Abstract
The methyl-CpG-binding domain (MBD) gene family was first linked to autism over a decade ago when Rett syndrome, which falls under the umbrella of autism spectrum disorders (ASDs), was revealed to be predominantly caused by MECP2 mutations. Since that time, MECP2 alterations have been recognized in idiopathic ASD patients by us and others. Individuals with deletions across the MBD5 gene also present with ASDs, impaired speech, intellectual difficulties, repetitive behaviors, and epilepsy. These findings suggest that further investigations of the MBD gene family may reveal additional associations related to autism. We now describe the first study evaluating individuals with ASD for rare variants in four autosomal MBD family members, MBD5, MBD6, SETDB1, and SETDB2, and expand our initial screening in the MECP2 gene. Each gene was sequenced over all coding exons and evaluated for copy number variations in 287 patients with ASD and an equal number of ethnically matched control individuals. We identified 186 alterations through sequencing, approximately half of which were novel (96 variants, 51.6%). We identified seventeen ASD specific, nonsynonymous variants, four of which were concordant in multiplex families: MBD5 Tyr1269Cys, MBD6 Arg883Trp, MECP2 Thr240Ser, and SETDB1 Pro1067del. Furthermore, a complex duplication spanning the MECP2 gene was identified in two brothers who presented with developmental delay and intellectual disability. From our studies, we provide the first examples of autistic patients carrying potentially detrimental alterations in MBD6 and SETDB1, thereby demonstrating that the MBD gene family potentially plays a significant role in rare and private genetic causes of autism.
Keywords: autism spectrum disorders (ASDs), copy number variation (CNV), methyl-CpG-binding domain (MBD), Rett syndrome, single nucleotide polymorphism (SNP)
Introduction
There is growing evidence of the involvement of the methyl-CpG-binding domain (MBD) genes in neurological disorders. To date, pathogenic mutations have been found in patients with clinical features along the autism continuum for two genes in this family, methyl-CpG-binding domain protein 5 (MBD5) and methyl-CpG-binding protein 2 (MECP2). Both genes carry an MBD domain, the unifying feature for the family that includes nine additional genes; BAZ1A, BAZ1B, MBD1, MBD2, MBD3, MBD4, MBD6, SETDB1 and SETDB2 (Roloff et al., 2003). The MBD genes are involved in a variety of functions, including chromatin remodeling (BAZ1A, BAZ1B, MBD1, MBD2, MBD3, and MECP2), DNA damage repair (BAZ1A and MBD4), histone methylation (SETBD1 and SETDB2), and X chromosome inactivation (MBD2, Roloff et al., 2003, Bogdanovic & Veenstra, 2009). There is also functional interplay among members of this family as they have been found to bind at the same promoter regions (MBD1, MBD2, MBD3, and MECP2), partner with each other in complexes (MBD1 and SETBD1), or act in the same complexes in a mutually exclusive manner (MBD2 and MBD3, Sarraf & I. Stancheva 2004; Ballestar et al., 2005; Le Guezennec et al., 2006; Matarazzo et al., 2007). Little is known thus far about the functions of MBD5 and MBD6; they each encode proteins that localize to chromatin but fail to bind methylated DNA (Laget et al., 2010).
One specific disorder in the autism spectrum, Rett syndrome, is caused almost exclusively by alterations in MECP2 (Amir et al., 1999). Due to the location of MECP2 on the X chromosome, mutations in females can lead to Rett syndrome while males with the same genetic changes typically present with neonatal encephalopathy (Moretti & Zoghbi 2006). Further investigations have demonstrated that MECP2 misregulation can lead to a wide range of clinical features including autism, Angelman-like symptoms, mental retardation with or without infantile seizures, mild learning disabilities, and schizophrenia (Watson et al., 2001; Klauck et al., 2002; Carney et al., 2003; Shibayama et al., 2004; Coutinho et al., 2007; Harvey et al., 2007; Lugtenberg et al., 2009). Our group previously evaluated the MECP2 gene in a dataset of female ASD patients and identified two mutations reported in classic Rett syndrome patients; an Arg294X mutation and a 41 base pair deletion (Leu386fs) predicted to generate a truncated protein (Carney et al., 2003). Furthermore, while point mutations in MECP2 were first recognized to result in abnormal clinical phenotypes, increased expression of the wild type protein due to gene duplication also results in neurodevelopmental disorders (Meins et al., 2005; Van Esch et al., 2005; del Gaudio et al., 2006; Ramocki et al., 2009).
A second gene in the MBD family, MBD5, was tied to neurodevelopmental disorders following the identification of microdeletions on chromosome 2q22–2q23 (Vissers et al., 2003; Koolen et al., 2004; de Vries et al., 2005; Wagenstaller et al., 2007; Jaillard et al., 2008; van Bon et al., 2009; Williams et al., 2009; Chung et al., 2011; Talkowski et al., 2011; Noh & J. M. Graham Jr 2012). The minimal region for these nonrecurrent deletions covers only a single gene, MBD5 (van Bon et al., 2009; Williams et al., 2009; Talkowski et al., 2011). This suggests that the common features of ASDs, delayed or impaired speech, intellectual disability, epilepsy, and stereotypic hand movements found across microdeletion patients manifest due to a decreased expression of this critical gene (van Bon et al., 2009; Williams et al., 2009; Talkowski et al., 2011). Notably, two cases of individuals with duplications across the critical MBD5 region also present with autistic features and developmental delay (Chung et al., 2012). This demonstrates that precise regulation of both MBD5 and MECP2 must be maintained as either increased or decreased expression of each gene can result in a range of neurodevelopmental disorders.
Supplementing clinical evidence, mouse models have reiterated the potential significance of the MBD family in autism etiology. Mbd1 and Mecp2 null models have abnormal neurobehavioral phenotypes including increased anxiety, and impaired social interactions and synaptic plasticity (Guy et al., 2001; Shahbazian et al., 2002b; Zhao et al., 2003; Allan et al., 2008). Furthermore, a transgenic Setdb1 model established a link between this gene and behavior (Jiang et al., 2010a). Additionally, Setdb1 plays a role in the repression of Grin2b, a gene linked to autism, bipolar disorder, intellectual disability, and schizophrenia (Avramopoulos et al., 2007; Allen et al., 2008; Endele et al., 2010; Jiang et al., 2010a; Myers et al., 2011; O'Roak et al., 2011).
Studies have demonstrated that each of the MBD genes are expressed in the brain, while their specific functions having only been determined for a subset of genes (Shahbazian et al., 2002a, Bogdanovic & Veenstra, 2009, Jiang et al., 2010b, Laget et al., 2010, Safran et al., 2010). MeCP2 is a transcriptional regulator believed to act in neuronal maturation as levels increase over time (Shahbazian et al., 2002a, Chahrour et al., 2008). Stable levels of MeCP2 are required through adulthood, as elimination of this protein in adult mice mimics features seen in knockout Mecp2 mice (McGraw et al., 2011). The H3K9 methyltransferase SETDB1 acts both in early development as well as later stages of life (Jiang et al., 2010a, Cho et al., 2012). Removal of Setdb1 in mice results in peri-implantation lethality (Dodge et al., 2004). Studies in the forebrain of transgenic Setdb1 mice demonstrate that it targets the NMDA receptors Grin2a and Grin2b as well as the glutamate receptor Grid2 (Yang et al., 2002, Jiang et al., 2010a).
While there is clinical evidence of MECP2 and MBD5 playing a role in autism, only two studies to date have evaluated patients with ASD for mutations in additional MBD family members (Li et al., 2005; Cukier et al., 2010). Previous work in our laboratory analyzed the coding regions of MBD1, MBD2, MBD3, and MBD4 in over 200 individuals with ASD of African and European ancestry and identified multiple variants that altered the amino acid sequence, were unique to patients with autism, and concordant with disease in multiplex families (Cukier et al., 2010). In contrast, a study by Li and colleagues was restricted to a dataset of 65 Japanese autistic patients and reported only a single variation that might be related to autism (Li et al., 2005). We now expand our initial study of MECP2 to a larger dataset that includes male patients and perform the first study evaluating patients with ASDfor alterations in four additional MBD family members: MBD5, MBD6, SETDB1 and SETDB2.
Materials and Methods
Ethics statement
We ascertained individuals at the John P. Hussman Institute for Human Genomics (HIHG) at the University of Miami, Miller School of Medicine (Miami, Florida), the University of South Carolina (Columbia, South Carolina), and the Center for Human Genetics Research at Vanderbilt University (Nashville, Tennessee). Written informed consent was obtained from parents for all minor children and those who were unable to give consent. In addition, we obtained assent from all participants of the appropriate developmental and chronological age. All participants were ascertained using the protocol approved by the appropriate Institutional Review Boards. Patients were collected for this study for over a decade, with protocols and amendments being approved at each stage. Oversight of the study at the University of Miami falls under the UM Medical IRB B committee, whose members include Ofelia Alvarez, M.D., Abdul Mian, Ph.D., Maria Alcaide, M.D., Michelle Dunaj Lucking, J.D., Jean Jose, D.O., Howard Landy, M.D., Bruce Nolan, M.D., FACP, FAASM, Emilio Weiss, Pharm.D., and Cecilia Grano de Oro, B.A.
Patient Ascertainment
Families of individuals with ASD were recruited from a number of clinical and educational settings throughout the United States and received informed consent in accordance with respective University institutional review boards. Core inclusion criteria for individuals with ASD included: (1) chronological age between 3 and 21 years of age, (2) a presumptive clinical diagnosis of ASD, (3) an expert clinical determination of ASD diagnosis using DSM-IV criteria (American Psychiatric Association, 1994) supported by the Autism Diagnostic Interview (ADI-R) (Rutter et al., 2003b) and (4) an IQ equivalent >35 or developmental level >18 months as determined by the Vineland Adaptive Behavior Scale (VABS) (Sparrow et al., 2005). ASD diagnosis was confirmed after review by a panel consisting of experienced clinical psychologists and a pediatric medical geneticist. In those instances where an ADI-R was not available, a best-estimate diagnosis was assigned using all available clinical information including clinician summaries, a caregiver report, and medical records. We excluded participants with severe sensory problems (e.g., visual impairment or hearing loss), significant motor impairments (e.g., failure to sit by 12 months or walk by 24 months), or identified metabolic, genetic (eg., chromosomal abnormalities, Fragile X mutations), or progressive neurological disorders. Family history and pedigree information was collected in a standard semi-structured interview from a knowledgeable informant, most frequently the mother. Additional clinical data were also collected by reviewing available medical and psychiatric records of affected individuals. DNA was isolated from whole blood collected from participants via venipuncture.
Healthy children between the ages of 4 and 21 years were recruited as controls. Participants were excluded if they had developmental, behavioral, or neurological conditions or first degree relatives with such disorders. We obtained consent from all participants or, in the case of minors, their parents. Participants provided a saliva sample with an Oragene DNA sample collection kit and a knowledgeable informant, usually the mother, completed the Social Communication Questionnaire (Rutter et al., 2003b) to screen for potential ASDs.
Two hundred and eighty-seven unrelated individuals with ASD were chosen from our dataset for sequencing. Two hundred and eighty-eight ethnically matched, healthy controls were also selected, for a total of 575 samples. Individuals were either of African (85 cases: 68 males, 17 females and 90 controls: 43 males, 47 females) or European ancestry (202 cases: 171 males, 31 females and 198 controls: 129 males, 69 females). Given our large dataset of European individuals, we hypothesized that MBD alterations would lead to more severe phenotypes and therefore enriched for these patients in the current study. We selected individuals with ASD of European ancestry with a VABS score less than or equal to 90. was determined by self-report and confirmed by comparing Illumina Human 1M BeadChip genotyping data with HapMap populations in the Eigenstrat program (Patterson et al., 2006; Price et al., 2006; Ma et al., 2009). Sixty-seven individuals evaluated in this study were included in our previous examination of the MBD1–4 genes (Cukier et al., 2010).
Sanger Sequencing and Genotyping
Five hundred and seventy-five samples were sequenced in one direction across all coding exons of MBD5, MBD6, MECP2, SETDB1, and SETDB2. Primer sets were designed to generate amplicons between 192 and 688 base pairs (bps) and cover each exon as well as 20 flanking bps (Supplemental Table 1). Primers were created using the Primer3 (v. 0.4.0) program (http://fokker.wi.mit.edu/primer3/input.htm) and the UCSC reference genome (GRCh37/hg19) for all genes except MECP2, for which previously published primers and conditions were used (Buyse et al., 2000; Quenard et al., 2006). Samples were placed in a 96 well format with cases and controls on each plate to control for batch effects. Sequencing reactions were performed with the Big Dye Terminator version 3.1 and run on an Applied Biosystems 3730xl DNA Analyzer (Life Technologies). Raw data were evaluated in the Sequencher program, version 4.10.1 (Gene Codes Corporation). Variations identified were confirmed by sequencing in the opposite direction.
When available, additional family members were sequenced to determine inheritance patterns and whether alterations were concordant with disease in multiplex families. Alterations were compared to the NCBI Database for Short Genetic Variations, dbSNP Build 134. Variations were run through the GERP (Genomic Evolutionary Rate Profiling, Cooper et al. 2005), PhastCons (Siepel et al., 2005), PolyPhen-2 (Polymorphism Phenotyping v2, Adzhubei et al., 2010), and SIFT (Sorting Intolerant From Tolerant, Kumar et al., 2009) programs to predict the level of damaging effect the variation might cause.
Evaluation of Copy Number Variations
To identify large structural variations, two TaqMan real-time polymerase chain reaction (PCR) copy number assays within each gene were selected from predesigned assays available from Applied Biosystems (Supplemental Table 2). All 575 samples were tested in quadruplicate according to the manufacturer’s standard protocol. Analysis was performed with the CopyCaller Software 1.0 (Applied Biosystems). Individuals having the same copy number change in both assays for a particular gene were further investigated using previously generated single nucleotide polymorphism (SNP) array data (Ma et al., 2009). Utilizing over 1 million SNPs genotyped on the Illumina Human 1M Beadchip, the PennCNV algorithm was used to detect deletions and duplications.
Cloning MBD5 Tyr1269Cys and Measuring Transcriptional Activation
We generated a plasmid (pMbd5-BD) that expresses the 1498 amino acid isoform of murine Mbd5 fused in front of a Gal4 binding domain and driven by a CMV promoter. Briefly, Mbd5 was isolated from a pooled cDNA library, cloned into the pGEMT-Easy plasmid (Promega) and then subcloned into pDsred-Express-N1 (Clontech). The Dsred-Express tag was replaced in frame with the Gal4 binding domain from pCMV-BD (Agilent). The human c.3806A>G (Tyr1269Cys) mutation was inserted into the corresponding murine Mbd5 c.3818 (p.1272) position using the QuikChange II XL Site-Directed Mutagenesis kit (Agilent). Sanger sequencing confirmed that each plasmid contained either wild type or Tyr1272Cys Mbd5 and that no other mutation had been introduced. HEK 293T cells were then transiently transfected with either the wild type or mutated pMbd5-BD plasmid along with pFR-Luc (Agilent) and pSV-β-Galactosidase (Promega) in quadruplicate using Lipofectamine 2000 (Invitrogen). Cells were permitted to grow for 48 hours and lysed. Activity was measured using the Stratagene Luciferase Assay Kit. To normalize, the lysates’ protein concentrations were measured using the Pierce BCA Protein Assay Kit (ThermoScientific) and β-galactosidase activity was measured by using the β-Galactosidase Assat Kit (Stratagene).
Results
Discovery of Novel Variations by Sequencing
Sequencing across the five MBD genes in 287 patients with ASD and 288 ethnically matched control individuals identified a total of 186 unique variations (Table 1, Supplemental Tables 3–7). These variants included 177 single nucleotide polymorphisms (SNPs), five deletions and four insertions. Ninety (48.4%) of the variations have been previously reported in either the dbSNP 134 database (http://www.ncbi.nlm.nih.gov/projects/SNP/) or RettBASE (http://mecp2.chw.edu.au/), while the remaining 96 variants (51.6%) are novel. Fifty-six variations are predicted to alter the amino acid sequence. Fifty-three of the changes were found solely in patients with ASD and absent from controls. To determine variants most likely to contribute to ASD susceptibility, we prioritized changes that were either unique to affected individuals or that had an increased frequency in cases when compared to controls. The 17 most interesting variants were nonsynonymous and unique to our ASD population (Table 1). We utilized four distinct programs to characterize the variants; GERP (Cooper et al., 2005) and PhastCons (Siepel et al., 2005) to measure the level of amino acid conservation across species and PolyPhen (Adzhubei et al., 2010) and SIFT (Kumar et al., 2009) to predict which alterations might have the damaging consequences to protein function.
Table 1.
ASD Unique, Nonsynonymous Variations
| ASD individuals | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Position | Hg19 Location |
Nucleotide | Amino Acid | SNP/Novel | African | European | Family | Inheritance | Concordant: Discordant |
GERP | Phast Cons |
Poly Phen-2 |
SIFT |
| MBD5 | exon 9 | 149,226,839 | c.1327G>A | Val443Met | rs137977565 | - | 1 | 37134 | maternal | N/A | 5.13 | 1 | 0.872 | tolerated |
| exon 12 | 149,247,640 | c.3740T>C | Ile1247Thr | novel | - | 1 | 37575 | paternal | N/A | 4.81 | 1 | 0.054 | damaging | |
| exon 12 | 149,247,706 | c.3806A>G | Tyr1269Cys | novel | - | 1 | 7763 | paternal | 3:0 | 5.64 | 1 | 0.234 | damaging | |
| exon 12 | 149,247,796 | c.3896G>A | Arg1299Gln | rs35934694 | 1 | - | 1072 | maternal | 1:1 | 5.54 | 1 | 0.997 | damaging | |
| MBD6 | exon 6 | 57,919,667 | c.916G>A | Gly306Arg | novel | - | 1 | 18019 | maternal | N/A | 3.04 | 1 | unknown | - |
| exon 6 | 57,920,130 | c.1379C>G | Ser460Cys | novel | - | 1 | 18006 | paternal | unknown | 3.2 | 1 | unknown | damaging | |
| exon 6 | 57,920,165 | c.1414G>T | Val472Leu | novel | - | 1 | 7935 | maternal | 2:1 | 3.2 | 1 | unknown | tolerated | |
| exon 10 | 57,921,969 | c.24446G>A | Glu816Lys | novel | - | 1 | 7987 | maternal | N/A | 3.51 | 0.992 | unknown | tolerated | |
| exon 10 | 57,922,170 | c.2647C>T | Arg883Trp | novel | - | 1 | 7979 | maternal | 2:0 | 4.46 | 1 | 0.998 | damaging | |
| exon 11 | 57,922,456 | c.2827C>G | Pro943Arg | novel | 1 | - | 7605 | paternal | N/A | 1.92 | 0.837 | 0.970 | damaging | |
| exon 12 | 57,922,754 | c.2899C>T | Arg967Cys | rs144794136 | - | 1 | 7933 | maternal | N/A | 4.39 | 1 | 0.989 | damaging | |
| MECP2 | exon 4 | 153,296,596 | c.719C>G | Thr240Ser | rs61749738 | 2 C/Y | - | 1072, 17130 | maternal | 2:0, N/A | 5.13 | 0.301 | 0.002 | tolerated |
| exon 4 | 153,296,207 | c.1108G>A | Ala370Thr | rs147017239 | 1 A/Y | - | 18024 | maternal | N/A | −0.673 | 0.392 | unknown | tolerated | |
| SETDB1 | exon 18 | 150,935,103–150,935,105 | c.3199delCTT | Pro1067del | novel | 1 | - | 17187 | maternal | 2:0 | - | - | - | - |
| SETDB2 | exon 10 | 50,056,954 | c.1274T>C | Ile425Thr | rs148270039 | - | 1 | 37575 | maternal | N/A | −5.09 | 0 | 0.001 | tolerated |
| exon 11 | 50,057,605 | c.1424C>T | Thr475Met | novel | - | 1 | 17548 | maternal | N/A | −0.596 | 0.937 | 0.000 | tolerated | |
| exon 12 | 50,059,890 | c.1607C>G^ | Pro536Arg^ | novel | - | 1 | 17212 | paternal | N/A | 0.54 | 0.004 | unknown | - | |
GERP score measures conservation between 34 mammalian species on a scale from −11.6 to 5.82
PhastCon score measures conservation between 17 vertebrate species on a scale from 0 to 1
PolyPhen-2 score measures the potential for an amino acid substitution to be damaging on a scale from 0 to 1
SETDB2 alternative isoform containing 548 amino acids
The mutational burden between cases and controls of African or European ancestry for each gene was not statistically significant by the chi-squared test (Supplemental Table 8). This was determined for the overall load of all variants as well as nonsynonymous alterations (Supplemental Table 9).
MBD5
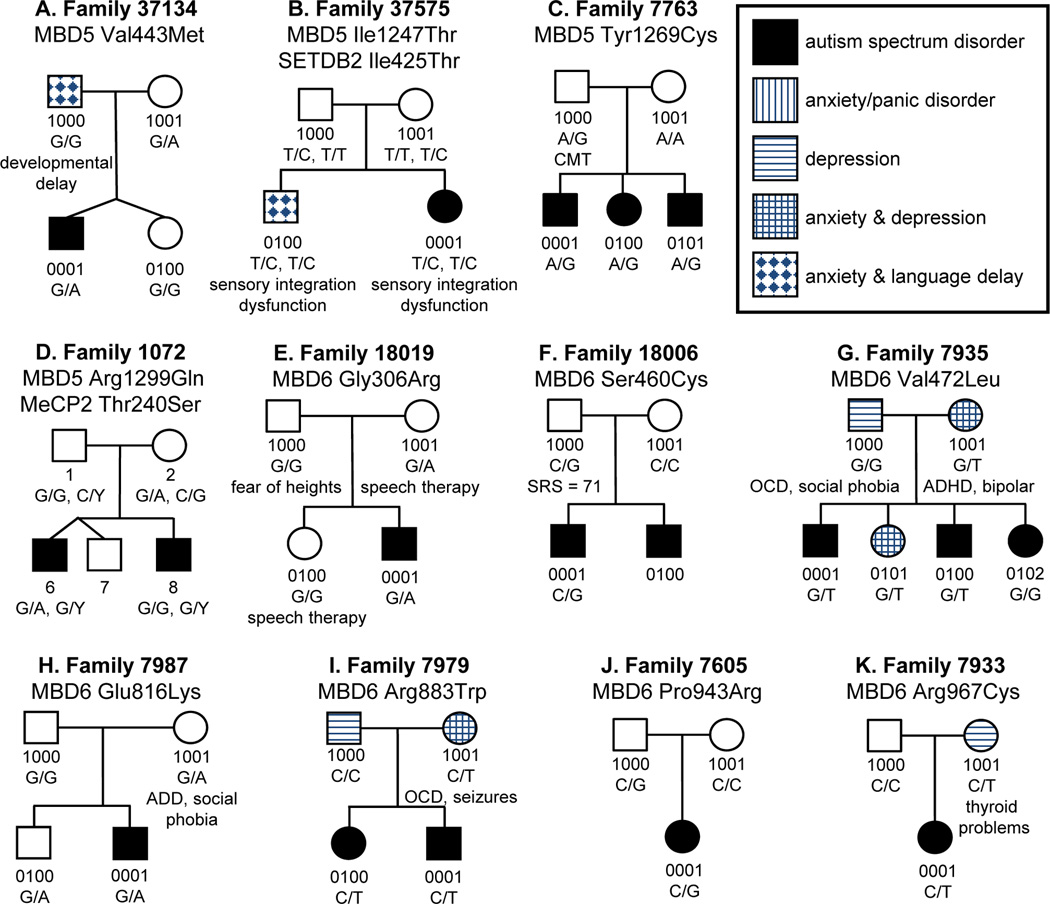
Thirty-two changes were identified in MBD5, 18 of which have been previously reported (Supplemental Table 3). A distinct set of 11 alterations were nonsynonymous, four of which were only identified in patients with ASD (Val443Met, Ile1247Thr, Tyr1269Cys, and Arg1299Gln, Figure 1A–D, Table 1). Three of these four alterations (75%) are predicted to be damaging by SIFT, as compared to only two of seven nonsynonymous variants (28.6%) identified solely in control individuals (Supplemental Table 3). One alteration of high interest, MBD5 Tyr1269Cys, was inherited paternally in all three ASD children in multiplex family 7763 (Figure 1C). Two of the affected individuals (0001 and 0100) were intellectually impaired with measured IQ in the moderate to severe range (Full Scale IQ: 40 and 50, respectively), while the remaining brother with autism (0101) had borderline intellectual functioning (Full Scale IQ=78). Furthermore, all three siblings had a delay in language and displayed self-injurious behaviors. Two individuals presented with macrocephaly (0100 and 0101), and individual 0100 has a history of epilepsy (recurrent non-febrile seizures).
Figure 1. Pedigrees of ASD families carrying alterations in MBD5 and MBD6.
Variants identified in MBD5 (A–D) and MBD6 (E–K) are shown. Each nucleotide change is marked under the individual in whom it occurs. Additional medical features in the family are noted. In two multiplex pedigrees (C, I), the variant is concordant with ASD.
MBD6
A total of 44 alterations were detected in MBD6, two being single base pair insertions and the remainder of which were SNPs (Supplemental Table 4). Sixteen of the single nucleotide changes have been previously reported and 28 are novel. A subset of 17 alterations was identified only in individuals with ASD, seven of which are predicted to cause missense changes (Table 1, Figure 1E–K). While each of these changes was only identified in a single proband, three of the alterations have high PolyPhen and SIFT scores and are novel (Arg883Trp, Pro943Arg and Arg967Cys), suggesting a strong functional consequence. Furthermore, one of these alterations, Arg883Trp, was identified in multiplex family 7979 and passed maternally to both affected children (Figure 1I). Individual 0001 has a diagnosis of autism and is nonverbal with moderate intellectual disability. His sister (0100) has a diagnosis of Pervasive Developmental Disorder-Not Otherwise Specified and mild intellectual disability, displaying some phrase speech. Both siblings have a history self-injurious behavior. Their mother (1001), who also carries the alteration, was diagnosed with anxiety/panic disorders, depression, obsessive compulsive disorder, and has a history of epilepsy (adolescent onset seizures).
Along with novel variations of interest in MBD6, we found that two known SNPs occur at a higher frequency within our affected population compared to our control population. The first variation, rs61741508 (c.-2C>A), was recognized in sixteen patients with ASD and five controls and is located just upstream of the ATG start site in the Kozak consensus sequence. This variation also has high conservation scores (Supplemental Table 4). The second SNP, rs117084250 (c.2407-64C>T), falls within intron nine and was found in twelve individuals with ASD but only four controls. However, the conservation scores were relatively low, thereby making this a variant of lesser interest (Supplemental Table 4).
MECP2
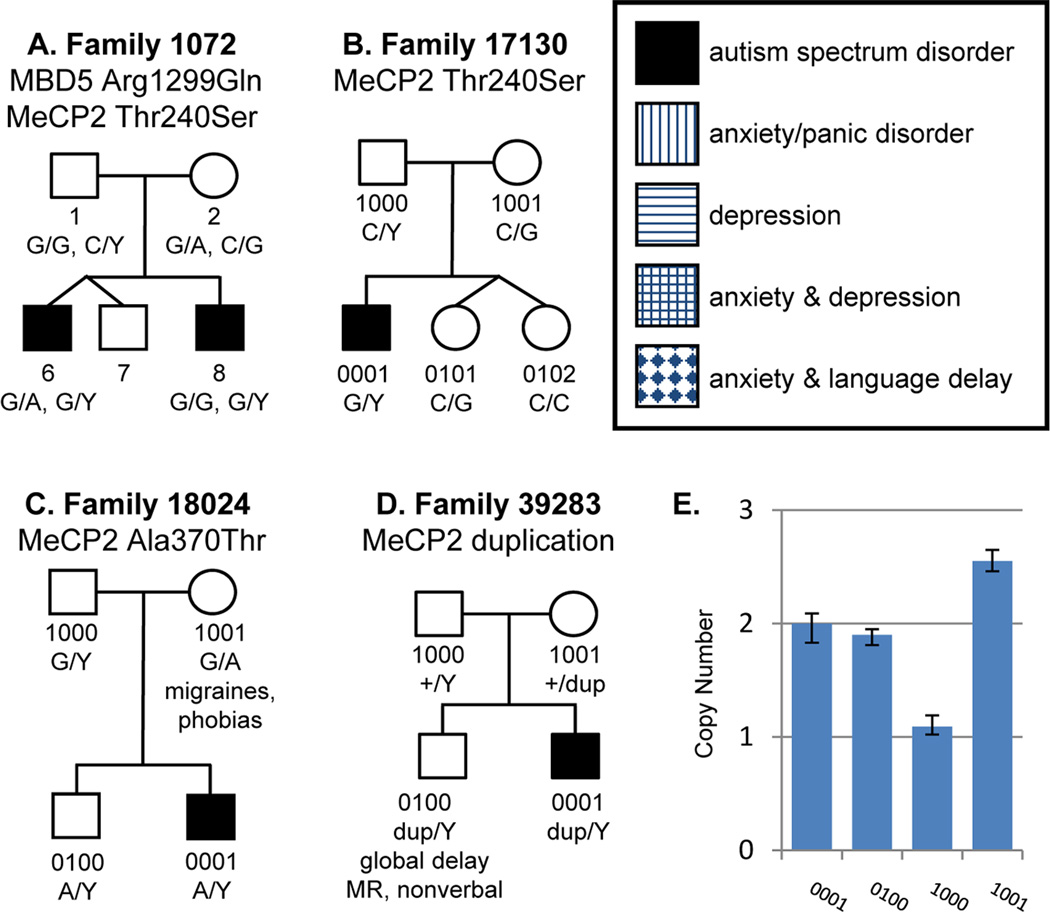
Twenty-eight alterations were identified in MECP2 (Supplemental Table 5). Sixteen of these are currently in the dbSNP database and another one has been previously reported in RettBASE, leaving 11 novel variations. While none of the frequently recurring, classic Rett syndrome variations were identified in this study, there are two previously reported MeCP2 alterations of undetermined pathogenicity (Thr240Ser and Ala370Thr) that may cause clinical phenotypes. This first variation, MeCP2 Thr240Ser (rs61749738), was identified in two families of African ancestry (1072 and 17130) and absent in control individuals (Figure 2A,B). Further investigation into additional family members showed that the variation was inherited maternally in both cases and concordant with disease in multiplex family 1072. The second alteration, Ala370Thr (rs147017239), was also inherited maternally in a single proband of African ancestry (family18024, Figure 2C).
Figure 2. Pedigrees of ASD families carrying alterations in MeCP2.
Point mutations (A–C) and a duplication (D) identified in MeCP2 are shown. Each nucleotide change is marked under the individual in whom it occurs. Additional medical features in the family are noted. (E) TaqMan real-time PCR CNV assay results for family 39283. The father (1000) carries a single copy of MeCP2 on his X chromosome, while both sons (0001 and 0100) harbor a duplication of MeCP2 on the X chromosome that they inherited from their mother (1001), who carries a total of three copies of MeCP2. Error bars represent the range (minimum to maximum) of the four replicates that were utilized to determine the copy number of each sample.
SETDB1
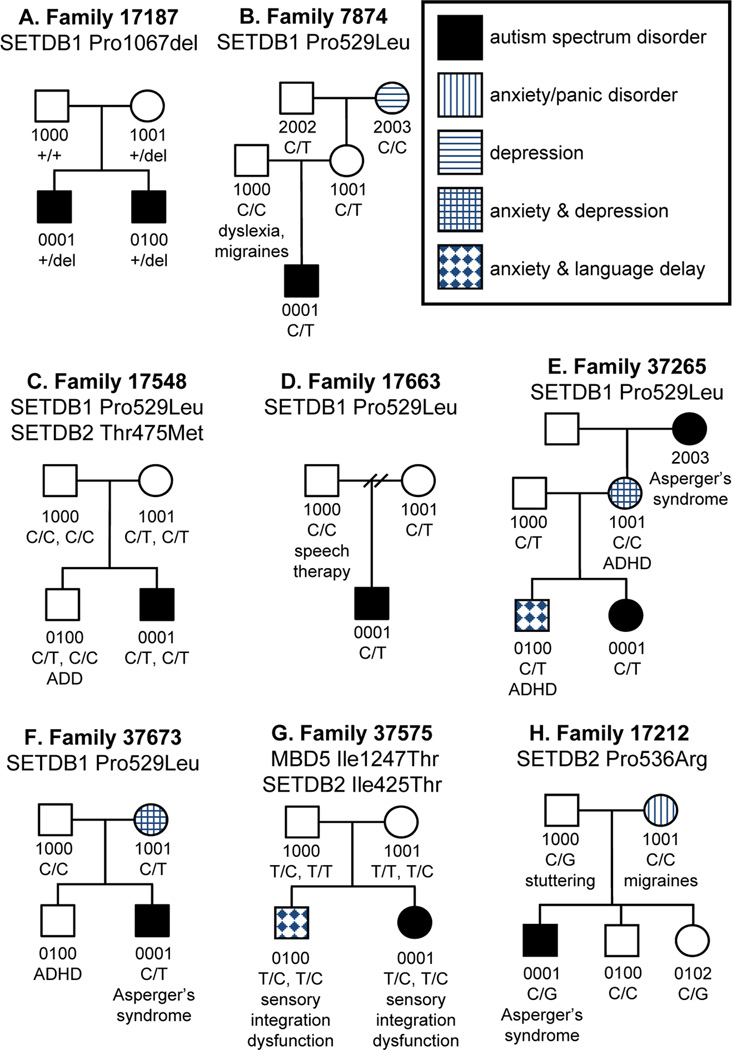
A total of 44 changes were found in SETDB1, comprised of 19 known and 25 novel alterations (Supplemental Table 6). Eight changes are predicted to be nonsynonymous, but only one of these, Pro1067del, was found solely in patients with ASD. This change is also the only ASD specific, nonsynonymous deletion identified in the entire study. The variant removes three nucleotides and predicts an in-frame deletion of a single amino acid. This deletion falls within the SET domain of the protein and was inherited maternally in both affected sons in family 17187 (Figure 3A).
Figure 3. Pedigrees of ASD families carrying alterations in SETDB1 and SETDB2.
Variants identified in SETDB1 (A–F) and SETDB2 (G–H) are shown. Each nucleotide change is marked under the individual in whom it occurs. Additional medical features in the family are noted. In one multiplex pedigree (A), the variant is concordant with ASD.
Another novel variation of interest in SETDB1 that we identified in a high proportion of cases versus controls, Pro529Leu, was identified in five ASD families of European ancestry and only a single control (Figure 3B–F). This variant was inherited paternally in one family and maternally in the remaining four families. In family 37265, the variation was passed from the father, who has dyslexia, to both the female proband with autism (0001) who was diagnosed with developmental and language delays as well as her brother (0100) who presented with ADHD, anxiety/panic disorder, language delay and macrocephaly (Figure 3E). In two of the families with maternal inheritance (17663 and 37673), the mothers presented with anxiety/panic disorder. In family 17663, the mother also presented with a history of seizures, sleep disorder and self-reported depression, while the mother in family 37673 reported history of adolescent onset Anorexia Nervosa. The increased incidence of this alteration in cases versus controls, along with neuropsychiatric and neurodevelopmental disorders in parents carrying the alteration, suggests that this variation may confer a variety of clinical consequences.
SETDB2
Thirty-eight single base pair alterations were identified in the SETDB2 gene, 21 of which have been previously reported and the remaining 17 are novel (Supplemental Table 7). Eight SNPs are predicted to alter amino acids and three of these were unique to affected individuals: Ile425Thr, Thr475Met and Pro536Arg (Table 1, Figure 3C,G,H). However, these alterations are not predicted to have a highly detrimental effect on the protein and occur within singleton families, making it difficult to determine whether they may play a pathogenic role in ASD.
Identification of a MECP2 Duplication
A single structural variation was identified in MECP2 by real-time PCR copy number assays and independently validated by SNP array data (Figure 2D,E). The duplication was passed maternally to both the proband (0001) as well as his brother (0100) in a family of Jamaican ancestry. The proband presented clinically with autism and mental retardation. He is nonverbal and has history of allergies, significant motor delays and non-febrile generalized seizures. His brother did not meet the criteria for autism, but presented with global developmental delay, intellectual disability, and allergies. The mother had two miscarriages between the birth of 0001 and 0100 and a subsequent miscarriage after the birth of 0100. Maternal family history is positive for infertility in three siblings and cystic fibrosis in two female siblings. This 91.1 kb duplication encompasses MECP2 and IRAK1 (chrX:152,931,687–153,022,805). Further evaluation of the region showed that the proband appears to also carry a second, nearby 294 kb duplication (chrX:152,641,759–152,895,758) that covers an additional 17 genes: BCAP31, ABCD1, KIAA1206, PLXNB3, SRPK3, IDH3G, SSR4, PDZD4, KIAA1444, L1CAM, LCAP, AVPR2, ARHGAP4, ARD1A, RENBP, HCFC1, and TMEM187.
Abnormal Function of a Novel MBD5 Alteration
To test whether one of the novel, ASD specific variations identified might have a quantifiable difference in protein function, we used a luciferase transcriptional activation assay. We selected the MBD5 Tyr1269Cys (c.3806A>G) variation because of its high conservation scores, moderate probability to affect function, and concordance in all three ASD siblings in family 7763. Due to the high homology between humans and mice (92% identical and 94% similar), we mutated murine Mbd5 at the corresponding amino acid, Tyr1272Cys (c.3818A>G). Six transient transfections were performed and there was an increase of 72.1% in the transcriptional activity of the mutated version of Mbd5 as compared to the wild type (p=5.06×10−9).
Discussion
Potentially Pathogenic Alterations in the MBD Genes
Along with isolating additional variations in MBD5 and MECP2 that may contribute to neuropsychiatric disease, this study is the first to report prospective pathogenic variations in MBD6 and SETDB1. These include two novel, nonsynonymous alterations in MBD6 (Arg883Trp and Pro943Arg) and one more in SETDB1 (Pro1067del). Furthermore, the MBD6 Arg883Trp and SetDB1 Pro1067del variations each segregated with ASD in the multiplex families. Potential for SETDB1 to play a role in neurobehavioral phenotypes is supported by results from transgenic Setdb1 mice demonstrating a role in mood behaviors (Jiang et al., 2010a).
To date, MBD5 mutations have been identified in individuals presenting a range of clinical phenotypes including ASD, developmental delay, intellectual disability, epilepsy, repetitive movements, and language impairments (Vissers et al., 2003; Koolen et al., 2004; de Vries et al., 2005; Wagenstaller et al., 2007; Jaillard et al., 2008; van Bon et al., 2009; Williams et al., 2009; Chung et al., 2011; Talkowski et al., 2011; Noh & Graham Jr 2012). These results suggest a significant role for the MBD5 isoform 1, which presents with increased expression in the brain (Laget et al., 2010). It has been estimated that between microdeletions and point mutations of MBD5, this gene may play a contributing genetic role in up to 1% of individuals with ASD (Talkowski et al., 2011). Of the nonsynonymous alterations identified in this study, ASD specific changes were more likely to be predicted to be damaging as compared to those variations found in control individuals (Supplemental Table 3). MBD5 Tyr1269Cys is a strong potentially pathogenic change due to its co-segregation with ASD in a multiplex family of three affected children, high conservation of this amino acid across species and altered function in the luciferase transcriptional activation assay. While this alteration does not fall in a known protein domain, it is specific to isoform 1, the isoform predominately expressed in brain (Laget et al., 2010). It seems likely that most alterations in MBD5 related to disease will be rare and unique, as the one alteration previously reported to have an increased frequency in patients with ASD, Gly79Glu, was only identified in a single control in the current study (Talkowski et al., 2011).
The role of MECP2 in developmental disorders is undisputed (Samaco & Neul 2011). Our study supports the possible pathogenicity of two specific MeCP2 alterations: Thr240Ser and Ala370Thr. The first variant, Thr240Ser was identified in two male probands from families of African ancestry, including the multiplex family 1072 where the variant segregated with ASD (Figure 2A,B). The maternal inheritance in family 17130 and presence of an unaffected carrier sister suggests that the variation may only present with a clinical phenotype in a hemizygous state. This variant falls within the transcriptional repression domain and has been previously reported in four studies; three cases of males with intellectual disability and one female with Rett syndrome (Yntema et al., 2002; Bourdon et al., 2003; Bienvenu & J. Chelly 2006; Campos et al., 2007; Bunyan & D. O. Robinson 2008). The second alteration, Ala370Thr, was identified in a singleton family of African ancestry and previously reported in three Chinese individuals: one female with Rett syndrome, her unaffected mother and a male presenting with epileptic encephalopathy (Figure 2C, Li et al., 2007; Wong & Li 2007). Both of these alterations must be further evaluated to isolate their potential functional consequences.
Finally, while we did identify variants of interest in four of the genes studied, SETDB2 alterations did not appear to be related to the occurrence of ASDs.
Complex Rearrangement on Xp28 Duplicating MECP2
To date, over one hundred Xp28 duplication patients have been described that share core clinical features including autism, developmental delay, infantile hypotonia, mental retardation, profound speech deficits, and recurrent infections (Meins et al., 2005; Van Esch et al., 2005; del Gaudio et al., 2006; Ramocki et al., 2009). The inheritance pattern found in family 39283, with transmission from a clinically normal mother to affected sons, is typical for alterations in this region (Figure 2D,E). The complex nature of the duplications found in this family is also fairly common due to the numerous low copy repeat sequences present in the local genomic architecture (Carvalho et al., 2009). While this structural variation may not be the only genetic component contributing to the neurodevelopmental features present in this family, it is most likely the predominant factor.
Interdependence of Members of the MBD Family
While there is evidence that some members of the MBD family either work together, in mutually exclusive complexes or bind the same genomic regions, the full extent of their interactions is still being revealed (Sarraf and Stancheva, 2004; Ballestar et al., 2005; Le Guezennec et al., 2006; Matarazzo et al., 2007). This interaction has also been demonstrated in animal models. The transgenic Setdb1 mouse was crossed to a Mecp2 null model to determine whether there was functional overlap; however, the Mecp2 phenotypes were not rescued and, in some cases, exacerbated (Jiang et al., 2010b). Therefore, it is interesting to note that family 1072 presented with ASD unique, nonsynonymous alterations at both MBD5 Arg1299Gln and MECP2 Thr240Ser in the proband. Furthermore, family 7605 has a novel, ASD specific alteration at MBD6 Pro943Arg and was previously identified with a potentially detrimental deletion in our prior study, MBD4 E314fsX316 (Cukier et al., 2010). It is not yet known whether these genes have any overlapping functions or cofactors, but these findings may hint at such a connection. Over time, we may discover that this is one example of the complex nature of autism genetics and how, in some cases, an accumulation of genetic insults must occur before a clinical phenotype manifests.
Aside from genes in the MBD family, there are numerous, additional genes and genomic regions that have been connected to ASDs (Betancur, 2011). Indeed, our laboratory has performed genome wide CNV studies to identify structural variations in our patients with ASDs (Cukier et al., 2011, Salyakina et al., 2011, Griswold et al., 2012). These studies identified deletions and duplications in regions previously linked to ASD such as 1q21.1, 15q13.1, and 16p11.2 (Griswold et al., 2012). However, individuals identified with ASD specific and unique, nonsynonymous MBD alterations were absent for such CNVs. Therefore, we feel that these alterations in MBD genes may play a role in ASD susceptibility and are not coincidentally occurring in patients with large, pathogenic genomic mutations.
This is the first study to evaluate the coding regions of MBD5, MBD6, SETDB1, and SETDB2 for rare alterations in individuals with ASD. We identified novel point mutations predicted to be damaging and concordant with disease in multiplex families, as well as a complex duplication encompassing MECP2. Additional studies, ideally both in patients and animal models, are required to determine the precise consequences of these alterations. The results described here compound the evidence of MECP2 and MBD5’s involvement in ASDs and neurodevelopmental disorders and provide the first examples of autistic patients carrying potentially detrimental alterations in MBD6 and SETDB1. This study demonstrates the expanding role MBD genes play in autism etiology.
Supplementary Material
Acknowledgements
We are extremely thankful to all participants and their family members, without whom this study would not be possible. We acknowledge Dr. Susan Folstein for her work in ascertaining a subset of the ASD families used in this study. Funding for this study was provided by the National Institute of Mental Health (R01 MH080647), the National Institute of Neurological Disorders and Stroke (P01 NS026630 and ARRA supplement), as well as a generous gift from the Hussman Foundation.
Footnotes
The authors declare that there is no conflict of interest.
Literature Cited
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan AM, Liang X, Luo Y, Pak C, Li X, Szulwach KE, et al. The loss of methyl-CpG binding protein 1 leads to autism-like behavioral deficits. Human Molecular Genetics. 2008;17:2047–2057. doi: 10.1093/hmg/ddn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nature Genetics. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genetics. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Avramopoulos D, Lasseter VK, Fallin MD, Wolyniec PS, McGrath JA, Nestadt G, et al. Stage II follow-up on a linkage scan for bipolar disorder in the Ashkenazim provides suggestive evidence for chromosome 12p and the GRIN2B gene. Genetics in Medicine : Official Journal of the American College of Medical Genetics. 2007;9:745–751. doi: 10.1097/gim.0b013e318159a37c. [DOI] [PubMed] [Google Scholar]
- Ballestar E, Ropero S, Alaminos M, Armstrong J, Setien F, Agrelo R, et al. The impact of MECP2 mutations in the expression patterns of Rett syndrome patients. Human Genetics. 2005;116:91–104. doi: 10.1007/s00439-004-1200-0. [DOI] [PubMed] [Google Scholar]
- Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Research. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nature Reviews.Genetics. 2006;7:415–426. doi: 10.1038/nrg1878. [DOI] [PubMed] [Google Scholar]
- Bogdanovic O, Veenstra GJC. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon V, Philippe C, Martin D, Verloes A, Grandemenge A, Jonveaux P. MECP2 mutations or polymorphisms in mentally retarded boys: diagnostic implications. Molecular Diagnosis : A Journal Devoted to the Understanding of Human Disease through the Clinical Application of Molecular Biology. 2003;7:3–7. doi: 10.1007/BF03260014. [DOI] [PubMed] [Google Scholar]
- Bunyan DJ, Robinson DO. Multiple de novo mutations in the MECP2 gene. Genetic Testing. 2008;12:373–375. doi: 10.1089/gte.2008.0012. [DOI] [PubMed] [Google Scholar]
- Buyse IM, Fang P, Hoon KT, Amir RE, Zoghbi HY, Roa BB. Diagnostic testing for Rett syndrome by DHPLC and direct sequencing analysis of the MECP2 gene: identification of several novel mutations and polymorphisms. American Journal of Human Genetics. 2000;67:1428–1436. doi: 10.1086/316913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Jr, Abdalla CB, Santos-Reboucas CB, dos Santos AV, Pestana CP, Domingues ML, et al. Low significance of MECP2 mutations as a cause of mental retardation in Brazilian males. Brain & Development. 2007;29:293–297. doi: 10.1016/j.braindev.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, et al. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatric Neurology. 2003;28:205–211. doi: 10.1016/s0887-8994(02)00624-0. [DOI] [PubMed] [Google Scholar]
- Carvalho CM, Zhang F, Liu P, Patel A, Sahoo T, Bacino CA, et al. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Human Molecular Genetics. 2009;18:2188–2203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Park JS, Kwon S, Kang YK. Dynamics of Setdb1 expression in early mouse development. Gene Expression Patterns. 2012;12:213–218. doi: 10.1016/j.gep.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Chung BH, Mullegama S, Marshall CR, Lionel AC, Weksberg R, Dupuis L, et al. Severe intellectual disability and autistic features associated with microduplication 2q23.1. European Journal of Human Genetics. 2012;20:398–403. doi: 10.1038/ejhg.2011.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BH, Stavropoulos J, Marshall CR, Weksberg R, Scherer SW, Yoon G. 2q23 de novo microdeletion involving the MBD5 gene in a patient with developmental delay, postnatal microcephaly and distinct facial features. American Journal of Medical Genetics.Part A. 2011;155A:424–429. doi: 10.1002/ajmg.a.33821. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, et al. NISC Comparative Sequencing Program. Distribution and intensity of constraint in mammalian genomic sequence. Genome Research. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho AM, Oliveira G, Katz C, Feng J, Yan J, Yang C, et al. MECP2 coding sequence and 3'UTR variation in 172 unrelated autistic patients. American Journal of Medical Genetics.Part B, Neuropsychiatric Genetics : The Official Publication of the International Society of Psychiatric Genetics. 2007;144:475–483. doi: 10.1002/ajmg.b.30490. [DOI] [PubMed] [Google Scholar]
- Cukier HN, Rabionet R, Konidari I, Rayner-Evans MY, Baltos ML, Wright HH, et al. Novel variants identified in methyl-CpG-binding domain genes in autistic individuals. Neurogenetics. 2010;11:291–303. doi: 10.1007/s10048-009-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukier HN, Salyakina D, Blankstein SF, Robinson JL, Sacharow S, Ma D, et al. Microduplications in an autism multiplex family narrow the region of susceptibility for developmental disorders on 15q24 and implicate 7p21. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2011;156B:493–501. doi: 10.1002/ajmg.b.31188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, Janssen IM, et al. Diagnostic genome profiling in mental retardation. American Journal of Human Genetics. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Gaudio D, Fang P, Scaglia F, Ward PA, Craigen WJ, Glaze DG, et al. Increased MECP2 gene copy number as the result of genomic duplication in neurodevelopmentally delayed males. Genetics in Medicine : Official Journal of the American College of Medical Genetics. 2006;8:784–792. doi: 10.1097/01.gim.0000250502.28516.3c. [DOI] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Molecular and Cellular Biology. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nature Genetics. 2010;42:1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, Chung RH, et al. Evaluation of Copy Number Variations Reveals Novel Candidate Genes in Autism Spectrum Disorder Associated Pathways. Human Molecular Genetics. 2012 doi: 10.1093/hmg/dds164. published online Apr 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genetic. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Harvey CG, Menon SD, Stachowiak B, Noor A, Proctor A, Mensah AK, et al. Sequence variants within exon 1 of MECP2 occur in females with mental retardation. American Journal of Medical Genetics.Part B, Neuropsychiatric Genetics : The Official Publication of the International Society of Psychiatric Genetics. 2007;144:355–360. doi: 10.1002/ajmg.b.30425. [DOI] [PubMed] [Google Scholar]
- Jaillard S, Dubourg C, Gerard-Blanluet M, Delahaye A, Pasquier L, Dupont C, et al. 2q23.1 microdeletion identified by array-CGH: an emerging phenotype with Angelman-like features? Journal of Medical Genetics. 2008;46:847–855. doi: 10.1136/jmg.2008.058156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Jakovcevski M, Bharadwaj R, Connor C, Schroeder FA, Lin CL, et al. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2010a;30:7152–7167. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Matevossian A, Guo Y, Akbarian S. Setdb1-mediated histone H3K9 hypermethylation in neurons worsens the neurological phenotype of Mecp2-deficient mice. Neuropharmacology. 2010b;60:1088–1097. doi: 10.1016/j.neuropharm.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauck SM, Lindsay S, Beyer KS, Splitt M, Burn J, Poustka A. A mutation hot spot for nonspecific X-linked mental retardation in the MECP2 gene causes the PPM-X syndrome. American Journal of Human Genetics. 2002;70:1034–1037. doi: 10.1086/339553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen DA, Vissers LE, Nillesen W, Smeets D, van Ravenswaaij CM, Sistermans EA, et al. A novel microdeletion, del(2)(q22.3q23.3) in a mentally retarded patient, detected by array-based comparative genomic hybridization. Clinical Genetics. 2004;65:429–432. doi: 10.1111/j.0009-9163.2004.00245.x. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Laget S, Joulie M, Le Masson F, Sasai N, Christians E, Pradhan S, et al. The human proteins MBD5 and MBD6 associate with heterochromatin but they do not bind methylated DNA. PLoS One. 2010;5(8):e11982. doi: 10.1371/journal.pone.0011982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Molecular and Cellular Biology. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yamagata T, Mori M, Yasuhara A, Momoi MY. Mutation analysis of methyl-CpG binding protein family genes in autistic patients. Brain & Development. 2005;27:321–325. doi: 10.1016/j.braindev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Li MR, Pan H, Bao XH, Zhang YZ, Wu XR. MECP2 and CDKL5 gene mutation analysis in Chinese patients with Rett syndrome. Journal of Human Genetics. 2007;52:38–47. doi: 10.1007/s10038-006-0079-0. [DOI] [PubMed] [Google Scholar]
- Lugtenberg D, Kleefstra T, Oudakker AR, Nillesen WM, Yntema HG, Tzschach A, et al. Structural variation in Xq28: MECP2 duplications in 1% of patients with unexplained XLMR and in 2% of male patients with severe encephalopathy. European Journal of Human Genetics : EJHG. 2009;17:444–453. doi: 10.1038/ejhg.2008.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Salyakina D, Jaworski JM, Konidari I, Whitehead PL, Andersen AN, et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Annals of Human Genetics. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo MR, De Bonis ML, Strazzullo M, Cerase A, Ferraro M, Vastarelli P, et al. Multiple binding of methyl-CpG and polycomb proteins in long-term gene silencing events. Journal of Cellular Physiology. 2007;210:711–719. doi: 10.1002/jcp.20879. [DOI] [PubMed] [Google Scholar]
- McGraw CM, Samaco RC, Zoghbi HY. Adult neuronal function requires MeCP2. Science. 2011;333:186. doi: 10.1126/science.1206593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meins M, Lehmann J, Gerresheim F, Herchenbach J, Hagedorn M, Hameister K, et al. Submicroscopic duplication in Xq28 causes increased expression of the MECP2 gene in a boy with severe mental retardation and features of Rett syndrome. Journal of Medical Genetics. 2005;42:e12. doi: 10.1136/jmg.2004.023804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Zoghbi HY. MeCP2 dysfunction in Rett syndrome and related disorders. Current Opinion in Genetics & Development. 2006;16:276–281. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Myers RA, Casals F, Gauthier J, Hamdan FF, Keebler J, Boyko AR, et al. A population genetic approach to mapping neurological disorder genes using deep resequencing. PLoS Genetics. 2011;7 doi: 10.1371/journal.pgen.1001318. e1001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh GJ, Graham JM., Jr 2q23.1 microdeletion of the MBD5 gene in a female with seizures, developmental delay and distinct dysmorphic features. European Journal of Medical Genetics. 2012;55:59–62. doi: 10.1016/j.ejmg.2011.10.001. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genetics. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Quenard A, Yilmaz S, Fontaine H, Bienvenu T, Moncla A, des Portes V, et al. Deleterious mutations in exon 1 of MECP2 in Rett syndrome. European Journal of Medical Genetics. 2006;49:313–322. doi: 10.1016/j.ejmg.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Ramocki MB, Peters SU, Tavyev YJ, Zhang F, Carvalho CM, Schaaf CP, et al. Autism and other neuropsychiatric symptoms are prevalent in individuals with MeCP2 duplication syndrome. Annals of Neurology. 2009;66:771–782. doi: 10.1002/ana.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roloff TC, Ropers HH, Nuber UA. Comparative study of methyl-CpG-binding domain proteins. BMC Genomics. 2003;4:1. doi: 10.1186/1471-2164-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, LeCouteur A, Lord C. Autism Diagnostic Interview, Revised (ADI-R) 2003a [Google Scholar]
- Rutter M, Bailey A, Berument SK, LeCouteur A, Lord C, Pickles A. Social Communication Questionnaire (SCQ) 2003b [Google Scholar]
- Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, Sirota-Madi A, Olender T, Golan Y, Stelzer G, Harel A, Lancet D. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010 doi: 10.1093/database/baq020. baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyakina D, Cukier HN, Lee JM, Sacharow S, Nations LD, Ma D, et al. Copy number variants in extended autism spectrum disorder families reveal candidates potentially involved in autism risk. PLoS One. 2011;6(10):e26049. doi: 10.1371/journal.pone.0026049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, Neul JL. Complexities of Rett Syndrome and MeCP2. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2011;31:7951–7959. doi: 10.1523/JNEUROSCI.0169-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Molecular Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Human Molecular Genetics. 2002a;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, et al. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002b;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Shibayama A, Cook EH, Jr, Feng J, Glanzmann C, Yan J, Craddock N, et al. MECP2 structural and 3'-UTR variants in schizophrenia, autism and other psychiatric diseases: a possible association with autism. American Journal of Medical Genetics.Part B, Neuropsychiatric Genetics : The Official Publication of the International Society of Psychiatric Genetics. 2004;128:50–53. doi: 10.1002/ajmg.b.30016. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Research. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow SS, Cicchetti DV, Balla D. Vineland Adaptive Behavior Scales--Second Edition. 2005 [Google Scholar]
- Talkowski ME, Mullegama SV, Rosenfeld JA, van Bon BW, Shen Y, Repnikova EA, et al. Assessment of 2q23.1 Microdeletion Syndrome Implicates MBD5 as a Single Causal Locus of Intellectual Disability, Epilepsy, and Autism Spectrum Disorder. American Journal of Human Genetics. 2011;89:551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bon BW, Koolen DA, Brueton L, McMullan D, Lichtenbelt KD, Ades LC, et al. The 2q23.1 microdeletion syndrome: clinical and behavioural phenotype. European Journal of Human Genetics : EJHG. 2009;18:163–170. doi: 10.1038/ejhg.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Esch H, Bauters M, Ignatius J, Jansen M, Raynaud M, Hollanders K, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. American Journal of Human Genetics. 2005;77:442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Vries BB, Osoegawa K, Janssen IM, Feuth T, Choy CO, et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. American Journal of Human Genetics. 2003;73:1261–1270. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenstaller J, Spranger S, Lorenz-Depiereux B, Kazmierczak B, Nathrath M, Wahl D, et al. Copy-number variations measured by single-nucleotide-polymorphism oligonucleotide arrays in patients with mental retardation. American Journal of Human Genetics. 2007;81:768–779. doi: 10.1086/521274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Black G, Ramsden S, Barrow M, Super M, Kerr B, et al. Angelman syndrome phenotype associated with mutations in MECP2, a gene encoding a methyl CpG binding protein. Journal of Medical Genetics. 2001;38:224–228. doi: 10.1136/jmg.38.4.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SR, Mullegama SV, Rosenfeld JA, Dagli AI, Hatchwell E, Allen WP, et al. Haploinsufficiency of MBD5 associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment, and seizures. European Journal of Human Genetics : EJHG. 2009;18:436–441. doi: 10.1038/ejhg.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VC, Li SY. Rett syndrome: prevalence among Chinese and a comparison of MECP2 mutations of classic Rett syndrome with other neurodevelopmental disorders. Journal of Child Neurology. 2007;22:1397–1400. doi: 10.1177/0883073807307091. [DOI] [PubMed] [Google Scholar]
- Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, et al. Molecular cloning of ESET, a novel histon H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene. 2002;21:148–152. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- Yntema HG, Oudakker AR, Kleefstra T, Hamel BC, van Bokhoven H, Chelly J, et al. In-frame deletion in MECP2 causes mild nonspecific mental retardation. American Journal of Medical Genetics. 2002;107:81–83. doi: 10.1002/ajmg.10085. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.