Abstract
Background
small B-cell neoplasms can show plasmacytic differentiation and may potentially progress to aggressive lymphoma (DLBCL). Epstein-Barr virus (EBV) infection may cause the transformation of malignant cells in vitro.
Design and Method
we established VR09 cell line with plasmacytic differentiation, obtained from a case of atypical, non-CLL B-cell chronic lymphoproliferative disease with plasmacytic features. We used flow cytometry, immunohistochemistry, polymerase chain reaction, cytogenetic analysis and florescence in situ hybridization in the attempt at thoroughly characterizing the cell line. We showed VR09 tumorigenic potential in vivo, leading to the development of activated DLBCL with plasmacytic features.
Results
VR09 cells displayed plasmacytic appearance and grew as spherical tumors when inoculated subcutaneously into immunodeficient Rag2−/− γ-chain−/− mice. VR09 cell line and tumors displayed the phenotype of activated stage of B cell maturation, with secretory differentiation (CD19+ CD20+ CD79a+ CD79b+/− CD138+ cyclin D1- Ki67 80% IgM+ IgD+ MUM1+ MNDA+ CD10- CD22+ CD23+ CD43+ K+, λ- Bcl2+ Bcl6-) and they presented episomal EBV genome, chromosome 12 trisomy, lack of c-MYC rearrangement and Myd88 gene mutation, presence of somatic hypermutation in the VH region, and wild-type p53.
Conclusion
This new EBV-positive cell line may be useful to further characterize in vivo activated DLBCL with plasmacytic features.
Introduction
Human lymphoma cell lines and animal models have contributed significantly to better understand the physiopathology of hematopoietic tumors. Cell lines are generally characterized by monoclonal origin, differentiation arrest and sustained proliferation in vitro, while maintaining most cellular features and specific genetic alterations of the primitive tumor [1]. However, primitive malignant cells may change some morphological and phenotypic features in culture, due to Epstein-Barr virus (EBV) infection. EBV is a B lymphotropic herpesvirus linked to several B cell malignancies and capable of transforming human B cell into permanent lymphoblastoid cell line in vitro [2]. Besides Hodgkin lymphoma, Burkitt lymphoma, human immunodeficiency virus-associated lymphomas and post-transplant lymphoproliferative disorders [3], EBV has been also detected in low-grade lymphoproliferative diseases such as chronic lymphocytic leukemia (CLL) where it has been shown to correlate with transformation to Richter syndrome in some cases [4], [5], [6].
Many types of small B lymphoid neoplasms can demonstrate plasmacytic differentiation and specific immunophenotypic or genetic markers for plasmacytic lymphoma; furthermore, small B-cell lymphomas with plasmacytic differentiation frequently represent a diagnostic challenge, and sometimes only a diagnosis of small B-cell lymphoma with plasmacytic differentiation can be done [7]. All of these disorders may potentially undergo transformation to large-cell lymphoma [8].
Diffuse large B-cell lymphoma (DLBCL) represents a very heterogeneous group of aggressive lymphomas. Following the introduction of gene expression profiling (GEP), it has become increasingly clear that DLBCL is a biologically and clinically heterogeneous disorder. Two major categories are now recognized, i.e. germinal center B-cell (GCB) and activated B-cell (ABC) types, which are different for the expression of more than 1000 genes. In the attempt at traslating GEP classification into daily practice, several immunohistochemical algorithms have been proposed as a surrogate for GEP analysis [9].
Some aggressive DLBCL show the phenotype of terminal B-cell differentiation and represent a continuum spectrum of lesions that ranges from conventional activated DLBCL to plasma cell disorders with specific molecular patterns [10], some of which, including Myd88, have been recently suggested to correlate with plasmacytic lymphoma histotype [11], [12].
We describe here the established VR09 cell line, an EBV-positive cell line with plasmacytic differentiation displaying tumorigenic potential in vivo and leading to the development of activated DLBCL with plasmacytic features when injected into Rag2−/− γ-chain−/− mice. Thus, VR09 xenotrasplantation can be used successfully as preclinical model for human DLBCL with plasmacytic differentiation to further characterize this disease.
Materials and Methods
Cell Collection and Culture
A 75-year old Caucasian man was admitted to hospital in September 2008 for fever, neutropenia and lymphocytosis (WBC 20.1×109/L, with neutrophils 0.8×109/L and lymphocytes 18.2×109/L), and moderate anemia and trombocytopenia (Hb 9 g/dl, PLTS 93×109/L). No significant superficial lymphoadenopathy or splenomegaly were present. Peripheral blood smear shows the predominance of small-medium sized mature lymphoid cells with abundant cytoplasm and compact chromatin ( Figure 1A ). A bone marrow sample was sent to our laboratory for first level-immunophenotyping by flow cytometry (FACSCanto, Becton Dickinson Biosciences, CA, USA), after written informed consent, as approved by the Ethics Committee of Azienda Ospedaliera Universitaria Integrata Verona (N. Prog. 1828, May 12, 2010 - ‘Institution of cell and tissue collection for biomedical research in Onco-Hematology’). Bone marrow smear appeared infiltrated (60%) by a cell population of lymphoplasmocytoid elements of small-medium size ( Figure 1B ). First level-immunophenotyping showed the presence of a cell population expressing CD19, CD20, CD22, CD138, surface immunoglobulins (sIg) at high level, and negative for CD5, CD10, ZAP-70, thus suggesting the diagnosis of non-CLL atypical B-cell chronic lymphoproliferative disease with plasmacytic features. No other exams could be performed, as the patient died of sepsis the following day.
Figure 1. Morphology of primary malignant cells and VR09 cell line.
Morphological features of patient’s primary cells and VR09 cells in suspension, as assessed by May-Grünwald-Giemsa staining. (A) Patient’s peripheral blood cells. (B) Patient’s bone marrow cells. (C) VR09 cells: small and round clumps in suspension. (D) VR09 cells: plasmacytoid appearance, with irregular nucleus, compact chromatin and abundant basophilic cytoplasm.
Mononuclear cells were purified from bone marrow sample by Ficoll-Paque centrifugation (Lymphoprep, Fresenius Kabi Norge AS for Axis-Shield Poc AS, Oslo, Norway), washed in phosphate-buffered saline solution (PBS) and resuspended at 1×106/mL concentration in RPMI 1640+ GlutaMAX 1X containing 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin (all from GIBCO, Invitrogen). Cells were cultured in 75 cm2 flask and incubated in humidified 5% CO2 atmosphere at 37°C. Half of culture medium was replaced every 3–4 days maintaining the same cell density of 1×106 cells/mL. To determine growth kinetics, cells were seeded at lower density (350,000/mL) and counted at 0, 24, 48 and 72 hours by flow-cytometry (FACSCanto, Becton Dickinson, Italy). No mitogens or growth factors were added during culture. Cells were maintained in culture up to one year. Cell morphology was evaluated on cytospins stained with May-Grunwald Giemsa dye.
Immunophenotypic Analysis
Cell vitality was assessed by acridine-orange/ethidium bromide staining and epifluorescent microscopy. Aliquots of 3×105 cells were incubated for 15 minutes at room temperature with three-color combinations of appropriate monoclonal antibodies anti-human CD3, CD19, CD20, CD22, CD23, CD25, CD38, CD43, CD45, FMC-7, CD79b 7AAD (Becton Dickinson, Italy), CD5, CD10, K, λ, IgG, IgM, IgD (Dako, Italy), CD103 (Beckman Coulter, Italy), CD138 (Cytognos, Italy), and isotype controls (Becton Dickinson, Italy). Samples were analyzed by FacsCANTO flow cytometer with BD FACSDiva software (Becton Dickinson, Italy).
Cell Cycle
For determination of the DNA content, 1.5×105 cells were incubated with 1 ml of staining solution including 5 ml of hypotonic solution, 50 µg of propidium iodide (Bender MedSystems) and 20 µg of RNAse for two hours at 4°C analyzed using appropriate settings by FacsCanto flow cytometer. Human peripheral mononuclear cells were used as control for the comparison of S fraction.
DNA and RNA Extraction, cDNA Synthesis
DNA and RNA were obtained from 107 cells by AllPrep DNA/RNA/Protein Mini Kit (Quiagen, Hilden, Germany). DNA quality was verified by spectrophotometry and RNA quality by the Agilent Bionanalyzer 2100; 1 µg of total RNA was reverse-transcribed by using SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, California) and cDNA was used as a template for reverse transcriptase polymerase chain reaction (RT-PCR) amplification using listed in Table 1 .
Table 1. Primers used to amplify and sequence P53, Card 11, CD79B, MYD88 AND RPMS1 genes.
| Primers | Sequence | Ta° | Amplicon lenght |
| P53-1-F | AAGTCTAGAGCCACCGTCCA | 55°C | 771 bp |
| P53-1-R | AAGTGTTTCTGTCATCCAAATACTC | ||
| P53-2-F | AGCCAAGTCTGTGACTTGCA | 55°C | 851 bp |
| P53-2-R | GGGGAACAAGAAGTGGAGAA | ||
| Card11-1-F | AGATGCAACGGGAGCCTGGC | 55°C | 631 pb |
| Card11-1-R | AGGTTAGCAGCTCCACGCGC | ||
| Card11-2-F | GGCCAAGGACCTGCAACGCT | 55°C | 703 pb |
| Card11-2-R | CCGCTCCACCTCCTCCAGCT | ||
| Card11-3-F | GAGGCCCTGGAGGACAGGCA | 55°C | 690 pb |
| Card11-3-R | TCCGCAGGAGCTAGGGCTGG | ||
| Card11-4-F | TCCTGCCCTACCATCCGCCC | 55°C | 671 pb |
| Card11-4-R | CAGCAGCTGGTGGCCCTCAC | ||
| Card11-5-F | TCCCAGCTCACCCTGCTGGG | 55°C | 739 pb |
| Card11-5-R | CCGAGATGATGCGGACCCGC | ||
| Card11-6-F | CCCGTCTCTCGCGAGCAAGC | 55°C | 928 pb |
| Card11-6-R | CGTCTGCTGGGGCAGCTCTG | ||
| CD79B-1-F | GCCTCGGACGTTGTCACGGG | 55°C | 858 pb* |
| CD79B-1-R | TGGGCCAGCTTCAGAGGCCA | ||
| MYD88-EX1F | CGCAGGAGAAAGAGGAAGC | 60°C | 491 bp |
| MYD88-EX1R | ATGGGAGACAGGATGCTGAG | ||
| MYD88-EX3F | TGGGTAAAGAGGTAGGCACTCCCAG | 60°C | 275 bp |
| MYD88-EX2R | GCCCATCTGCTTCAAACACCCATGC | ||
| MYD88-EX3F | AAGCCTTCCCATGGAGCTCTGACCAC | 60°C | 311 bp |
| MYD88-EX3R | GCTAGGAGGAGATGCCCAGTATCTG | ||
| MYD88-EX3F | ACTAAGTTGCCACAGGACCTGCAGC | 60°C | 194 bp |
| MYD88-EX3R | ATCCAGAGGCCCCACCTACACATTC | ||
| MYD88-EX3F | GTTGTTAACCCTGGGGTTGAAG | 60°C | 297 bp |
| MYD88-EX3R | GCAGAAGTACATGGACAGGCAGACAGATAC | ||
| RPMS1F | GGATGGGAGAGGGTGATCTT | 60°C | 151 bp |
| RPMS1R | ACGTGGAGTTTGCAGTCCTC |
Isoform 1: 858 pb; Isoform 2: 546 pb; Isoform 3: 861 pb.
Analysis of VH Rearrangement, p53, CD79B, Card 11 and MYD88 Mutation
To verify the identity between cells from the patient and the cell line, rearranged VH genes were analyzed as previously described [13] in frozen DNA from both the patient and the cell line TP53, CD79B and CARD11 genes were amplified and sequenced by using the primers reported in Table 1 . MYD88 exons 1 to 5 were sequenced as previously described [11], [12]. VH and CD79B PCR bands were excised from agarose gels and purified by using spin columns (PureLink Quick Gel Extraction and PCR Purification Combo Kit, Invitrogen). PCR products were sequenced by dye terminator reaction (Big Dye Terminator Cycle Sequencing Kit v.3.1, Applied Biosystems, Warrington, UK) on AB3130XL automated sequencer (Applied Biosystems). TP53, CARD11 and MYD88 PCR products were directly purified with magnetic beads (Agencourt AMPure XP, Beckman Coulter Genomics, Beverly, Massachussets, USA) and sequenced as reported above and by outsourcing to Base Clear BV (Leiden, The Netherlands).
VH genes sequences were compared to published germline sequences using IgBLAST (http://www.ncbi.nlm.nih.gov/igblast/). Sequences with 2% or less deviation from any germline IgVH sequence were considered unmutated. TP53, CD79B, CARD11 and MYD88 amplified sequences were compared to reference sequences by using Geneious software (Biomatters Ltd., Auckland, New Zealand).
Preparation of Formalin-fixed Paraffin-embedded (FFPE) Cell Block and Immunohistochemical Staining
Briefly, 12×106 cells were centrifuged, washed once in PBS and then incubated for 15 minutes in 4% buffered formalin. Cells were centrifuged again and resuspended in 1 ml of 1% low melting point agarose at 37°C; then, they were poured into disposable plastic moulds and cooled for ten minutes at +4°C. Solid agarose blocks were put inside a histological cassette, with 4% buffered formalin at room temperature, and then processed routinely as tissue block; 4 µm-thick sections were cut from the block and stained with ematoxilin-eosin to verify the quality of the cell inclusion. Many sections from the block were immunolabeled with the broad panel of antibodies described in Table 2 . A 25% cut-off was used to distinguish positive from negative. All samples were processed by using a sensitive ‘Bond polymer Refine’ detection system in automated Bond immunostainer (BondMax, Vision-Biosystem, Menarini, Florence, Italy).
Table 2. Marker expression by VR09 cell line and tumor masses, as assessed by immunohistochemistry performed on FFPE cell block.
| Antibody | Species | Clone | Source | Diluition | VR09 cell line | Tumor masses |
| Annexin a1 | mouse | B01P | Abnova | 100 | neg | neg |
| Bcl-2 | mouse | 124 | Dako | 40 | pos | pos |
| Bcl-6 | mouse | LN22 | Novocastra | 20 | neg | neg |
| Cyclin D1 | rabbit | SP4 | Neomarkers | 10 | neg | neg |
| CD03 | rabbit | SP7 | Labvision | 150 | neg | neg |
| CD05 | mouse | 4C7 | Novocastra | 200 | neg | neg |
| CD10 | mouse | 56C6 | Novocastra | 50 | neg | neg |
| CD19 | mouse | LE-CD19 | Serotec | 200 | pos | pos |
| CD20 | mouse | L26 | Novocastra | 100 | pos | pos |
| CD22 | mouse | FPC1 | Novocastra | 20 | pos | pos |
| CD23 | mouse | 1B12 | Novocastra | 100 | pos | pos |
| CD25 IL2R | mouse | 4C9 | Novocastra | 100 | pos/neg | pos/neg |
| CD38 | mouse | 38C03 | Neomarkers | 50 | neg | pos/neg |
| CD43 (T cell) | mouse | MT1 | Novocastra | 30 | pos | pos |
| CD79alfa | mouse | JCB117 | Dako | 100 | pos | pos |
| CD79beta | mouse | JS01 | Novocastra | 50 | pos/neg | pos/neg |
| CD123 | mouse | 7G3 | Bd | 100 | neg | pos/neg |
| CD138 (syndecan-1) | mouse | BB4 | Serotec | 50 | pos/neg | pos/neg |
| DBA44 hairy cell | mouse | DBA.44 | Dako | 20 | neg | neg |
| FOXP1 | mouse | JC12 | Abcam | 500 | neg | pos/neg |
| EBV (EBER) | mRNA probe* | Vision Byosistems | pos | pos | ||
| GCET1 | mouse | RAM341 | Abcam | 500 | neg | neg |
| HHV-8 | mouse | 13B10 | Novocastra | 50 | neg | neg |
| IgD | rabbit | polyclonal | Dako | 20 | pos | pos/neg |
| IgG | rabbit | polyclonal | Dako | 10000 | neg | neg |
| IgM | rabbit | polyclonal | Dako | 5000 | pos/neg | pos/neg |
| Ki-67 | mouse | MM1 | Novocastra | 20000(15′ incubation) | pos(40%) | pos(80%) |
| ZAP-70 | mouse | 2F3.2 | Upstate | 200 | neg | neg |
| κ chains | rabbit | polyclonal | Dako | 50 | pos | pos |
| λ chains | rabbit | Dako | 30000 | neg | neg | |
| MNDA | mouse | 235 | provided by CNIO** | 2 | pos | pos |
| MUM1 Protein | mouse | MUM1p | Dako | 50 | pos | pos |
| PAX 5 | mouse | 1EW | Novocastra | 50 | pos/neg | pos/neg |
| P53 Protein | mouse | DO-7 | Novocastra | 20 | pos/neg | pos/neg |
| Sox11 | rabbit | polyclonal | Atlas Sigma | 50 | pos/neg | pos/neg |
| TCL1A | mouse | 1–21 | Santa Cruz | 100 | pos/neg | pos/neg |
| TdT | rabbit | polyclonal | Dako | 30 | neg | neg |
| TRAcP | mouse | 26E5 | Neomarkers | 30 | neg | neg |
Establishment of the Mouse in vivo Model
Three month-old immunodeficient Rag2−/− γ-chain−/− mice (Taconic animal models, New York’s River Valley, NY USA) were injected either subcutaneously (s.c.) (n = 6) with 5×106 VR09 cells, resuspended in 0.2 ml PBS, or intravenously (i.v) (n = 6) with 0.5×106 VR09 cells resuspended in 0.1 ml PBS. Animals were checked three times a week for the development of tumors. When s.c. tumors developed up to approximately 2 cm of diameter, corresponding to a spheric volume of approximately 4.187 cm3, mice were killed. Half of the tumor mass was frozen and half included in paraffin according to standard procedures. Sections of 5–8 µm from each tumoral mass were prepared and analyzed for morphology, immunohistochemistry and FISH. Paraffin sections for immunohistochemistry were stained with a panel of antibodies, as described above ( Table 2 ). Moreover, a little portion of the mass from two of six s.c injected mice, was dissected and mechanically dissociated into single-cell suspension; 1.5×106 cells from each tumor were cultured in RPMI +10% FBS and incubated as described above. Immunophenotipic analysis was performed on secondary cell culture after two months of culture, as previously described.
Three of six mice injected i.v. were euthanized at +30 days, while the others were sacrificed after four months from the treatment. Spleen, liver, femurs, lymph nodes, lungs and bowel of the i.v. injected animals were collected and included in paraffine. Tissue sections were immunostained with antibodies against CD20 and CD138 to verify the presence on tumor infiltration. In addition, immunophenotyping with anti-human CD19 and CD45 monoclonal antibodies and isotype controls was performed by standard procedures on peripheral blood mononuclear cells of all mice injected both s.c. and i.v.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The in vivo studies were approved by Verona University Ethical Committee for experimentation on animals (Prot. n°51 del 16/06/11, D.lgs 116/92).
EBV Status Evaluation
The presence of EBV was assessed on both cells cultured in vitro and cells obtained from the tumoral mass developing after in vivo injection of VR09 cell line. Epstein-Barr virus-encoded RNA (EBER) hybridization was performed on cytoinclusion by means of specific fluorescein-conjugated EBER probes (Bond ISH EBER Probe, Vision-Biosystem, Menarini, Florence, Italy). EBER sequence was detected by anti-fluorescin antibody associated with a sensitive ‘Bond polymer Refine’ chromogenic detection system in an automated immunostainer (Bondmax, Vision-Biosystem, Menarini, Florence, Italy).
To verify that EBV infection of VR09 derived directly from the patient, the presence of EBV gene RPMS1 was detected by PCR amplification on both DNA from the patient and VR09 cell line. The reaction was performed in a final volume of 50 ml, containing 5 ml Taq Buffer Advanced (5 Prime, Hamburg, Germany), 5 pmol of each primer (listed in Table 1), 200 mM each dNTP and 1.25 U of Taq DNA Polymerase (5 Prime). Cycling conditions were 2 minutes at 94°C, 20 seconds at 60°C, 40 seconds at 68°C, then a final elongation of 5 minutes at 68°C. PCR products were analyzed on a 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany), using the Agilent DNA1000 chip and reagents. Electronic data were analyzed using the manufacturer’s software (Agilent 2100 Expert).
Karyotyping and Fluorescence in situ Hybridization (FISH)
Cytogenetic analysis was performed with standard methods on both cells cultured in vitro and cell suspensions from tumors developing after in vivo injection of VR09 cell line: 10–20×106 cells were centrifuged, resuspended in RPMI 1640 medium containing 20% fetal bovine serum (FBS, Lonza, Verviers, Belgium) and cultured for 24 hours. Before harvest, 0.15 µg/ml colchicin (Eur-Clone, Italy) was added. Chromosome preparation and staining using QFQ technique was performed according to standard protocols. Karyotypes were scored according to the International System for Human Cytogenetic Nomenclature (ISCN) [14]. Images were captured with a ZEISS Axioplan microscope (ZEISS, Jena, Germany) and evaluated by Cytovision applied imaging system (Molecular Devices, New Milton).
Interphase cytogenetic FISH was performed on nuclei from VR09 cell suspension and formalin-fixed and paraffin-embedded tissue sections from in vivo tumor masses developing after in vivo injection of VR09 cell line. The following kits were used: LSI BCL-2 dual color probes for 14q32;18q21 (Abbott-Vysis, Olympus), LSI CCND1 (11q13) break-apart probes (Abbott-Vysis, Olympus), LSI IGH/CCND1 probe for 11q13-14q32.3 (Abbott-Vysis, Olympus), LSI MYC dual color break-apart probe for 8q24 (Abbott-Vysis, Olympus), LSI C-MYC dual color probes for 8q24-14q32 (Abbott-Vysis, Olympus) and CEP (centromeric) mapping 12p11.1-q11.1 (Abbott-Vysis, Olympus). Chromosome 9 probes from human painting box kit (Spectral Imaging) were also used to identify whole chromosome 9 on metaphases of cell derived from in vivo tumor masses. The procedure was performed according to the methods described elsewhere [15].
FISH slides were examined by using either Axioplan (Zeiss, Germany) or Olympus BX61 (Olympus, Hamburg, Germany) epifluorescent automated microscopes. The signals were recorded using a CCD camera (Axiocam HRm, Zeiss and Digital Camera Olympus), and score was assigned according to manufacturer’s instructions, available in each commercial FISH kit.
Results
Establishment and Characterization of VR09 Cell Line
Primary cells proliferated very slowly during the first six weeks of culture, with a consequent decline in cell number. Remaining cells formed a few clusters of proliferation and started growing in suspension forming small round clumps ( Figure 1C ). At ten weeks of culture a stable proliferating cell line (designed VR09) was established, with a doubling time of approximately 84 hours. May-Grünwald-Giemsa staining showed a wide cellular size spectrum, from medium to large cells, with plasmacytoid features and occasional bizarre shapes: cells had a round-ovoid, often eccentric or moderately irregular nucleus, with compact chromatin and abundant basophilic cytoplasm ( Figure 1D ). At present, VR09 cells grow in suspension in RPMI 1640 medium supplemented with 10% heat-inactivated FBS without requiring any other supplements, and can be vitally stored in a medium consisting of 60% RPMI 1640 medium, 30% heat-inactivated FBS and 10% DMSO. Once thawed and put in culture, VR09 cell line grows rapidly and maintains the same doubling time of the early culture. Cells are optimally maintained at 500,000 and 1,000,000/mL cell density and may be split 1∶5 every 72 hours.
Immunophenotype and Immunohistochemistry
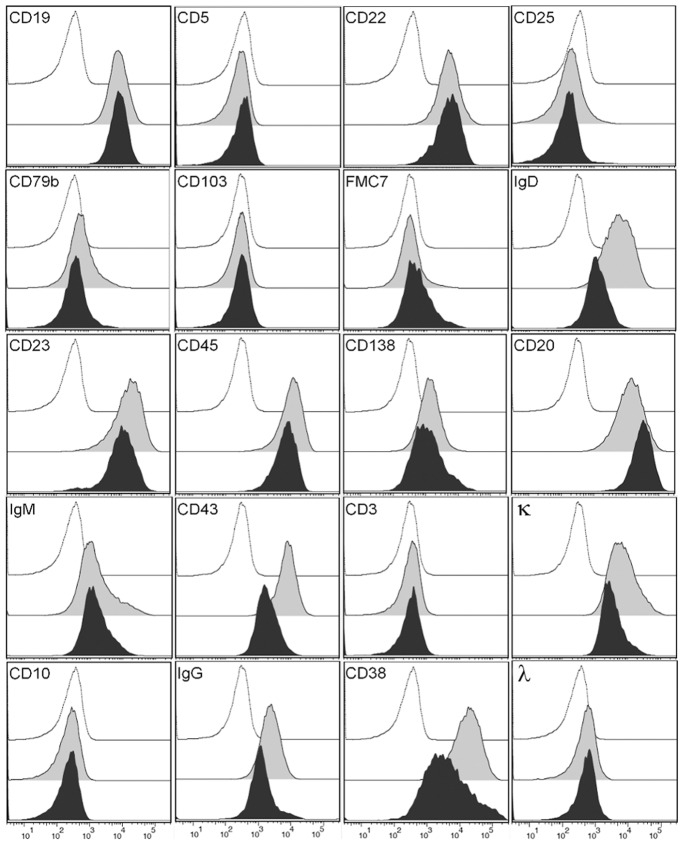
Immunophenotyping by flow cytometry was performed on VR09 cell line at 10 weeks and one year after continuous culture, resembling the phenotype of atypical B-cell chronic lymphoproliferative disease with plasmacytic features found in the patient. In particular, cells were CD45+, CD19+, CD20+, CD22+, CD23+, CD43+, CD38+, CD138+, IgD+, IgM+, IgG+ and kappa chains+, and negative for CD3, CD5, CD10, CD25, CD79b, CD103, FMC7 and lambda chains ( Figure 2 ); Zap-70, as expected, was not expressed. VR09 surface markers remained unchanged over time, with the exception of CD20, whose expression had a 1 log increase as compared to starting culture.
Figure 2. Immunophenotype.
Immunophenotyping of VR09 cell line (grey and filled curves) and cell suspensions from in vivo tumor mass (black and filled curves), as compared with isotype control (white curves).
Results of immunohistochemistry performed on FFPE cell block are summarized in Table 2: all the markers shown by flow cytometry were confirmed, with the exception of IgG and CD38 expression that resulted negative by immunohistochemistry; proliferation index (Ki-67+ cells) was about 40%. In addition, cells were positive for Bcl-2, MNDA [16] and MUM1, and negative for Bcl-6, Cyclin D1, Annexin A1, DBA44, GCET1, ZAP-70, TdT and TRAP; other markers, such as PAX-5, Sox11, FOXP1 and TCL1, were weakly and variably expressed. In summary, neoplastic cells displayed a late B-cell phenotype (MNDA+, FOXP1+, IRF4/MUM1+, cyK+, CD138+), while germinal center markers (Bcl-6, CD10, TCL1, GCET1) and immature B-cell markers (TdT) were negative. Cyclin D1 was included to exclude the remote possibility of a blastoid variant of mantle cell lymphoma, and also because plasma cell myeloma can be cyclin D1-positive. Hairy cell leukemia markers (Annexin A1, DBA44, TRAP) were investigated to exclude the very remote possibility of evolution from hairy cell leukemia (in consideration of the clinical setting).
Cell Cycle
DNA content of VR09 cell line was analyzed at six months and one year of culture. High S-phase rate was observed (diploid S 22.66%), in absence of tetraploid or aneuploid peak, as compared to the control (diploid S: 1.64%).
IVGH, TP53, CARD11, CD79B and MYD88 Mutation Analysis
VR09 cell line and the DNA from the patient showed the same VH3-7/D4-23/JH4 gene rearrangement. The comparison of the sequence with published germline sequences showed 95.7% identity in both VR09 cell line and DNA from the patient, with evidence of somatic hypermutation (SHM) in the VH region, defined as more than 2% mutations as compared to germline sequences. The analysis of the sequences obtained from both strands of cDNA and compared with the published TP53 sequence (U94788.1) showed that this sequence in VR09 cell line was wild-type (data not shown). CARD11 sequence was compared to the published one (NM_032415): VR09 cell line showed a silent heterozygous mutation leading to D533D variation in the protein linker region. Finally, VR09 cell line expressed all the three isoforms of the CD79B gene (NM_000626.2; NM_021602.2; NM_001039933.1): cDNA sequence analysis showed that variants 1 and 3 had an homozygous silent mutation in exon 3 leading to C122C variation in the Ig-like V-type protein domain, involving the disulfide bond; moreover, variant 2 had an Alanine insertion in the exon 2b corresponding to the protein signal peptide region. The analysis of exons 1 to 5 of MYD88 showed no mutations compared to the reference sequence (NG_016964).
Mouse Model
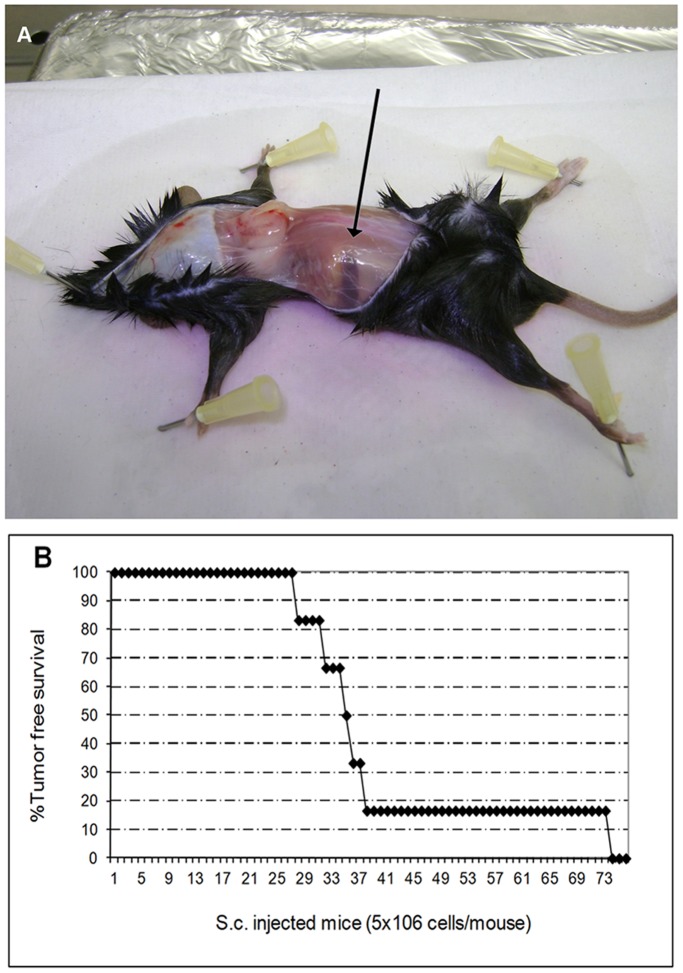
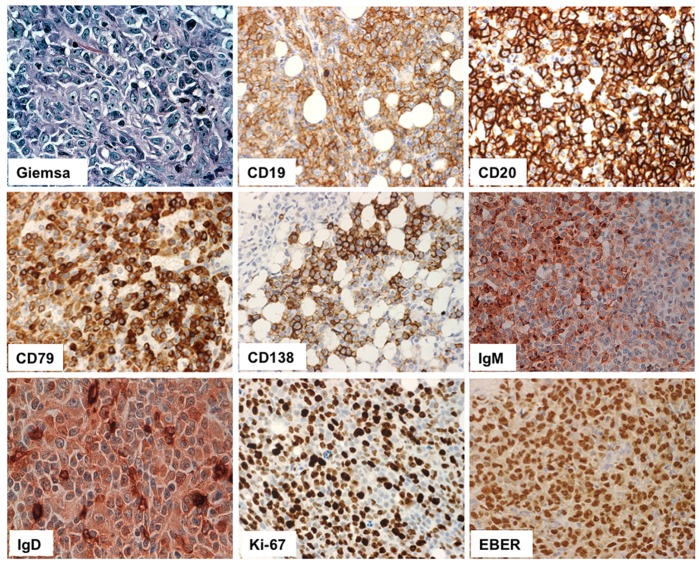
All six mice injected s.c. with VR09 cell line developed spherical tumor masses around the site of injection by 34–74 days after treatment and were sacrificed 10 days later. Subcutaneous mass and tumor incidence in vivo are shown in Figure 3 . Cells from subcutaneous masses were evaluated by H&E and Giemsa staining: cells ranged from medium to large size, with irregular nuclei, condensed chromatin without nucleoli and basophilic cytoplasm. Immunohistochemistry showed no significant differences in tumor cell immunophenotype as compared to VR09 cell line in culture ( Table 2 and Figure 4 ).
Figure 3. Cell line growth in vivo. (.
A) Spherical subcutaneous mass (arrow) in Rag-2−/− γ-chain−/− mice 36 days after VR09 cell line injection (representative case). (B) Timing of tumor development in vivo (6 mice s.c. injected with 5×106 cells/mouse).
Figure 4. Representative histological markers detected on tumors.
High magnification (20X) of histological sections of tumors developing after in vivo injection of VR09 cell line. Lymphoid infiltrates display large size, plasmablastic-plasmacytic features and high Ki-67 index. Cells express CD19, CD20, CD138, CD79a, IgM, IgG and EBV protein (EBER).
Cells suspensions obtained by disaggregation of in vivo tumor masses showed proliferative capacity when cultured with the same procedure used for primary VR09 cell line. Again, cells appeared as small and round clumps with plasmacytoid features and bizarre shape: immunophenotyping on these cells after two months of continuous secondary culture did not show any significant change of cell surface marker expression as compared to primary VR09 cell line ( Figure 2 ).
Of the six i.v. injected mice, three were euthanized after 30 days from injection and the other 3 after four months: histopathologic examination of spleen, liver, femurs, lymph nodes and lungs did not show any evidence of tumor infiltration. In addition, no circulating human CD45/CD19-positive cells were detected in peripheral blood of both s.c. and i.v. injected mice.
EBV Status
EBV RNA evaluation was performed on FFPE cell blocks and tumoral masses by using a fluorescein-conjugated probe coupled to chromogenic detection (Bond ISH EBER Probe, Vision-Biosystem, Menarini, Florence, Italy). In situ hybridization for EBV-encoded RNA (EBER) showed a clear nuclear signal in both VR09 cell line and tumors ( Table 2 and Figure 4 ).
Analysis of the RPMS1 gene PCR products, performed on both DNA from patient and VR09 cell line, showed the same 151 bp specific amplicon ( Figure 5 ), thus showing that EBV infection was already present at the beginning of culture.
Figure 5. EBV positivity in the original tumor and in the VR09 cell line.

PCR products analysis by Agilent 2100 Bioanalyzer showed the presence of the same 151 bp specific amplicon for EBV RPMS1 gene, thus demonstrating that EBV infection was present in the original cells from patient. A normal DNA from pancreas was used as negative control.
Karyotyping
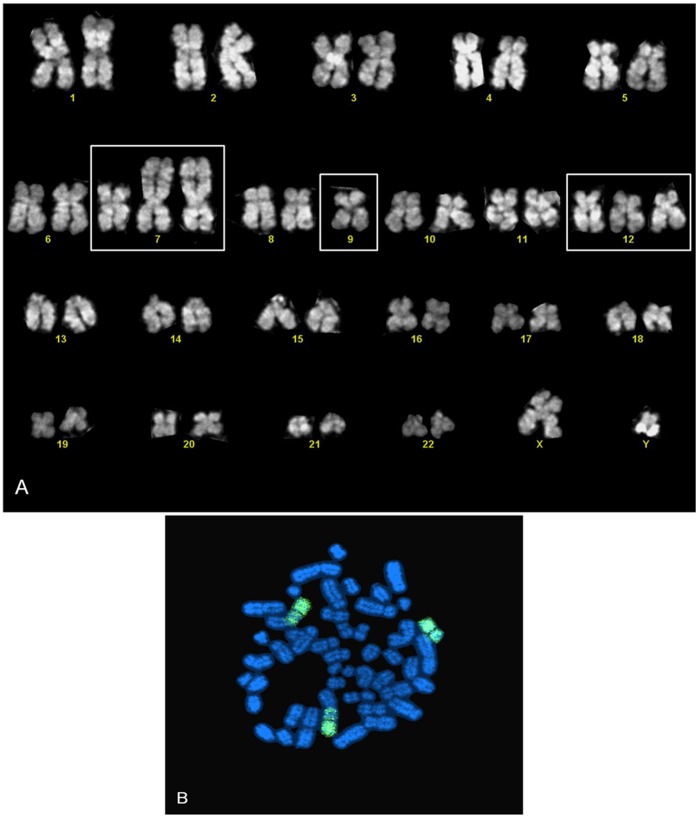
Chromosome analysis performed at 10 weeks and one year of continuous culture showed a male karyotype with 47 chromosomes. All metaphases exhibited trisomy of chromosome 12. Karyotype was repeated on secondary cell cultures obtained by disaggregation of in vivo tumor masses, thus revealing the presence of additional structural chromosome aberration involving chromosomes 7 and 9, i.e. 47, XY, der(7)(9qter->9p23::7p13->7qter), +der(7)(9qter->9p23::7p13->7qter), −9, +12 ( Figure 6A ).
Figure 6. Karyotyping and FISH of secondary culture.
(A) Trisomy of chromosome 12 and structural chromosome aberration involving chromosomes 7 and 9 on secondary culture detected by karyotype; (B) Chromosome 9 staining by FISH of secondary culture.
Fluorescence in situ Hybridization (FISH)
Adjacent or fused fluorescent signals (LSI BCL-2) were found in 5% of nuclei on tissue sections; this percentage is below the established 10% signal threshold for considering a case positive (adjacent signals are generated by random overlapping of genomic regions). No break-apart fluorescent signals were found for LSI CCND1 (11q13) and LSI MYC (8q) probes on tissue sections. No adjacent or fused fluorescent signals were found for LSI IGH/CCND and LSI C-MYC dual color probes (4% of nuclei, below threshold). Trisomy of chromosome 12 was found in 45% of nuclei. The rearrangement involving chromosomes 9 and 7 detected by karyotyping on cell suspension from in vivo tumor mass was confirmed by fluorescent signals derived from hybridization of chromosome 9 DNA probes to target chromosome 9 (chromosome 9 paint) ( Figure 6B ). The main features of VR09 cell line are summarized in Table 3 .
Table 3. Summary of VR09 cell line features.
| Parameter | Features |
| Cell Line | |
| Name of cell line | VR09 |
| Cell line type | Lymphoid |
| Cell phenotype | CD19+ CD20+ CD79a+ CD79b+/− CD138+ cyclin D1- Ki67 80% IgM+ IgD+ MUM1+ MNDA+ CD10− CD22+ CD23+ CD43+ K+, λ− Bcl2+ Bcl6− |
| Cytogenetic karyotype | Trisomy of chromosome 12 |
| Mutation analysis | VH3-7/D4-23/JH4 |
| Tumorigenic capacity | Subcutaneously growth into immunodeficient Rag2−/− γ-chain−/− mice |
| Clinical data | |
| Primary disease of patient | Atypical B-cell chronic lymphoproliferative disease with plasmacytic features |
| Disease Status | At diagnosis |
| Patient data (age, race, sex) | 75-year old Caucasian man |
| Source of material | Bone marrow |
| Year of establishment | 2008 |
| Cell culture data | |
| Culture Medium | RPMI 10% FBS |
| Subcultivation routine | Maintained at 0.5–1×1.06 cells/mL; 1∶5 split every 72 hours |
| Minimum cell density | 0.5×106 cells/mL |
| Maximum cell density | 1.0×106 cells/mL |
| Doubling time | 84 hours |
| Cell storage condition | 60% RPMI 1640 medium, 30% heat-inactivated FBS and 10% DMSO |
| In situ morphology | medium-to-large sized cells, plasmacytoid features and occasional bizarre shapes; round clumps in suspension |
| Viral status | EBV-positive |
Discussion
We describe here the establishment and molecular characterization of novel human EBV-positive DLBCL cell line with plasmacytic differentiation obtained from a patient with atypical B-cell chronic lymphoproliferative disease with plasmacytic features.
Human B-lymphoid cell lines are extensively used world wide as models in studies of various aspects of B cell biology and as a tools in research on the pathogenesis of leukemia and lymphoma. Nevertheless, the efficiency establishment of new leukemia and lymphoma cell line is rather low and remains a random process [1]. Sometimes, the spontaneous proliferation and immortalization of normal or malignant cells in vitro is due to EBV infection. It is well known for several decades that infection of normal B cell with EBV leads to establishment of lymphoblastoid cell line, thus confirming its oncogenic potential [17]. EBV is associated to several malignancies in vivo and may lead to spontaneous growth and transformation of malignant cells in vitro [2]; nevertheless, lymphoproliferative cell lines from CLL-like chronic lymphoproliferative disorders have never been reported, unless infected by EBV or in prolymphocytoid transformation [18]. The establishment of VR09 cell line was likely due to EBV infection; in fact, as EBV was already present in patient’s B cells, we can reasonably assume that the infection drove the transformation into DLBCL once cultured in vitro and injected into mice, thus excluding a culture-dependent contamination by EBV. Indeed, the Association between EBV infection and indolent B-cell neoplasms, such as chronic lymphocytic leukemia, marginal zone lymphoma, lymphoplasmacytic lymphoma has been reported [4], [19], [20] Thus, the positivity of EBV in VR09 cell line is not surprising. Moreover, some studies showed that cases of DLBCL developed from low-grade lymphoproliferative diseases are EBV-positive and that EBV may have a potential role, although not yet completed defined, in the progression of the indolent disease [21], [22].
Unfortunately, we lack much information about the patient; however, bone marrow cell morphology and immunophenotype are in agreement with the diagnosis of atypical, non-CLL B-cell chronic lymphoproliferative disease with plasmacytic features. Low-grade B-cell lymphoproliferative disorders, such as CLL/small lymphocytic lymphoma and marginal zone B-cell lymphoma may have overlapping features, thus making the differential diagnosis sometimes difficult. Furthermore, many types of small B lymphoid neoplasms can display plasmacytic differentiation and a phenotype resembling lymphoplasmacytic lymphoma; the latter consists of small lymphocytes, plasmacytoid lymphocytes and plasma cells expressing IgM and pan-B-cell antigens, such as CD19, CD20 and CD22, and usually negative for CD5, CD10 and CD23. As we lack information about serum IgM paraprotein, we concluded as a case of atypical B-chronic lymphoproliferative disease with plasmacytic differentiation, rather than a plasmacytic lymphoma.
Among lymphoproliferative disorders, VR09 cell line presents features of DLBCL with plasmacytic differentiation, with medium to large sized and plasmablastic/plasmacytic-like cells, high Ki-67 index and activated phenotype according to Hans’ and new immunostaining algorithm [9]. Moreover, cells are positive for MUM1, CD38, and CD138, thus suggesting a terminal-B cell differentiation.
Although lymphoplasmacytic lymphoma and other lymphoproliferative disease with plasmacytic differentiation may develop into diffuse large B cell lymphoma, similarly to CLL, the frequency of transformation is low [22]. Consequently, VR09 cell line can be useful as a model of DLBCL variant corresponding to a very late or activated stage of B cell maturation [23], i.e. the early post-germinal stage differentiation, not entirely terminally differentiated.
Despite the aggressive transformation of indolent disease is not always clonally related to original disease, some data are indirectly consistent with the hypothesis of clonal evolution: in fact, VR09 cell line maintains the immunophenotype and plasmacytoid features of the primary cells at diagnosis, displays the presence of the same clonal gene rearrangement found in patient’s DNA and has a tumorigenic capacity in vivo; the latter feature strongly suggests its malignant behaviour. VR09 cell line grow subcutaneously without dissemination in peripheral blood and no engraftment when it is administrated i.v. This strange peculiarity could be simply due to the in vivo model used, i.e. Rag2−/− γ-chain−/− mice. They are immunodeficient mice with C57B16/10 background devoid of functional B and T cells because of the complete lack of function of the V(D)J recombinase enzyme system, and displaying deficient innate immunity due to the absence of γ chain of interleukin-2-receptor [24]. This model has the advantage to show a stable phenotype without developing spontaneous tumors, as compared to SCID mice [25]. Indeed, VR09 cell line injection led to 100% incidence of developing tumors in vivo, always maintaining the same features.
We found trisomy of chromosome 12 in our model. This aberration is often detected in chronic lymphocytic leukemia [26], [27] and in other chronic B cell malignancies, including sometimes lymphoplasmacytic lymphoma. We also found an additional chromosomal abnormality involving chromosomes 7 and 9 after secondary culture: genetic alterations of cell lines are usually stable over the time, but it is debated whether cell line may acquire additional rearrangements in culture [1].
We also found a synonymous variant of CD79B involving the insertion of Alanine in the exon 2b of variant 2. This variant may be physiological since it has been detected in other isoforms; however, it has never been described before. Moreover, a recent study suggested that also synonymous variants may be involved in the pathogenesis of some diseases [28]; therefore, one could speculate that this novel variant of CD79B gene could be involved in the pathogenesis of lymphomas. VR09 cell line did not display the common missense mutations of Card11, usually involved in the constitutive activation of NF-kB pathway in some cases of human activated-B-cell like DLBCL [29]. However, the detection of synonymous mutations of Card11 reveals the accuracy of sequencing method. Interestingly, VR09 cell line showed unmutated Myd88 gene, differently from what described in recents reports [11], [12], thus suggesting that lymphomas with plasmacytic features may have different gene patterns.
In conclusion, VR09 is a new cell line of activated DLBCL with plasmacytic differentiation that grow as solitary tumor once injected s.c. in immunodeficient mice. This model could be useful for further studies about the development of DLBCL in patients with low-grade B-cell lymphoproliferative disorders with plasmacytic differentiation, which is a rare but possible event in clinical practice.
Acknowledgments
The authors thank Irene Dalai for her contribution in genetic analysis.
Funding Statement
This work was supported by Italian Ministry of University and Scientific Research (PRIN 2005, PRIN 2007), Fondazione CARIVERONA, (Bando 2003 and 2008) and Fondazione AIRC (MFAG 2008). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Drexler HG, Matsuo AY, MacLeod RA (2000) Continuous hematopoietic cell lines as model systems for leukemia-lymphoma research. Leuk Res 24: 881–911. [DOI] [PubMed] [Google Scholar]
- 2. Altmann M, Hammerschmidt W (2005) Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol 3(12): e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen MR (2011) Epstein-barr virus, the immune system, and associated diseases. Front Microbiol 2:5 Jan 26. [DOI] [PMC free article] [PubMed]
- 4. Tarrand JJ, Keating MJ, Tsimberidou AM, O'Brien S, LaSala RP, et al. (2010) Epstein-Barr virus latent membrane protein 1 mRNA is expressed in a significant proportion of patients with chronic lymphocytic leukemia. Cancer 116(4): 880–7. [DOI] [PubMed] [Google Scholar]
- 5. Rossi D, Gaidano G (2009) Richter syndrome: molecular insights and clinical perspectives. Hematol Oncol 27: 1–10. [DOI] [PubMed] [Google Scholar]
- 6. Ansell SM, Li CY, Lloyd RV, Phyliky RL (1999) Epstein-Barr virus infection in Richter’s transformation. Am J Hematol 60: 99–104. [DOI] [PubMed] [Google Scholar]
- 7. Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, et al. (2011) The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood 117: 5019–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pangalis GA, Angelopoulou MK, Vassilakopoulos TP, Siakantaris MP, Kittas C (1999) B-chronic lymphocytic leukemia, small lymphocytic lymphoma, and lymphoplasmacytic lymphoma, including Waldenström's macroglobulinemia: a clinical, morphologic, and biologic spectrum of similar disorders. Semin Hematol 36: 104–14. [PubMed] [Google Scholar]
- 9. Choi WW, Weisenburger DD, Greiner TC, Piris MA, Banham AH, et al. (2009) A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res 15: 5494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Montes-Moreno S, Gonzalez-Medina AR, Rodriguez-Pinilla SM, Maestre L, Sanchez-Verde L, et al. (2010) Aggressive large B-cell lymphoma with plasma cell differentiation: immunohistochemical characterization of plasmablastic lymphoma and diffuse large B-cell lymphoma with partial plasmablastic phenotype. Haematologica 95: 1342–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, et al. (2011) Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475: 101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, et al. (2011) Oncogenically active MYD88 mutations in human lymphoma. 470(7332): 115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zamò A, Ott G, Katzenberger T, Adam P, Parolini C, et al. (2006) Establishment of the MAVER-1 cell line, a model for leukemic and aggressive mantle cell lymphoma. Haematologica 91: 40–7. [PubMed] [Google Scholar]
- 14. Brothman AR, Persons DL, Shaffer LG (2009) Nomenclature evolution: Changes in the ISCN from the 2005 to the 2009 edition. Cytogenet Genome Res 127: 1–4. [DOI] [PubMed] [Google Scholar]
- 15. Brunelli M, Eccher A, Gobbo S, Ficarra V, Novara G, et al. (2008) Loss of chromosome 9p is an independent prognostic factor in patients with clear cell renal cell carcinoma. Mod Pathol 21: 1–6. [DOI] [PubMed] [Google Scholar]
- 16. Kanellis G, Roncador G, Arribas A, Mollejo M, Montes-Moreno S, et al. (2009) Identification of MNDA as a new marker for Nodal Marginal Zone Lymphoma. Leukemia 23: 1847–57. [DOI] [PubMed] [Google Scholar]
- 17. Nilsson K (1999) Human B-lymphoid cell lines. Hum Cell 5: 25–41. [PubMed] [Google Scholar]
- 18. Stacchini A, Aragno M, Vallario A, Alfarano A, Circosta P, et al. (1999) MEC1 and MEC2: two new cell lines derived from B-chronic lymphocytic leukaemia in prolymphocytoid transformation. Leuk Res 23: 127–36. [DOI] [PubMed] [Google Scholar]
- 19. Varghese AM, Sayala H, Evans PA, O’Connor SJ, Patmore R, et al. (2008) Development of EBV-associated diffuse large B-cell lymphoma in Waldenstrom macroglobulinemia and mantle cell lymphoma. Leuk Lymphoma 49: 1618–9. [DOI] [PubMed] [Google Scholar]
- 20. Kojima M, Nakamura N, Itoh H, Shimizu K, Shimizu K, et al. (2010) Epstein-Barr virus-related atypical lymphoproliferative disorders in Waldeyer's ring: a clinicopathological study of 9 cases. Pathobiology 77: 218–24. [DOI] [PubMed] [Google Scholar]
- 21. Tsimberidou AM, Keating MJ (2005) Richter syndrome: biology, incidence, and therapeutic strategies. Cancer 103: 216–28. [DOI] [PubMed] [Google Scholar]
- 22. Lin P, Mansoor A, Bueso-Ramos C, Hao S, Lai R, et al. (2003) Diffuse large B-cell lymphoma occurring in patients with lymphoplasmacytic lymphoma/Waldenström macroglobulinemia. Clinicopathologic features of 12 cases. Am J Clin Pathol 120: 246–53. [DOI] [PubMed] [Google Scholar]
- 23. Falini B, Fizzotti M, Pucciarini A, Bigerna B, Marafioti T, et al. (2000) A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood 95: 2084–92. [PubMed] [Google Scholar]
- 24. Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, et al. (1992) RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68: 855–67. [DOI] [PubMed] [Google Scholar]
- 25. Van Rijn RS, Simonetti ER, Hagenbeek A, Bonyhadi M, Storm G, et al. (2007) Quantitative assessment of human T lymphocytes in RAG2(−/−)gammac(−/−) mice: the impact of ex vivo manipulation on in vivo functionality. Exp Hematol 35: 117–27. [DOI] [PubMed] [Google Scholar]
- 26. Stilgenbauer S, Döhner K, Bentz M, Lichter P, Döhner H (1998) Molecular cytogenetic analysis of B-cell chronic lymphocytic leukemia. Ann Hematol 76: 101–10. [DOI] [PubMed] [Google Scholar]
- 27. Hjalmar V, Kimby E, Matutes E, Sundström C, Jacobsson B, et al. (1998) Trisomy 12 and lymphoplasmacytoid lymphocytes in chronic leukemic B-cell disorders. Haematologica 83: 602–9. [PubMed] [Google Scholar]
- 28. Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, et al. (2011) A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet 43: 242–5. [DOI] [PubMed] [Google Scholar]
- 29. Lenz G, Davis RE, Ngo VN, Lam L, George TC, et al. (2008) Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319(5870): 1676–9. [DOI] [PubMed] [Google Scholar]