provides broad coverage of the known problem strains. It would be an advantage if these were T4-related phages, about which so much is known. Ideally, component phage identification and detailed characterization and maintenance of this bank should be supported by WHO and/or the European and American CDCs [24]. The O104:H4 outbreak caused considerable suffering and resulted in a strain on healthcare and public health systems [25], which could provide an incentive for competent authorities and physicians to use phages, as additional tools, in the prevention and treatment of otherwise virtually untreatable infections. Meanwhile, preventive microbiology remains crucial for early detection of major health threats caused by infectious diseases [26].
Supporting Information
Comparison of ORFs/genes of phage GEC-3S with other representatives of the RB49 group.
(XLSX)
Acknowledgments
We thank Lieutenant Colonel Serge Jennes, Colonel Pierre Neirinckx and Major General Geert Laire for their continuous support.
Funding Statement
This work was suported by the Royal Higher Institute for Defense (grant MED 12). The authors would like to acknowledge the Research community “Phagebiotics” (WO.022.09) grant from the FWO Vlaanderen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Frank C, Werber D, Cramer JP, Askar M, Faber M, et al. (2011) Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365: 1771–1780. [DOI] [PubMed] [Google Scholar]
- 2. Casey PG, Hill C, Gahan CG (2011) E. coli O104:H4: social media and the characterization of an emerging pathogen. Bioeng Bugs 2: 189–193. [DOI] [PubMed] [Google Scholar]
- 3. Bielaszewska M, Mellmann A, Zhang W, Köck R, Fruth A, et al. (2011) Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 11: 671–676. [DOI] [PubMed] [Google Scholar]
- 4. Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, et al. (2011) Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS ONE 6: e22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Altmann M, Spode A, Altmann D, Wadl M, Benzler J, et al. (2011) Timeliness of surveillance during outbreak of Shiga Toxin-producing Escherichia coli infection, Germany, 2011. Emerg Infect Dis 17: 1906–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buchholz U, Bernard H, Werber D, Böhmer MM, Remschmidt C, et al. (2011) German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med 365: 1763–1770. [DOI] [PubMed] [Google Scholar]
- 7.Michael M, Elliott EJ, Ridley GF, Hodson EM, Craig JC (2009) Interventions for haemolytic uraemic syndrome and thrombotic thrombocytopenic purpura. Cochrane Database Syst Rev CD003595. [DOI] [PMC free article] [PubMed]
- 8. Kimmitt PT, Harwood CR, Barer MR (2000) Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg Infect Dis 6: 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pirnay JP, Verbeken G, Rose T, Jennes S, Zizi M, et al. (2012) Introducing yesterday’s phage therapy in today’s medicine. Future Virol 7: 379–390. [Google Scholar]
- 10. Faruque SM, Bin Naser I, Islam MJ, Faruque ASG, Ghosh AN, et al. (2005) Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc Natl Acad Sci U S A 102: 1702–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bruttin A, Brüssow H (2005) Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob Agents Chemother 49: 2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodridge LD, Bisha B (2011) Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage 1: 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muniesa M, Hammerl JA, Hertwig S, Appel B, Brüssow H (2012) Shiga toxin-producing Escherichia coli O104:H4: a new challenge for microbiology. Appl Environ Microbiol 78: 4065–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maura D, Morello E, du Merle L, Bomme P, Le Bouguénec C, et al. (2011) Intestinal colonization by enteroaggregative Escherichia coli supports long-term bacteriophage replication in mice. Environ Microbiol 14: 1844–1854. [DOI] [PubMed] [Google Scholar]
- 15. Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, et al. (2009) Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS ONE 4: e4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kropinski AM, Van den Bossche A, Lavigne R, Noben JP, Babinger P, et al. (2012) Genome and proteome analysis of 7-7-1, a flagellotropic phage infecting Agrobacterium sp H13-3. Virol J 9: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vandersteegen K, Mattheus W, Ceyssens PJ, Bilocq F, De Vos D, et al. (2011) Microbiological and molecular assessment of bacteriophage ISP for the control of Staphylococcus aureus . PLoS ONE 6: e24418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 20. Myers E, Miller W (1988) Optimal alignments in linear space. CABIOS 4: 11–17. [DOI] [PubMed] [Google Scholar]
- 21. Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 22. Edgell DR, Gibb EA, Belfort M (2010) Mobile DNA elements in T4 and related phages. Virology Journal 7: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bezuidt O, Pierneef R, Mncube K, Lima-Mendez G, Reva ON (2011) Mainstreams of horizontal gene exchange in enterobacteria: consideration of the outbreak of enterohemorrhagic E. coli O104:H4 in Germany in 2011. PLoS ONE 6: e25702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brüssow H (2012) What is needed for phage therapy to become a reality in Western medicine? Virology [Epub ahead of print]. [DOI] [PubMed]
- 25. Struelens MJ, Palm D, Takkinen J (2011) Enteroaggregative, Shiga toxin-producing Escherichia coli O104:H4 outbreak: new microbiological findings boost coordinated investigations by European public health laboratories. Euro Surveill 16: 19890. [DOI] [PubMed] [Google Scholar]
- 26. Friedrich A (2011) Enterohaemorrhagic Escherichia coli O104:H4: are we prepared now? Euro Surveill 16: 19938. [PubMed] [Google Scholar]
- 27. Imberechts H, Wild P, Charlier G, De Greve H, Lintermans P, et al. (1996) Characterization of F18 fimbrial genes fedE and fedF involved in adhesion and length of enterotoxemic Escherichia coli strain 107/86. Microb Pathog 21: 183–192. [DOI] [PubMed] [Google Scholar]
- 28. Arbiol C, Comeau AM, Kutateladze M, Adamia R, Krisch HM (2010) Mobile regulatory cassettes mediate modular shuffling in T4-type phage genomes. Genome Biol Evol 2: 140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of ORFs/genes of phage GEC-3S with other representatives of the RB49 group.
(XLSX)